Phosphorylation of the Cap-Binding Protein Eukaryotic Translation Initiation Factor 4E by Protein Kinase Mnk1 In Vivo (original) (raw)
Abstract
Eukaryotic translation initiation factor 4E (eIF4E) binds to the mRNA 5′ cap and brings the mRNA into a complex with other protein synthesis initiation factors and ribosomes. The activity of mammalian eIF4E is important for the translation of capped mRNAs and is thought to be regulated by two mechanisms. First, eIF4E is sequestered by binding proteins, such as 4EBP1, in quiescent cells. Mitogens induce the release of eIF4E by stimulating the phosphorylation of 4EBP1. Second, mitogens and stresses induce the phosphorylation of eIF4E at Ser 209, increasing the affinity of eIF4E for capped mRNA and for an associated scaffolding protein, eIF4G. We previously showed that a mitogen- and stress-activated kinase, Mnk1, phosphorylates eIF4E in vitro at the physiological site. Here we show that Mnk1 regulates eIF4E phosphorylation in vivo. Mnk1 binds directly to eIF4G and copurifies with eIF4G and eIF4E. We identified activating phosphorylation sites in Mnk1 and developed dominant-negative and activated mutants. Expression of dominant-negative Mnk1 reduces mitogen-induced eIF4E phosphorylation, while expression of activated Mnk1 increases basal eIF4E phosphorylation. Activated mutant Mnk1 also induces extensive phosphorylation of eIF4E in cells overexpressing 4EBP1. This suggests that phosphorylation of eIF4E is catalyzed by Mnk1 or a very similar kinase in cells and is independent of other mitogenic signals that release eIF4E from 4EBP1.
Mitogens stimulate protein and RNA synthesis (56, 65). The increase in protein synthesis is partly due to increased initiation on preexisting mRNAs, with the result that those mRNAs are recruited into larger polysomes. In addition to an increase in basal translation, specific mRNAs are preferentially upregulated, suggesting that mitogenic signal transduction pathways impinge on the components of the translation machinery that interact with the mRNA.
mRNAs are brought to the ribosome by eukaryotic initiation factor (eIF4F) (for reviews, see references 58, 60, and 61). eIF4F is a multiprotein complex formed from 25-, 46- and 220-kDa subunits, called eIF4E, eIF4A, and eIF4G, respectively. eIF4E, also known as cap-binding protein, is responsible for binding the 5′-terminal 7-methyl-GTP (m7GTP) cap found on all eukaryotic mRNAs. eIF4A is a subunit of an RNA helicase that seems to unwind secondary structure in the mRNA. eIF4G is the scaffolding subunit, to which the other subunits bind. It also has a binding site for eIF3, which links the eIF4F-mRNA complex to the 40S ribosomal subunit. In yeast, eIF4G has an additional functional region, to which the poly(A)-binding protein and the 3′ end of the mRNA bind (64). Besides serving as a passive scaffold, eIF4G plays a regulatory role, stimulating the binding of capped mRNA to eIF4E (24). Thus, the eIF4F complex promotes interactions between the 5′ end of the mRNA, the ribosome, and an RNA helicase.
As the main mRNA-binding component of the translation machinery, the eIF4F complex has the potential to distinguish between mRNAs for differential translation in mitogen-treated cells. The eIF4E subunit is thought to be the main regulatory component since it is present in limiting molar amounts (11, 26, 53), and eIF4E availability in quiescent cells is further restricted by eIF4E-binding proteins, including 4EBP1 or PHAS-I (61). 4EBPs prevent eIF4E binding to eIF4G without altering mRNA binding to eIF4E (23, 44, 52). Mitogenic stimuli induce the phosphorylation of 4EBPs and the release of eIF4E, allowing eIF4E to associate with eIF4G and participate in translation. In addition, mitogens stimulate the phosphorylation of ribosomal protein S6 and several translation initiation factors, including eIF4E. Phosphorylation of 40S ribosomal protein S6 specifically stimulates the translation of a class of mRNAs that have pyrimidine-rich 5′ ends (29). However, cells lacking the S6 protein kinase still exhibit mitogen-induced increases in basal translation (32), indicating that other mechanisms, such as 4EBP1 and eIF4E phosphorylation, also mediate mitogenic stimulation of translation.
Several lines of evidence suggest that phosphorylation of eIF4E stimulates translation initiation. Mitogen-enhanced eIF4E phosphorylation usually correlates with increased protein synthesis (36), and phosphorylation increases the binding of eIF4E to capped mRNA and to eIF4G in vitro (5, 39, 48). The location of Ser 209 adjacent to the cap-binding pocket is consistent with an effect of phosphorylation on mRNA binding (45, 46). Significantly, overexpression of wild-type eIF4E, but not of a substitution mutant that is not phosphorylated, leads to malignant transformation and high rates of protein synthesis (10, 41). Studies of cells transformed by eIF4E overexpression show an increased ability to translate mRNAs with increased cap-proximal secondary structure (37, 55, 59), consistent with increased binding of the eIF4F complex and its associated helicase activity. While some other reports indicate that changes in eIF4E phosphorylation do not invariably correlate with increased translation (34, 49, 51, 54, 57, 70), it seems likely that eIF4E phosphorylation modulates translation initiation in cells.
Different signal transduction pathways control the phosphorylation of 4EBP1, S6, and eIF4E. In fibroblasts, the phosphorylation of 4EBP1 and S6 occurs by an extracellular signal-regulated kinase (ERK)/mitogen-activated protein (MAP) kinase-independent signaling pathway that is sensitive to a specific inhibitor, rapamycin (4, 6, 18, 25, 68, 69). In contrast, mitogen-stimulated eIF4E phosphorylation occurs via a rapamycin-insensitive, Ras- and ERK/MAP kinase-dependent pathway (14–16). This means that phosphorylation of eIF4E is independent of phosphorylation of 4EBPs. eIF4E is also phosphorylated in response to stresses, including anisomycin and hypertonicity, dependent on the p38 MAP kinase (49, 51, 70). This suggests that the MAP kinases ERK and p38, or a protein kinase activated by ERK or p38, may phosphorylate eIF4E in cells. However, neither ERK nor p38 is a candidate to phosphorylate eIF4E directly, because the residue phosphorylated, Ser 209 (13, 31), lacks proline residues required for MAP kinase recognition (1, 7). Therefore, eIF4E is more likely to be phosphorylated by one or more MAP kinase-dependent protein kinases.
A number of MAP kinase-dependent kinases are now known, and of these, Mnk1, MAPKAPK-3 (3pK), and Msk1 are activated by both ERK and p38 (9, 17, 43, 71). Direct in vitro phosphorylation assays suggest that MAPKAPK-3 and Mnk1 can both phosphorylate eIF4E directly, while certain other MAP kinase-dependent protein kinases, including Rsk and MAPKAPK-2, cannot (51, 52a, 71). Under a variety of stimulation and inhibitor conditions, the in vivo phosphorylation of eIF4E correlates with the activity of Mnk1 (70). This correlative evidence suggests that Mnk1, or a similarly activated protein kinase, phosphorylates eIF4E in cells. In view of the potential importance of eIF4E phosphorylation in regulating translation initiation, we have investigated whether Mnk1 phosphorylates eIF4E in vivo. In this report, we demonstrate that Mnk1 is a member of the eIF4F complex, binding directly to eIF4G. We have identified activating phosphorylation sites in Mnk1 and created activated and dominant negative mutants. Coexpression of a dominant negative mutant Mnk1 inhibits mitogen-induced and basal phosphorylation of eIF4E, while expression of an activated mutant Mnk1 results in constitutive phosphorylation of eIF4E. Phosphorylation of eIF4E by Mnk1 also occurs in the presence of excess 4EBP1. We suggest that Mnk1 or a protein kinase with similar binding properties and enzymatic specificity phosphorylates eIF4E in mitogen- and stress-stimulated cells.
MATERIALS AND METHODS
Plasmids and mutagenesis.
The plasmids pLexA-Mnk1, pEBG-Mnk1, and pEBG-T2A2 (Mnk1 containing Thr197Ala/Thr202Ala mutations) have been described previously (71). Untagged Mnk1 was expressed from pCS2+ (66), and myc-tagged (MT-) Mnk1 from pCS3+MT (66a) (this vector encodes six myc tags at the N terminus of the fusion protein). The mutations Thr197Ser (T197S), Thr202Ser (T202S), Thr332Ser (T332S), Thr197Asp/Thr202Asp (T2D2), Thr332Asp (T332D), Thr197Asp/Thr202Asp/Thr332Asp (T3D3), and Thr197Ala/Thr202Ala/Thr332Ala (T3A3) were created by _Pfu_-mediated mutagenesis (Quickchange, Stratagene) and confirmed by nucleotide sequencing. ΔN Mnk1 was created by _Taq_-mediated PCR (Perkin-Elmer), resulting in a 23-residue deletion from the N terminus, and ΔC Mnk1 was made similarly to delete 85 residues from the C terminus. The human eIF4E open reading frame was PCR amplified from a bacterial expression clone (62) and inserted into the _Bgl_II site of pcDL-SRα456 (63) to encode eIF4E with a single HA tag at the N terminus. eIF4E-209Ala was subcloned into the same vector by using a mutant C-terminal oligonucleotide and Taq-mediated PCR. Its sequence was confirmed. The eIF4E open reading frame was also cloned into pCS3+MT for expression of MT-eIF4E. Vectors expressing HA-tagged and Flag-tagged 4EBP1 were the kind gifts of Anne-Claude Gingras and Nahum Sonenberg (18).
Two-hybrid screens.
Strain L40 yeast cells (67) expressing pLexA-Mnk1 were transformed with a human HeLa cDNA library (Clontech). From an estimated 5 × 106 primary transformants, 51 _HIS3_-positive colonies were selected. Of 25 which possessed strong β-galactosidase activity, 12 were specific for the original bait plasmid and did not interact with ΔN Mnk1. Three of these clones encoded p220-2 or eIF4GII (22) (human EST Z34918), and nine of them encoded human hRch1, a form of α-importin (19, 21).
In vitro binding to eIF4G.
Recombinant eIF4GI was made in Sf9 cells by using the baculovirus expression system and purified as described for eIF2 (33). Glutathione _S_-transferase (GST)-Mnk1 or GST-T2A2 proteins were made in Escherichia coli and purified on glutathione-Sepharose. Beads were mixed with eIF4G in a buffer containing 100 mM CH3COOK, 2 mM (CH3COO)2Mg, 1 mM phenylmethylsulfonyl fluoride, 7 mM 2-mercaptoethanol, and 50 mM HEPES (pH 7.4). Samples were mixed at 4°C for 1 h and washed three times by centrifugation with the same buffer containing 0.1% Triton X-100. Bound proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting with rabbit antiserum to eIF4G, raised as described previously (35).
Copurification assays.
293 cells in 50-mm dishes were transiently transfected (Lipofectamine; Gibco-BRL) with pEBG-Mnk1 or pCS3+MT-Mnk1 DNA (4 μg) and grown for 48 h after the addition of DNA. The cells were then serum starved overnight with 0.1 to 0.5% fetal bovine serum. They were lysed in buffer containing 1% Triton X-100 and immunoprecipitated (anti-myc monoclonal antibody 9E10 for MT-Mnk1) or purified with glutathione-Sepharose (GST-Mnk1) (71). The glutathione-Sepharose was washed three times in the same buffer before SDS-PAGE and Western blotting were performed. Anti-HA monoclonal antibody 12CA5 was from D. Kremer and L. Breeden. Anti-Flag monoclonal antibody M2 was from Kodak. Rabbit antiserum to the 9E10 epitope, used for Western blotting, was from Babco. Rabbit antiserum to GST was raised by standard procedures.
Immunolocalization.
NIH 3T3 cells were transiently transfected with pSRα-eIF4E, fixed with 4% paraformaldehyde in phosphate-buffered saline, permeabilized with 0.1% Triton X-100, and stained with 12CA5 anti-HA antibody or chicken anti-Mnk1 antiserum. Antiserum was raised to GST-Mnk1, made in E. coli (71), by immunizing chickens. Immunoglobulin was purified from eggs (Aves Inc., Tigard, Oregon) and affinity purified with GST-Mnk1, coupled to CNBr-Sepharose, and eluted with urea. Secondary antibodies were fluorescein-conjugated anti-chicken antibody and Texas red-conjugated anti-mouse antibodies (Jackson Immunoresearch Laboratories). Images were obtained with a Deltavision image-deconvoluting microscope and represent optical sections.
Phosphopeptide analysis.
For labeling, 50-mm dishes of 293 cells were transfected with pEBG-Mnk1 and starved as described above; at 4.5 h before lysis, the cells were washed once and incubated for 30 min in phosphate-free Dulbecco modified Eagle medium containing 1 mM pyruvate, 1 mg of fatty acid-free bovine serum albumin per ml, and 20 mM HEPES (pH 7.4). The fluids were replaced with the same mixture containing 0.5 mCi of [32P]orthophosphate (NEN) per ml, and incubation was continued for 4 h. The cells were stimulated as needed, and GST-Mnk1 was isolated by using glutathione-Sepharose and SDS-PAGE. The protein was eluted from the dried gel, oxidized with performic acid, and digested with trypsin followed by thermolysin as described previously (3). Phosphopeptides were resolved by electrophoresis in cellulose thin layers at pH 1.9 followed by ascending chromatography in a buffer containing isobutyric acid (3), with the anode at the left. Individual phosphopeptides were extracted from the cellulose, partially hydrolyzed in 6 M HCl at 110°C for 2 h, mixed with nonradioactive phosphoserine, phosphothreonine, and phosphotyrosine, and analyzed by electrophoresis at pH 3.5 (3).
In vitro kinase assays.
GST-Mnk1 was purified from starved or tetradecanoyl phorbol acetate (TPA)-stimulated 293 cells as described above, except that a wash in 0.5 M LiCl was used to remove associated MAP kinases, as described previously (71). Protein kinase assays were performed (71) with 5 μg of recombinant eIF4E substrate and 10 μCi of [γ-32P]ATP. Recombinant eIF4E was prepared as described previously (62) and was the kind gift of C. G. Proud (University of Dundee).
Analysis of the eIF4E phosphorylation state.
293 cells were transiently transfected with the following plasmids as needed (the quantity of DNA per 50-mm dish is given in parentheses): wild-type or mutant Mnk1 in vector pEBG (4 μg), HA-eIF4E in vector pSRα (1 μg), and Flag-4EBP1 in vector pcDNA3 (4 μg). Vector DNA was added as needed. Transfected cells were serum starved between 48 and 60 h after DNA addition), labeled with [32P]orthophosphate between 60 and 64 h as described above, and treated with 0 or 100 nM TPA for the last 15 min before lysis. [32P]HA-eIF4E was recovered from cells lysed in 1% Triton buffer by using immunoprecipitation with 12CA5 anti-HA antibody and analyzed by SDS-PAGE and autoradiography. Samples for isoelectric focusing were prepared by lysing nonradioactive cells (in 100-mm dishes) by Dounce homogenization in 4E buffer (50 mM β-glycerophosphate, 1 mM phenylmethylsulfonyl fluoride, 2 mM Na3 VO4, 1 μM microcystine [Calbiochem]). Protein concentrations were equalized, and samples were removed for immunoprecipitation and for eIF4E purification as needed. Immunoprecipitations were performed as above, and the immunoprecipitates were washed with 1% Triton buffer. eIF4E was purified with m7GTP-Sepharose (Pharmacia). Samples were washed in four times in 4E buffer, eluted in buffer containing urea and ampholytes, and separated on a one-dimensional denaturing isoelectric focusing gel containing equal parts of pH 4 to 6 and pH 6 to 8 ampholytes, as described previously (13).
RESULTS
Mnk1 binds to eIF4G and other components of the eIF4F initiation complex.
The Mnk1 primary sequence predicts a protein kinase domain flanked by an N-terminal region of unknown function and a C-terminal region that binds the MAP kinases ERK1, ERK2, and p38 (17, 71). To identify potential substrates for Mnk1, we used the yeast two-hybrid system (67, 71). From 5 × 106 transformants, we isolated 2 cDNAs that encoded proteins that interacted with full-length Mnk1 but not with control proteins. The cDNAs encoded portions of an α-importin relative (20, 21) and of eIF4GII, one of two known forms of eIF4G (22). Both fragments bind to full-length Mnk1 and to a mutant which lacks the C-terminal 85 amino acids but do not bind to a mutant in which the N-terminal 23 amino acids are deleted (Fig. 1A and data not shown). Since the C terminus is needed for ERK or p38 MAP kinase binding (71), this suggests that the binding of α-importin and eIF4GII is not mediated by MAP kinase. The α-importin clone encodes residues 43 to 239, which contain the region hypothesized to interact with nuclear localization signal peptides. Since the N terminus of Mnk1 is rich in basic residues, it may be recognized by α-importin as a nuclear localization signal. However, we have not detected a significant portion of Mnk1 within the nucleus (Fig. 2), suggesting that Mnk1 may not bind functionally to α-importin in mammalian cells. The two-hybrid isolate of eIF4GII encodes the C-terminal 192 residues, implying that this region of eIF4GII is sufficient to confer Mnk1 binding. The amino acid sequences of eIF4GI and eIF4GII are 68% identical in this region, suggesting that Mnk1 may bind to both forms of eIF4G.
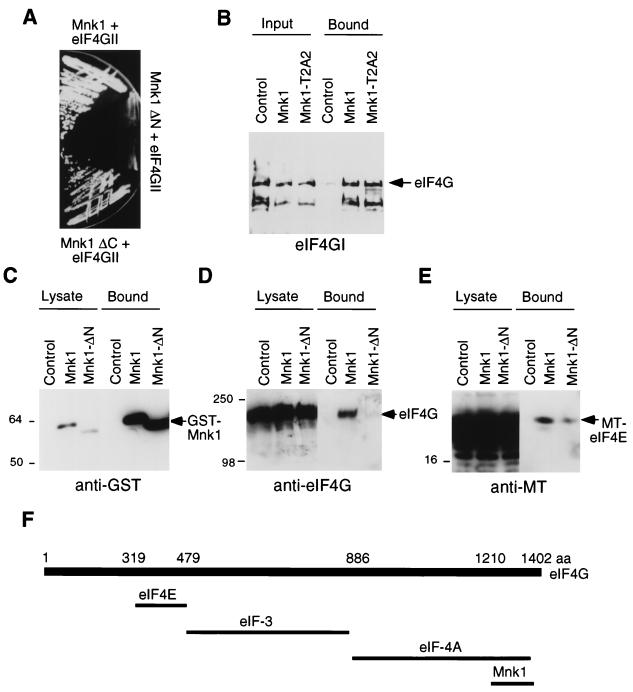
FIG. 1.
(A) Growth of yeast strain L40 expressing LexA-Mnk1 and VP16-eIF4GII (top), LexA-Mnk1-ΔN and VP16-eIF4GII (middle), and LexA-Mnk1-ΔC and VP16-eIF4GII (bottom) on medium lacking histidine. Growth indicates transactivation of the HIS3 reporter gene. (B) GST-Mnk1 and GST-Mnk1-T2A2 (T197A/T202A) were purified from E. coli and incubated with purified mammalian eIF4G. GST fusion proteins were isolated, and bound eIF4G was eluted and analyzed by SDS-PAGE and Western blotting. Input lanes represent one-fifth of the sample used for binding. (C to E) 293 cells were transiently transfected with pEBG, pEBG-Mnk1, or pEBG-Mnk1ΔN, together with pCS3+eIF4E. GST-Mnk1 was purified with glutathione-Sepharose, and bound proteins were visualized with GST antibody (C), eIF4G antibody (D), or 9E10 anti-MT antibody (E). Lysate lanes represent 1/10 of the sample used for purification. (F) Diagram of characterized eIF4G binding sites. See the text for details. aa, amino acids.
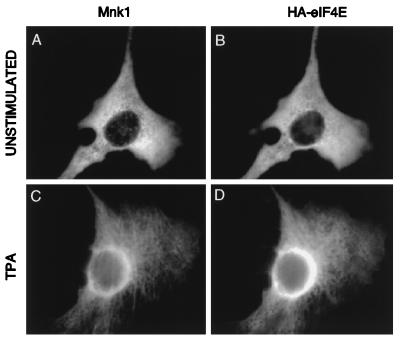
FIG. 2.
Subcellular localization of Mnk1 and eIF4E. NIH 3T3 cells were transfected with pCS2-Mnk1 and pSRα-eIF4E. Untagged Mnk1 and HA-eIF4E were visualized with an affinity-purified chicken-anti-Mnk1 antibody and 12CA5 (anti-HA) ascites fluid, followed by fluorescein-conjugated anti-chicken antibody and Texas red-conjugated anti-mouse antibody. (A and C) Mnk1 immunofluorescence; (B and D) HA-eIF4E. Cells were either serum starved (A and B) or treated with phorbol ester for 60 min (C and D).
To test whether eIF4GI, like eIF4GII, would bind Mnk1 and to test whether the binding is direct, we mixed purified eIF4GI with a GST fusion protein encoding full-length Mnk1. In this experiment, full-length eIF4G and a breakdown product bound to GST-Mnk1 and to a GST-Mnk1 mutant lacking two predicted phosphorylation sites (T197A/T202A, denoted T2A2 [see below]) (Fig. 1B). To determine whether Mnk1 binds to eIF4G in cells, we transiently transfected 293-HEK cells with a plasmid encoding GST-Mnk1. GST-Mnk1 was recovered with glutathione-Sepharose (Fig. 1C), and associated eIF4GI was detected by immunoblotting (Fig. 1D). In cells overexpressing Mnk1, 5% of the endogenous eIF4GI copurified with Mnk1. Deletion of the N-terminal 23 residues abrogated this interaction, as expected from the yeast two-hybrid experiment (Fig. 1D).
The binding sites on eIF4G for eIF4E, eIF4A, and eIF3 have been mapped (27, 38, 44, 50). eIF4E binds residues 319 to 479, eIF3 binds 480 to 886, and eIF4A binds 887 to 1402 (numbering from rabbit eIF4GI) (Fig. 1F). A Mnk1-binding site is contained in the two-hybrid isolate, residues 1210 to 1402. This implies that Mnk1 may bind to eIF4G without displacing eIF4E or eIF3, although it may interfere with eIF4A binding. To test whether Mnk1 complexes contain eIF4E, we cotransfected cells with vectors encoding GST-Mnk1 and myc-tagged (MT)-eIF4E. A small fraction of MT-eIF4E was found associated with GST-Mnk1 (Fig. 1E). This eIF4E-Mnk1 interaction is likely to be partly mediated by eIF4G, since the N-terminal deletion mutant of Mnk1 which does not bind eIF4G has reduced binding of eIF4E (Fig. 1E). However, there is also eIF4G-independent binding of eIF4E to the N-terminal mutant of Mnk1. This alternative mode of Mnk1-eIF4E binding may occur via another eIF4F component, because Mnk1 and eIF4E do not interact in the yeast two-hybrid system (data not shown). It remains to be determined whether Mnk1 also interacts with eIF4A and eIF3.
Subcellular localization of Mnk1.
The initiation factors eIF4E and eIF4G are localized predominantly in the cytoplasm, with 12 to 25% of eIF4E being found in the nucleus (40, 42). To determine the subcellular distribution of Mnk1 within cells, we expressed untagged Mnk1 and HA-tagged eIF4E in NIH 3T3 cells and examined their localization by indirect immunofluorescence. Anti-Mnk1 antiserum did not stain untransfected cells significantly. When Mnk1 was overexpressed, the bulk of Mnk1 localized in the cytoplasm in serum-starved cells and a small population was found within the nucleus (Fig. 2A). Upon stimulation with TPA, Mnk1 shifted to a perinuclear ring (Fig. 2C). HA-eIF4E was also mostly cytoplasmic and shifted to a perinuclear ring in a similar way after TPA stimulation (Fig. 2B and D). The shift in localization of both Mnk1 and eIF4E after TPA treatment could reflect a general reorganization of the cytoplasm or a change in the localization of a subset of proteins. Under both unstimulated and stimulated conditions, the majority of Mnk1 and eIF4E colocalize, consistent with the binding of Mnk1 to eIF4G and eIF4E. To confirm these data, we fractionated extracts of cells expressing Mnk1 or HA-eIF4E. Both Mnk1 and eIF4E were present predominantly within the cytosolic compartment, consistent with our immunofluorescence data (data not shown).
Phosphorylation site substitution mutants of Mnk1.
The binding of Mnk1 to eIF4G and eIF4E and the in vitro phosphorylation of eIF4E by Mnk1 suggested that Mnk1 may phosphorylate eIF4E in cells. To test this, we designed dominant negative and activated mutants of Mnk1. Because Mnk1 is activated by phosphorylation by ERK or p38 MAP kinases, we reasoned that replacement of regulatory phosphorylation sites by nonphosphorylated or acidic residues might inhibit or activate Mnk1, respectively. We examined the Mnk1 sequence for likely MAP kinase phosphorylation sites. The MAP kinase consensus, ψX[S/T]P, where ψ is proline or aliphatic (1, 7), occurs three times in Mnk1, at Thr 197, Thr 202, and Thr 332. Thr 197 and Thr 202 are located within the T-loop or activation loop (30) of Mnk1, and Thr 332 is close to the COOH terminus. We tested whether these residues are phosphorylated in vivo by peptide mapping.
GST-Mnk1 was expressed by transient transfection of 293 cells, which were then serum starved and labeled with [32P]orthophosphate. Before cell lysis, cultures were treated with mitogenic (TPA) or stress (NaCl) stimuli. GST-Mnk1 was recovered on glutathione-Sepharose and digested with proteases. To distinguish between phosphorylation at Thr 197 and Thr 202, which are contained in the same predicted tryptic peptide, trypsin digests were further treated with thermolysin. Phosphopeptides were separated by thin-layer electrophoresis and chromatography and detected by autoradiography.
Wild-type Mnk1 is phosphorylated on two major and several minor tryptic and thermolytic phosphopeptides (Fig. 3A). The major peptides (peptides 2 and 7 in Fig. 3F) contained phosphothreonine and phosphoserine, respectively (Fig. 3E, top). Stimulation of cells with either TPA or NaCl resulted in the increased labeling of phosphopeptides 1, 3, 4, 5, and 6 (Fig. 3B, C, and F). Phosphoamino acid analysis of individual phosphopeptides showed that peptides 1 to 3 contained phosphothreonine while peptides 4 to 7 contained phosphoserine (Fig. 3E, top).
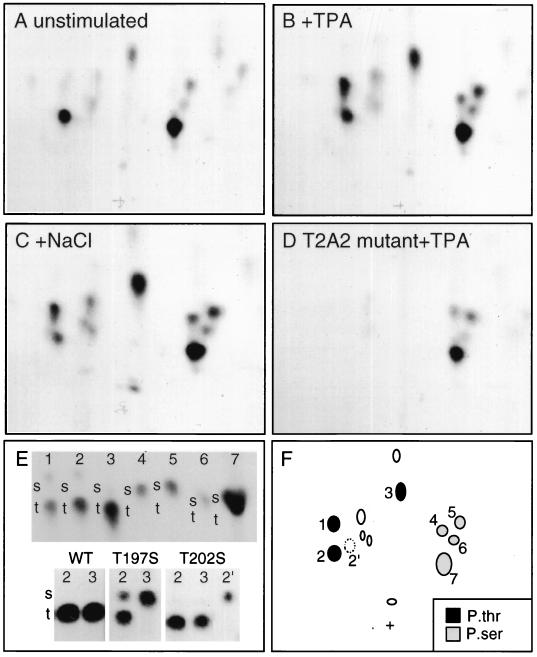
FIG. 3.
Phosphopeptide analysis of Mnk1. 293 cells were transfected with wild-type Mnk1 (A to C and E), T197A/T202A (T2A2) mutant Mnk1 (D), or various mutants (E) of EBG-Mnk1 and metabolically labeled with [32P]orthophosphate. The cells were serum starved (A and E) or stimulated with 100 nM TPA for 15 min (B, D, and E) or 0.4 M NaCl (C) for 30 min. GST-Mnk1 was purified by glutathione-Sepharose and SDS-PAGE and digested with trypsin and thermolysin. Phosphopeptides were resolved by electrophoresis and chromatography and detected by autoradiography (A to D). (E) The Phosphoamino acid content of individual phosphopeptides was determined. The top panel shows phosphoamino acid analysis of phosphopeptides 1 to 7; the bottom panel shows phosphoamino acid analysis of phosphopeptides 2 and 3 from wild-type (WT), T197S, and T202S mutants of Mnk1. Radioactive phosphoamino acids were detected by autoradiography, and the positions of nonradioactive internal standards, phosphoserine (s), and phosphothreonine (t) were detected with ninhydrin. (F) Schematic showing phosphoserine-containing (grey symbols), phosphothreonine-containing (black symbols), and unanalyzed (open symbols) phosphopeptides. Phosphopeptide 2′ was detected in maps of T202S mutant only.
To identify which phosphopeptides correspond to the MAP kinase consensus sites, we created GST-Mnk1 mutants with Thr 197, Thr 202, or Thr 332 individually or combinatorially replaced with alanine or serine and labeled the proteins with [32P]orthophosphate in 293 cells. A mutant (T2A2), containing both Thr 197 and Thr 202 replaced with Ala, was not phosphorylated at peptides 1, 2, or 3 in control or TPA- or NaCl-treated cells (Fig. 3D and data not shown). This suggests that peptides 1 to 3 either contain Thr 197 or Thr 202 or are dependent on them for their phosphorylation. In contrast, all phosphopeptides were detected in a T332A mutant (data not shown). When Thr 197 or Thr 332 was individually replaced with Ser, the phosphopeptide maps were indistinguishable from that of wild-type Mnk1 (data not shown). Phosphopeptides 1 to 3 were phosphorylated only on threonine in the T332S mutant (data not shown). This suggests that residue 332 is not a phosphorylation site. In contrast, phosphoamino acid analysis of individual phosphopeptides showed that replacement of Thr 197 with Ser (T197S) resulted in an increased phosphoserine content in peptide 2 and a total shift from phosphothreonine to phosphoserine in peptide 3 (Fig. 3E, bottom). This suggests that residue 197 is a phosphorylation site in Mnk1. Peptide 2 may actually be a mixture of two peptides, one phosphorylated at serine and one phosphorylated at threonine in the T197S mutant. Similar analysis of a T202S mutant showed a novel peptide 2′ (Fig. 3F) that was exclusively phosphorylated on serine (Fig. 3E, bottom). This suggests that the tryptic or thermolytic peptide 2 is composed of two distinct, comigrating peptides, one containing Thr 197 and one containing Thr 202. Peptide 3 contains only Thr 197. These results indicate that Mnk1 is phosphorylated at Thr 197 and Thr 202 but is probably not phosphorylated at Thr 332.
Effects of phosphorylation site mutants on Mnk1 kinase activity.
The kinase activities of substitution mutants of Mnk1 were assessed by expressing wild-type or mutant GST-Mnk1 in 293 cells. Cells were unstimulated or stimulated with TPA before purification of GST-Mnk1 and assay with recombinant eIF4E substrate (Fig. 4B). Expression levels were assessed by Western blotting and found to be comparable (Fig. 4A). When incubated with radioactive ATP, all mutants were autophosphorylated or phosphorylated by a contaminating kinase, but the mutants differed in their ability to phosphorylate eIF4E (Fig. 4B). Wild-type GST-Mnk1 exhibits TPA-stimulated eIF4E phosphorylating activity, whereas two mutants lacking the T-loop threonines, T197A/T202A (T2A2) and T197A/T202A/T332A (T3A3), were unable to phosphorylate eIF4E in vitro (Fig. 4B) (71). Consistent with our negative evidence for phosphorylation at Thr 332, replacement of Thr 332 with Ala did not inactivate Mnk1 and actually caused partial activation (T332A) (Fig. 4B).
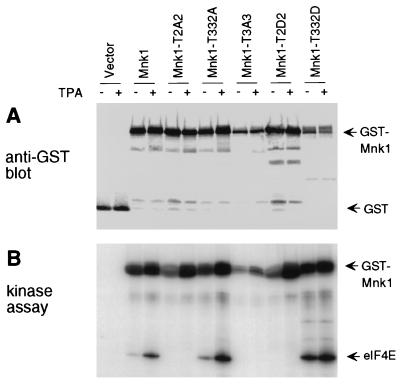
FIG. 4.
Expression and kinase activity of Mnk1 mutants. GST-Mnk1 and mutants T2A2 (T197A/T202A), T332A, T3A3 (T197A/T202A/T332A), T2D2 (T197D/T202D), and T332D were synthesized in 293 cells. Transfected cells were serum starved and treated with TPA or left untreated. Mnk1 was purified with glutathione-Sepharose. (A) Samples were immunoblotted with polyclonal anti-GST antibody. (B) The same samples were incubated with radiolabeled ATP and eIF4E. Products were detected by SDS-PAGE and autoradiography.
In an attempt to activate GST-Mnk1 further, the T-loop phosphorylation sites, Thr 197 and Thr 202, were replaced with Asp. However, this mutant, T197D/T202D (T2D2), was inactive in phosphorylation of eIF4E in vitro (Fig. 4B), suggesting that introduction of acidic residues does not functionally substitute for phosphorylation of these sites. Surprisingly, altering Thr 332 to Asp stimulated both basal and TPA-dependent eIF4E phosphorylating activity (T332D) (Fig. 4). This unexpected activation may be a result of a posttranslational modification, such as phosphorylation, because GST-Mnk1-T332D expressed in E. coli is inactive unless incubated with ERK (data not shown). Thus, the activation of the T332D mutant is probably due to structural changes that alter the access of other sites for phosphorylation. Whatever the explanation of the increased activity of the T332D mutant, this mutant provides a useful tool to study the effects of deregulated Mnk1 activity in cells.
Mnk1 regulates eIF4E phosphorylation in cells.
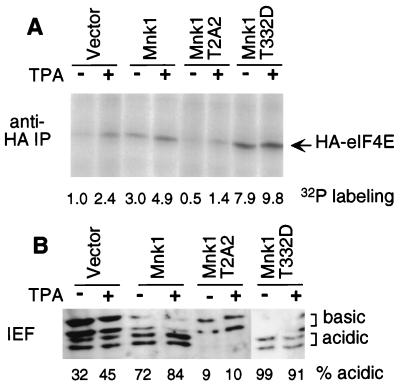
To determine the ability of Mnk1 to phosphorylate eIF4E in vivo, we cotransfected 293 cells with HA-tagged eIF4E and wild-type or mutant GST-Mnk1. The phosphorylation status of eIF4E was analyzed either by metabolic labeling with [32P]orthophosphate labeling or by isoelectric focusing and Western blotting (Fig. 5).
FIG. 5.
Phosphorylation of eIF4E in cells overexpressing Mnk1. 293 cells were transfected with pSRα-eIF4E and either vector pEBG, wild-type Mnk1, Mnk1-T2A2 (T197A/T202A), or Mnk1-T332D. (A) Cells were serum starved, metabolically labeled with 32P, and treated with TPA or left untreated. HA-eIF4E was isolated by immunoprecipitation (IP) and detected by SDS-PAGE and autoradiography. Incorporation was quantified with a PhosphorImager and normalized to incorporation in unstimulated cells expressing vector DNA. (B) Cells were serum starved and treated with TPA or left untreated. HA-eIF4E was purified with m7GTP-Sepharose and subjected to one-dimensional isoelectric focusing (IEF) and Western blotting with anti-HA antibodies. The amounts of the two basic and two acidic species were quantified, yielding a percentage value for acidic species for each sample (indicated below the gel). The inhibition of HA-eIF4E phosphorylation by T2A2 mutant Mnk1 was confirmed by using the pCS3+MT vector, which expresses MT-Mnk1-T2A2 (data not shown).
Epitope-tagged HA-eIF4E was recovered from [32P]orthophosphate-labeled cells by immunoprecipitation, resolved by SDS-PAGE, and quantified (Fig. 5A). Control experiments showed that the expression of HA-eIF4E was not affected by coexpression of wild-type or mutant GST-Mnk1 (data not shown; see also Fig. 6). Phosphate incorporation into HA-eIF4E was enhanced 2.4-fold by TPA treatment of cells that were not overexpressing Mnk1 (Fig. 5A). Expression of either wild-type or activated T332D mutant GST-Mnk1 led to a three- to eightfold increase in eIF4E labeling in the absence of TPA stimulation, suggesting that increased basal or constitutive mutant Mnk1 activity stimulates eIF4E phosphorylation in vivo. These increases were approximately additive with the increase due to TPA stimulation, suggesting that overexpression of Mnk1 does not interfere with the endogenous activation process. Expression of the inactive T197A/T202A (T2A2) mutant Mnk1 suppressed both the basal and TPA-stimulated phosphate incorporation into eIF4E, suggesting that this mutant is dominant negative and interferes with the normal mechanism of eIF4E phosphorylation in vivo.
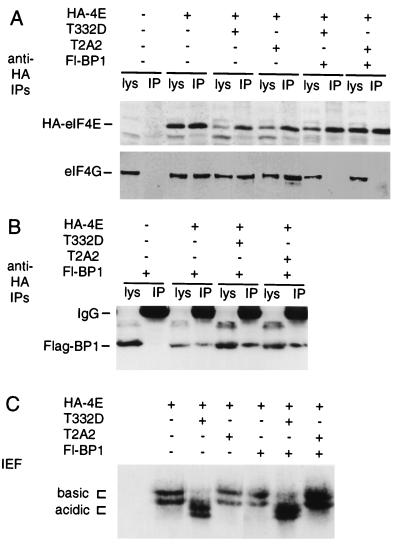
FIG. 6.
Effect of Mnk1 overexpression on complex formation and phosphorylation of eIF4E. 293 cells were transfected with vectors encoding HA-eIF4E, Flag-4EBP1, and GST-Mnk1-T2A2 (T197A/T202A) or GST-Mnk1-T332D. Proteins were purified by immunoprecipitation (IP) with antibody to HA (12CA5 monoclonal antibody) or with m7GTP-Sepharose and subjected to SDS-PAGE (A and B) or isoelectric focusing (IEF) (C) and Western blotting with antibodies to HA (A and C), eIF4G (A), or Flag (B). All samples are from the same experiment, which was repeated with similar results. Note that basal eIF4E phosphorylation was low, so that inhibitory effects of GST-Mnk1-T2A2 were not detectable. lys, lysate.
To determine whether this increase in eIF4E kinase activity resulted in increased steady-state levels of eIF4E phosphorylation, transfected HA-eIF4E was purified from unlabeled cells by using m7GTP-Sepharose and analyzed by isoelectric focusing followed by Western blotting (Fig. 5B). This method resolved two species of endogenous eIF4E, an acidic phosphorylated form and a basic nonphosphorylated form (data not shown). When anti-HA antibodies were used, two acidic and two basic species, corresponding to phosphorylated and nonphosphorylated forms of HA-eIF4E, were detected (Fig. 5B). We assume that the splitting of each form into two is an artifact of the HA tag. In unstimulated cells, approximately one-third of HA-eIF4E was acidic, and this increased to 45% following TPA stimulation. Coexpression of wild-type Mnk1 increased the stoichiometry of eIF4E acidic isoforms to approximately 70%, while the activated T332D mutant caused an increase to 95%. The expression of the dominant interfering mutant, T197A/T202A (T2A2), reduced the stoichiometry to approximately 10% and inhibited the response to TPA.
These results show that overexpression of wild-type or activated mutant Mnk1 increases eIF4E phosphorylation stoichiometry as well as phosphate incorporation. The nonproportionality between phosphorylation stoichiometry and phosphate incorporation assays is consistent with non-steady-state phosphate labeling and an increase in dephosphorylation as well as phosphorylation when Mnk1 is overexpressed (12). Overexpression of the inactive mutant of Mnk1 decreases the rate and stoichiometry of eIF4E phosphorylation, suggesting a dominant negative effect on the endogenous eIF4E kinase.
Effects of overexpressed activated mutant Mnk1 in cells overexpressing 4EBP1.
The preceding results indicate that increased Mnk1 activity leads to increased phosphorylation of eIF4E in transfected cells. However, these experiments had made use of overexpressed, epitope-tagged eIF4E to assay the phosphorylation state of eIF4E in the cells that were overexpressing Mnk1. Thus, the normal balance between eIF4E and 4EBP1 levels was not maintained in the transfected cells. To determine whether increased Mnk1 activity would also increase eIF4E phosphorylation in cells where eIF4E is associated with 4EBP1 and to test whether eIF4E phosphorylation altered the binding of eIF4E to either eIF4G or 4EBP1, we cotransfected 293 cells with various combinations of plasmids expressing mutant GST-Mnk1, HA-tagged eIF4E, and Flag-tagged 4EBP1 (18) and analyzed HA-eIF4E complexes and phosphorylation state (Fig. 6).
As expected, immunoprecipitation with anti-HA antibodies efficiently recovered HA-eIF4E (Fig. 6A, top; compare lysate and immunoprecipitate). Endogenous eIF4G was recovered in the anti-HA immunoprecipitates only when cells were co-transfected with HA-eIF4E (Fig. 6A, bottom). Coexpression of dominant negative (T2A2) or activated (T332D) mutant GST-Mnk1 did not significantly alter the coprecipitation of eIF4G with HA-eIF4E. However, as expected (61), overexpression of 4EBP1 competed with eIF4G for binding to HA-eIF4E, so that eIF4G was not present in anti-HA immunoprecipitates when Flag-4EBP1 was overexpressed. This shows that the expression level of Flag-4EBP1 was sufficient to sequester most of the HA-eIF4E in inactive complexes. Flag-4EBP1 was efficiently recovered in anti-HA immunoprecipitates only when cells were co-transfected with HA-eIF4E (Fig. 6B, bottom). Neither the binding of 4EBP1 nor the displacement of eIF4G from HA-eIF4E was significantly altered by coexpression of dominant negative (T2A2) or activated (T332D) mutant GST-Mnk1 (Fig. 6A and B).
Isoelectric focusing and anti-HA Western blotting showed that phosphorylation of HA-eIF4E was strongly stimulated by overexpression of activated mutant GST-Mnk1 T332D (Fig. 6C), even in the presence of 4EBP1 in amounts sufficient to displace all detectable eIF4G from HA-eIF4E. This suggests that activated Mnk1 can drive high-level phosphorylation of eIF4E even in cells where most eIF4E is present in complexes with 4EBP1. Thus, Mnk1 activation may lead to eIF4E phosphorylation independent of the release of eIF4E from 4EBP1 complexes. Furthermore, changes in the eIF4E phosphorylation state do not alter the stability of eIF4E-4EBP1 or eIF4E-eIF4G complexes.
DISCUSSION
Previous studies have shown that the cap-binding protein eIF4E is phosphorylated at a single site, Ser 209, when cells are stimulated with a variety of mitogens or stresses, but the kinase responsible has not been identified. The ERK and p38 MAP kinase-activated protein kinase, Mnk1, is able to phosphorylate eIF4E in vitro at Ser 209 (71). We have now found that Mnk1 is bound to the eIF4E-associated scaffold eIF4G and regulates the phosphorylation state of eIF4E in transfected cells. This suggests that endogenous Mnk1 may phosphorylate eIF4E in response to mitogen and stress stimuli. We have also found that Mnk1 can lead to extensive eIF4E phosphorylation under conditions where most eIF4E is associated with the inhibitor, 4EBP1. This suggests that Mnk1 can operate independently of 4EBP1 phosphorylation and may explain how eIF4E becomes phosphorylated in stressed cells, where 4EBP1 is not phosphorylated (51, 70).
The detection of the C-terminal region of eIF4G as a binding partner for Mnk1 in an unbiased yeast two-hybrid screen suggests that Mnk1 has a high specificity for binding eIF4G in cells. eIF4E could be copurified with Mnk1 and eIF4G from transfected cells. The proximity of Mnk1 to eIF4E bound to eIF4G suggests that Mnk1 is likely to phosphorylate eIF4E even when not overexpressed. Mnk1 binds to eIF4G independently of the C terminus of Mnk1, to which ERK and p38 MAP kinases bind. Thus, it seems likely that a complex of Mnk1 and MAP kinase is associated with eIF4G in cells. Together with the specificity of Mnk1 for the physiological phosphorylation site in eIF4E in vitro (71) and the effects of overexpressing Mnk1 mutants, it is highly likely that Mnk1 phosphorylates eIF4E in vivo when ERK or p38 is activated. In addition, Mnk1 may phosphorylate other components of the eIF4F complex. This possibility remains to be explored.
The second protein found to bind to Mnk1 in the yeast two-hybrid screen was α-importin. α-Importin binding requires the N-terminal basic region of Mnk1, which resembles a nuclear localization signal. However, our immunofluorescence experiments suggest that the large majority of Mnk1 is cytosolic in fibroblasts, under unstimulated and stimulated conditions. Mnk1 redistributed to the perinuclear region in TPA-stimulated cells but still appeared to be outside the nucleus (Fig. 2). It remains to be determined whether Mnk1 enters the nucleus under certain conditions or perhaps shuttles rapidly out of the nucleus once it enters. We also found that the majority of epitope-tagged eIF4E is in the cytoplasm, either by viewing immunofluorescence or by cell fractionation. While the cytoplasmic localization of eIF4E is consistent with an Mnk1-eIF4G-eIF4E complex, it is not fully consistent with the results of a previous study of eIF4E localization (42). Those authors found that a monoclonal antibody to eIF4E stained the nucleus more intensely than it stained the cytoplasm. In addition, biochemical fractionation showed that about 12 to 25% of the protein was nuclear and the remainder was split equally between perinuclear membranes and the cytosol (42). It is possible that perinuclear eIF4E appears to be inside the nucleus under some conditions. Newly synthesized capped RNA is bound to a distinct cap-binding protein inside the nucleus (28) and may be transferred to perinuclear eIF4E during export.
Biochemical and mutational analysis shows that phosphorylation of Mnk1 occurs in mitogen- and stress-stimulated cells at a common set of serine and threonine residues. We found that Thr 197 and probably Thr 202, both in the T loop of the Mnk1 kinase domain, are phosphorylated at increased levels in mitogen- or stress-stimulated cells. These sites both lie in the known MAP kinase phosphorylation consensus, ψX[S/T]P, and thus are likely to be phosphorylated directly by ERK and p38 MAP kinases. The MAP kinase-activated protein kinases, Rsk and MAPKAPK-2, are similarly phosphorylated in their T loops by ERK and p38, respectively (2, 8). T-loop phosphorylation is critical for activation of a variety of protein kinases, and in some cases, replacement of T-loop phosphorylation site residues with acidic residues is sufficient for activation (30). However, replacement of Thr 197 and Thr 202 with aspartate did not activate Mnk1. Instead, replacement of a C-terminal residue, Thr 332, which does not appear to be phosphorylated, is sufficient for Mnk1 activation when expressed in mammalian cells. This residue may be critical for maintaining an inhibitory conformation, since small C-terminal deletions also activate Mnk1 (61a). It is likely that the activation of Mnk1 mutant T332D involves phosphorylation, since this mutant is activated further by mitogens and is not activated when made in bacteria. Detailed analysis of Rsk and MAPKAPK-2 has shown that phosphorylation at several sites in the C- and N-terminal noncatalytic regions contributes to activity (2, 8). Additional phosphorylation sites in Mnk1 may well regulate activity and remain to be identified.
Previous evidence has indicated that eIF4E phosphorylation depends on ERK MAP kinase in mitogen-stimulated cells and on p38 MAP kinase in stressed cells (14–16, 49, 51, 70). Mnk1 binds to and is activated by the ERK and p38 MAP kinases in vitro and in transfected cells (17, 71). Of the MAP kinase-activated protein kinases tested, Mnk1, its relative Mnk2, and MAPKAPK-3 can specifically phosphorylate eIF4E at Ser 209 in vitro (51, 70a, 71). MAPKAPK-3 was reported to be activated by the ERK, p38, and Jun N-terminal kinase (JNK) classes of MAP kinases (43). Since JNK is activated by stresses and since blockade of p38 but not JNK inhibits eIF4E phosphorylation in stressed cells (51, 70), it is unlikely that a JNK-stimulated protein kinase such as MAPKAPK-3 phosphorylates eIF4E in vivo. Our new results indicate that Mnk1 can phosphorylate eIF4E in cells when overexpressed, and they implicate Mnk1 in stimulating eIF4E phosphorylation in mitogen-stimulated 293 cells. However, it is also possible that a kinase other than Mnk1 is the physiological eIF4E kinase in other cell types or under other stimulation conditions. By binding to the C terminus of eIF4G, the kinase-defective T197A/T202A (T2A2) mutant Mnk1 may displace the physiological eIF4E kinase and inhibit eIF4E phosphorylation. One obvious candidate for another in vivo eIF4E kinase is Mnk2. However, it is unlikely that Mnk2 is responsible for stress activation of eIF4E phosphorylation, since Mnk2 is not activated by p38 MAP kinase (71).
The relationship between eIF4E phosphorylation and 4EBP1 phosphorylation is complex. As discussed above, inhibitor studies indicate that different signal transduction pathways activate the kinases that phosphorylate eIF4E and 4EBP1. In particular, rapamycin inhibits 4EBP1 phosphorylation (61). However, Mendez et al. (47) showed that insulin-stimulated phosphorylation of eIF4E is inhibited by rapamycin and Morley and McKendrick (51) showed that rapamycin inhibited serum-induced eIF4E phosphorylation. On the other hand, the latter authors also showed that rapamycin did not inhibit stress (anisomycin)-induced eIF4E phosphorylation (51). Wang et al. (70) found that 4EBP1 reduced eIF4E phosphorylation by Mnk1 in vitro, although they used large quantities of 4EBP1. We have now found that excess 4EBP1 did not prevent eIF4E phosphorylation by overexpressed Mnk1. This suggests either that Mnk1 can phosphorylate the eIF4E-4EBP1 complex or that free eIF4E is phosphorylated by Mnk1 and exchanges with unphosphorylated eIF4E in the 4EBP1 complex. Either way, it is clear that eIF4E can be phosphorylated to high stoichiometry when Mnk1 is activated in the absence of 4EBP1 phosphorylation. Our experiments do not address the possibility that eIF4E bound to the eIF4G-Mnk1 complex is phosphorylated faster than the free eIF4E or eIF4E bound to 4EBP1.
In conclusion, we have shown that Mnk1 binds to the C terminus of eIF4G and that overexpressed activated Mnk1 increases the stoichiometry of eIF4E phosphorylation in transfected cells, even in the presence of excess 4EBP1. Mnk1 activation correlates with stress- and mitogen-stimulated eIF4E phosphorylation, and dominant-negative Mnk1 interferes with mitogen-stimulated and basal eIF4E phosphorylation, suggesting that Mnk1 regulates eIF4E phosphorylation independently of 4EBP1 phosphorylation in vivo.
ACKNOWLEDGMENTS
We are very grateful to A.-C. Gingras, A. Vojtek, D. Turner, Y. Gotoh, C. Proud, L. Breeden, I. Scheffler, L. Shantz, E. Foulstone, and A. Geballe for reagents; R. Fukunaga, T. Hunter, N. Sonenberg, and C. Proud for interesting discussions; and B. Howell, S. Morris, and C. Sachsenmaier for their comments on the manuscript.
This work was supported by NIH grants R01-CA-73987 (to J.A.C.) and R01-DK-13499 (to S.R.K.).
REFERENCES
- 1.Alvarez E, Northwood I C, Gonzalez E A, Latour D A, Seth A, Abate C, Curran T. Pro-Leu-Ser/Thr-Pro is a consensus primary sequence for substrate phosphorylation. Characterization of the phosphorylation of c-myc and c-jun proteins by an epidermal growth factor receptor threonine 669 kinase. J Biol Chem. 1991;266:15277–15285. [PubMed] [Google Scholar]
- 2.Ben-Levy R, Leighton I A, Doza Y N, Attwood P, Morrice N, Marshall C J, Cohen P. Identification of novel phosphorylation sites required for activation of MAPKAP kinase-2. EMBO J. 1995;14:5920–5930. doi: 10.1002/j.1460-2075.1995.tb00280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyle W J, van der Geer P, Hunter T. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 1991;201:110–148. doi: 10.1016/0076-6879(91)01013-r. [DOI] [PubMed] [Google Scholar]
- 4.Brunn G J, Hudson C C, Sekulic A, Williams J M, Hosoi H, Houghton P J, Lawrence J C, Jr, Abraham R T. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science. 1997;277:99–101. doi: 10.1126/science.277.5322.99. [DOI] [PubMed] [Google Scholar]
- 5.Bu X, Haas D W, Hagedorn C H. Novel phosphorylation sites of eukaryotic initiation factor-4F and evidence that phosphorylation stabilizes interactions of the p25 and p220 subunits. J Biol Chem. 1993;268:4975–4978. [PubMed] [Google Scholar]
- 6.Burnett P E, Barrow R K, Cohen N A, Snyder S H, Sabatini D M. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc Natl Acad Sci USA. 1998;95:1432–1437. doi: 10.1073/pnas.95.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark-Lewis I, Sanghera J S, Pelech S L. Definition of a consensus sequence for peptide substrate recognition by p44mpk, the meiosis-activated myelin basic protein kinase. J Biol Chem. 1991;266:15180–15184. [PubMed] [Google Scholar]
- 8.Dalby K N, Morrice N, Caudwell F B, Avruch J, Cohen P. Identification of regulatory phosphorylation sites in mitogen-activated protein kinase (MAPK)-activated protein kinase-1a/p90rsk that are inducible by MAPK. J Biol Chem. 1998;273:1496–1505. doi: 10.1074/jbc.273.3.1496. [DOI] [PubMed] [Google Scholar]
- 9.Deak M, Clifton A D, Lucocq J M, Alessi D R. Mitogen- and stress-activated protein kinase-1 (MSK1) is directly activated by MAPK and SAPK2/p38, and may mediate activation of CREB. EMBO J. 1998;17:4426–4441. doi: 10.1093/emboj/17.15.4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Benedetti A, Rhoads R E. Overexpression of eukaryotic protein synthesis initiation factor 4E in HeLa cells results in aberrant growth and morphology. Proc Natl Acad Sci USA. 1990;87:8212–8216. doi: 10.1073/pnas.87.21.8212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duncan R, Milburn S C, Hershey J W. Regulated phosphorylation and low abundance of HeLa cell initiation factor eIF-4F suggest a role in translational control. Heat shock effects on eIF-4F. J Biol Chem. 1987;262:380–388. [PubMed] [Google Scholar]
- 12.Ferrell J E., Jr Tripping the switch fantastic: how a protein kinase cascade can convert graded inputs into switch-like outputs. Trends Biochem Sci. 1996;21:460–466. doi: 10.1016/s0968-0004(96)20026-x. [DOI] [PubMed] [Google Scholar]
- 13.Flynn A, Proud C G. Serine 209, not serine 53, is the major site of phosphorylation in initiation factor eIF-4E in serum-treated Chinese hamster ovary cells. J Biol Chem. 1995;270:21684–21688. doi: 10.1074/jbc.270.37.21684. [DOI] [PubMed] [Google Scholar]
- 14.Flynn A, Proud C G. Insulin and phorbol ester stimulate initiation factor eIF-4E phosphorylation by distinct pathways in Chinese hamster ovary cells overexpressing the insulin receptor. Eur J Biochem. 1996;236:40–47. doi: 10.1111/j.1432-1033.1996.00040.x. [DOI] [PubMed] [Google Scholar]
- 15.Flynn A, Proud C G. Insulin-stimulated phosphorylation of initiation factor 4E is mediated by the MAP kinase pathway. FEBS Lett. 1996;389:162–166. doi: 10.1016/0014-5793(96)00564-9. [DOI] [PubMed] [Google Scholar]
- 16.Frederickson R M, Mushynski W E, Sonenberg N. Phosphorylation of translation initiation factor eIF-4E is induced in a ras-dependent manner during nerve growth factor-mediated PC12 cell differentiation. Mol Cell Biol. 1992;12:1239–1247. doi: 10.1128/mcb.12.3.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fukunaga R, Hunter T. MNK1, a new MAP kinase-activated protein kinase, isolated by a novel expression screening method for identifying protein kinase substrates. EMBO J. 1997;16:1921–1933. doi: 10.1093/emboj/16.8.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gingras A C, Kennedy S G, Ma O L, Sonenberg N, Hay N. 4E-BP1, a repressor of mRNA translation, is phosphorylated and inactivated by the Akt(PKB) signaling pathway. Genes Dev. 1998;12:502–513. doi: 10.1101/gad.12.4.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorlich D, Kostka S, Kraft R, Dingwall C, Laskey R A, Hartmann E, Prehn S. Two different subunits of importin cooperate to recognize the nuclear localization signals and bind them to the nuclear envelope. Curr Biol. 1995;5:383–392. doi: 10.1016/s0960-9822(95)00079-0. [DOI] [PubMed] [Google Scholar]
- 20.Gorlich D, Mattaj I W. Nucleocytoplasmic transport. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 21.Gorlich D, Prehn S, Laskey R A, Hartmann E. Isolation of a protein that is essential for the first step of nuclear protein import. Cell. 1994;79:767–778. doi: 10.1016/0092-8674(94)90067-1. [DOI] [PubMed] [Google Scholar]
- 22.Gradi A, Imataka H, Svitkin Y V, Rom E, Raught B, Morino S, Sonenberg N. A novel functional human eukaryotic translation initiation factor 4G. Mol Cell Biol. 1998;18:334–342. doi: 10.1128/mcb.18.1.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haghighat A, Mader S, Pause A, Sonenberg N. Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J. 1995;14:5701–5709. doi: 10.1002/j.1460-2075.1995.tb00257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haghighat A, Sonenberg N. eIF4G dramatically enhances the binding of eIF4E to the mRNA 5′-cap structure. J Biol Chem. 1997;272:21677–21680. doi: 10.1074/jbc.272.35.21677. [DOI] [PubMed] [Google Scholar]
- 25.Hara K, Yonezawa K, Kozlowski M T, Sugimoto T, Andrabi K, Weng Q P, Kasuga M, Nishimoto I, Avruch J. Regulation of eIF-4E BP1 phosphorylation by mTOR. J Biol Chem. 1997;272:26457–26463. doi: 10.1074/jbc.272.42.26457. [DOI] [PubMed] [Google Scholar]
- 26.Hiremath L S, Webb N R, Rhoads R E. Immunological detection of the messenger RNA cap-binding protein. J Biol Chem. 1985;260:7843–7849. [PubMed] [Google Scholar]
- 27.Imataka H, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol Cell Biol. 1997;17:6940–6947. doi: 10.1128/mcb.17.12.6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Izaurralde E, Lewis J, McGuigan C, Jankowska M, Darzynkiewicz E, Mattaj I W. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell. 1994;78:657–668. doi: 10.1016/0092-8674(94)90530-4. [DOI] [PubMed] [Google Scholar]
- 29.Jefferies H B J, Fumagalli S, Dennis P B, Reinhard C, Pearson R B, Thomas G. Rapamycin suppresses 5′TOP mRNA translation through inhibition of p70S6K. EMBO J. 1997;16:3693–3704. doi: 10.1093/emboj/16.12.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson L N, Noble M E M, Owen D J. Active and inactive protein kinases: structural basis for regulation. Cell. 1996;85:149–158. doi: 10.1016/s0092-8674(00)81092-2. [DOI] [PubMed] [Google Scholar]
- 31.Joshi B, Cai A L, Keiper B D, Minich W B, Mendez R, Beach C M, Stepinski J, Stolarski R, Darzynkiewicz E, Rhoads R E. Phosphorylation of eukaryotic protein synthesis initiation factor 4E at Ser-209. J Biol Chem. 1995;270:14597–14603. doi: 10.1074/jbc.270.24.14597. [DOI] [PubMed] [Google Scholar]
- 32.Kawasome H, Papst P, Webb S, Keller G M, Johnson G L, Gelfand E W, Terada N. Targeted disruption of p70S6K defines its role in protein synthesis and rapamycin sensitivity. Proc Natl Acad Sci USA. 1998;95:5033–5038. doi: 10.1073/pnas.95.9.5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kimball S R, Horetsky R L, Jagus R, Jefferson L S. Expression and purification of the α subunit of eukaryotic initiation factor eIF2: use as a kinase substrate. Protein Expression Purif. 1998;12:415–419. doi: 10.1006/prep.1998.0863. [DOI] [PubMed] [Google Scholar]
- 34.Kimball S R, Horetsky R L, Jefferson L S. Signal transduction pathways involved in the regulation of protein synthesis by insulin in L6 myoblasts. Am J Physiol. 1998;274:C221–C228. doi: 10.1152/ajpcell.1998.274.1.C221. [DOI] [PubMed] [Google Scholar]
- 35.Kimball S R, Rannels S L, Elensky M B, Jefferson L S. Quantitation of proteins by dot-blot immunoassay: a comparison of visualization methods using eukaryotic initiation factor 2 and a monospecific antibody. J Immunol Methods. 1988;106:217–223. doi: 10.1016/0022-1759(88)90200-1. [DOI] [PubMed] [Google Scholar]
- 36.Kleijn M, Scheper G C, Voorma H O, Thomas A A M. Regulation of translation initiation factors by signal transduction. Eur J Biochem. 1998;253:531–544. doi: 10.1046/j.1432-1327.1998.2530531.x. [DOI] [PubMed] [Google Scholar]
- 37.Koromalis A E, Lazaris-Karatzas A, Sonenberg N. mRNAs containing extensive secondary structure in their 5′ noncoding region translate efficiently in cells overexpressing initiation factor eIF-4E. EMBO J. 1992;11:4153–4158. doi: 10.1002/j.1460-2075.1992.tb05508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lamphear B J, Kirchweger R, Skern T, Rhoads R E. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J Biol Chem. 1995;270:21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- 39.Lamphear B J, Panniers R. Cap binding protein complex that restores protein synthesis in heat-shocked Ehrlich cell lysates contains highly phosphorylated eIF-4E. J Biol Chem. 1990;265:5333–5336. [PubMed] [Google Scholar]
- 40.Lang V, Zanchin N I, Lunsdorf H, Tuite M, McCarthy J E. Initiation factor eIF-4E of Saccharomyces cerevisiae. Distribution within the cell, binding to mRNA, and consequences of its overproduction. J Biol Chem. 1994;269:6117–6123. [PubMed] [Google Scholar]
- 41.Lazaris-Karatzas A, Montine K S, Sonenberg N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature. 1990;345:544–547. doi: 10.1038/345544a0. [DOI] [PubMed] [Google Scholar]
- 42.Lejbkowicz F, Goyer C, Darveau A, Neron S, Lemieux R, Sonenberg N. A fraction of the mRNA 5′ cap-binding protein, eukaryotic initiation factor 4E, localizes to the nucleus. Proc Natl Acad Sci USA. 1992;89:9612–9616. doi: 10.1073/pnas.89.20.9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ludwig S, Engel K, Hoffmeyer A, Sithanandam G, Neufeld B, Palm D, Gaetsel M, Rapp U R. 3pK, a novel mitogen-activated protein (MAP) kinase-activated protein kinase, is targeted by three MAP kinase pathways. Mol Cell Biol. 1996;16:6687–6697. doi: 10.1128/mcb.16.12.6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mader S, Lee H, Pause A, Sonenberg N. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4 gamma and the translational repressors 4E-binding proteins. Mol Cell Biol. 1995;15:4990–4997. doi: 10.1128/mcb.15.9.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marcotrigiano J, Gingras A C, Sonenberg N, Burley S K. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell. 1997;89:951–961. doi: 10.1016/s0092-8674(00)80280-9. [DOI] [PubMed] [Google Scholar]
- 46.Matsuo H, Li H, McGuire A M, Fletcher C M, Gingras A C, Sonenberg N, Wagner G. Structure of translation factor eIF4E bound to m7GDP and interaction with 4E-binding protein. Nat Struct Biol. 1997;4:717–724. doi: 10.1038/nsb0997-717. [DOI] [PubMed] [Google Scholar]
- 47.Mendez R, Myers Jr M G, White M F, Rhoads R E. Stimulation of protein synthesis, eukaryotic translation initiation factor 4E phosphorylation, and PHAS-I phosphorylation by insulin requires insulin receptor substrate-1 and phosphatidylinositol 3-kinase. Mol Cell Biol. 1996;16:2857–2864. doi: 10.1128/mcb.16.6.2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Minich W B, Balasta M L, Goss D J, Rhoads R E. Chromatographic resolution of in vivo phosphorylated and nonphosphorylated eukaryotic translation initiation factor eIF-4E: increased cap affinity of the phosphorylated form. Proc Natl Acad Sci USA. 1994;91:7668–7672. doi: 10.1073/pnas.91.16.7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morley S J. Signalling through either the p38 or ERK mitogen-activated protein (MAP) kinase pathway is obligatory for phorbol ester and T cell receptor complex (TCR-CD3)-stimulated phosphorylation of initiation factor (eIF) 4E in Jurkat T cells. FEBS Lett. 1997;418:327–332. doi: 10.1016/s0014-5793(97)01405-1. [DOI] [PubMed] [Google Scholar]
- 50.Morley S J, Curtis P S, Pain V M. eIF4G: Translation’s mystery factor begins to yield its secrets. RNA. 1997;3:1085–1104. [PMC free article] [PubMed] [Google Scholar]
- 51.Morley S J, McKendrick L. Involvement of stress-activated protein kinase and p38/RK mitogen-activated protein kinase signaling pathways in the enhanced phosphorylation of initiation factor 4E in NIH 3T3 cells. J Biol Chem. 1997;272:17887–17893. doi: 10.1074/jbc.272.28.17887. [DOI] [PubMed] [Google Scholar]
- 52.Nishi N, Morino S, Tomoo K, Youtani T, Ishida T. Expression of a synthetic gene for initiation factor 4E-binding protein 1 in Escherichia coli and its interaction with eIF-4E and eIF-4E × m7GTP complex. J Biochem. 1998;123:157–161. doi: 10.1093/oxfordjournals.jbchem.a021903. [DOI] [PubMed] [Google Scholar]
- 52a.Proud, C. Personal communication.
- 53.Rau M, Ohlmann T, Morley S J, Pain V M. A reevaluation of the cap-binding protein, eIF4E, as a rate-limiting factor for initiation of translation in reticulocyte lysate. J Biol Chem. 1996;271:8983–8990. doi: 10.1074/jbc.271.15.8983. [DOI] [PubMed] [Google Scholar]
- 54.Rinker-Schaeffer C W, Austin V, Zimmer S, Rhoads R E. Ras transformation of cloned rat embryo fibroblasts results in increased rates of protein synthesis and phosphorylation of eukaryotic initiation factor 4E. J Biol Chem. 1992;267:10659–10664. [PubMed] [Google Scholar]
- 55.Rousseau D, Kaspar R, Rosenwald I, Gehrke L, Sonenberg N. Translation initiation of ornithine decarboxylase and nucleocytoplasmic transport of cyclin D1 mRNA are increased in cells overexpressing eukaryotic initiation factor 4E. Proc Natl Acad Sci USA. 1996;93:1065–1070. doi: 10.1073/pnas.93.3.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rudland P S, Jimenez de Asua L. Action of growth factors in the cell cycle. Biochim Biophys Acta. 1979;560:91–133. doi: 10.1016/0304-419x(79)90004-0. [DOI] [PubMed] [Google Scholar]
- 57.Rychlik W, Rush J S, Rhoads R E, Waechter C J. Increased rate of phosphorylation-dephosphorylation of the translational initiation factor eIF-4E correlates with the induction of protein and glycoprotein biosynthesis in activated B lymphocytes. J Biol Chem. 1990;265:19467–19471. [PubMed] [Google Scholar]
- 58.Sachs A B, Sarnow P, Hentze M W. Starting at the beginning, middle and end: translation initiation in eukaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 59.Shantz L M, Hu R H, Pegg A E. Regulation of ornithine decarboxylase in a transformed cell line that overexpresses translation initiation factor eIF-4E. Cancer Res. 1996;56:3265–3269. [PubMed] [Google Scholar]
- 60.Sonenberg N. mRNA 5′ cap-binding protein eIF4E and control of cell growth. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 245–269. [Google Scholar]
- 61.Sonenberg N, Gingras A C. The mRNA 5′ cap-binding protein eIF4E and control of cell growth. Curr Opin Cell Biol. 1998;10:268–275. doi: 10.1016/s0955-0674(98)80150-6. [DOI] [PubMed] [Google Scholar]
- 61a.Stear, J., and A. J. Waskiewicz. Unpublished results.
- 62.Stern B D, Wilson M, Jagus R. Use of nonreducing SDS-PAGE for monitoring renaturation of recombinant protein synthesis initiation factor, eIF-4 alpha. Protein Expression Purif. 1993;4:320–327. doi: 10.1006/prep.1993.1041. [DOI] [PubMed] [Google Scholar]
- 63.Takebe Y, Seiki M, Fujisawa J-I, Hoy P, Yokota K, Arai K-I, Yoshida M, Arai N. SRα promoter: an efficient and versatile mammalian cDNA expression system composed of an SV40 early promoter and the R-U5 segment of human T-cell leukemia virus type I long terminal repeat. Mol Cell Biol. 1988;8:466–472. doi: 10.1128/mcb.8.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tarun S Z, Jr, Sachs A B. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 65.Thomas G, Thomas G, Luther H. Transcriptional and translational control of cytoplasmic proteins after serum stimulation of quiescent Swiss 3T3 cells. Proc Natl Acad Sci USA. 1981;78:5712–5716. doi: 10.1073/pnas.78.9.5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Turner D L, Weintraub H. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev. 1994;8:1434–1447. doi: 10.1101/gad.8.12.1434. [DOI] [PubMed] [Google Scholar]
- 66a.Vojtek, A. Personal communication.
- 67.Vojtek A B, Hollenberg S M, Cooper J A. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 68.von Manteuffel S R, Dennis P B, Pullen N, Gingras A C, Sonenberg N, Thomas G. The insulin-induced signalling pathway leading to S6 and initiation factor 4E binding protein 1 phosphorylation bifurcates at a rapamycin-sensitive point immediately upstream of p70s6k. Mol Cell Biol. 1997;17:5426–5436. doi: 10.1128/mcb.17.9.5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.von Manteuffel S R, Gingras A C, Ming X F, Sonenberg N, Thomas G. 4E-BP1 phosphorylation is mediated by the FRAP-p70s6k pathway and is independent of mitogen-activated protein kinase. Proc Natl Acad Sci USA. 1996;93:4076–4080. doi: 10.1073/pnas.93.9.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang X, Flynn A, Waskiewicz A J, Webb B L, Vries R G, Baines I A, Cooper J A. The phosphorylation of eukaryotic initiation factor eIF4E in response to phorbol esters, cell stresses, and cytokines is mediated by distinct MAP kinase pathways. J Biol Chem. 1998;273:9373–9377. doi: 10.1074/jbc.273.16.9373. [DOI] [PubMed] [Google Scholar]
- 70a.Waskiewicz, A. J. Unpublished results.
- 71.Waskiewicz A J, Flynn A, Proud C G, Cooper J A. Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J. 1997;16:1909–1920. doi: 10.1093/emboj/16.8.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]