Regulation of Fibronectin EDA Exon Alternative Splicing: Possible Role of RNA Secondary Structure for Enhancer Display (original) (raw)
Abstract
The fibronectin primary transcript undergoes alternative splicing in three noncoordinated sites: the cassette-type EDA and EDB exons and the more complex IIICS region. We have shown previously that an 81-nucleotide region within the EDA exon is necessary for exon recognition and that this region contains at least two splicing-regulatory elements: a polypurinic enhancer (exonic splicing enhancer [ESE]) and a nearby silencer element (exonic splicing silencer [ESS]). Here, we have analyzed the function of both elements in different cell types. We have mapped the ESS to the nucleotide level, showing that a single base change is sufficient to abolish its function. Testing of the ESE and ESS elements in heterologous exons, individually or as part of the complete EDA regulatory region, showed that only the ESE element is active in different contexts. Functional studies coupled to secondary-structure enzymatic analysis of the EDA exon sequence variants suggest that the role of the ESS element may be exclusively to ensure the proper RNA conformation and raise the possibility that the display of the ESE element in a loop position may represent a significant feature of the exon splicing-regulatory region.
Alternative splicing is a widespread process used in higher eucaryotes to regulate gene expression. A single primary transcript can generate multiple proteins with distinct functions in a tissue- and/or development-specific manner. Differential pre-mRNA processing occurs via several pathways, including skipped exons, included introns, alternative 5′ and 3′ splice sites, mutually exclusive exons, alternative promoters, and different polyadenylation sites. The intron-exon junction sequences in complex organisms are degenerate, many pre-mRNAs contain multiple introns, and large introns may contain several cryptic splicing sites (16). The selection of splicing sites in constitutively or alternatively spliced pre-mRNAs is determined by parameters such as exon length (3, 32, 41), the presence of splicing enhancer and silencer elements (23), and the strength of splicing signals (16). In some cases the formation of secondary structure participates in the regulation of splice site selection by modifying the physical distances within introns or by being involved in the definition of exons (2, 7, 8, 12, 14, 25, 34, 40).
Considerable progress has been made in recent years in identifying the sequences and the cellular factors that through protein-RNA and protein-protein interactions regulate the alternative splice site selection. However, less attention was given to the RNA structural aspects. The _cis_-acting elements of alternatively spliced exons must be exposed in order to be recognized by the _trans_-acting cellular factors. In the case of the chicken β-tropomyosin gene, the secondary structure of the primary transcript regulates alternative splicing by completely hiding the muscle-specific exon (8). It was recently shown that the Drosophila SR proteins Tra/Tra2 and B52 recognize specific RNA hairpin-loop structures in their target RNAs (17, 33). This type of recognition may be widespread. However, the definition of the structural features of the RNA substrates has not progressed at the same pace as the study of the protein partners of the system.
The fibronectin (FN) gene is a classical example to illustrate genome evolution by exon shuffling (20), generation of protein diversity by alternative splicing (20), and topological coordination between transcription and splicing (9). The FN primary transcript undergoes alternative splicing in three noncoordinated sites (5) that gives rise to 20 polypeptides in humans (for recent reviews, see references 22 and 15). One of the alternatively spliced regions, the EDA exon (also called EIIIA or EDI), is a cassette-type alternatively spliced exon. It is selectively excluded in the FN mRNAs produced by hepatocytes and is included to various extents by other cell types. Mardon et al. (28) found that the inclusion of the EDA exon was totally dependent on the presence of an 81-nucleotide sequence located within the central region of the exon. This region is also active in promoting EDA inclusion in an in vitro splicing assay (24). We have previously reported that this 81-nucleotide region contains a bipartite structure consisting of _cis_-acting exonic sequences that control the alternative splicing of the EDA exon (6). This region is composed of a purine-rich exonic splicing enhancer (ESE) (5′GAAGAAGA3′) plus a second exonic element (5′CAAGG3′) located 13 nucleotides downstream of the ESE, which acts as an exonic splicing silencer (ESS). In fact, deletion of the entire ESS results in constitutive inclusion of the EDA exon. This linear bipartite structure may be an oversimplification, and higher-order RNA structures may be implicated, as was shown for a different region of the EDA exon (35).
In this work we show that both the ESE and ESS regulate splicing of the EDA exon by a common mechanism in different cell types. The mapping of the silencer element has been extended to the nucleotide level, and both the ESE and ESS elements have been tested in heterologous exons. Enzymatic mapping has shown a peculiar secondary structure of the EDA exon that localizes the ESE element in an exposed position as part of a loop region. The ESS seems to be determinant for the RNA conformation, as it lies in a stem structure. We suggest that the secondary structure characterized here mediates proper recognition of the EDA exon by the splicing machinery.
MATERIALS AND METHODS
Plasmid construction.
Constructs pSVED-A Tot, pSVED-A Sac-Stu, pSVED-A Δ2E, pSVED-A Δ4, and pSVED-A Δ3 have been previously described (6).
(i) Constructs pSVED-A ES.1, ES.2, ES.3U, ES.3A, ESE-Mut, and ESE-ESS-Mut.
The _Sal_I/_Sac_I fragment obtained from the vector pSVED-A Tot Sac (6) was cloned into the pUC18 vector, creating the pUC-SS3.9 construct. ES.1, ES.2, ES.3U, ES.3A, ESE-Mut, and ESE-ESS-Mut mutations were obtained by annealing the degenerated primers containing the desired mutations. The mutated synthetic fragments were cloned in _Sac_I-_Stu_I-digested pUC-SS3.9 to obtain intermediate constructs, from which the _Sal_I/_Sac_I fragment was excised and ligated into the previously _Sal_I-_Sac_I-digested pSVED-A Tot Sac vector.
(ii) Constructs pSVED-A ES.4, ES.5, ES.6, and ES.7.
The fragments ES.4, ES.5, ES.6, and ES.7 were obtained by PCR-directed mutagenesis and cloned in the _Stu_I-_Bam_HI-digested M13mp18 EDA Tot (6). From these constructs the _Sal_I/_Bam_HI fragment was excised, blunt ended, and cloned in _Bst_EII-digested pSVα1W vector (18) to obtain the final constructs.
(iii) Constructs pSVαESS+S, ESS+AS, ESS+2cS, ESS−S, and ESS−AS.
pSVαBstINS was obtained by digestion of pSVα1W with _Bst_EII, blunt ending, and insertion of the synthetic fragment SpeINS. The pSVαESS+S, ESS+AS, ESS+2cS, and ESS−S, ESS−AS variants were obtained by insertion of the fragments ESS+ and ESS− inside the _Pml_I-digested pSVαBstINS vector in different orientations (sense and antisense) and in tandem (+3cS). The ESS sequence (5′AGCTGCAAGGCCTCAGACCGG3′) was inserted in sense and antisense orientations.
(iv) B series constructs.
The EDB hybrid minigene construct B1 (see Fig. 3A), which is analogous to the pSVED-A Tot FN–α-globin chimeric minigene, contains the genomic EDB exon region, its flanking introns, and part of the −1 and +1 flanking exons. This region was amplified from human genomic DNA cloned in the pUC18 vector, creating pUC.EDBTot. The insert was fully sequenced, excised, and cloned in the _Bst_EII site of the pSVα1W vector, creating the B1 plasmid. A new _Xho_I site was introduced by PCR-directed mutagenesis 98 bases downstream of the _Kpn_I site of the EDB exon, generating the B2 construct. The B4 plasmid was created by deleting the EDB sequence between the _Kpn_I and _Xho_I sites.
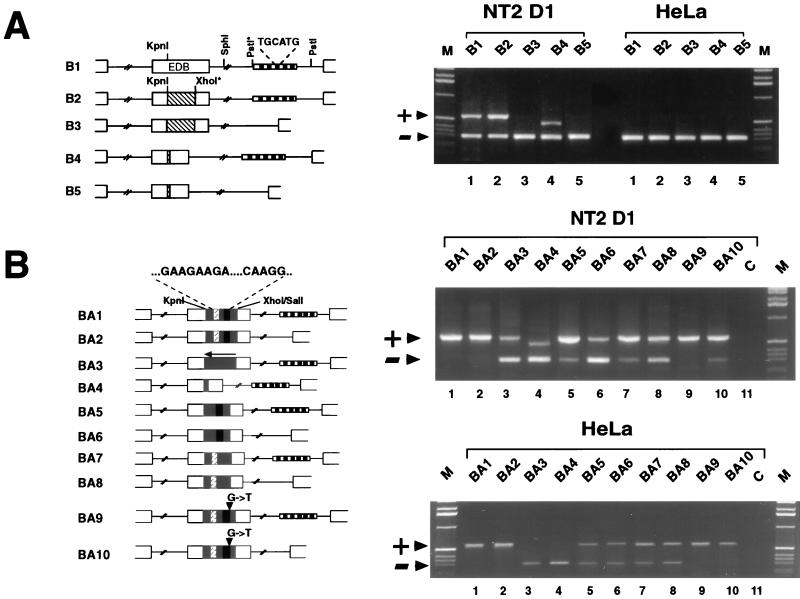
FIG. 3.
The ESS element is not functional in the EDB context even when inserted as a part of the entire EDA splicing-regulatory region. (A) The ISE elements are necessary for EDB inclusion. A diagrammatic representation of the EDB hybrid minigene constructs is on the left. The hatched box inside the EDB exon represents the region of the exon deleted in B4. The boxed region in the intron represents the region deleted in order to remove the ISEs. Restriction enzyme sites marked by asterisks were introduced by means of PCR primers. RT-PCR analysis from cells transfected with EDB hybrid minigenes is shown on the right. The upper band of 560 bp represents inclusion of the EDB exon, while the lower 287-bp band represents skipping of the EDB exon. B4 gives an upper band of 460 bp concordant with the 100-bp _Kpn_I-_Xho_I deletion in the EDB exon. (B) The EDA splicing-regulatory region overrides the ISE requirement of the EDB exon. A schematic representation of the chimeric EDB-EDA hybrid minigene constructs with and without the intron enhancer repeats is shown on the left. The shaded space inside the EDB exon indicates the part of the EDA exon inserted in the EDB exon. The white, hatched box inside this shaded space represents the positive element, and the black box represents the negative element. The arrow on BA3 indicates that the EDA fragment is cloned in the antisense direction. Restriction sites utilized for cloning are indicated. Agarose gel electrophoresis of RT-PCR products from cells transfected with chimeric EDB-EDA hybrid minigenes is shown on the right. The upper 560-bp band represents inclusion of the EDB exon, and the lower 287-bp band represents skipping of the EDB exon. Arrowheads and lanes M and C are as defined for Fig. 1.
(v) BA series constructs.
To prepare the chimeric EDB-EDA constructs (BA series), the respective EDA fragments were amplified with primers bearing _Kpn_I and _Sal_I sites and cloned in _Kpn_I-_Xho_I-digested B4. For the preparation of constructs lacking intron repeats, the 269-bp _Pst_I/_Sph_I fragment in the +1 intron was excised from the construct pUCEDBTot, creating pUCEDBTotΔir. The _Kpn_I/_Bam_HI fragment from this construct was excised and cloned in B1 and B4, creating B3 and B5, respectively. The chimeric BA constructs with intron deletions (BA2, BA6, BA8, and BA10) were prepared from B5 by inserting the respective EDA fragments amplified with primers EDA Kpn (5′TCCAGGTACCGGGTGACCTA3′) and EDA Sal (5′TGACTGTCGACTCAGAACC3′) in the _Kpn_I-_Xho_I cassette.
(vi) EDB-ESE and -ESS series constructs.
To make the EDB-B2-X-ESE, EDB-B2-K-ESE, and EDB-B4-ESE constructs, the oligonucleotides hESEdir (5′CCTGATGGTGAAGAAGACACTGCA3′) and hESErev (5′GTGACTTCTTCACCATCAGGTGCA3′) were annealed and cloned at the _Xho_I, _Kpn_I (_Asp_718), and _Xho_I-_Asp_718 sites (previously made blunt), respectively, of each vector. Orientation and copy number were verified by PCR and DNA sequencing. A similar approach was utilized to obtain the EDB-B2-K-ESS, EDB-B2-X-ESS, and EDB-B4-ESS plasmids by annealing the hESSdir (5′GCAAGGCCTCAGACCGAGCT3′) and hESSrev (5′CGGTCTGAGGCCTTGCAGCT3′) oligonucleotides.
Cell culture and transfection.
HeLa (human cervical carcinoma), Hep3B (human hepatocarcinoma), CaCo2 (human colon carcinoma), SK-N-SH (human neuroblastoma), WI38 Va13 (simian virus 40 [SV40]-transformed human fibroblast), MA104 (simian kidney), and COS1 (SV40-transformed simian kidney) cells were maintained in Dulbecco’s modified Eagle medium supplemented with 10% fetal calf serum, 50 μg of gentamicin per ml, and 4 mM l-glutamine. NT2D1 (human embryonal teratocarcinoma) cells were maintained in Dulbecco’s modified Eagle medium supplemented with 4.5 g of glucose per liter, 10% fetal calf serum, 50 μg of gentamicin per ml, and 4 mM l-glutamine. About 1.6 × 106 cells were transfected with 5 μg of specific plasmid purified by CsCl gradient centrifugation and 3 μg of T-antigen expression plasmid (pβ5′sVBglII) by means of a modified calcium phosphate precipitation method (21) or DOTAP reagent (Boehringer Mannheim) as suggested by the manufacturers. Cells were harvested 36 h after transfection.
RNA preparation and RT-PCR analysis.
Total RNA was prepared from the transfected cells by a single-step extraction method with RNAzol B (TEL-TEST Inc., Friendswood, Tex.). A standard reverse transcription (RT) protocol utilizing Moloney murine leukemia virus supplied by Gibco/BRL was used.
(i) RT-PCR assay for analysis of pSVED-A construct transfection.
cDNA was synthesized by utilizing the specific primer pSVcDNA (5′GGTATTTGGAGGTCAGCA3′). Each cDNA was then amplified by PCR with the primers PSV5′J and PSV3′J as previously described (6).
(ii) RT-PCR assay for analysis of pSVαESS construct transfection.
cDNA from HeLa cells and HepG2 total RNA were synthesized by using the specific primer αglocDNA (5′GTATTTGGAGGTCAGCA3′). The cDNA was amplified by PCR through 28 cycles of 93°C for 45 s, 59°C for 1 min, and 72°C for 30 s. The primers αgloE1-5′ (5′CGCACGCTGGCGAGTATGGT3′) and αgloE3-3′ (5′TCACAGAAGCCAGGAACT3′) were used to discriminate the transfected sequences from the endogenous α-globin messengers.
(iii) RT-PCR assay for analysis of pSVα-B (EDB) and BA (EDA-EDA) construct transfection.
About 2 μg of RNA samples was reverse transcribed in 25-μl reaction mixtures as described above with 25 pM Pharmacia random hexanucleotide primer. FN message derived from the transiently transfected constructs was specifically amplified by the combination of sense primer in the −1 FN EDB exon (F1edb, ACTGCCTGCTGGTGACCTG) and antisense primer in the third exon of α-globin (Glob Rev, GAAGCCAGGAACTTGTCCA). The amplifications were performed in a total volume of 50 μl for a total of 30 cycles of amplification (30 s at 94°C, 30 s at 56°C, and 30 s at 72°C), and 5 μl of the PCR products was analyzed on 1.5% agarose–ethidium bromide gels.
(iv) Quantification of the PCR results.
For quantification purposes, 3 μCi of [α-32P]dCTP was added to the basic PCR mixture, and reactions were carried through 24 cycles. Ten milliliters of each reaction product was then run on a 6% acrylamide gel, dried, and exposed with an A202400 Instant Imager (Camberra Packard Instruments). The counts from each splicing band were corrected by the number of C/G nucleotides in the product sequence. Four independent transfection RT-PCR assays were carried out for each sample. The data analysis was performed with the StatView (Abacus Concept, Inc.) program. The unpaired Student t test was used to determine significance. A P value of <0.05 was considered significant.
Enzymatic analysis of RNA secondary structure.
Single-strand-specific (S1 nuclease and T1 RNase) and double-strand-specific (V1 RNase) enzymes were used to analyze the secondary structures of the in vitro-transcribed RNAs. The EDA sequences originally inserted in the pSVED-A Tot Sac construct (6) or in the Δ4, Δ2e, ES.3A, ES.3U, ESE-Mut, BA1, or BA7 mutant constructs were cloned in the _Sma_I site of the pBSKSII(+) plasmid and transcribed with T7 RNA polymerase. Enzymatic digestion was performed essentially as described previously (4). Briefly, reaction mixes contained 1 μg of RNA and 0.02 U of RNase V1 (Pharmacia Biotech), 0.5 U of RNase T1 (Sigma), or 20 U of S1 nuclease (Pharmacia Biotech) and were incubated at 30°C for 15 min. A control aliquot of RNA without the addition of RNases was processed simultaneously with the digested samples. The RNase cleavage sites were identified by primer extension with a 32P-end-labelled oligonucleotide primer (5′CTGTGGACTGGGTTCCAATC3′), and the RT reaction products were loaded on a 6% polyacrylamide gel and exposed to Kodak X-Omat AR films for 12 to 24 h.
RESULTS
The EDA splicing-regulatory mechanism is similar in a variety of cell types.
To study the alternative splicing regulation of the FN EDA exon, we used an FN–α-globin hybrid minigene, named pSVED-A Tot, which contains the EDA exon, fragments of the flanking exons, and the complete flanking introns. We have previously established that the EDA expression is controlled by a bipartite splicing-regulatory region constituted by the ESE and ESS. Deletion of the polypurinic ESE sequence in the pSVED-A Δ2e plasmid causes complete exon skipping, while deletion of the ESS sequence in the pSVED-A Δ4 construct causes total exon inclusion (Fig. 1A and B, lanes 2 and 4, respectively) (6). Since the previous studies were performed exclusively with HeLa cells, we wanted to analyze the cell type specificity of the two elements that constitute this bipartite splicing-regulatory region by transfecting a variety of cell types derived from different tissues.
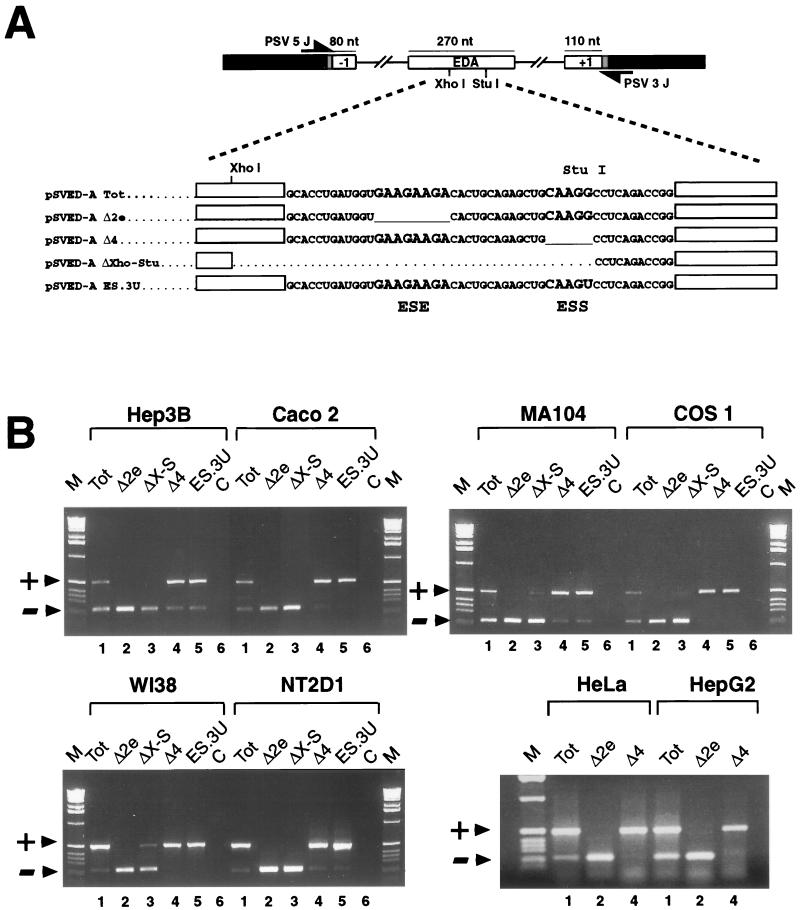
FIG. 1.
Expression of the EDA minigene variants in different cell types. (A) Schematic representation of the pSVED-A minigenes. FN exons and introns are shown by white boxes and lines, linker sequences are shaded, and α-globin exon 3 is indicated by black boxes. Locations of the primers used in the RT-PCR assay are shown. The sequence of the exonic region involved in EDA splicing regulation is reported for the wild-type minigene (pSVED-A Tot) and for the mutants carrying the deletions of the ESE (pSVED-A Δ2e) and ESS (pSVED-A Δ4), the complete deleted region (pSVED-A ΔXho-Stu), and the ESS point mutation (pSVED-A ES.3U). ESE and ESS sequences are in larger type. nt, nucleotides. (B) RT-PCR analysis of total RNA from cells expressing each of the indicated constructs in Hep3B (human hepatocarcinoma), CaCo2 (human colon carcinoma), MA104 (simian kidney), COS1 (SV40-transformed simian kidney), WI38 Va13 (SV40-transformed human fibroblast), NT2D1 (human embryonal teratocarcinoma), HeLa (human cervical carcinoma), and HepG2 (human hepatocarcinoma) cell lines. Arrowheads indicate PCR products either containing (+) or lacking (−) the FN EDA exon in the messenger transcribed from the transfected minigene. Lane M, molecular size markers (1 kb; Life Technologies); lane C, PCR controls.
The splicing patterns of the mRNAs generated from the different EDA hybrid minigene constructs (Fig. 1A) were analyzed by RT-PCR amplifications with specific primers, generating 231- and 501-bp bands for the EDA skipped and included products, respectively (Fig. 1B). The wild-type EDA minigene (Tot) produced EDA-positive and EDA-negative RNAs matching the endogenous pattern of EDA splicing in the respective cell types (data not shown). In all of the cell lines tested, the deletion of the 81-bp central sequence of the exon (ΔXho-Stu), as well as the deletion of the positive polypurine element (Δ2e), abolished alternative splicing of EDA, producing exclusively EDA− RNA (Fig. 1B, lanes 2 and 3). In contrast, the deletion of the negative element (Δ4) resulted in complete or almost complete inclusion of the EDA exon (Fig. 1B, lanes 4), as seen previously in HeLa cells (6, 28, 39). In HeLa, HepG2, COS1, NT2D1, CaCo2, and WI38 Va13 cells, the EDA exon is completely included (Fig. 1B, lanes 4), while in MA104, Hep3B, and SK-N-SH cells (not shown), a small amount (5 to 15%) of mRNA lacking the exon is sometimes evident.
These results show that the exonic splicing-regulatory region containing the ESE and ESS controls EDA splicing ubiquitously and not in a cell-type-specific manner.
Single point mutations in the ESS element can promote complete EDA exon inclusion.
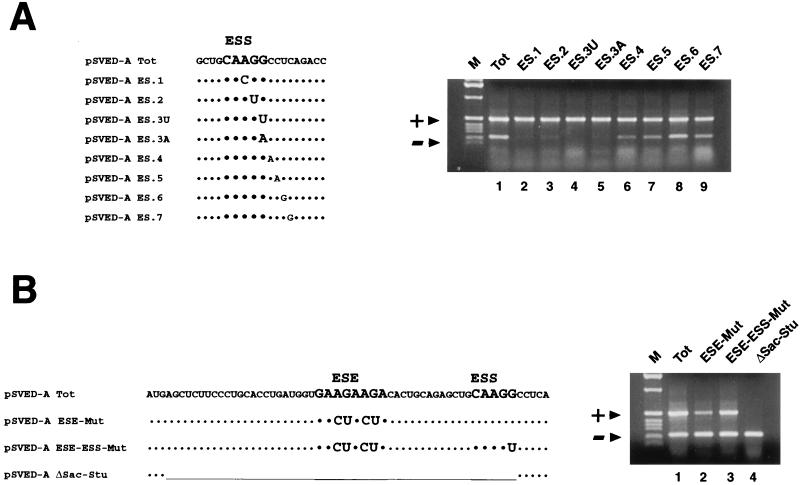
The ESS sequence (5′CAAGG3′) has been previously characterized by deletion analysis, and it was desirable to finely map the element by introducing single and paired point mutations in the sequence previously defined as the ESS and in its nearby 3′ flanking sequence. Since the EDA is predominantly included in HeLa cells, slight variations towards an increased exon inclusion are difficult to detect. RT-PCR analysis of the expression of the pSVED-A Tot plasmid showed that in the HepG2 cell line the EDA exon is included in 60% of the endogenous FN messengers (Fig. 1B). As all of the different cell lines had a common mechanism that regulates the expression of the FN–α-globin minigene, we performed the following experiments with the HepG2 cell line, where small variations in the splicing inclusion/exclusion ratios of the various FN minigenes are easier to observe. Minigenes carrying single and combined point mutations were transiently transfected in HepG2 cells, and the processed messenger was analyzed by RT-PCR. Single point mutations of the ESS AGG core sequence were as efficient as the original 5-nucleotide deletion in increasing the EDA inclusion to 95 to 100% of the mRNA product (Fig. 2A, lanes 2 to 5). Increased exon inclusion was also observed after the substitution of the two cytosine nucleotides immediately downstream from the ESS element (82 and 79% in the ES.4 and ES.5 constructs, respectively [Fig. 2A, lanes 6 and 7]), while mutations further downstream did not produce any significant effect (63 and 59% in the ES.6 and ES.7 constructs, respectively [Fig. 2A, lanes 8 and 9]). We did not analyze the nucleotides at the 5′ end, since previous deletion analysis indicated that they were not primarily involved in the EDA regulation (Δ3 construct in reference 6). Moreover, the disruption of the silencing effect by a single point mutation was also observed in all of the cell types tested (Fig. 1A and B, lane 5). In addition, the expression of the double or triple base substitutions reflected the behavior of the single point mutations showed in Fig. 2A (data not shown). Single point mutations can produce the same effect of the original CAAGG deletion, and these data indicate that the core sequence of this element must be extended to CAAGGCC.
FIG. 2.
(A) Single point mutations promote complete EDA exon inclusion in the expressed pSVED-A minigenes. The left panel shows the sequence of the EDA region surrounding the ESS. Single nucleotides in the EDA region containing the characterized ESS have been mutated. Nucleotides involved in the deletion that previously defined the ESS are in larger type. The right panel shows the RT-PCR analysis of the ESS point mutant series transiently expressed in HepG2 cells. (B) Polypurinic ESE point mutation and ESE-ESS deletion. pSVED-A Tot and pSVED-A ESE-Mut, ESE-ESS-Mut, and ΔSac-Stu sequences are shown on the left. In the pSVED-A ΔSac-Stu construct the whole regulatory region has been deleted. RT-PCR assays of the ESE and ESE-ESS combined mutations in HepG2 cell line are shown at the right. ESE and ESS sequences are in larger type. Symbols and abbreviations are as in Fig. 1.
Nonpurine exonic sequences flanking the polypurinic ESE sequence are also involved in EDA exon inclusion.
Most ESE sequences are polypurine stretches with a general sequence (GAR)n, where R is either A or G (23). We have introduced four point mutations completely disrupting the polypurine arrangement in the pSVED-A ESE-Mut construct (the GAAGAAGA sequence was replaced by GACTACTA). In both HeLa and HepG2 cells, pSVED-A ESE-Mut expression failed to produce complete exon skipping. In fact, about 10% of exon inclusion occurred (Fig. 2B, lane 2), while it was completely skipped in the Δ2e construct (Fig. 1B, lane 2), where the entire element was deleted. The polypurine is therefore necessary for efficient exon inclusion, although its sequence can be disrupted without total loss of the splicing enhancement activity. This suggests that the purine stretch is likely to be part of a more complex sequence and/or secondary structure. More evidence that supports the hypothesis that sequences flanking the defined polypurine ESE are involved in the inclusion of the EDA exon was obtained with the constructs pSVED-A ESE-ESS-Mut and pSVED-A ΔSac-Stu. In pSVED-A ESE-ESS-Mut both the ESE and the ESS were mutated, and the splicing patterns obtained showed that the mutation of the ESS element could partially rescue the loss of splicing enhancement activity due to the purine mutation (34 versus 61% of the activity of the Tot plasmid [Fig. 2B, lanes 1 to 3]). A strikingly different effect is observed upon expression and analysis of the construct pSVED-A ΔSac-Stu, where a 34-nucleotide region including both the ESE and the ESS region has been deleted. The EDA exon was totally skipped (Fig. 2B, lane 4), suggesting that exonic sequences other than the polypurine ESE exert a positive control on the EDA exon splicing. Therefore, it would be more correct to refer to the FN ESE as an exonic region that contains the characterized polypurine sequence.
Function of the ESE and ESS elements in a heterologous exonic context.
To test whether the ESE and ESS could modify splicing of constitutive and alternatively spliced heterologous exons, a series of plasmids was created. The ESS sequence was inserted into the constitutively spliced second exon of the α-globin gene, and both the ESS and ESE were inserted into the FN EDB alternatively spliced exon.
The EDB exon was used because it is homologous to the EDA and itself undergoes alternative splicing. Since the distance between the exon enhancers and the 3′ splice site seems to be critical for the interactions favored by binding of protein factors on the enhancer sequences (24), the manipulations were done in such a way that the corresponding EDA distance was kept. A preliminary characterization of the human EDB behavior was necessary to confirm that its splicing regulation was the same as that in rat (19). The wild-type human EDB exon (B1 construct) was transfected into NT2D1 cells, which are in some ways equivalent to the rat F9 cell line used by Huh et al. (19). The pattern of splicing seen was a 1:1 ratio between EDB+ and EDB− mRNA forms. On the other hand the human EDB exon was completely excluded in HeLa cells (Fig. 3A, lane 1). A similar splicing efficiency was observed with the B2 plasmid, where an _Xho_I site was introduced to facilitate further cloning (Fig. 3A, lane 2). The deletion of the central portion of the EDB exon (B4) caused no modification in the alternative splicing pattern (Fig. 3A, lane 4). Deletion of five of the intronic splicing enhancer (ISE) elements out of the total seven repeats present in the human gene (B3 and B5 constructs) completely abolished the inclusion of the EDB exon. These results confirm those observed with the rat ISE elements (19, 26). Transfection into HeLa cells, whose endogenous pattern is EDB minus, showed no difference between the different constructs (Fig. 3A).
Our results indicate that the ISEs composed of TGCATG repeats are important for the cell-type-specific inclusion of the human EDB exon (Fig. 3A). This is consistent with the recent analysis done by Lim and Sharp (26), who showed the cell specificity of the rat elements. The experiments described above also demonstrate the dominance of the ISE sequences over the putative exonic regulators present in the human EDB exon, as their deletion produced complete exon skipping, as previously seen for the rat FN EDB exon (19).
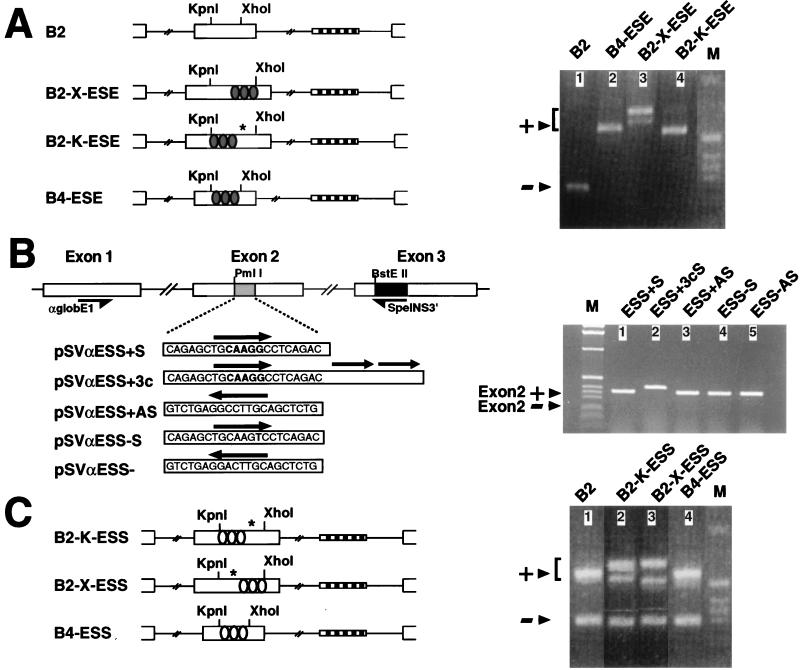
The function of the human EDA ESE and ESS elements in a heterologous context was analyzed by introducing the individual elements into α-globin- and FN EDB-bearing plasmids. The EDA polypurinic enhancer element notably enhanced the EDB exon inclusion in a series of EDA-EDB chimeric constructs. The stimulatory effect was better seen with the Hep3B cell line, where the EDB exon is normally skipped (Fig. 4A, lane 1). When the ESE element plus its flanking sequences was inserted at the place of the deleted _Kpn_I/_Xho_I fragment of the B4 plasmid or at the _Xho_I site of the B2 construct, constitutive inclusion of the EDB exon was observed (B4-ESE and B2-X-ESE plasmids, respectively [Fig. 4A, lanes 2 and 3]). The same splicing-stimulatory effect was observed when the ESE was placed at the _Kpn_I site of the B2 construct (B2-K-ESE plasmid), although a cryptic 5′ splicing site located 30 bases downstream of the _Kpn_I site was utilized (Fig. 4A, lane 4). The inclusion of the ESE element in the B2 and B4 constructs also produced constitutive EDB exon inclusion in different cell types (not shown). A secondary band close to the EDB plus product was observed in the case of the B2-X-ESE construct (Fig. 4A, lane 3), and direct sequencing of the PCR product revealed that a 5′ cryptic splicing site located near the newly generated _Xho_I site was utilized in this case by a minority of the mRNAs.
FIG. 4.
Insertion of the ESE and ESS sequences in heterologous exons. (A) ESE element in the FN EDB alternatively spliced exon. Schemes representing the insertion of the ESE sequences into the B2 and B4 constructs (left) and RT-PCR analysis of transient transfection in Hep3B cells (right) are shown. The shaded ovals represent ESE sequences (5′CCTGATGGTGAAGAAGACACTGCA3′). (B) ESS element in the α-globin constitutively spliced exon 2. A graphic representation of the pSVαESS minigene series is shown on the left. The α-globin region of the pSVαESS constructs is indicated by white boxes. The black box indicates the linker sequence inserted in the _Bst_EII restriction site. The shaded box indicates the FN sequences inserted in the second exons. Primers SpeINS3′ and αglobE15′ used for the RT-PCR analysis are indicated. The sequences of the insertions are indicated, together with their orientations and number of tandem repeats (black arrows). RT-PCR analysis of the pSVαESS series expression in HeLa cells is shown on the right. The bands corresponding to mRNAs including exon 2 and to the expected mRNAs excluding it are indicated by arrows. (C) ESS element in the FN EDB alternatively spliced exon. The EDB ESS construct scheme is shown on the left. The RT-PCR analysis of transient transfection in NT2D1 cells is shown on the right. NT2D1 cells were used because they have a high inclusion/exclusion ratio of the EDB exon, increasing the sensitivity of the assay. The white ovals represent ESS sequences (5′AGCTGCAAGGCCTCAGACCGAGCT3′). The EDB excluded and included bands are indicated by arrows. Different included bands are present due to the use of cryptic splicing sites. The positions of the cryptic sites are indicated by asterisks. Arrowheads and lane M are as defined for Fig. 1.
The expression of the ESS inserted into the α-globin gene constructs produced no modification in the constitutive globin exon 2 inclusion, even when added in multiple copies (Fig. 4B, lanes 1 and 2). This result indicates that the splicing-inhibitory activity of the ESS sequence is not significant in a constitutive exon and that the inhibitory mechanism may depend on other factors such as weak splicing sites and/or the adjacent ESE. To test this possibility, we prepared a group of constructs by inserting the ESS element, in one or multiple copies, in different positions inside the EDB exon or by replacing the central part of the exon (Fig. 4C). Surprisingly, the ESS had no inhibitory activity when placed in the alternatively spliced EDB exon, even in multiple copies and at different distances from the 5′ splicing site (Fig. 4C). In fact, no reduction in the inclusion ratio was observed after transient transfection of the NT2D1 cells with the constructs bearing the silencer element. In lanes 2 and 3 of Fig. 4C, a second band can be seen and is the result of the activation of a 5′ cryptic splice site, probably due to the excessive length of the chimeric exon.
The EDA ESS element activity depends exclusively on the complete EDA context.
The transfection experiments with the ESS inserted into the EDB exon context presented in the previous sections introduced the possibility for the requirement of a more complex secondary structure and/or a crosstalk between the positive and negative _cis_-acting elements present in the bipartite enhancer. To elucidate this possibility we made a series of constructs in which the entire EDA splicing-regulatory region was inserted into the FN EDB heterologous exon. The _Kpn_I-_Xho_I region in the hybrid minigene B2 was replaced with the wild-type regulatory region of the EDA exon (Fig. 3B, BA1 to BA4 constructs) or with individual mutations of the positive or negative elements in the chimeric EDB-EDA hybrid minigenes, with or without the ISE elements (Fig. 3B, BA5 to BA10 constructs). These chimeric constructs were transfected in two distinct classes of cell lines representing predominantly the EDB+ or EDB− phenotype (NT2D1 or WI38 Va13 [not shown] and HeLa or Hep3B [not shown], respectively), and the splicing pattern was analyzed by RT-PCR (Fig. 3B).
The BA1 chimeric construct containing the entire bipartite enhancer from the EDA exon showed constitutive inclusion of the EDB exon in both groups of cells regardless of the endogenous pattern of EDB splicing in these cells. The deletion of the intron repeats, which resulted in complete exclusion of the exon in the wild-type EDB construct (B3 construct [Fig. 3A]), had no effect on the splicing pattern in either group of cells when the EDA sequence was present (BA2 construct [Fig. 3B]). The insertion of the EDA sequence in the opposite orientation (BA3), as well as the combined deletion of the core region containing the GAAGAAGA and CAAGGCC elements (BA4), shows a partial exon inclusion in NT2D1 cells and no inclusion in HeLa cells. As previously seen with the EDA ΔSac-Stu construct, the enhancer effect of the EDA splicing-regulatory region elements is lost when the ESE and ESS elements are deleted, as in the case of the BA3 and BA4 EDB-EDA hybrid constructs (compare BA3 to B1 and BA4 to B4). These results indicate that the bipartite splicing enhancer from the EDA exon, when transferred to another alternatively spliced exon regulated mainly by ISEs, can override the necessity for ISEs. In addition, this effect is not cell type specific as is true for the behavior of this bipartite enhancer in the context of the EDA exon.
The construct BA5 has a deletion of the positive ESE element, but the intronic enhancers have been preserved. There is up to 90% inclusion of the EDB exon in NT2D1 cells (Fig. 3B, lane 5) and WI38 Va13 cells (not shown). The deletion of both exonic and intronic positive elements (BA6) considerably reduced the percentage of EDB inclusion in these cells (30%), without producing complete exon skipping, confirming the previous hypothesis that nonpurinic exonic sequences are involved in the function of the EDA composite enhancer region.
Unexpectedly, the deletion of the negative element (BA7 plasmid) produced the opposite effect from that observed for the Δ4 construct (Fig. 1) (6). In fact, a reduction in exon inclusion was observed in BA7 when compared with the BA1, which contains the entire EDA region. Moreover, in the absence of both the negative ESS element and the ISE elements (BA8 construct), the percentage of exon inclusion was surprisingly lower than that for the products of the constructs BA2 and BA7 (which contain the intronic positive elements).
The combination of the G/T point mutation in the ESS element and the absence of the ISE elements (BA10 plasmid) produced a minor increase in EDB exon skipping in NT2D1 cells (compare BA2 with BA10 in Fig. 3B). This unexpected result, analogous to the one observed for the BA7 and BA8 constructs, was in contrast with the stimulatory effect seen in the EDA context for the same mutations (Fig. 2A). The presence of the intron enhancers in the BA9 construct may have masked the G/T mutation effect, as no difference in splicing could be seen (compare BA1 with BA9 in Fig. 3B). The splicing pattern was unaffected when the same constructs were tested in HeLa cells. Deletion of the positive or negative elements (BA5 and BA7, respectively) had a more pronounced effect in HeLa cells, reducing the EDB inclusion to 60%. There was no further reduction when the intron repeats were deleted (BA6 and BA8).
These results indicate that the positive and negative elements from the EDA exon behave as parts of a composite enhancer in the context of the EDB exon and that the EDB exon is recognized independently of the presence or absence of the ISE elements when the EDA regulatory region is inserted in the place of its exonic regulatory region (polypurinic enhancer plus downstream silencer elements [13]).
The paradoxical behavior of the ESS element in the BA7 to BA10 plasmids and its loss of function after its inclusion in a heterologous alternatively spliced exon (B4-ESS, B2-X-ESS, and B2-K-ESS constructs [Fig. 4C]) strongly suggest that the ESS is part of an EDA-specific splicing-regulatory region. This may indicate a possible structural role for the ESS element that depends exclusively on a complete EDA context. This result is in contrast to the full function of the nearby enhancer element in different heterologous contexts.
The EDA presents a secondary structure and the ESE is located in a terminal loop.
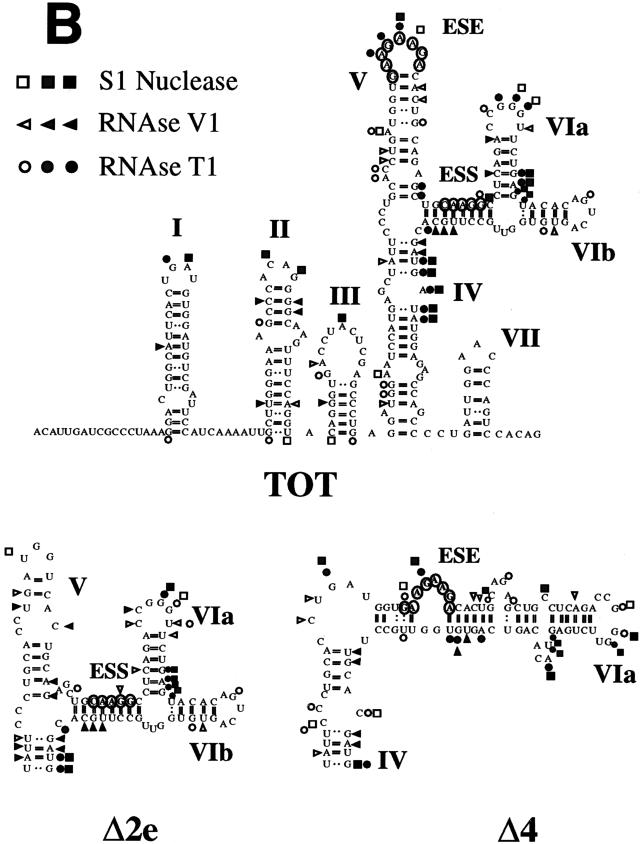
To evaluate the structure of the EDA-specific splicing-regulatory region, we have investigated the RNA secondary structures of the wild-type EDA exon and some mutants. The conformations of the in vitro-transcribed RNAs were mapped by enzymatic reactions. Both RNase T1 (cleaves after a guanosine residue) and S1 nuclease (cleaves single-stranded nucleic acids) were used to identify single-stranded regions. Double-stranded or stacked regions were investigated with RNase V1, which has no apparent base specificity. The cleaved bases in the RNA molecules were then identified by primer extension analysis. To help with the interpretation of the mapping experiments, a systematic search of potential base pairing possibilities was assisted by use of the Zuker M-Fold computer program with free energy minimization (42).
Using computer predictions and mutational analysis, Staffa et al. (35) had already suggested the existence of a conserved RNA secondary structure within the first 118 nucleotides of the EDA exon. Our enzymatic cleavage analysis not only confirms the presence of the proposed structure in the first 118 nucleotides of the exon but also extends the analysis of the EDA secondary structure to the whole exon, focusing on the polypurine enhancer element. In fact, confirmatory cleavages in the stem-loop I, II, and III structures proposed by Staffa et al. (35) were observed with both single- and double-strand-specific nucleases (Fig. 5).
FIG. 5.
Enzymatic determination of the RNA secondary structure of the EDA exon. (A) Enzymatic analysis of RNA templates of the Δ4, Tot, and Δ2e constructs. In vitro-transcribed RNAs were enzymatically digested with S1 nuclease and T1 and V1 RNases and reverse transcribed, and the RT products were separated on a polyacrylamide sequencing gel as described in Materials and Methods. A sequencing reaction with the same primer was run in parallel to precisely determine the cleavage sites. Squares, circles, and triangles indicate S1 nuclease and RNase T1 and V1 cleavage sites, respectively. Black, shaded, and white symbols indicate high, medium, and low cleavage intensities, respectively. No enzyme was added to the reaction mixture in lanes C. (B) Secondary-structure models of the wild-type human EDA exon (TOT) and of the variants bearing deletions of the ESE and ESS elements (Δ2e and Δ4 constructs). The regions flanking the ESE and ESS elements are shown for the Δ2e and Δ4 constructs. The structures of the other regions are similar to the one presented for the Tot construct. The data was optimized by computer-assisted RNA modelling (42). The ESE and ESS elements are circled.
The proposed secondary structure of the entire exon is composed of the already-reported stem-loops I, II, and III (35); stem IV, ending in an internal loop; stem-loop V, which contains the ESE in its terminal loop; and stem-loop VI, which ends in two minor stem-loop structures (stem-loops VIa and VIb). The experimental data shown in Fig. 5A supports the presence of these structures and in particular the stem-loop V structure. RNase V1 digested paired regions of the stem, and both S1 and T1 nucleases showed cuts in the GAAGAAGA enhancer element, clearly suggesting its localization in a loop structure (Fig. 5A). There is good correlation between the thermodynamically most stable structure and the experimental data for the enzymatic specific cleavage.
In the case of the Δ2e construct, where the ESE was deleted, the overall secondary structure is well conserved, with a shortening and modification of region V (Fig. 5). In any case, the absence of the ESE from loop V completely abolished EDA exon recognition, as observed in Fig. 1. The other stem-loop structures remained invariable.
Deletion or single-base mutation of the ESS element increases single-strand-specific digestion in the ESE region.
Probing of the mutant Δ4 RNA lacking the silencer element showed the conservation of the stem-loop I, II, III, IV, and VII structures and a partial conservation of the stem-loop VI structure (Fig. 5). Although there is a modification of the stem-loop V structure as seen by computer prediction analysis, the enhancer element is still digested by S1 and T1 nucleases, indicating its positioning in an exposed and single-stranded region. Significantly, there is an increase of single-strand-specific digestion in the ESE-ESS area, indicating an overall change in the stem-loop V and VI structures that is consistent with the enhanced in vivo splicing activity of the Δ4 construct (Fig. 1).
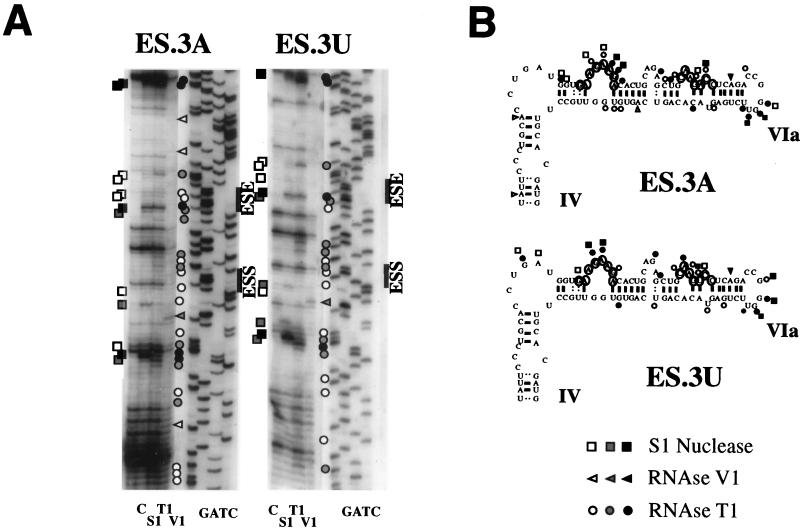
Likewise, enzymatic analysis of the ES.3A and ES.3U constructs clearly showed the localization of the ESE element also in an exposed position (Fig. 6A). The secondary structure shown in Fig. 6B revealed for ES.3A and ES.3U an RNA conformation analogous to the one described for the Δ4 construct. In fact, both RNase T1 and S1 nuclease produced cleavages within the ESE sequence, suggesting its localization in a single-stranded region (Fig. 6), as already observed for the pSVED-A Tot and Δ4 plasmids (Fig. 5). The digestion pattern also suggests a more open structure in the ESE flanking area, similar to the observed results for the Δ4 construct. The ESS element region shows single-strand-specific enzymatic digestion, suggesting that the overall structure of the region is modified in a similar manner in the ES.3A, ES.3U, and Δ4 constructs. This is consistent with in vivo experiments that indicate complete, or even enhanced, function of the ESE in the Δ4, ES.3A, and ES.3U constructs (Fig. 1 and 2A). The enhanced activity of the ESE could be explained by a different conformation of the RNA, as seen in Fig. 5. This suggestion is supported by the fact that the digestion pattern surrounding the enhancer element indicates changes towards a more open structure that may facilitate both protein-RNA and protein-protein interactions between the basic splicing machinery and the SR proteins recognizing the region (24).
FIG. 6.
Secondary-structure analysis of the ES.3A and ES.3U constructs. (A) Enzymatic analysis of RNA templates of the ES.3A and ES.3U constructs. In vitro-transcribed RNAs were enzymatically analyzed as described in the legend to Fig. 5. Symbols are as described in the legend to Fig. 5. (B) Secondary-structure model of the ES.3A and ES.3U constructs. Only the regions flanking the ESE and ESS elements are shown. The structures of the other regions are similar to the one presented for the Tot construct in Fig. 5. The data was optimized by computer-assisted RNA modelling (42).
Extension of the sequence and structural analyses to templates containing the flanking introns by computer-assisted modelling showed a conservation of the proposed structure and did not show any particular configuration that could hide the splicing sites in any of the wild-type or mutant RNAs (not shown).
EDB splicing efficiency is reduced when the ESE element is not exposed in the loop of a hairpin structure.
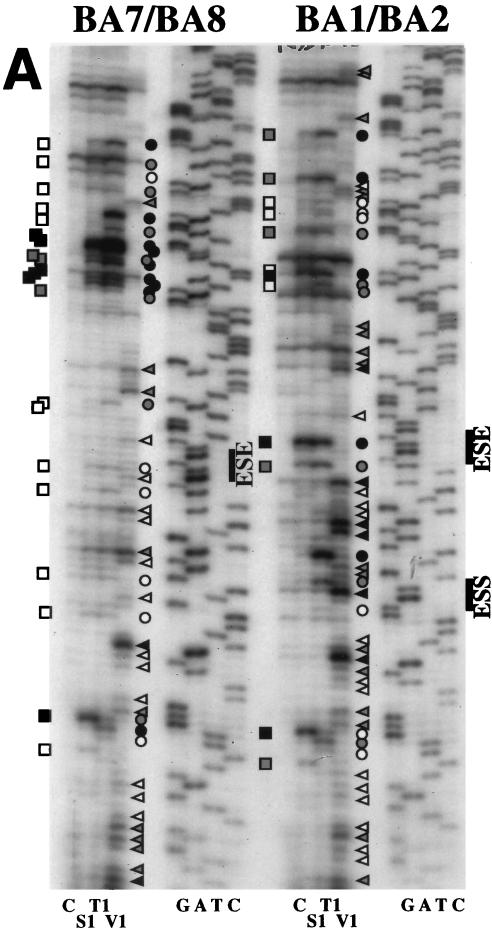
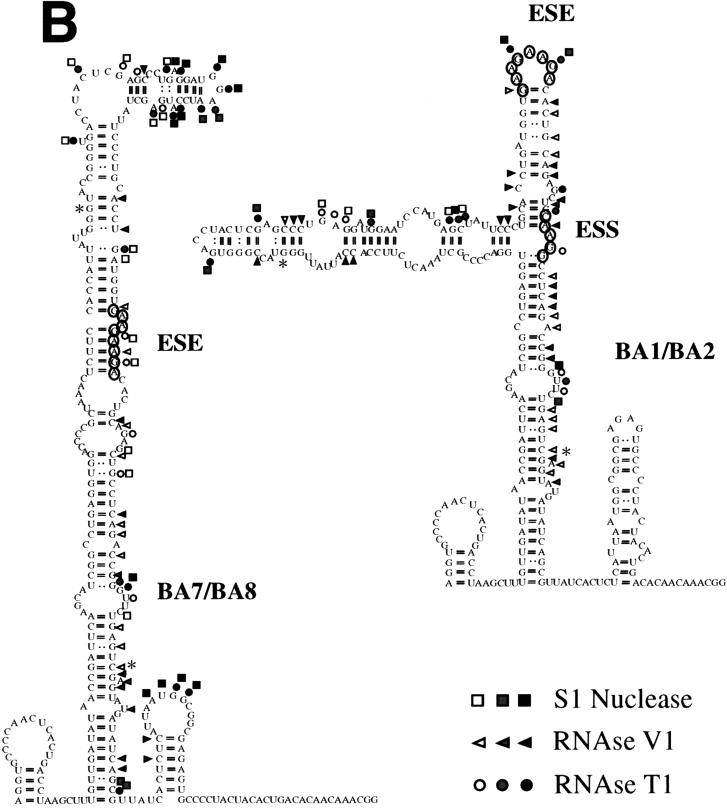
We had particular interest in the unexpected reduction of splicing activity obtained with the chimeric BA7 and BA8 constructs (which contain a deletion in the ESS element) when compared with the BA1 and BA2 constructs (bearing the complete EDA splicing regulatory region). This result is contradictory if the ESS element was acting through interactions mediated by a linear sequence, and this difference in splicing activity may be the consequence of a particular secondary structure.
RNase and S1 nuclease digestion, assisted by computer modelling of the EDB-EDA hybrid exons (BA1 and BA2 constructs [Fig. 3]) showed essentially a conservation of the EDA structure with the ESE in a loop (Fig. 7). Strong S1 nuclease and T1 RNase cleavages can be observed at the ESE position, suggesting, as already observed for the EDA Tot construct, that the ESE resides in an exposed position in a loop region. The absence of the ESS sequence resulted in a different digestion pattern in the BA7 and BA8 constructs, confirming the structural role of the ESS element. However, in contrast with the Δ4, ES.3A, and ES.3U structures (Fig. 5 and 6), the BA7 and BA8 constructs show a clear reduction, if not complete absence, of single-strand-specific digestion in the ESE region (Fig. 7A). In fact, deletion of the ESS sequence results in a different secondary structure of the EDA region, where the ESE polypurinic sequence is no longer displayed in a loop but is masked in a stem (Fig. 7B), providing a clear explanation for the decrease in exon inclusion observed when the ESS element is deleted (Fig. 3B).
FIG. 7.
RNA secondary-structure determination by enzymatic digestion of the BA1/BA2 and BA7/BA8 chimeric constructs. (A) Enzymatic analysis of RNA templates of the BA1/BA2 and BA7/BA8 constructs. The digestion reaction conditions were the same as for Fig. 5. Symbols are as described in the legend to Fig. 5. (B) The digestion data obtained from the digestion reactions shown in panel A was optimized by computer-assisted modelling (42), and the digestion sites are indicated. The asterisks indicate the junctions between EDB and EDA sequences.
The analysis of the wild-type and mutant RNAs indicates the presence of a defined and complex RNA secondary structure of the EDA exon, suggesting that _cis_-acting element and _trans_-acting factor interactions are mediated by the RNA conformation.
DISCUSSION
We have previously shown that an 81-nucleotide region in the FN EDA exon regulates its alternative splicing in HeLa cells (28). More recently, the sequence elements responsible for this regulation have been mapped to a GAAGAAGA element that functions as a positive modulator of splicing and to a CAAGG element that functions as a negative modulator of splicing in HeLa cells (6).
Since FN alternative splicing is subjected to tissue-specific and developmental stage-specific regulation (29), the effect of these positive and negative elements on EDA splicing could change in different cell types. We have tested a series of cell lines derived from a variety of tissues, including liver, kidney, intestine, and nervous system, by transfecting EDA hybrid minigene constructs. The analysis of the splicing patterns showed that both the positive and negative elements of the EDA exon are functional in all of these cell types and that the minigene splicing is modulated in the same fashion as the endogenous FN gene in all cell lines tested. This result indicates that each cell type carries a specific combination of _trans_-acting factors that determine the degree of inclusion or exclusion of the alternatively spliced exon.
The silencer activity can be abolished by single base mutations.
The EDA exonic silencer element was the first reported mammalian sequence to act as a negative modulator of splicing. Deletion of the previously described ESS element, a 5-nucleotide stretch (5′CAAGG3′), results in total EDA inclusion (6). Here we have shown that single nucleotide substitutions can induce the same effect and that the mutation of the two cytosine residues downstream also increases exon inclusion, redefining the ESS element as a CAAGGCC sequence. This sequence does not have common features with the human immunodeficiency virus type 1 tat exon 2 and 3 ESS (1, 36) or with the splicing silencer present in the K-SAM alternative exon of fibroblast growth factor 2 (10), but it is remarkably similar to a CAGACCT ESS in the bovine growth hormone exon 5 (37). Nevertheless, when the EDA ESS was mutated to match the core sequence of the latter repressor in the construct pSVED-A ES.3A (Fig. 2A, lane 5), the EDA exon was included in 100% of the final messengers, diminishing the significance of the sequence homology and pointing more to structural elements as the basis of the observed functions.
Extra sequences besides the polypurine enhancer are required for efficient exon recognition.
The FN ESE element sequence (GAAGAAGA) contains the consensus obtained for the ASF/SF2 splicing factor (38). It has been previously shown that there is an interaction of the proteins recognizing the ESE with the splicing machinery recognizing the EDA splicing sites (24, 30). However, the interaction with other _cis_- and _trans_-acting regulatory elements and factors is not yet well understood. When the EDA polypurine GAAGAAGA is mutated to GACUACUA, the exon is still included in 15% of the mRNA molecules, while deletion of the purine stretch causes total exon exclusion (Fig. 2B). Further evidence that the ESE sequence is not limited to the purine stretch is given by the comparison of the results obtained with the minigenes pSVED-A ΔSac-Stu and pSVED-A ESE-ESS-Mut (Fig. 2B) and with the BA5 and BA6 constructs (Fig. 3). In the first case, the complete deletion of the EDA regulatory region leads to complete exclusion of the EDA exon, while the inactivation of the ESS can partially overcome the effect due to the purine mutations in the ESE (15% of inclusion in pSVED-A ESE-Mut and 35% in pSVED-A ESE-ESS-Mut). In the second case, a stimulatory effect in splicing was observed with the BA5 and BA6 constructs despite the absence of the polypurinic enhancer element in the chimeric EDB-EDA exon. Thus, the splicing enhancement activity of the EDA splicing-regulatory region is also exerted by sequences other than the GAAGAAGA polypurinic enhancer. This is consistent with the observation that overlapping SRp40, ASF/SF2, and SRp55 binding sites (as defined by the systematic evolution of ligands by exponential enrichment [SELEX] methodology by Liu et al. [27]) can be found in the region between the ESE and ESS elements. An increase in the EDA exon inclusion efficiency in the Δ4, ES.3A, and ES.3U constructs may be the consequence of a two- and three-dimensional structural modification. This change may improve the exposure of the positive _cis_-acting elements to the splicing machinery and may generate an increase in the EDA exon inclusion rate. Furthermore, the complete regulatory region is dominant over the ESS inhibition, as we can see from the ESE-Mut and ESE-ESS double-mutant transfection experiments.
The splicing-regulatory region of the EDA exon is active in the EDB exon and dominates over the EDB ISE and ESE elements.
The cell-type-specific regulation of EDB splicing was altered when the EDA regulatory element was introduced inside the EDB exon. This chimeric construct showed 100% exon inclusion in both NT2D1 and HeLa cells, irrespective of the wild-type pattern and independently of the presence or absence of the ISEs. The insertion of the 81-bp region from the EDA exon in the antisense direction within the EDB exon (BA3) did not have a considerable effect on the splicing pattern, confirming that the enhancer effect is sequence specific. Similar dominant effects of exonic splicing modulators were also obtained when the negative TAGG element from the FGFR2 K-SAM exon was inserted inside the rat FN EDB exon (11). In spite of the presence of the enhancer positive elements in the downstream intron, the EDB exon was skipped when the TAGG silencer element was inserted in the exon, suggesting that the exonic splicing-regulatory elements present in the FN EDA exon and in the K-SAM exon of the FGFR-2 gene are dominant over the intronic regulatory elements present in the EDB downstream intron. In contrast, the activity of the exonic splicing-regulatory elements present within the EDB exon is dependent on the presence of the ISEs in the downstream intron.
The ESS function strictly depends on the EDA exonic context.
The insertion of the ESS element in either a constitutively or alternatively spliced exon (as a single or multiple element or as a part of the entire EDA region) showed no effect on splicing. These results suggest that the silencer element function depends exclusively on the complete EDA exonic context. On the other hand, we have shown that the ESE is functional in different exonic contexts, confirming the results reported by Staffa and Chochrane (36), who utilized a two-exon, one-intron minigene system. This observation may be the consequence of a peculiar characteristic of the enhancer sequence (a polypurine) that may facilitate its display in an RNA structure. It should be noted that when the ESS element was deleted (BA7 and BA8 constructs [Fig. 3]), the proportion of exon inclusion paradoxically diminished. This effect is particularly visible in the absence of the ISE elements (BA8 construct). Again, this result suggests that the recognition motifs are not only linear nucleotide sequences and that there may be structural features whose conservation is essential for optimal exon recognition.
The EDA exon has a secondary structure that may mediate its recognition by _trans_-acting factors.
The secondary structures derived from computer modelling supported the data obtained by enzymatic digestion of RNA templates within the exonic region of the EDA exon. The presence of three stem-loop structures in the first 118 bases of the exon has already been suggested by Staffa et al. (35), who utilized computer predictions and mutational analysis. We have confirmed these structures, and we have further identified the presence of other stem-loop structures in the rest of the exon. In our model, the GAAGAAGA enhancer sequence resides within a terminal loop in domain V of the exon (Fig. 5), a situation that may facilitate interactions with _trans_-acting factors. It was recently shown that the Drosophila SR proteins Tra/Tra2 and B52 interact with hairpin-loop structures containing the ESE elements (17, 33) and that the RRM domain of the RNA binding protein U1A, which is similar to that present in the SR proteins, specifically interacts with the single-stranded region of a hairpin structure formed by U1A snRNA (31). Furthermore, Lavigueur et al. (24) demonstrated that SR proteins interact with the polypurine element of the EDA exon. We propose that the stem-loop structure where the enhancer resides may be necessary for this interaction. Furthermore, the efficient function of the ESE element depends on the maintenance of the single-stranded configuration. This hypothesis is supported by the in vivo data obtained by transfecting the BA series of constructs and the ESS mutants. An unexpected decrease in EDB exon inclusion was observed when the ESS was absent from the EDB-EDA chimeric construct (compare results for the BA1 and BA2 constructs with those for the BA7 and BA8 constructs). Detailed analysis of the RNA secondary structure showed that the ESE element was displayed in a single-stranded region when the wild-type EDA sequence was inserted into the EDB exon (Fig. 7). In the case of the BA7 and BA8 constructs, the ESE element was partially masked in a stem (Fig. 7), providing an explanation for the paradoxical behavior of the hybrid minigene, which, notwithstanding the absence of the ESS silencer, decreases significantly the inclusion of the exon. In the case of the ESS mutants, there is an overall increase in single-stranded cleavages in the ESE-ESS region, suggesting an enhanced ability to interact with _trans_-acting factors. The overlapping SRp40, ASF/SF2, and SRp55 binding sites found in this region (see above) may be more accessible, and therefore there is an increase of exon inclusion.
The display of the ESE in a loop may be the main feature of the exon identifier. The function of the silencer sequences, which are so context dependent, may be to ensure the proper structure that facilitates display of the SR binding sequences. The understanding of this regulatory mechanism will be complete after further characterization of the _trans_-acting factors that participate in EDA exon recognition. Experiments to identify the proteins involved in the modulation of the EDA exon splicing are under way.
ACKNOWLEDGMENTS
A.F.M., M.C., and R.P. contributed equally to the manuscript and should be considered joint first authors.
We thank A. Iaconcig and C. Stuani for technical help.
REFERENCES
- 1.Amendt B A, Si Z H, Stoltzfus C M. Presence of exon splicing silencers within human immunodeficiency virus type 1 tat exon 2 and tat-rev exon 3: evidence for inhibition mediated by cellular factors. Mol Cell Biol. 1995;15:4606–4615. doi: 10.1128/mcb.15.8.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balvay L, Libri D, Fiszman M Y. Pre-mRNA secondary structure and the regulation of splicing. Bioessays. 1993;15:165–169. doi: 10.1002/bies.950150304. [DOI] [PubMed] [Google Scholar]
- 3.Black D L. Does steric interference between splice sites block the splicing of a short c-src neuron-specific exon in non-neuronal cells? Genes Dev. 1991;5:389–402. doi: 10.1101/gad.5.3.389. [DOI] [PubMed] [Google Scholar]
- 4.Brown E A, Day S P, Jansen R W, Lemon S M. The 5′ nontranslated region of hepatitis A virus RNA: secondary structure and elements required for translation in vitro. J Virol. 1991;65:5828–5838. doi: 10.1128/jvi.65.11.5828-5838.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caputi M, Baralle F E, Melo C A. Analysis of the linkage between fibronectin alternative spliced sites during ageing in rat tissues. Biochim Biophys Acta. 1995;1263:53–59. doi: 10.1016/0167-4781(95)00067-q. [DOI] [PubMed] [Google Scholar]
- 6.Caputi M, Casari G, Guenzi S, Tagliabue R, Sidoli A, Melo C A, Baralle F E. A novel bipartite splicing enhancer modulates the differential processing of the human fibronectin EDA exon. Nucleic Acids Res. 1994;22:1018–1022. doi: 10.1093/nar/22.6.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chebli K, Gattoni R, Schmitt P, Hildwein G, Stevenin J. The 216-nucleotide intron of the E1A pre-mRNA contains a hairpin structure that permits utilization of unusually distant branch acceptors. Mol Cell Biol. 1989;9:4852–4861. doi: 10.1128/mcb.9.11.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clouet d’Orval B, d’Aubenton Carafa Y, Sirand-Pugnet P, Gallego M, Brody E, Marie J. RNA secondary structure repression of a muscle-specific exon in HeLa cell nuclear extracts. Science. 1991;252:1823–1828. doi: 10.1126/science.2063195. [DOI] [PubMed] [Google Scholar]
- 9.Cramer P, Pesce C G, Baralle F E, Kornblihtt A R. Functional association between promoter structure and transcript alternative splicing. Proc Natl Acad Sci USA. 1997;94:11456–11460. doi: 10.1073/pnas.94.21.11456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Del Gatto F, Breathnach R. Exon and intron sequences, respectively, repress and activate splicing of a fibroblast growth factor receptor 2 alternative exon. Mol Cell Biol. 1995;15:4825–4834. doi: 10.1128/mcb.15.9.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Del Gatto F, Gesnel M C, Breathnach R. The exon sequence TAGG can inhibit splicing. Nucleic Acids Res. 1996;24:2017–2021. doi: 10.1093/nar/24.11.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Domenjoud L, Gallinaro H, Kister L, Meyer S, Jacob M. Identification of a specific exon sequence that is a major determinant in the selection between a natural and a cryptic 5′ splice site. Mol Cell Biol. 1991;11:4581–4590. doi: 10.1128/mcb.11.9.4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du K, Peng Y, Greenbaum L E, Haber B A, Taub R. HRS/SRp40-mediated inclusion of the fibronectin EIIIB exon, a possible cause of increased EIIIB expression in proliferating liver. Mol Cell Biol. 1997;17:4096–4104. doi: 10.1128/mcb.17.7.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Estes P A, Cooke N E, Liebhaber S A. A native RNA secondary structure controls alternative splice-site selection and generates two human growth hormone isoforms. J Biol Chem. 1992;267:14902–14908. [PubMed] [Google Scholar]
- 15.ffrench-Constant C. Alternative splicing of fibronectin—many different proteins but few different functions. Exp Cell Res. 1995;221:261–271. doi: 10.1006/excr.1995.1374. [DOI] [PubMed] [Google Scholar]
- 16.Green M R. Biochemical mechanisms of constitutive and regulated pre-mRNA splicing. Annu Rev Cell Biol. 1991;7:559–599. doi: 10.1146/annurev.cb.07.110191.003015. [DOI] [PubMed] [Google Scholar]
- 17.Hertel K J, Lynch K W, Hsiao E C, Liu E H, Maniatis T. Structural and functional conservation of the Drosophila doublesex splicing enhancer repeat elements. RNA. 1996;2:969–981. [PMC free article] [PubMed] [Google Scholar]
- 18.Higgs D R, Goodbourn S E, Lamb J, Clegg J B, Weatherall D J, Proudfoot N J. Alpha-thalassaemia caused by a polyadenylation signal mutation. Nature. 1983;306:398–400. doi: 10.1038/306398a0. [DOI] [PubMed] [Google Scholar]
- 19.Huh G S, Hynes R O. Regulation of alternative pre-mRNA splicing by a novel repeated hexanucleotide element. Genes Dev. 1994;8:1561–1574. doi: 10.1101/gad.8.13.1561. [DOI] [PubMed] [Google Scholar]
- 20.Hynes R. Fibronectins. New York, N.Y: Springer-Verlag; 1990. [Google Scholar]
- 21.Jordan M, Schallhorn A, Wurm F M. Transfecting mammalian cells: optimization of critical parameters affecting calcium-phosphate precipitate formation. Nucleic Acids Res. 1996;24:596–601. doi: 10.1093/nar/24.4.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kornblihtt A R, Pesce C G, Alonso C R, Cramer P, Srebrow A, Werbajh S, Muro A F. The fibronectin gene as a model for splicing and transcription studies. FASEB J. 1996;10:248–257. doi: 10.1096/fasebj.10.2.8641558. [DOI] [PubMed] [Google Scholar]
- 23.Krainer A R. Eukaryotic mRNA processing. New York, N.Y: Oxford University Press Inc; 1997. [Google Scholar]
- 24.Lavigueur A, La Branche H, Kornblihtt A R, Chabot B. A splicing enhancer in the human fibronectin alternate ED1 exon interacts with SR proteins and stimulates U2 snRNP binding. Genes Dev. 1993;7:2405–2417. doi: 10.1101/gad.7.12a.2405. [DOI] [PubMed] [Google Scholar]
- 25.Libri D, Piseri A, Fiszman M Y. Tissue-specific splicing in vivo of the beta-tropomyosin gene: dependence on an RNA secondary structure. Science. 1991;252:1842–1845. doi: 10.1126/science.2063196. [DOI] [PubMed] [Google Scholar]
- 26.Lim L P, Sharp P A. Alternative splicing of the fibronectin EIIIB exon depends on specific TGCATG repeats. Mol Cell Biol. 1998;18:3900–3906. doi: 10.1128/mcb.18.7.3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu H X, Zhang M, Krainer A R. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev. 1998;12:1998–2012. doi: 10.1101/gad.12.13.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mardon H J, Sebastio G, Baralle F E. A role for exon sequences in alternative splicing of the human fibronectin gene. Nucleic Acids Res. 1987;15:7725–7733. doi: 10.1093/nar/15.19.7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKeown M. Alternative mRNA splicing. Annu Rev Cell Biol. 1992;8:133–155. doi: 10.1146/annurev.cb.08.110192.001025. [DOI] [PubMed] [Google Scholar]
- 30.Muro A F, Iaconcig A, Baralle F E. Regulation of the fibronectin EDA exon alternative splicing. Cooperative role of the exonic enhancer element and the 5′ splice site. FEBS Lett. 1998;437:137–141. doi: 10.1016/s0014-5793(98)01201-0. [DOI] [PubMed] [Google Scholar]
- 31.Oubridge C, Ito N, Evans P R, Teo C H, Nagai K. Crystal structure at 1.92 A resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature. 1994;372:432–438. doi: 10.1038/372432a0. [DOI] [PubMed] [Google Scholar]
- 32.Robberson B L, Cote G J, Berget S M. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 1990;10:84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi H, Hoffman B E, Lis J T. A specific RNA hairpin loop structure binds the RNA recognition motifs of the Drosophila SR protein B52. Mol Cell Biol. 1997;17:2649–2657. doi: 10.1128/mcb.17.5.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Solnick D. Alternative splicing caused by RNA secondary structure. Cell. 1985;43:667–676. doi: 10.1016/0092-8674(85)90239-9. [DOI] [PubMed] [Google Scholar]
- 35.Staffa A, Acheson N H, Cochrane A. Novel exonic elements that modulate splicing of the human fibronectin EDA exon. J Biol Chem. 1997;272:33394–33401. doi: 10.1074/jbc.272.52.33394. [DOI] [PubMed] [Google Scholar]
- 36.Staffa A, Cochrane A. Identification of positive and negative splicing regulatory elements within the terminal tat-rev exon of human immunodeficiency virus type 1. Mol Cell Biol. 1995;15:4597–4605. doi: 10.1128/mcb.15.8.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun Q, Dirksen W P, Rottman F M. RNA Processing Meeting—1995. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1995. An exonic splicing repressor in bovine growth hormone pre mRNA; p. 311. [Google Scholar]
- 38.Tacke R, Manley J L. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J. 1995;14:3540–3551. doi: 10.1002/j.1460-2075.1995.tb07360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vibe-Pedersen K, Kornblihtt A R, Baralle F E. Expression of a human alpha-globin/fibronectin gene hybrid generates two mRNAs by alternative splicing. EMBO J. 1984;3:2511–2516. doi: 10.1002/j.1460-2075.1984.tb02165.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watakabe A, Inoue K, Sakamoto H, Shimura Y. A secondary structure at the 3′ splice site affects the in vitro splicing reaction of mouse immunoglobulin mu chain pre-mRNAs. Nucleic Acids Res. 1989;17:8159–8169. doi: 10.1093/nar/17.20.8159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu R, Teng J, Cooper T A. The cardiac troponin T alternative exon contains a novel purine-rich positive splicing element. Mol Cell Biol. 1993;13:3660–3674. doi: 10.1128/mcb.13.6.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zuker M. Computer prediction of RNA structure. Methods Enzymol. 1989;180:262–288. doi: 10.1016/0076-6879(89)80106-5. [DOI] [PubMed] [Google Scholar]