The Linker Domain of Stat1 Is Required for Gamma Interferon-Driven Transcription (original) (raw)
Abstract
Upon binding of gamma interferon (IFN-γ) to its receptor, the latent transcription factor Stat1 becomes phosphorylated, dimerizes, and enters the nucleus to activate transcription. In response to IFN-α, Stat1 binds to Stat2 in a heterodimer that recruits p48, an IRF family member, to activate transcription. A number of functional domains of the STATs, including a C-terminal transactivation domain, a dimerization domain, and an SH2 domain, are known. However, the highly conserved residues between the DNA binding and SH2 domains (463 to 566), recently christened the linker domain on the basis of crystallographic studies, have remained without a known function. In the present study, we report that KE544-545AA point mutants in Stat1 abolish transcriptional responses to IFN-γ but not to IFN-α. We further show that this mutant Stat1 undergoes normal phosphorylation, nuclear translocation, and DNA binding. Taken together with recent structural evidence, these results suggest that the linker domain acts as a critical contact point during the construction of a Stat1-driven transcriptional complex.
The STATs are a family of transcription factors that are latent in the cytoplasm until activated in response to the occupation of a cell surface receptor by a polypeptide ligand (5, 23). They become activated by phosphorylation on a single tyrosine by either a JAK kinase bound to a cytokine receptor (e.g., gamma interferon [IFN-γ]) or a receptor tyrosine kinase (e.g., the epidermal growth factor receptor). The phosphorylated STATs dimerize, translocate to the nucleus, and initiate specific transcriptional programs. The highly conserved nature of STATs from humans to Drosophila has aided in identifying a number of functional domains in these proteins. For example, the carboxyl terminus (40 to 50 amino acids long) is required for transcriptional activation and can independently transactivate when coupled to a Gal4 binding element (3, 16, 17, 28, 32, 33). The region between residues ∼550 and 625 comprises an SH2 domain, which binds the single phosphotyrosine (around residue 700) of the opposing Stat monomer (22). STATs show slightly different preferences in DNA binding sites (21), and the specificity for DNA site selection can be transferred between STATs by swapping amino acids in the region between residues 400 and 500, implicating this region in DNA binding (12). STAT dimer-dimer interaction has recently been mapped to the amino-terminal 60 to 130 amino acids (25, 26, 30).
Additionally, interactions between STATs and several other DNA binding proteins or coactivators have been recently described and in some cases mapped. For example, the formation of ISGF3, the IFN-α-induced transcription factor, requires interaction between the IRF family member p48 and Stat1, an event critically dependent on K161 of Stat1 (11). STATs have also been shown to interact with the transcription factors USF-1, Sp1, and c-Jun and with the glucocorticoid receptor (15, 18, 19, 24). The coactivator CREB binding protein (CBP)/p300 has been shown to bind both the amino and carboxyl termini of Stat1 and Stat2 (9, 33). Recently, the replication factor MCM5 was also shown to interact with the carboxyl terminus of Stat1 (32).
One of the STAT regions which has eluded functional description is the linker domain (LD). The LD was originally described as an SH3 homology region prior to the resolution of the STAT and SH3 structures (7); it is now apparent that the LD consists of a highly helical, novel protein fold (2, 4). The LD spans residues 463 to 566 of Stat1 and sits between the DNA binding domain and the SH2 domain. This region constitutes one of the most highly conserved regions in the STAT molecules. In the present series of experiments, four highly conserved residues in the LD of Stat1 were mutated to alanine. Full-length Stat1 bearing mutations of K and E at residues 544 and 545 was found to lack the ability to induce transcriptional responses to IFN-γ. Phosphorylation, DNA binding, and nuclear translocation occurred normally, and the mutant protein retained the ability to participate in responses to IFN-α. The phenotype of the KE544-545 mutant closely resembles that of the C-terminally truncated isoform Stat1β, which also fails to support IFN-γ transcriptional responses but does participate in the IFN-α response. Thus, the LD may contain previously unrecognized contact points for the interaction of at least some STAT homodimers with the transcriptional machinery of the cell.
MATERIALS AND METHODS
Cell culture.
Human U3A cells deficient in Stat1 (gift of George Stark, Cleveland Clinic, Cleveland, Ohio) were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% bovine calf serum (Cosmic serum; HyClone) at 37°C and 5% CO2. Transient transfections were performed by the calcium phosphate method (1). Following 8 h of exposure to precipitate, cells were washed with phosphate-buffered saline and allowed to recover for 14 to 16 h before stimulation. Stable cell lines were constructed by selecting for and maintaining transformants in 400 μg of G418 per ml. A stable cell line expressing wild-type Stat1 was a gift of Curt Horvath (Mount Sinai School of Medicine). Clones were screened by Western blotting with Stat1c antibody. IFN-γ (gift of Amgen) was used at a concentration of 5 ng/ml, and IFN-α (gift of Hoffmann-La Roche) was used at a concentration of 500 IU/ml unless stated otherwise.
Plasmids.
Stat1 mammalian expression vectors were constructed by the insertion of cDNAs into the RC/CMV expression vector (Invitrogen). The luciferase reporter construct 3×Ly6E GAS-LUC was cloned as previously described (29). The Stat1 point mutations were generated according to a protocol by Ausubel et al. (1). Mutagenesis was confirmed by standard dideoxy termination sequencing. pCMVβ, a beta-galactosidase control vector, was purchased from Invitrogen.
Transcript analysis.
Luciferase assay was performed by transient transfection of pCMVβ (2 μg), Stat1 expression construct (8 μg), and 3×Ly6E GAS-LUC (8 μg) on semi-confluent 10-cm-diameter plates of U3A cells. Specifics of the protocol are described elsewhere (29). Each sample was assayed in triplicate for each experiment and normalized to beta-galactosidase activity. Similar results were obtained on three occasions. Reverse transcriptase (RT)-PCR was performed on stable cell lines following cytokine activation as described previously (11). Briefly, total RNA was isolated from confluent six-well plates by using Trizol reagent (Gibco BRL) and digested with DNase I, followed by reverse transcription. An aliquot of the cDNA was then used as a template for a PCR spiked with 0.1 μl of [α-32P]dATP per 25 μl of reaction mixture. Following a 25-cycle PCR, bands were resolved on a 5% acrylamide gel. Primers for the target genes were as follows: ISG54s, 5′-AATGCCATTTCACCTGGAACTTG-3′; ISG54a, 5′-GTGATAGTAGACCCAGGCATAGT-3′; GBPs, 5′-TGAGCAGCACCTTCGTGTACAAT-3′; GBPa, 5′-TAGGAACAGAAGTCTGCTACTTG-3′; IRF-1s, 5′-ATGAGACCCTGGCTAGAG-3′; IRF-1a, 5′-AAGCATCCGGTACACTCG-3′; ISG15s, 5′-CAACGAATTCCAGGTGTC-3′; ISG15a, 5′-CCCTTGTTATTCCTCACC-3′; GAPDHs, 5′-GTGAAGGTCGGAGTCAAC-3′; and GAPDHa, 5′-TGGAATTTGCCATGGGTG-3′.
Immunoblotting.
Whole-cell and fractionated-cell extracts were obtained by previously described procedures (22, 29). Western blotting was performed under standard conditions (1). Anti-Stat1c antibody has been previously described (20) and was used at a 1:1,000 dilution for blotting. Covalent modifications were detected by anti-phosphotyrosine 701 (New England Biolabs) and anti-phosphoserine 727 (Upstate Biotechnology) antibodies used at a 1:1,000 dilution. Nitrocellulose membranes were stripped for 30 min at 55°C in a solution containing 0.7% β-mercaptoethanol, 65 mM Tris (pH 6.5), and 2% sodium dodecyl sulfate (SDS).
Electrophoretic mobility shift assay (EMSA).
Fractionated- and whole-cell extracts (5 to 8 μg) were assayed for DNA binding activity with 32P-labeled m67 SIE oligonucleotide (27). DNA-protein complexes were then resolved on a 4% (29:1) polyacrylamide-bisacrylamide gel at 4°C (400 V, 0.25× Tris-borate-EDTA). Quantitation and analysis were performed on a Molecular Dynamics Storm PhosphorImager as previously described (8).
Interferon cytopathic-effect assay.
Assays for interferon-mediated protection from viral infection have been described extensively elsewhere (10). Briefly, three 12-well clusters of a given cell line were grown to confluence on 96-well plates. After treatment with 1,000 IU of IFN-α per ml or 10 ng of IFN-γ per ml, the cell culture medium was aspirated off, and the medium was replaced with prewarmed, serum-free Dulbecco’s modified Eagle’s medium containing encephalomyocarditis virus (EMCV). After 24 h, the medium was removed and the viable cell population was visualized by staining with 2% methylene blue in 50% ethanol.
GST protein-protein association assay.
The glutathione _S_-transferase (GST) reagents were produced by a 3-h isopropylthiogalactoside induction of bacteria carrying the respective constructs at 29°C. Purification was performed according to the manufacturer’s instructions (Amersham Pharmacia). The GSTMCM5(251-400) and GSTMCM5(351-400) constructs were generated by PCR subcloning of the appropriate fragments into the _Bam_HI and _Hin_dIII sites in pRSETA (Invitrogen) and were then further subcloned into pGEX-2TK (Pharmacia) by using the _Bam_HI sites and the filled-in _Hin_dIII site with the _Sma_I site. The CBP CREB binding domain (CBD) fusion protein was produced as described previously (33). The pull-down assay was performed by preclearing a radioimmunoprecipitation assay (RIPA) buffer extract containing IFN-γ-activated cells with glutathione-Sepharose 4B beads. The extracts were then rocked overnight at 4°C in the presence of 10 to 15 μg of fusion protein bound to glutathione-Sepharose beads. Following three 10-min washes (two RIPA and one whole cell), the precipitated extract was separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by Western blotting with anti-Stat1n monoclonal antibody (Transduction Laboratories).
RESULTS
Analysis of target gene induction with full-length Stat1 point mutants in the LD.
The LD of the STATs contains several residues that are highly conserved throughout STATs from mammals and lower organisms (Fig. 1). Reasoning that functionally important residues would be the most conserved, we prepared four alanine mutants with sequence identity throughout the human STATs: F506, R512, W539, and KE544-545 (the KE mutant consisted of a double mutation to AA544-545). In addition, when these experiments began, the three-dimensional structure was not available, and the hypothesis that the region from ∼500 to 580 was an SH3 domain was still entertained. Along these lines, sequence alignment suggested that W539 might be the critical tryptophan which lines the SH3 binding pocket and is required for polyproline helix binding activity (6).
FIG. 1.
Sequence alignment of human and Drosophila STAT LDs. The residues having sequence identity with Stat1 are in boxes. Homologous residues are indicated by shading. In the present experiments, four residues were mutated to alanine, as indicated by the asterisks: F506, R512, W539, and KE544-545. Sequence alignment was performed according to the ClustalW method (24a) and rendered with SeqVu 1.1.
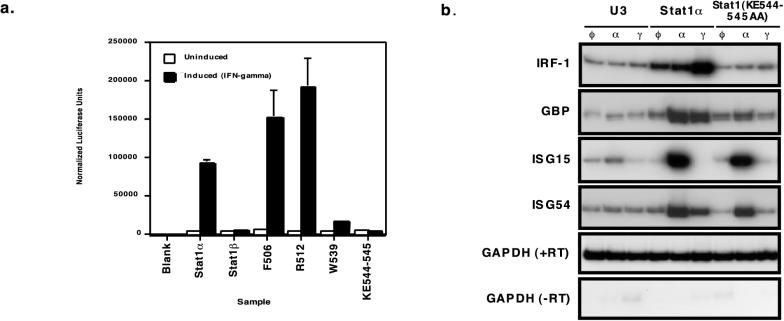
As an initial screen for the altered phenotype of these point mutants, the ability to induce gene expression upon stimulation with IFN-γ was tested. A luciferase reporter construct with three Stat1 binding sites (3×Ly6E GAS-LUC) was transiently cotransfected into U3A cells which lack Stat1 with vectors expressing the full-length Stat1 in either wild-type (Stat1α) or point mutant form. Transfected cells were then stimulated with IFN-γ for 4 h or left untreated. Upon normalization with a cotransfected beta-galactosidase control plasmid, the results showed the expected ∼25-fold induction for wild-type Stat1α and no induction with transcriptionally inactive Stat1β. While the F506 and R511 mutants exhibited slightly better-than-normal gene induction, the W539 and KE544-545 mutants were markedly attenuated (Fig. 2a). Subsequent studies showed that the Stat1(KE544-545AA) mutants failed to induce transcription at 2, 4, and 6 h and that the wild-type Stat1 induced transcription at all these time points (31).
FIG. 2.
Transcription activity of Stat1 point mutants. (a) Transient transfections were carried out with the U3A (no Stat1) cell line by using the indicated Stat1 expression vectors. Transcriptional responses to IFN-γ were monitored by the 3×Ly6E GAS-LUC reporter plasmid after 4 h of IFN-γ stimulation. This experiment was performed three times, each time with triplicate samples. A representative experiment is shown with the standard errors for the three samples. (b) RT-PCR analysis was performed by using wild-type-complemented (Stat1α) or mutant-complemented [Stat1(KE544-545AA)] U3A-derived stable cell lines. Cells were treated with the specified interferon for 4 h or left untreated, followed by RNA harvesting and reverse transcription. PCR amplification of the resulting cDNAs with radioactive nucleotides was performed with a primer pair representing the mRNAs indicated on the left. The products were resolved by acrylamide electrophoresis and autoradiography.
To examine the effect of the mutant Stat1 on the induction of physiologically relevant target genes, we generated U3A-derived stable transfectants of the full-length Stat1(KE544-545AA) mutant and analyzed interferon-induced transcripts by RT-PCR. Because response to IFN-α requires Stat2-Stat1 heterodimers but not the Stat1 transactivation domain, we tested responses to both IFN-α and IFN-γ to obtain information regarding the structural and functional integrity of the Stat1(KE544-545AA) mutant protein. A representative experiment is shown in Fig. 2b. The IRF-1 and guanylate binding protein genes were induced much more strongly by IFN-γ than IFN-α, whereas ISG15 and ISG54 were activated only by IFN-α. These differences in gene activation reflect the differing specificities of Stat1-Stat1 and Stat2-Stat1 dimers. While the cells containing wild-type Stat1 exhibited marked and appropriate gene induction in response to both interferons, the Stat1(KE544-545AA) mutant cell line failed to activate transcription of IFN-γ-inducible genes while retaining responsiveness to IFN-α. The W539A mutant exhibited a similar pattern of responsiveness (31).
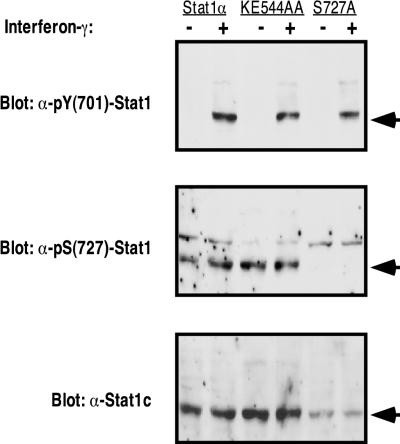
Stat1(KE544-545AA) mutants undergo phosphorylation at tyrosine 701 and serine 727.
Impaired phosphorylation, at either tyrosine 701 or serine 727, was one possible explanation for the mutant phenotype. To determine the status of the two known phosphorylation sites, stable cell lines expressing wild-type Stat1α, the Stat1(KE544-545AA) mutant, or the Stat1(S727A) mutant were treated with IFN-γ, and the resulting extracts were probed with antibodies specific for tyrosine- or serine-phosphorylated Stat1 (Fig. 3). As shown in Fig. 3, treatment with IFN-γ clearly induced tyrosine phosphorylation of Stat1 in all three cell lines. In addition, both wild-type and mutant Stat1 had constitutive phosphorylation at serine 727 despite a 24-h serum starvation period prior to the experiment. The specificity of the pS(727)-Stat1 antibody is confirmed by its failure to stain the lanes with extract from an S727A mutant cell line. Constitutive S727 phosphorylation in U3A cell line derivatives has been noted before (29, 35) and makes it difficult to assess regulation of serine phosphorylation in these U3A-derived cells. Nonetheless, it is clear that the absence of S727 phosphorylation does not contribute to the observed phenotype of the Stat1(KE544-545AA) mutants.
FIG. 3.
Stat1(KE544-545AA) mutants undergo normal posttranslational modification. Stable cell lines complemented with Stat1α, the Stat1(KE544-545AA) mutant, or the Stat1(S727A) mutant were grown to ∼20% confluence, serum starved for 24 h, and activated for 1 h with IFN-γ. Cells were lysed in whole-cell extract buffer, and the extracts were resolved by SDS-PAGE. Forty micrograms of extract was used for the Stat1α and S727A cell lines, but 20 μg of protein was used for the Stat1(KE544-545AA) cell line to compensate for differences in protein expression levels. Western blotting was first performed with the pY(701)-Stat1 antibody. The nitrocellulose membrane was then stripped and reprobed with the other two antibodies listed. The expected band position of Stat1 is indicated by the arrow to the right of each panel.
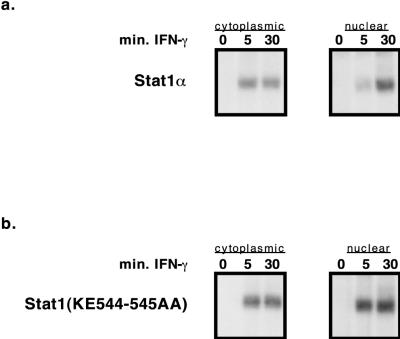
The Stat1(KE544-545AA) mutant translocates to the nucleus and can bind DNA.
To examine whether the Stat1(KE544-545AA) mutant protein entered the nucleus and was competent to bind DNA, Stat1-complemented U3A stable cell lines were treated with IFN-γ, and fractionated-cell extracts were prepared. DNA binding Stat1 molecules were then detected by using EMSA with a radioactively labeled m67 SIE deoxyoligonucleotide probe. Though the Stat1(KE544-545AA) mutant’s DNA binding activity consistently exhibited slightly faster accumulation in the nucleus than wild-type Stat1’s (Fig. 4), wild-type Stat1 and mutant Stat1 were clearly capable of entering the nucleus and binding the oligonucleotide probe to approximately the same extent.
FIG. 4.
Stat1(KE544-545AA) mutants bind DNA and enter the nucleus. Wild-type and Stat1(KE544-545AA) mutant cell lines were activated for 0, 5, or 30 min with IFN-γ. Following extract fractionation, the DNA binding activity of each extract was assayed by EMSA with a radiolabeled m67 SIE oligonucleotide.
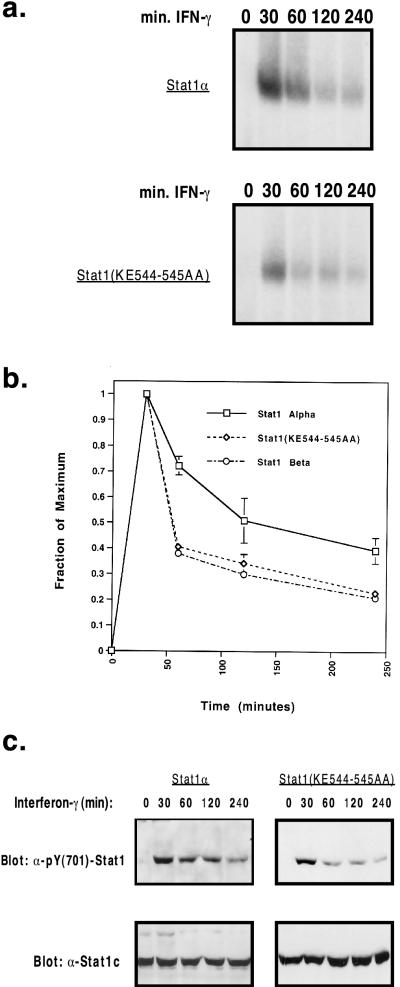
Phosphorylated Stat1(KE544-545AA) persists in the cell over a 4-h period.
Given the otherwise normal biochemical profile of the Stat1(KE544-545AA) mutant, we sought to see if there were any differences between the activation-inactivation cycles of mutant and wild-type Stat1 over a 4-h course of IFN-γ treatment (8). As shown in Fig. 5a, whole-cell extracts of both mutant and wild-type cell lines showed similar DNA binding activities over the entire 4-h period. These results were consistent with those of a corresponding Western blot for phosphotyrosine in a similar experiment (Fig. 5c). We then quantitated and averaged the EMSA signals of at least two of these time course experiments (Fig. 5b). By referencing any signal to the peak binding activity at 30 min, the relative DNA binding activity at each time point can be determined. The residual signals at 4 h were found to be slightly different: 40% for the wild type and 23% for the Stat1(KE544-545AA) mutant. This discrepancy can be attributed largely to differences in the rate of decay between 30 and 60 min, with the rates of decay being similar between 60 and 240 min. Interestingly, a set of kinetic experiments comparing the decay of Stat1β to that of Stat1α revealed a similar pattern of small differences in signal decay. Given the severity of the phenotype of the Stat1(KE544-545AA) mutant, the slightly faster inactivation of the protein seems an unlikely explanation for the complete failure to activate target genes in response to IFN-γ.
FIG. 5.
Kinetics of the response to IFN-γ by Stat1α and Stat1(KE544-545AA). (a) EMSA of whole-cell extracts from interferon-stimulated cell lines. Stat1α and Stat1(KE544-545AA) mutants were exposed to IFN-γ for the indicated periods and were assayed as described in the legend to Fig. 4. (b) Autoradiograms from time course experiments were quantitated by a PhosphorImager. Normalization to the maximum signal intensity (30 min) yielded the decay pattern shown. The graph represents the averages of two to three independent experiments. (c) Western blots of a time course experiment comparing phosphorylations of Stat1α and Stat1(KE544-545AA). Following interferon stimulation for the indicated periods, 30 μg of whole-cell extract from each experimental condition was resolved by SDS-PAGE and probed with the pY(701)-Stat1 antibody. Membranes were then stripped and reprobed with the Stat1c antibody.
Biochemical characteristics of wild-type Stat1 and mutant Stat1 parallel their ability to confer an antiviral response to the interferons.
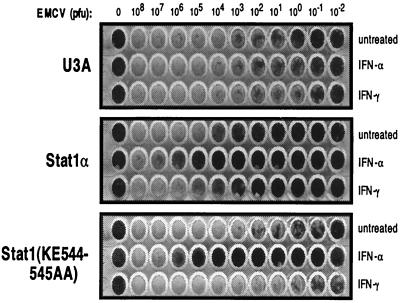
The interferons were first identified on the basis of their ability to induce a protective antiviral state. This observed antiviral activity requires Stat1-driven gene induction in response to the interferons (10). For example, IFN-γ-mediated resistance to EMCV, a picornavirus, is highly dependent on IRF-1 induction (13). To determine the effects of the Stat1(KE544-545AA) mutant on the induction of a physiologic pathway, we treated U3A, Stat1α-complemented U3A, and Stat1(KE544-545AA)-complemented U3A cell lines with interferon for 14 to 16 h, followed by infection with serially diluted EMCV (Fig. 6). IFN-α treatment protected both mutant- and wild-type-complemented cell lines, in accord with our previous observations of target gene responses by RT-PCR (Fig. 2b). However, IFN-γ protection was seen only with the wild-type Stat1α cell line. This cell line had a much higher resistance to infection than the other two cell lines when left untreated, a result which may be due to the high IRF-1 background of this cell line (Fig. 2b). In any case, wild-type-complemented cells were protected by both interferons, while the Stat1(KE544-545AA) mutant cell line was protected only by IFN-α. Thus, the resistance to viral infection correlated with the biochemical assays of the Stat1(KE544-545AA) mutant.
FIG. 6.
Antiviral response of cells expressing Stat1α and Stat1(KE544-545AA). The indicated cell lines were grown to confluence and then treated with the indicated cytokine for 14 to 16 h. Serum-free stocks of EMCV were then prepared and distributed to the appropriate wells. Following 24 h of incubation, the remaining viable cells were visualized by methylene blue staining.
The Stat1(KE544-545AA) mutant interacts normally with two proteins implicated in Stat1 transactivation.
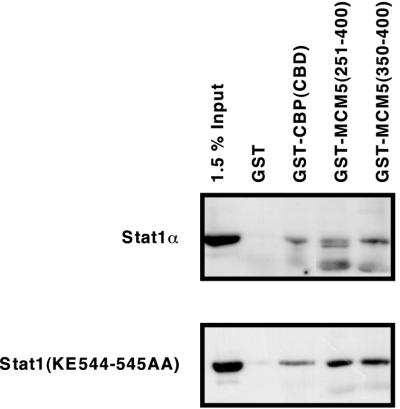
Although all known protein interactions with Stat1 map outside of the LD, we decided to test whether our Stat1(KE544-545AA) mutant could disrupt any such known interactions. Using GST fusion proteins to precipitate cell extracts from wild-type- or mutant-complemented U3A cell lines, we assessed the interaction of Stat1 with the CBP CBD and two overlapping regions of MCM5. As shown in Fig. 7, both wild-type and mutant Stat1 interacted specifically with the regions of CBP and MCM5 tested. Therefore, it is unlikely that disruption of the interaction with CBP or MCM5 accounts for the observed phenotype.
FIG. 7.
Stat1(KE544-545AA) interacts normally with CBP and MCM5 fusion proteins. RIPA extracts of IFN-γ-activated, mutant- and wild-type-complemented U3A cell lines were precipitated overnight with GST, GSTCBP(CBD), GSTMCM5(251-400), or GSTMCM5(350-400). After washing and separation by SDS-PAGE (7% acrylamide), the precipitates were visualized by a monoclonal antibody to the Stat1 N terminus.
DISCUSSION
The determination of the three-dimensional structure of the cores of Stat1 and Stat3 ended speculation that the region of the STATs from ∼500 to 580 was an SH3 domain: the SH3 structure is characterized by β sheets (14), whereas the STAT domain from ∼500 to 580 is entirely composed of α-helical elements (2, 4). On structural grounds, the role of the region from 500 to 580 is to serve as a linker between the SH2 domain and the section of the STAT proteins which contains DNA contacts, the DNA binding domain. However, the structural analyses do not suggest an obvious biochemical function of the LD.
When it was initially found that Stat1(KE544-545AA) mutants affected IFN-γ transcriptional induction in the commonly used luciferase assay, it was not clear whether the protein was somehow destabilized by the mutations. However, in response to IFN-γ, tyrosine and serine phosphorylation, dimerization, nuclear translocation, and DNA binding of the Stat1(KE544-545AA) protein all occurred normally. Moreover, the mutant protein very clearly can continue to function normally in the IFN-α response pathway, suggesting that the mutant phenotype is due to a highly specific loss of function rather than a global problem with protein stability or folding. GST pull-down assays suggest that Stat1(KE544-545AA) also interacts normally with MCM5 and CBP.
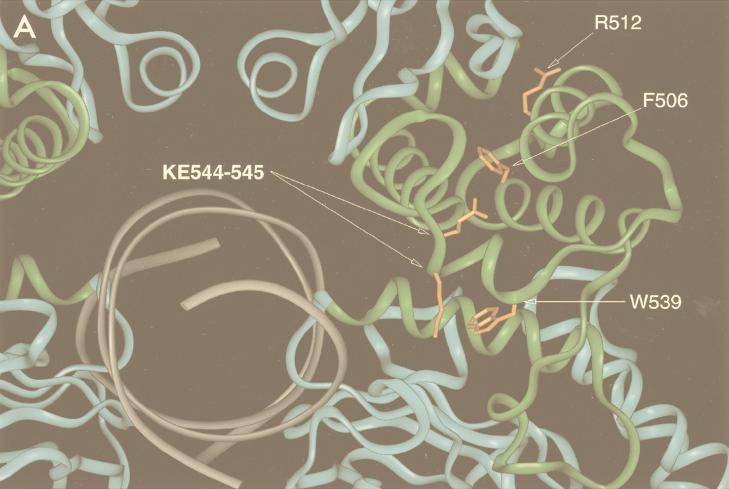
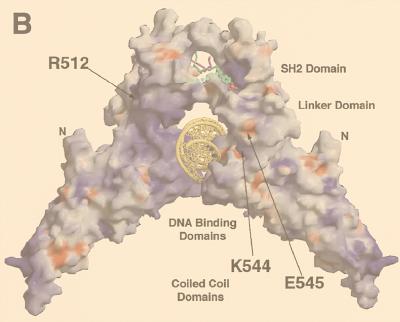
The locations of the mutations in this study also merit reconsideration in light of the Stat1 crystal structure. Of the four mutations made, two are completely or partially buried (W539 and F506) and two (KE544-545 and R512) fall on the charged surface of the protein (Fig. 8A). In the case of KE544-545, the residues create two charged pits on the surface of the molecule proximal to the major groove of the bound DNA element (Fig. 8B). Mutation of the residues to alanine in our experiments effectively removed these charges, which we found to be necessary for Stat1-driven transcriptional activation. Proteins which bind to this region are being sought but have not yet been found (31).
FIG. 8.
Structure-function relationships in the Stat1 LD. Both panels depict a Stat1 dimer core (residues 130 to 712) bound to an m67 SIE oligonucleotide as reported by Chen et al. (4). (A) Depiction of the residues mutated to alanine in this study. The oligonucleotide is indicated in gray, with the DNA binding and SH2 domains located at the bottom and top, respectively. The LD is highlighted in green, and the mutated residues are orange. (B) Mutated residues in the context of the exposed surface of Stat1. The positive surface charge is blue, the negative surface charge is red, and the neutral charge is white-gray. The figure is adapted from reference 4 with permission.
As for the specific inability of the Stat1(KE544-545AA) mutant to function in the IFN-γ pathway, there are several plausible explanations. The most obvious is that the LD is by itself an independent transactivation domain. This idea was tested with LD-Gal4 fusions by Gal4 luciferase assay, but no transactivation activity was observed. There is, of course, no guarantee of proper folding of the LD under such circumstances, and so no firm conclusion is yet possible (31). A second, perhaps more likely, possibility is that the LD cooperates in an obligatory manner with the known COOH-terminal transactivation domain. The core structure presently available lacks the COOH-terminal 38 amino acids of Stat1. It seems that with such a short run of amino acids an actual physical interaction between the LD and the terminal 38 amino acids is unlikely. But perhaps dual contacts between the two STAT domains and coactivators or general transcription factors of the basal transcription machinery can occur. There is precedence for such dual contacts with the STATs. For example, the histone acetyltransferase CBP contacts Stat1 twice, at both the COOH-terminal and the amino-terminal regions, which are quite far apart (33). Finally, it is possible that the C terminus of Stat1 attracts one set of coactivators and that the LD attracts another, independent set of coactivators. Regardless of which model proves to be true, any interaction with the LD may be essential to transcriptional responses, given the complete loss of transactivation by the KE544-545AA mutant.
ADDENDUM
Another protein was recently implicated in Stat1-mediated transcriptional activation (34). The Nmi protein was shown to boost transcriptional responses to IFN-γ and to interact with Stat1. Additionally, Stat5 was shown to interact with Nmi via its coiled-coil domain. We are currently attempting to characterize the interaction of Stat1(KE544-545AA) with Nmi.
ACKNOWLEDGMENTS
We thank Yuhong Shen for the gift of titered EMCV, amplified from a stock provided by Robert H. Silverman (Cleveland Clinic). We also thank members of the Darnell lab for helpful discussions and Lois Cousseau for manuscript preparation.
This work was supported by NIH grants AI32489 and AI34420 to J.E.D. E.Y. is supported by MSTP grant GM07739, and R.H. is supported by NIH training grant CA09673.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1994. [Google Scholar]
- 2.Becker S, Groner B, Mueller C W. Three-dimensional structure of the Stat3β homodimer bound to DNA. Nature. 1998;394:145–151. doi: 10.1038/28101. [DOI] [PubMed] [Google Scholar]
- 3.Bhattacharya S, Eckner R, Grossman S, Oldread E, Arany Z, D’Andrea A, Livingston D M. Cooperation of Stat2 and p300/CBP in signalling induced by interferon-α. Nature. 1996;383:344–347. doi: 10.1038/383344a0. [DOI] [PubMed] [Google Scholar]
- 4.Chen X, Vinkemeier U, Zhao Y, Jeruzalmi D, Darnell J E, Jr, Kuriyan J. Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell. 1998;93:827–839. doi: 10.1016/s0092-8674(00)81443-9. [DOI] [PubMed] [Google Scholar]
- 5.Darnell J E., Jr STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 6.Erpel T, Superti-Furga G, Courtneidge S A. Mutational analysis of the Src SH3 domain: the same residues of the ligand binding surface are important for intra- and intermolecular interactions. EMBO J. 1995;14:963–975. doi: 10.1002/j.1460-2075.1995.tb07077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu X-Y. A transcription factor with SH2 and SH3 domains is directly activated by an interferon α-induced cytoplasmic protein tyrosine kinase(s) Cell. 1992;70:323–335. doi: 10.1016/0092-8674(92)90106-m. [DOI] [PubMed] [Google Scholar]
- 8.Haspel R L, Salditt-Georgieff M, Darnell J E., Jr The rapid inactivation of nuclear tyrosine phosphorylated Stat1 depends upon a protein tyrosine phosphatase. EMBO J. 1996;15:6262–6268. [PMC free article] [PubMed] [Google Scholar]
- 9.Horvai A E, Xu L, Korzus E, Brard G, Kalafus D, Mullen T-M, Rose D W, Rosenfield M G, Glass C K. Nuclear integration of JAK/STAT and Ras/AP-1 signaling by CBP and p300. Proc Natl Acad Sci USA. 1997;94:1074–1079. doi: 10.1073/pnas.94.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horvath C M, Darnell J E., Jr The antiviral state induced by alpha interferon and gamma interferon requires transcriptionally active Stat1 protein. J Virol. 1996;70:647–650. doi: 10.1128/jvi.70.1.647-650.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horvath C M, Stark G R, Kerr I M, Darnell J E., Jr Interactions between STAT and non-STAT proteins in the interferon-stimulated gene factor 3 transcription complex. Mol Cell Biol. 1996;16:6957–6964. doi: 10.1128/mcb.16.12.6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horvath C M, Wen Z, Darnell J E., Jr A STAT protein domain that determines DNA sequence recognition suggests a novel DNA-binding domain. Genes Dev. 1995;9:984–994. doi: 10.1101/gad.9.8.984. [DOI] [PubMed] [Google Scholar]
- 13.Kimura T, Nakayama K, Penninger J, Kitagawa M, Harada H, Matsuyama T, Tanaka N, Kamijo R, Vilcek J, Mak T W, Taniguchi T. Involvement of the IRF-1 transcription factor in antiviral responses to interferons. Science. 1994;264:1921–1924. doi: 10.1126/science.8009222. [DOI] [PubMed] [Google Scholar]
- 14.Kuriyan J, Cowburn D. Modular peptide recognition domains in eukaryotic signalling. Annu Rev Biophys Biomol Struct. 1997;26:259–288. doi: 10.1146/annurev.biophys.26.1.259. [DOI] [PubMed] [Google Scholar]
- 15.Look D C, Pelletier M R, Tidwell R M, Roswit W T, Holtzman M J. Stat 1 depends on transcriptional synergy with Sp1. J Biol Chem. 1995;270:30264–30267. doi: 10.1074/jbc.270.51.30264. [DOI] [PubMed] [Google Scholar]
- 16.Lu B, Reichel M, Fisher D, Smith J, Rothman P. Identification of a STAT6 domain required for IL-4 induced activation of transcription. J Immunol. 1997;159:1255–1264. [PubMed] [Google Scholar]
- 17.Moriggl R, Berchtold S, Friedrich K, Standke G J R, Kammer W, Heim M, Wissler M, Stöcklin E, Gouilleux F, Groner B. Comparison of the transactivation domains of Stat5 and Stat6 in lymphoid cells and mammary epithelial cells. Mol Cell Biol. 1997;17:3663–3678. doi: 10.1128/mcb.17.7.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muhelthaler-Mottet A, Di Berardino W, Otten L A, Mach B. Activation of the MHC class II transactivator CIITA by interferon-gamma requires cooperative interaction between Stat1 and USF-1. Immunity. 1998;8:157–166. doi: 10.1016/s1074-7613(00)80468-9. [DOI] [PubMed] [Google Scholar]
- 19.Schaefer T S, Sanders L K, Nathans D. Cooperative transcriptional activity of Jun and Stat3β, a short form of Stat3. Proc Natl Acad Sci USA. 1995;92:9097–9101. doi: 10.1073/pnas.92.20.9097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schindler C, Shuai K, Prezioso V R, Darnell J E., Jr Interferon-dependent tyrosine phosphorylation of a latent cytoplasmic transcription factor. Science. 1992;257:809–813. doi: 10.1126/science.1496401. [DOI] [PubMed] [Google Scholar]
- 21.Seidel H M, Milocco L H, Lamb P, Darnell J E, Jr, Stein R B, Rosen J. Spacing of palindromic half sites as a determinant of selective STAT (signal transducers and activators of transcription) DNA binding and transcriptional activity. Proc Natl Acad Sci USA. 1995;92:3041–3045. doi: 10.1073/pnas.92.7.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shuai K, Horvath C M, Tsai-Huang L H, Qureshi S, Cowburn D, Darnell J E., Jr Interferon activation of the transcription factor Stat91 involves dimerization through SH2-phosphotyrosyl peptide interactions. Cell. 1994;76:821–828. doi: 10.1016/0092-8674(94)90357-3. [DOI] [PubMed] [Google Scholar]
- 23.Stark G R, Kerr I M, Williams B R G, Silverman R H, Schreiber R D. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 24.Stocklin E, Wissler M, Gouilleux F, Groner B. Functional interactions between Stat5 and the glucocorticoid receptor. Nature. 1996;383:726–728. doi: 10.1038/383726a0. [DOI] [PubMed] [Google Scholar]
- 24a.Transfac-Team. 1998, copyright date. Sequence alignment. [Online.] http://transfac.gbf.de/dbsearch/clustalw.html. [19 May 1999, last date accessed.]
- 25.Vinkemeier U, Cohen S L, Moarefi I, Chait B T, Kuriyan J, Darnell J E., Jr DNA binding of in vitro activated Stat1α, Stat1β, and truncated Stat1: interaction between NH2-terminal domains stabilizes binding of two dimers to tandem DNA sites. EMBO J. 1996;15:5616–5626. [PMC free article] [PubMed] [Google Scholar]
- 26.Vinkemeier U, Moarefi I, Darnell J E, Jr, Kuriyan J. Structure of the amino-terminal protein interaction domain of STAT-4. Science. 1998;279:1048–1052. doi: 10.1126/science.279.5353.1048. [DOI] [PubMed] [Google Scholar]
- 27.Wagner B J, Hayes T E, Hoban C J, Cochran B H. The SIF binding element confers sis/PDGF inducibility onto the c-fos promoter. EMBO J. 1990;9:4477–4484. doi: 10.1002/j.1460-2075.1990.tb07898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang D, Stravopodis D, Teglund S, Kitazawa J, Ihle J N. Naturally occurring dominant negative variants of Stat5. Mol Cell Biol. 1996;16:6141–6148. doi: 10.1128/mcb.16.11.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wen Z, Zhong Z, Darnell J E., Jr Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 30.Xu X, Sun Y-L, Hoey T. Cooperative DNA binding and sequence selective recognition conferred by the STAT amino-terminal domain. Science. 1996;273:794–797. doi: 10.1126/science.273.5276.794. [DOI] [PubMed] [Google Scholar]
- 31.Yang, E., and J. E. Darnell. Unpublished observations.
- 32.Zhang J J, Zhao Y, Chait B T, Lathem W W, Ritzi M, Knippers R, Darnell J E., Jr Ser727-dependent recruitment of MCM5 by Stat1α in IFN-γ-induced transcriptional activation. EMBO J. 1998;17:6963–6971. doi: 10.1093/emboj/17.23.6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J J, Vinkemeier U, Gu W, Chakravarti D, Darnell J E., Jr Two contact regions between Stat1 and CBP/p300 in interferon γ signalling. Proc Natl Acad Sci USA. 1996;93:15092–15096. doi: 10.1073/pnas.93.26.15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu M H, John S, Berg M, Leonard W J. Functional association of Nmi with Stat5 and Stat1 in IL-2 and IFN-γ-mediated signalling. Cell. 1999;96:121–130. doi: 10.1016/s0092-8674(00)80965-4. [DOI] [PubMed] [Google Scholar]
- 35.Zhu X, Wen Z, Xu L Z, Darnell J E., Jr Stat1 serine phosphorylation occurs independently of tyrosine phosphorylation and requires an activated Jak2 kinase. Mol Cell Biol. 1997;17:6618–6623. doi: 10.1128/mcb.17.11.6618. [DOI] [PMC free article] [PubMed] [Google Scholar]