Massive Apoptosis of Thymocytes in T-Cell-Deficient Id1 Transgenic Mice (original) (raw)
Abstract
Id1 is an inhibitor of a group of basic helix-loop-helix transcription factors, collectively called E proteins, which includes E12, E47, E2-2, and HEB. We have generated transgenic mice in which Id1 is specifically expressed in T cells. The total number of thymocytes in these mice is less than 4% of that in wild-type mice. The majority of the transgenic thymocytes are CD4 and CD8 double negative and bear the cell surface markers of multipotent progenitor cells. A small number of thymocytes, however, differentiate into CD4 or CD8 single-positive T cells, which also display different characteristics from their wild-type counterparts. More importantly, apoptotic cells constitute about 50% of the total thymocytes. These apoptotic thymocytes have rearranged their T-cell receptor genes, suggesting that they are differentiating T cells. This finding has raised the possibility that the T-cell deficiency in Id1 transgenic mice is the result of a massive apoptosis of differentiating T cells triggered by Id1 expression as opposed to a developmental block at the earliest progenitor stage. The progenitor cells accumulated in the transgenic mice might have survived because they are not susceptible to the apoptotic signals. Despite the massive cell death of the thymocytes at young ages, Id1 transgenic mice frequently develop T-cell lymphoma later in their life span, and lymphomagenesis appears to occur at different stages of T-cell development. Taken together, our data suggest that E proteins, being the targets of Id1, are essential regulators for normal T-cell differentiation and tumor suppression.
A subclass of the basic helix-loop-helix family of transcription factors includes E12, E47, E2-2, and HEB proteins (24, 25, 40), which are collectively called E proteins. E12 and E47 are encoded by the E2A gene as a result of alternative splicing (40, 55), whereas E2-2 and HEB are products of their respective genes. Although encoded by different genes, these E proteins are highly homologous in their DNA binding, dimerization and _trans_-activation domains (36, 50). Consequently, they have similar functions; i.e., they bind to E-box sequences and activate transcription. In lymphoid cells, the E proteins exist predominantly as homo- or heterodimers among themselves (4), even though they can form heterodimers with other tissue-specific basic helix-loop-helix proteins such as MyoD and NeuroD/BETA2 in muscle, neuronal, and pancreatic tissues (29, 41, 42). The critical role of E proteins in B-lymphocyte development has been clearly demonstrated by the characterization of E2A-deficient mice (3, 64). Disruption of the E2A gene leads to a block of B-cell differentiation at the earliest stage (fraction A stage; B220+ HSA− BP1− CD43−) (4, 22). In these mice, no rearrangement of the immunoglobulin genes takes place and most of the B-cell-specific genes are not expressed. In comparison, null mutation of the E2-2 or HEB gene causes a modest impairment of B-cell development (65), probably due to the relatively low abundance of the E2-2 and HEB proteins expressed in the B-cell lineage. HEB is able to rescue B-cell development in E2A-deficient mice when its cDNA is inserted in the E2A locus so that HEB can be translated through an internal ribosomal entry site (65). Therefore, HEB can functionally complement E2A. Similar to strategies of disrupting the E2A gene, we have previously generated transgenic mice in which Id1 is specifically expressed in B cells (57). Because Id1 forms heterodimers with E proteins and prevents them from binding to DNA, Id1 acts as a naturally occurring dominant negative inhibitor of E proteins. In these transgenic mice, B-cell development is also blocked at the earliest stage (57), a stage when endogenous Id1 is expressed at a high level (32). Taken together, these findings suggest that E proteins are essential for B-cell development.
Analogous to B-cell development, T-cell differentiation is also a stepwise process that involves the sequential rearrangement and expression of T-cell receptor (TCR) genes as well as the expression of other T-cell-specific genes. Rather than developing in the bone marrow as B cells do, the primary site for T-cell differentiation is the thymus. The developmental stages of T cells are well characterized by the expression of cell surface markers. In general, T cells in the thymus can be divided into four populations: CD4 and CD8 double negative (DN), double positive (DP), and CD4 (CD4+) or CD8 (CD8+) single positive (26). T cells in the thymus develop from the DN to the DP stage through an intermediate stage, termed the immature CD8 single-positive (ISP) stage. The DP cells then differentiate into single-positive cells before exiting the thymus. Early T-cell development at the DN stage can be further characterized by the expression of CD44 and CD25 cell surface markers. Multipotent progenitor cells marked as CD44+ CD25− commit to the T-cell lineage and differentiate through the CD44+ CD25+, CD44− CD25+, and then CD44− CD25− stages. In the process, the TCR β gene is rearranged and expressed. The β chain pairs with the pre-T α chain to form a pre-TCR, which is essential for proliferation and differentiation of immature T cells. Data from gene disruption experiments have demonstrated that lack of any of the components that lead to the formation of pre-TCRs, as well as the CD3 signal-transducing molecules, results in the arrest of T-cell differentiation at the CD44− CD25+ stage (20, 35, 38, 53). However, the mechanisms that dictate the differentiation of progenitor cells from the CD44+ CD25− stage to the CD44− CD25+ stage are less well understood. Parallel with the progression of the developmental program are the apoptosis and antiapoptosis events that ensure the survival of properly developed T cells and the elimination of unwanted T cells. These events are intimately related to the processes of TCR rearrangement and mitogenic stimulation through TCR-mediated signaling pathways as well as other signaling pathways. Shifting the balance between apoptosis and antiapoptosis has direct consequences on T-cell development.
In contrast to the well-defined role of E2A gene products in B-cell development, the role of E proteins in T-cell ontogeny is not entirely clear. Homozygous E2A or HEB-deficient mice display moderate defects in T-cell development (45, 63). The E2A-deficient mice have reduced numbers of T cells in both the thymus and the spleen. The percentage of DP thymocytes decreases by 50%, while the percentage of DN cells and CD8+ cells increases significantly in either E2A or HEB-deficient mice. These modest effects of E2A or HEB mutation raise the question whether the E proteins are merely facilitators of T-cell development or whether they are actually crucial regulators in T-cell development but their role has not been revealed due to the redundant function of E2A and HEB genes. Because of the neonatal death of E2A and HEB deficient mice respectively, it has not been possible to generate E2A and HEB double-knockout mice through simple genetic crosses. We thus created transgenic mice in which the Id1 gene (7), encoding the inhibitor of both E2A and HEB, is specifically expressed in T cells. Strikingly, T-cell development in these mice appear to be arrested at the earliest progenitor stage and massive apoptosis of thymocytes is detected, resulting in mice with rudimentary thymuses. This data, together with the previous findings with B cells, thus prompts us to propose that E proteins, being the targets of the Id1 inhibitor, act as important regulators in lymphocyte commitment and differentiation in both the B- and T-cell lineages. Similar to the E2A-deficient mice (5, 63), Id1 transgenic mice also develop T-cell lymphoma, suggesting an oncogenic potential for the Id1 protein.
MATERIALS AND METHODS
Id1 transgenic mice.
The Id1 transgenic construct was generated by inserting mouse Id1 cDNA into the _Bam_HI site of the vector plck-hGH (17), which contains the T-cell-specific lck proximal promoter and the human growth hormone (hGH) gene with introns and a polyadenylation signal. The Id1 cDNA was modified by including a Kozak translation initiation sequence at the ATG codon and by fusing the sequence encoding the influenza virus HA epitope tag with the 3′ end of the Id1 coding sequence. Transgenic founders were identified by Southern blot analysis of the tail genomic DNA. Transgenic offspring were determined by PCR of the tail genomic DNA with the transgene-specific primers: 5′-hGH (CGAACCACTCAGGGTCCTGTGG) and 3′-hGH (GGATTTCTGTTGTGTTTCCTCCCTG).
Flow cytometry.
Cell suspensions were prepared from the thymus, spleen, and lymph nodes. Spleen cells were purified on Ficoll cushions by a 30-min centrifugation at 4°C, and cells in the supernatant were collected by centrifugation. Thymocytes were also purified similarly. The cells were stained with antibodies for two-color or three-color fluorescence-activated cell sorter (FACS) analysis on a FACScan-II (Becton-Dickinson, Franklin Lakes, N.J.). The following antibodies were purchased from Caltag Laboratories (Burlingame, Calif.): phycoerythrin (PE)-conjugated anti-CD4 (PE-CD4), Tri-color (TC)-CD4, fluorescein isothiocyanate (FITC)-CD8, TC-CD8, FITC-CD3, FITC-TCRβ (H57), FITC-CD24, and FITC-c-kit. FITC-TCRγδ (GL3), FITC-CD25, and PE-CD44 were from Pharmingen (San Diego, Calif.).
PCR for TCR rearrangement.
Thymic genomic DNA was prepared from 106 unpurified cells by lysis at 55°C for 1 h in 200 μl of buffer containing 10 mM Tris (pH 8.4), 50 mM KCl, 2 mM MgCl2, 0.45% Nonidet P-40, 0.45% Tween 20, and 60 μg of proteinase K per ml. A 1-μl volume of the DNA was subjected to PCR in a 50-μl reaction mixture for 25 cycles (for the Id2 gene) or 30 cycles (for other genes) by denaturing at 94°C for 1 min, annealing at 62°C for 30 s, and elongating at 72°C for 1.5 min. One-tenth of the reaction mixture was analyzed by Southern blot hybridization. Prehybridization was performed for 6 h at 37°C in a buffer containing 6× SSC (pH 7.0) (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 5× Denhardt solution, 0.05% sodium pyrophosphate, 0.1% sodium dodecyl sulfate, and 100 μg of sheared and denatured salmon sperm DNA per ml. End-labeled oligonucleotide probe was added subsequently for hybridization for 18 h at 37°C. The filters were washed three times for 10 min each at 37°C in 6× SSC–0.05% sodium pyrophosphate–0.1% sodium dodecyl sulfate. The final wash was for 30 min at 37°C in 6× SSC–0.05% sodium pyrophosphate. Quantitation was performed with a PhosphorImager (Molecular Dynamics, Inc., Sunnyvale, Calif.).
The oligonucleotides used for TCR gene rearrangement assays were as follows (unless specified, 3′ primers were used as probes): Vβ3-5′ (CCTTGCAGCCTAGAAATTCAGTCC) (12), Dβ2-5′ (GTAGGCACCTGTGGGGAAGAAACT), Jβ2-3′ (TGAGAGCTGTCTCCTACTATCGATT) (2), Jβ2 (probe) (GTCTACTCCAAAC TAC TC), Vα2C-5′ (ACTGTCTCTGAAGGAGCCTCTCTG), VαF3-5′ (ACCCAGACAGAAGGCCTGGTCACT), VαH-5′ (CAGAAGGTGCAGCAGAGCCCAGAA), JαTT11-3′ (GACCCTATTACTCACATACTTGGCTTG), JαTT11 (probe) (GAAAGCAGAGTCCCAATTCCAAAG) (30), Vδ1-5′ (GGGGGATCCTGCCTCCTTCTAC), Jδ1-3′ (AAAAAGCTTACTCAACACGACTGGA), JδH (probe) (GGAAGCTTACTTCCAACCTCTTTAGGT) (11); Id2-5′ (GAACCGAGCCTGGTGCCGCGCAGTCAGCTC), and Id2-3′ (GGCGGATCCTTATTTAGCCACAGAGTAC) (57).
RT-PCR for gene expression.
Thymic total RNAs were prepared with Trizol (Life Technologies, Gaithersburg, Md.) as specified by the manufacturer. First-strand cDNAs were synthesized from 10 μg of total RNA with the oligo(dT) primer and Moloney murine leukemia virus reverse transcriptase (RT) (Life Technologies). One-fortieth of the first-strand cDNA reaction product was used for PCR with a reaction volume of 25 μl. Serial dilution at 1:10 and 1:100 were also performed to establish the linearity of the amplification. Amplification was performed for 25 cycles (for glyceraldehyde-3-phosphate dehydrogenase [GAPDH]) and 30 cycles (for other genes) by denaturing at 94°C for 45 s, annealing at 60°C for 20 s, and elongating at 72°C for 1 m. One-fifth of each reaction product was analyzed and quantitated by Southern blot hybridization and PhosphorImager measurement. GAPDH signals were used for normalization.
The oligonucleotides used for RT-PCR were as follows (unless specified, 3′ primers were used as probes): GAPDH-5′ (ATGGTGAAGGTCGGTGTGAACGGATTTGGC), GAPDH-3′ (GCATCGAAGGTGGAAGAGTGGGAGTTGCTG) (57), pTα-5′ (CACACTGCTGGTAGATGGAAGGC), pTα-3′ (GTCAGGAGCACATCGAGCAGAAG) (43), Vβ8-5′ (ATGTACTGGTATCGGCAGGACACGG), Cβ-5′ (GGATCTGAGAAATGTGACTC), Cβ-3′ (CTGACCAGCACAGCATATAG), Cβ (probe) (GTCACACAGAACATCAGTGCAG) (27), CD3δ-5′ (GAAGATGGAACACAGCGGGATTCTG), CD3δ-3′ (CTTAAGATTTCTTGTTCCGGGGCCAGT), CD3ɛ-5′ (GTGGAACACTTTCTGGGGCATCCTG), CD3ɛ-3′ (GTCAGACTGCTCTCTGATTCAGGCCA), CD3γ-5′ (CATGGAGCAGAGGAAGGGTCTGGCTG), CD3γ-3′ (GTTCACTTCTTCCTCAGTTGGTTTC), CD3ζ-5′ (GATGAAGTGGAAAGTGTCTGTTCTC), CD3ζ-3′ (CTGTTAGCGAGGGGCCAGGGTCTGC), RAG1-5′ (CCAAGCTGCAGACATTCTAGCACTC), RAG1-3′ (CAACATCTGCCTTCACGTCGATCC), RAG2-5′ (CACATCCACAAGCAGGAAGTACAC), RAG2-3′ (GGTTCAGGGACATCTCCTACTAAG) (13), TdT-5′ (GAAGATGGGAACAACTCGAAGAG), TdT-3′ (CAGGTGCTGGAACATTCTGGGAG) (31), Ku70-5′ (TGGAGAAGAAGGTCATAGCAGTGTG), Ku70-3′ (TGGGCTTCTGAGCTTTAGTCAGTTC), Ku80-5′ (CAAGGTTGGAAGTGTGAATCCTGTTG), Ku80-3′ (TCCTTATGGTCACTCTGTAGAGACC), IL7Rα-5′ (AGCTGTTTCTGGAGAAAGTGG), IL7Rα-3′ (AACGACTTTCAGGTCAGAGGG) (33), Ikaros-5′ (GATAGATCTATGGATGTCGATGAGGGTCAAGAC), Ikaros-3′ (GATGAATTCTTAGCTCAGGTGGTAACGATGCTC) (19), c-kit-5′ (GTGTATTCACAGAGATTTGGCAGCC), and c-kit-3′ (CTGCGTAGAAGAGGCGCTGCTGC). For endogenous Id1 expression, Id1-5′ (CCAGTGGCAGTGCCGCAGCCGCTGCAGGC) and Id1-3′ (GTAGTGTCTTTCCCAGAGATCCCCTGG) were used. Oligonucleotide GGCTGGAGTCCATCTGGTCCCTCAGTGC was used as a probe. For transgenic Id1 expression, Id1-5′ and hGH-3′ (CCACAGGACCCTGAGTGGTTCG) were used as primers.
RESULTS
Dramatic reduction of the number of thymocytes in the Id1 transgenic mice.
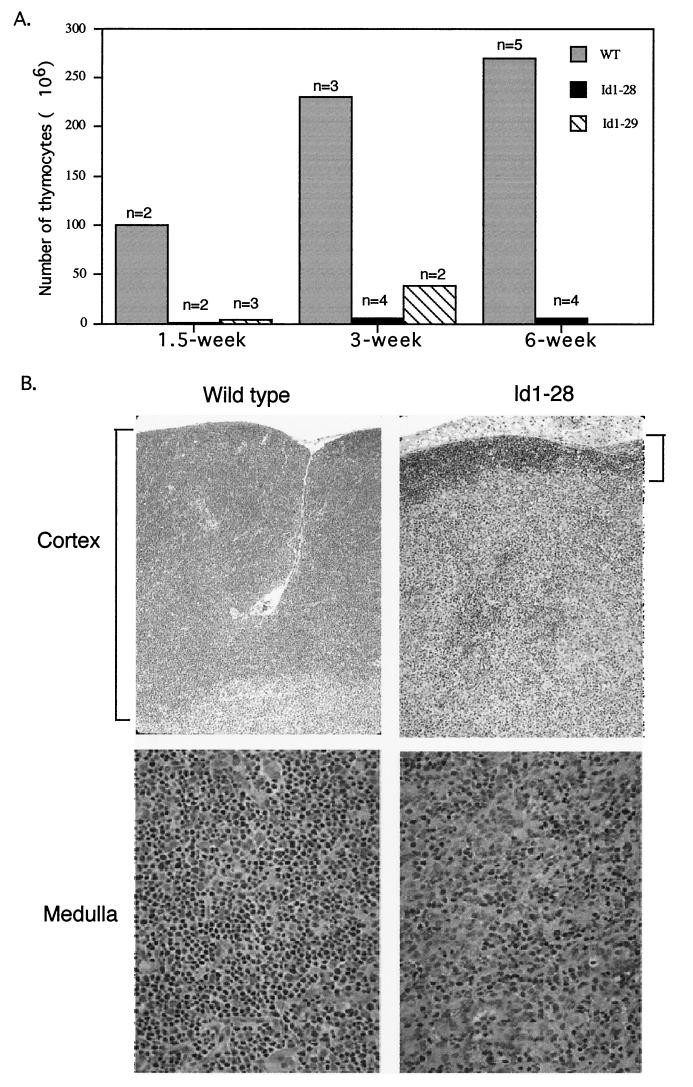
To inhibit the activities of all E proteins in T cells, we have generated transgenic mice in which the Id1 transgene is specifically expressed in T cells. Id1 forms heterodimers with the E proteins, but the resulting heterodimers cannot bind to DNA. We inserted the Id1 cDNA into the plck-hGH transgenic vector (17), which contains the proximal promoter of the lck gene for directing T-cell specific transcription and a portion of the hGH gene for the proper processing of mRNA by splicing and polyadenylation. The Id1 transgenic fragment was microinjected into pronuclei of the oocytes of FVB/N mice. Expression of the Id1 transgene was detected in two independent founder lines (Id1-28 and Id1-29) by RT-PCR assays with primers annealing to the Id1 and hGH sequences (data not shown). Three other founder lines died in a non-pathogen-free facility prior to rederivation, possibly due to their immunodeficiency. The total numbers of thymocytes in the Id1-28 and Id1-29 mice were less than 4% of the numbers in their wild-type littermates in the first 1.5 weeks after birth (Fig. 1A). While this low thymocyte count in the Id1-28 transgenic mice persisted throughout adulthood, the deficit in the Id1-29 transgenic mice was subsequently alleviated, suggesting a leakier block, probably as a result of a lower level of Id1 expression. Therefore, further examination was carried out by using the Id1-28 transgenic mice. Consistent with the low thymocyte counts, the thymuses of the Id1-28 transgenic mice were rudimentary. Histological examination revealed that the thymuses of Id1 transgenic mice were almost devoid of the cortex, where developing T-cells are normally found (Fig. 1B). In addition, the medulla of the thymus showed a significant reduction in the number of lymphocytes.
FIG. 1.
(A) Reduced cellularity in the thymuses of Id-1 transgenic mice. Unpurified viable thymocytes were counted with a hemocytometer. The numbers of cells per thymus are the average for n wild-type (WT) or Id1-28 and Id1-29 transgenic mice at the indicated ages. (B). Hematoxylin and eosin stain of wild-type and Id1-28 transgenic thymus sections.
T-cell developmental blocks in the Id1 transgenic mice.
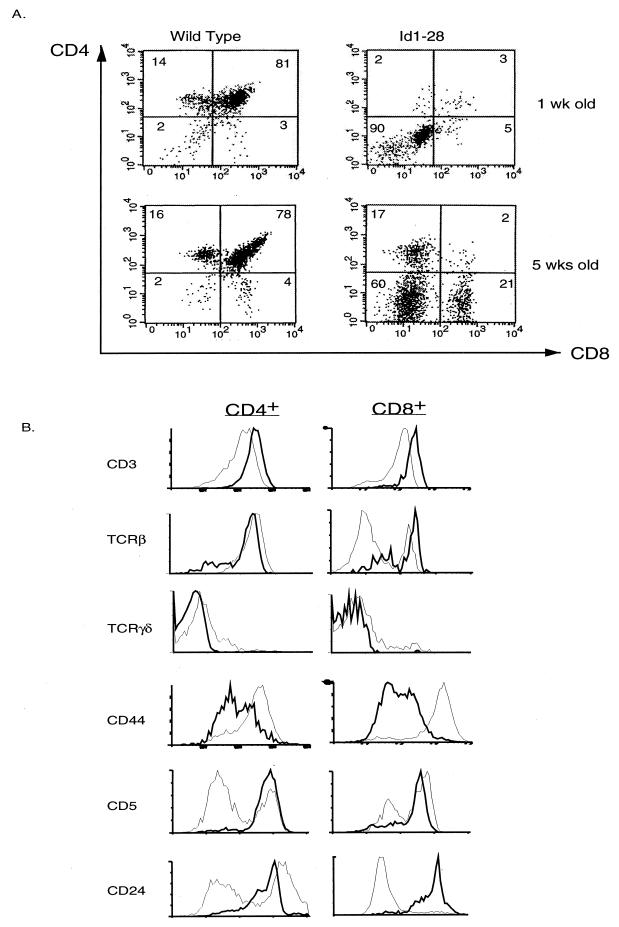
To examine intrathymic T-cell development, thymocytes from 1- and 5-week-old Id1-28 transgenic mice and their wild-type littermates were first analyzed by using flow cytometry with antibodies against CD4 and CD8 (Fig. 2A). In contrast to the predominance of DP cells in wild-type thymuses at both ages, the 1-week-old transgenic thymuses contained primarily DN cells, suggesting that the initial defect is at the DN stage. At the age of 5 weeks, CD4+ and CD8+ single-positive cells appeared in the transgenic thymuses while the DP cells were still lacking. Moreover, the CD8+ population in the transgenic mice constitutes 20% of the thymocytes, compared to 2.6% in wild-type mice. The appearance of these CD4+ and CD8+ cells is not dependent on the age of the animals but on the degree of inhibition of E-protein function. For example, in 5-week-old homozygous transgenic mice, fewer single positive cells were detected (data not shown), suggesting that higher levels of Id1 expression lead to a greater inhibition of E-protein function and a more complete block of T-cell development.
FIG. 2.
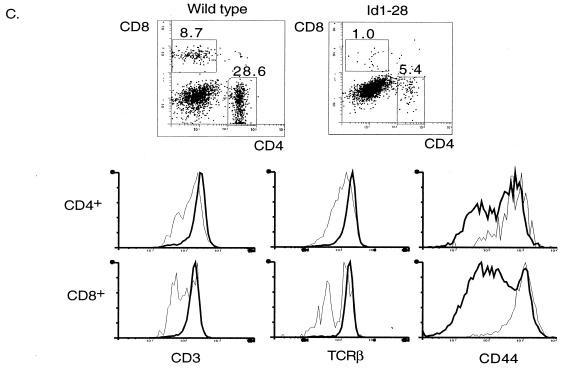
FACS analyses of cells from Id1 transgenic mice. (A) CD4 and CD8 profiles of the wild-type and transgenic littermates at the indicated ages. The percentage of each cell population is shown in each quadrant. (B) Further analysis of CD4+ and CD8+ single-positive thymocytes. Thymocytes from 5-week-old mice were stained with TC-CD4 and PE-CD8 together with FITC-conjugated antibodies as indicated. The CD4+ and CD8+ populations were defined as shown in panel A. The expression of each indicated cell surface marker is plotted as fluorescence intensity versus cell number (different scales were used for wild-type and transgenic cells). The profiles of the wild-type mice are shown as thick lines, and those of the Id1 transgenic mice are shown as thin lines. (C) Analyses of Ficoll-purified spleen cells from 5-week-old mice as described in panel B. The percentage of CD4+ and CD8+ single-positive cells are shown on top of the boxes.
We have further characterized the CD4+ and CD8+ single-positive thymocytes that emerged in the 5-week-old transgenic mice. In the CD4+ cells, expression of CD3 was somewhat decreased while expression of CD44 was dramatically increased compared to the expression in their wild-type counterparts (Fig. 2B). The majority of the CD4+ cells appeared to be αβ T cells rather than γδ T cells. Staining for CD5 and CD24 markers revealed heterogeneous populations in CD4+ cells; i.e., some cells are CD5+ or CD24−, characteristic of more mature T cells, whereas others appear as CD5− or CD24+, suggesting an immature phenotype. The CD8+ cells, which represent an unusually large population of thymocytes, also express lower levels of CD3 and extremely high level of CD44. Unlike CD4+ cells, only 30% of the CD8+ cells were identified as αβ T cells and 10% were identified as γδ T cells. The remainder of the CD8+ cells did not carry high levels of any surface TCR and were thus suspected to be ISP cells. However, the majority of these CD8+ cells appeared as CD5+ and CD24− cells, which would tend to classify the cells as mature T cells rather than ISP cells. Therefore, the expression patterns of TCR and CD5 CD24 in the CD8+ cells seem uncoupled, and the mechanisms remain to be investigated. Nevertheless, our FACS data described above would suggest that T-cell development in the Id1 transgenic mice is impaired primarily at the DN stage. However, the single-positive cells that have developed also display obvious abnormalities, which would imply that Id1 expression continues to affect the developmental program beyond the DN stage. Compared to the total number of αβ T cells in Id1 transgenic mice, which is 100-fold smaller than the wild-type count, the number of γδ T cells is reduced by only fivefold. Moreover, some of the γδ T cells appear to be CD8+, which is extremely rare in wild-type mice. This finding is generally in agreement with that found by Blom et al., where Id3 blocks the development of αβ but not γδ T cells differentiated from committed progenitor T cells (9).
Consistent with the findings in the transgenic thymus, the number of peripheral T cells in the spleen was also dramatically decreased (about eightfold) (Fig. 2C). Similar to thymic CD4+ and CD8+ single-positive cells, splenic T cells also exhibited low levels of CD3 and TCR β and a high level of CD44 surface expression. Moreover, there was also a small but distinct population of CD8+ cells that had much lower levels of cell surface TCR β, perhaps corresponding to γδ T cells. Despite the unusually high percentage of CD8+ cells in the thymus, the ratio of CD4+ and CD8+ cells in the spleens of transgenic mice appeared similar to that in their wild-type littermates.
It has been shown that overexpression of Id3 in bipotential progenitor cells from human thymus inhibits the differentiation of the T-cell lineage under appropriate culture conditions while stimulating the differentiation of the NK cell lineage (23). However, we have not detected any significant increase in the number of NK cells in either the thymuses or spleens of the Id1 transgenic mice (data not shown). This may be due to the different mechanisms used in human and mouse hematopoiesis, different functions of Id1 and Id3, different promoters for Id expression, or different experimental systems, i.e., in fetal thymic organ culture and in mice.
Further characterization of the DN cells in the transgenic mice.
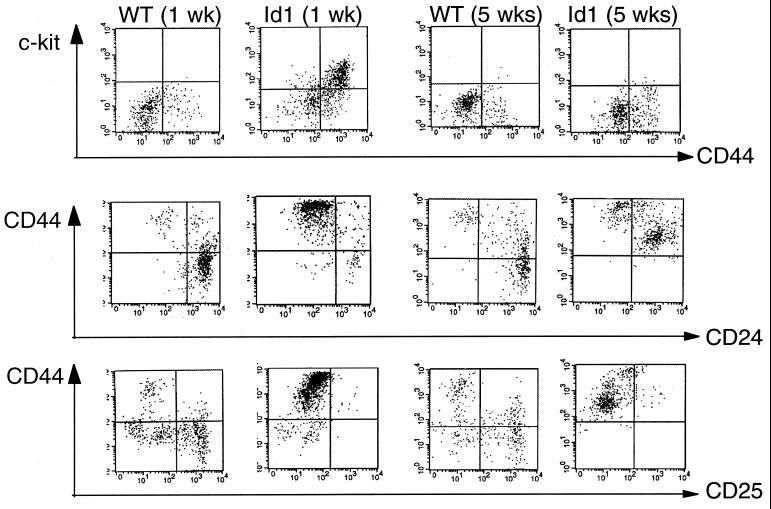
The FACS analysis in Fig. 2A suggested that the T-cell developmental defect in the transgenic mice occurred primarily at the DN stage. To further define the developmental stages of the DN transgenic cells, thymocytes that were negative for anti-CD4 and anti-CD8 staining were examined for c-kit, CD44, CD24, and CD25 expression (Fig. 3). In wild-type mice, the DN cells were characterized predominantly as c-kit− CD44− CD24+, with CD25 being positive or negative. In contrast, the majority of DN cells in 1-week-old transgenic mice appeared to be c-kit+ CD44+ CD24− CD25−, which marks multipotent progenitor cells (51, 62). At 4 weeks later, while the wild-type cells exhibited indistinguishable characteristics from the 1-week-old cells, the transgenic DN cells progressed to a stage displaying cell surface markers as c-kit− CD44+ CD24+ CD25− and remained at this stage throughout adulthood. It thus appears that the major rate-limiting steps in T-cell development in the Id1 transgenic mice might involve the commitment of lymphoid progenitor cells to the T-cell lineage and the initial differentiation of committed T cells.
FIG. 3.
Further characterization of CD4 and CD8 DN cells. Thymocytes from wild-type (WT) and transgenic littermates at the indicated ages were stained with TC-CD4 and TC-CD8 together with PE- and FITC-conjugated antibodies as indicated. The TC-negative population was gated for these analyses.
TCR rearrangement in the transgenic mice.
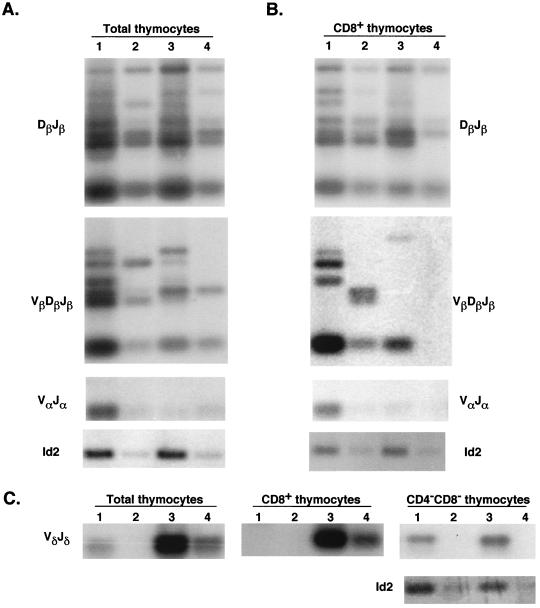
The V(D)J recombination of TCR genes is a crucial process during T-cell differentiation. The majority of thymic T cells rearrange and express the TCR αβ, while a minor population of T cells bear TCR γδ. The orderly events in TCR rearrangement begin with the joining of the D and J regions of the β chain gene of TCR followed by the V-to-DJ recombination. The productive rearrangement of the β-chain gene, together with the pre-T α chain, results in the formation of a pre-TCR, which then prompts the V-to-J recombination of the α-chain gene. Therefore, these step-by step events of TCR rearrangement have been used as indicators for the developmental stages of T cells. The γδ T cells arise from progenitor T cells by rearranging TCR δ and γ genes prior to TCR α gene rearrangement. We have examined the rearrangement of TCR genes in the thymocytes from the Id1 transgenic mice to further assess their developmental status (Fig. 4).
FIG. 4.
TCR rearrangement. Total and sorted thymocytes (labeled at the top of each panel) from wild-type and transgenic littermates at the indicated ages were used to prepare DNA for PCR analyses. Thymocytes from wild-type and transgenic (lanes 1 and 3) mice and their respective 10-fold-diluted samples (lanes 2 and 4) were used in PCRs to detect TCR rearrangement events as indicated. The PCR product of the Id2 gene served as a control for the amount of DNA present in each sample. The PCR products were analyzed by Southern blotting.
In the total thymocytes of 5-week-old Id1 transgenic mice, the Dβ-to-Jβ rearrangement, which is not believed to be a rate-limiting step, occurred at a similar efficiency to that in the wild-type mice. However, the events of the Vβ-to-DβJβ recombination were significantly reduced (Fig. 4), most probably as a result of the lack of T cells that had reached the developmental stage necessary to undergo V-to-DJ rearrangement. Surprisingly, although the CD4+ or CD8+ single-positive T cells constituted about 40% of the thymocytes in the 5-week-old transgenic mice, a very low frequency of rearrangement in the α locus was detected by using three 5′ primers binding to different Vα regions and two 3′ primers corresponding to different Jα regions (Fig. 4 and data not shown). It remains to be verified whether this low frequency is due to the intrinsic defect in TCR α rearrangement, or because the single-positive cells in the transgenic mice are not all αβ T cells but also include γδ and immature T cells.
To further examine the CD8 single-positive cells which represent an abnormally large population in the transgenic mice, we have isolated these cells from wild-type and transgenic mice by using a cell sorter and compared the gene rearrangement in various TCR loci (Fig. 4B). While the frequency of Dβ-Jβ rearrangement was comparable between wild-type and transgenic cells, that of Vβ-DβJβ rearrangement in the transgenic cells was much lower than in the wild-type cells. Like the finding in total thymocytes, rearrangement at the α locus in the transgenic cells was detected at a very low frequency compared to that in the wild-type cells. These low frequencies of TCR α and β rearrangement might be explained by the small fraction (about 30%) of αβ T cells in the CD8+ population (Fig. 2A).
In contrast to the α and β loci, rearrangement of the δ locus in the transgenic mice did not appear to be inhibited. Compared to the total thymocytes in their wild-type littermates, the frequency of Vδ-to-DδJδ recombination was increased dramatically in the transgenic mice. Since purified DN cells from wild-type and transgenic mice showed similar efficiencies of Vδ-to-Jδ rearrangement (Fig. 4C), the relative increase in the rearrangement events in the δ locus may be partially explained by the much higher percentage of DN cells in the transgenic mice. Moreover, in the CD8+ cells of transgenic mice, Vδ-Jδ rearrangement was readily detectable in the transgenic sample but not the wild-type sample, most probably due to the presence of γδ T cells as well as immature single-positive cells in the CD8+ population (Fig. 2B).
Massive cell death in the thymuses of the Id1 transgenic mice.
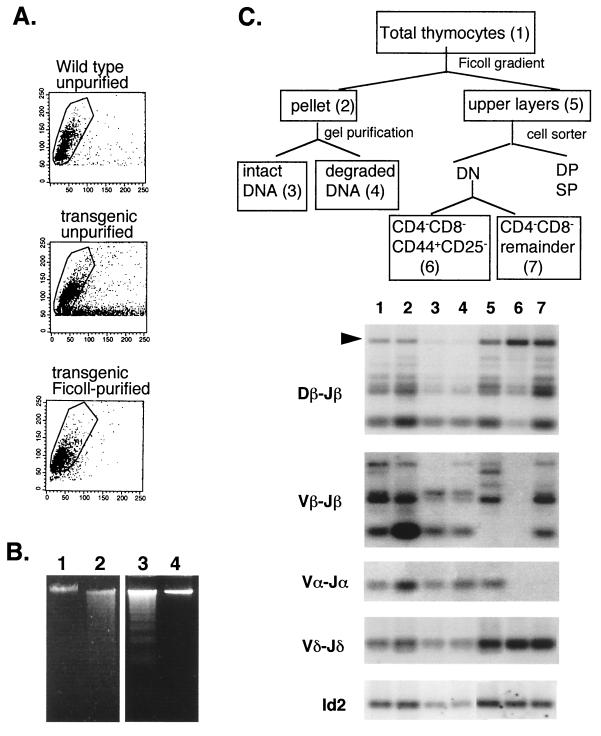
Another striking feature of the Id1 transgenic mice was the presence of a large number of dead cells in the thymus as determined microscopically and by forward- and side-scatter flow cytometry analysis (Fig. 5A). The dead cells, most probably apoptotic cells, constituted about 50% of the total thymocytes. To further characterize the dead cells, we have separated the dead cells from live ones by centrifugation of total thymocytes on a Ficoll cushion. The resulting pellet contained dead cells as well as contaminating live cells, while the supernatant had mostly live cells as evidenced by the scatter analysis (Fig. 5A). Moreover, gel electrophoresis of DNA isolated from total thymocytes of the transgenic mice but not the wild-type mice showed DNA fragmentation, a characteristic of apoptotic cells (Fig. 5B, lanes 1 and 2). The fragmented DNA was contributed by the cells partitioned to the pellet after centrifugation on the Ficoll cushion, whereas the DNA prepared from cells in the supernatant appeared intact (lanes 3 and 4). To assess the developmental status of the apoptotic cells, genomic DNA was isolated from different fractions of cells for analysis of TCR rearrangement. As diagrammed in Fig. 5C, total thymocytes (sample 1) were first separated into pellet (sample 2) and supernatant (sample 5) components. The DNA isolated from the pelleted cells was further fractionated by gel electrophoresis into intact DNA (sample 3), which was probably derived from contaminating live cells, and fragmented DNA, which presumably originated from the apoptotic cells (sample 4). Cells in the supernatant were then stained with TC-CD4, TC-CD8, PE-CD44, and FITC-CD25. By using a cell sorter, DN cells, which constituted 70% of the thymocyte, were gated, and CD44+ CD25− cells (sample 6) and the remainder of the DN cells (sample 7) were collected. These samples were then used to isolate DNA for PCR analysis of TCR rearrangement.
FIG. 5.
Apoptosis in the Id1 transgenic thymus. (A) Forward- and side-scatter analysis of thymocytes with or without Ficoll purification as indicated. Live cells are circled. (B) Electrophoresis of DNA from wild-type and transgenic unpurified thymocytes (lanes 1 and 2), transgenic thymocytes in the pellet after centrifugation on Ficoll (lane 3), and transgenic thymocytes in the supernatant (lane 4). (C) TCR rearrangement in different fractions of thymocytes in 10-day-old transgenic mice. The fractionation procedure is diagrammed at the top. The different fractions are numbered 1 through 7, corresponding to the lane numbers. The PCR product representing the germ line configuration in Dβ-Jβ region is marked by an arrowhead.
Although the frequencies of TCR rearrangement at the α and β loci were lower in the transgenic mice than in wild-type mice, as shown in Fig. 4, these rearrangement events were detectable in both the live and dead cells in the transgenic mice when PCR amplified through additional cycles (Fig. 5C, lanes 1 to 5). Of particular significance was that in the fragmented DNA (lane 4), the TCR β locus was predominantly in the rearranged configuration. Rearrangement of the TCR α locus was also detectable. This would imply that the cells undergoing apoptosis have committed to the T-cell lineage and reached certain stages in T-cell development. In contrast to these apoptotic cells, the majority of CD44+ CD25− DN progenitor cells, which constituted at least 50% of the transgenic thymocytes, had not undergone rearrangement in the TCR α or β locus (lane 6). The weak signals of Dβ-Jβ rearrangement detected in this sample might come from a subpopulation of cells that have aberrantly initiated rearrangement or from the γδ T cells (10) or from contaminating DN cells that belong to later stages (lane 7). Whether TCR gene rearrangement triggers apoptosis in the transgenic thymocytes or whether apoptosis occurs only in differentiating T cells remains to be determined.
Since a large number of Vδ-Jδ rearrangement events were detected in the CD44+ CD25− DN cells (Fig. 5C, lane 6), it would appear that some of the γδ T cells display a surface phenotype as CD44+ CD25−. FACS analysis with antibodies against TCR γδ together with antibodies against CD44 or CD25 had confirmed that the γδ-positive cells were CD44+ CD25− in both wild-type and transgenic mice (data not shown). While this surface phenotype seems to be consistent with the notion that rearrangement of the TCR γδ loci precedes that of the αβ loci (42), TCR δ rearrangement was detected in the CD25− cells of the Id1 transgenic mice, where TCRδ rearrangement was not believed to take place until the cells become CD25+ (62). Whether this discrepancy is due to the expression of Id1 transgene remains to be determined. Similar to αβ T cells, γδ T cells also undergo apoptosis, since Vδ-Jδ rearrangement was also detected in the apoptotic cells. This may explain the fivefold reduction in the total number of γδ T cells in the thymuses of transgenic mice based on FACS analyses (data not shown).
Gene expression in the Id1 transgenic mice.
Since the E proteins are potent transcription activators, it is important to examine gene expression which might be altered as a result of the loss of E-protein activity. By using RT-PCR analysis, we compared the expression levels of various genes important for T-cell differentiation between transgenic mice and their wild-type littermates at the ages of 1 and 5 weeks (Table 1). We found that expression of the genes encoding components of the pre-TCR complexes (pre-Tα, CD3δ, CD3γ, CD3ɛ, and CD3ζ) was not dramatically inhibited except for Vβ expression. However, genes involved in the rearrangement of TCR, such as the RAG1, RAG2, TdT, and Ku70 genes, were expressed at less than 15% of the wild-type levels in 1-week-old mice and their expression increased in 5-week-old mice. Ku80 expression, on the other hand, did not change dramatically in either the 1- or 5-week-old mice. The mRNA level of Ikaros transcription factor also decreased modestly in the 1-week-old transgenic mice but increased in older mice (Table 1 and data not shown). These results may be interpreted to mean that the lower levels of gene expression are due to the lack of T cells which have reached the stages when these genes are up-regulated (51). Alternatively, these genes may be the direct targets of the E proteins, and inhibition of E protein activities leads to lower levels of their expression. By the same token, the genes encoding c-kit and interleukin-7 (IL-7) receptor α, both of which are important for the survival of progenitor T cells, appeared to be expressed at higher-than-wild-type levels. This may also be explained by the enrichment for the T cells at early stages in the transgenic mice.
TABLE 1.
Gene expression in Id1 transgenic micea
| Gene | Expression (%) in mice agesb: | |
|---|---|---|
| 1 wk | 5 wk | |
| pTα | 48 | NDc |
| Cβ | 75 | 74 |
| Vβ | 0 | 24 |
| CD3δ | 54 | ND |
| CD3γ | 89 | ND |
| CD3ɛ | 70 | ND |
| CD3ζ | 43 | ND |
| RAG1 | 4 | 62 |
| RAG2 | 3 | 12 |
| TdT | 1 | 111 |
| Ku70 | 15 | 70 |
| Ku80 | 50 | 254 |
| Ikaros | 30 | ND |
| IL-7R | 671 | 255 |
| c-kit | 8,507 | ND |
| Id-1 | 1,722 | 53 |
One remarkable finding from the gene expression analysis is that the endogenous Id1 transcript in the 1-week-old transgenic mice was present at a level 17-fold higher than that in their wild-type littermates (Table 1). Since the majority of thymocytes in the transgenic mice at this age belong to a population characterized as CD4− CD8− and c-kit+ CD44+ CD24− CD25−, it is possible that Id1 is normally expressed at a high level in this population of progenitor T cells, which are present as an extremely minor population in the wild-type littermates (Fig. 2A and 3). In 5-week-old mice, this population in the transgenic mice progressed to a stage displaying cell surface markers such as CD4− CD8− and c-kit− CD44+ CD24+ CD25− (Fig. 3) and Id1 expression decreased to a level comparable to that in their wild-type littermates. Based on these results, we therefore propose that the endogenous Id1 gene is expressed in the thymus at the progenitor stage marked as CD4− CD8− and c-kit+ CD44+ CD24− CD25− and that its expression is down-regulated as differentiation proceeds. Interestingly, when Id1 down-regulation is not possible due to ectopic Id1 overexpression, as in the transgenic mice, T-cell development appears to be impaired at this progenitor stage due to the failure of further differentiation of the progenitor cells or the survival of differentiated T cells.
T-cell lymphoma in the Id1 transgenic mice.
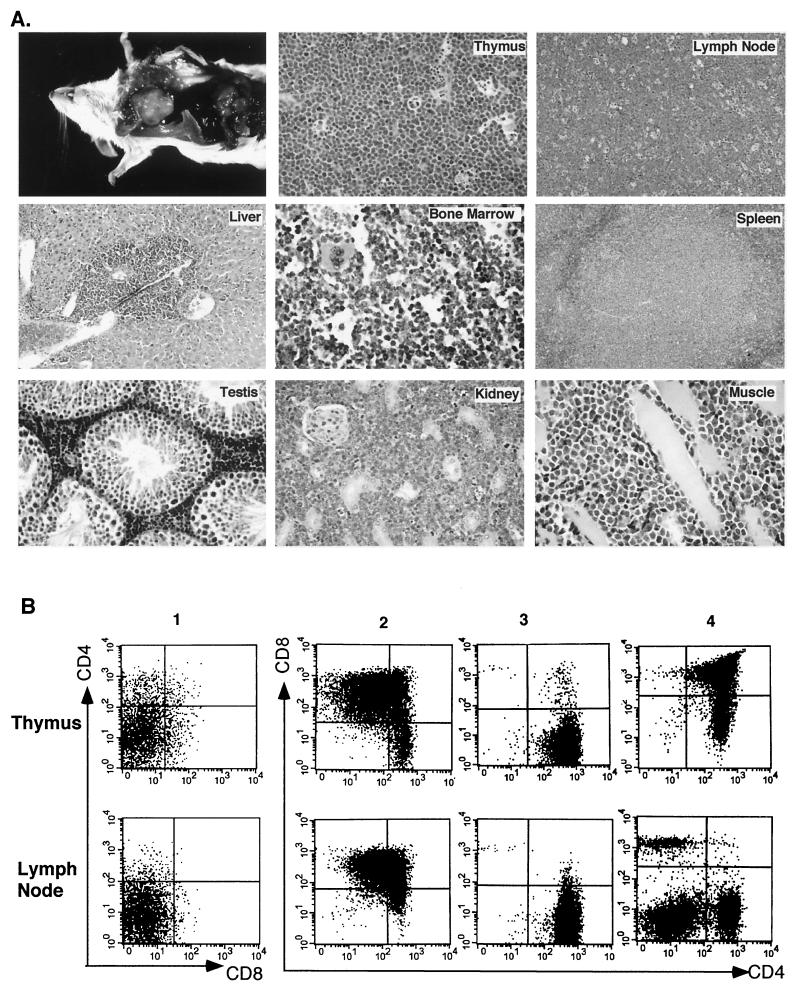
Since disruption of the E2A gene was found to cause T-cell lymphoma in E2A-deficient mice (5, 63), we were interested in determining if overexpression of Id1 could also lead to tumor formation in the transgenic mice. Despite the severe block of T-cell development at an early progenitor stage, adult transgenic mice developed T-cell lymphoma at a high frequency and at an age as early as 2.5 months. Figure 6A shows the physical appearance of one such thymic lymphoma and the histological examination of the lymphoma and its metastatic sites. The normal histological structure of the thymus was completely effaced and replaced by a monotonous population of intermediate-size neoplastic lymphocytes with abundant mitotic figures and apoptotic cells. Morphologically, this tumor resembled Burkitt’s lymphoma in humans. Although upon gross examination only the lymph nodes and spleen appeared affected by the tumor, microscopically all sampled organs demonstrated interstitial infiltration of neoplastic lymphocytes.
FIG. 6.
T-cell lymphoma in Id1 transgenic mice. (A) The appearance of a thymic lymphoma in a 3.5-month-old mouse and histological examination of the tumor and its metastatic sites. (B) FACS analysis of the thymus and lymph nodes of four 4-month-old transgenic mice numbered 1 to 4. The size of each thymus was similar to that shown in panel A. Lymph nodes were enlarged to various degrees. Cells were stained with antibodies against CD4 and CD8 as labeled.
Cohorts of wild-type (n = 14) and Id1 transgenic (n = 12) mice were sacrificed at the age of 4 months to examine tumor formation. While all of the wild-type thymuses had involuted by this age, 6 of the 12 transgenic thymuses exhibited various degree of enlargement, indicative of tumor formation. Four of the transgenic mice had thymuses as large as that shown in Fig. 6A, and some of them displayed apparently enlarged lymph nodes and spleens. To further characterize the tumor cells, cells from these thymic tumors as well as the affected lymph nodes and spleen were stained with antibodies against CD4 and CD8 for FACS analysis. Surprisingly, the analysis revealed profound heterogeneity in their CD4 and CD8 profiles. Figure 6B shows several examples of such profiles of the enlarged thymus and lymph nodes, which range from uniformly DN to CD4+ or CD8+ single positive (Fig. 6B and data not shown). Sometimes, the tumors appeared to consist of a mixture of cells with different CD4 and CD8 profiles, as exemplified by mouse 2 (Fig. 6B). CD3 and CD44 expression on the surface of these cells has also been examined, but no correlation between the expression levels and tumor type or size was observed (data not shown). In most instances, the surface phenotypes of cells from the thymuses and lymph nodes are similar (mice 1 to 3, Fig. 6B), suggesting that the tumor cells originating from the thymus may have migrated to the lymph nodes and continued to expand there. Interestingly, although the thymus and lymph nodes were extremely enlarged in mouse 4, the CD4 and CD8 profiles of these organs appeared more or less normal. This would suggest that while T cells maintain their ability to differentiate normally, preneoplastic hyperproliferation might have been occurring in the thymus, resulting in a large number of mature T cells being poured out to the periphery including the lymph nodes. It would be interesting to investigate whether this preneoplastic proliferation precedes all tumor formation or whether this condition represents a different form of cell transformation as a result of Id1 overexpression.
Finally, one significant observation is that the developmental stages of the neoplastic or preneoplastic cells are all beyond the initial developmental block. The majority of thymocytes found in the Id1 transgenic mice that had not developed tumors were at the CD44+ CD25− DN stage. However, even the tumor with a DN phenotype appeared as CD44− CD25+. This brings up the question whether only cells that have escaped the initial block can be transformed or whether the neoplastic cells are derived from the developmentally arrested cells and continue to differentiate during tumorigenesis.
DISCUSSION
Role of E and Id proteins in T-cell development.
Id1 overexpression arrests T-cell development at the progenitor stage in the Id1 transgenic mice, a defect similar to but much more severe than that in the E2A- or HEB-deficient mice (5, 65). Since Id1 is an inhibitor of all E proteins (56), it is likely that it exerts its dramatic effect by interfering with the function of multiple E proteins which are expressed simultaneously in T cells. In fact, we have found that expression of E47, a product of the E2A gene, can rescue the T-cell deficiency in Id1 transgenic mice (data not shown). Therefore, findings with the Id1 transgenic mice have demonstrated that the E proteins collectively are as crucial for T-cell development as they are for B-cell development. The thymocytes that accumulated in the Id1 transgenic mice were identified as c-kit+ CD44+ CD24− CD25− or c-kit− CD44+ CD24+ CD25− progenitor cells that have the potential to differentiate into T, B, and NK cells (15, 51). Interestingly, the developmental stages of these cells appear to parallel the stage known as fraction A (B220+ CD43+ CD24− BP-1−) in bone marrow (22), the stage at which B-cell development is blocked in the E2A-deficient mice (4). Fraction A cells can be further characterized based on their surface expression of the AA4.1 marker. AA4.1+ cells are considered prepro-B cells, while AA4.1− cells include cells expressing the NK1.1 markers and a subset of T cells (32). Because B and T cells have common strategies in their differentiation, the similar effect on B- and T-cell development as a result of E-protein ablation either by gene disruption or by overexpression of their inhibitor would suggest that E proteins might control B- and T-cell development through similar mechanisms.
In the Id1 transgenic mice, abundant expression of the endogenous Id1 gene most probably occurs in c-kit+ CD44++ CD24− CD25− progenitor cells. When this population of cells disappear in adult mice, Id1 expression decreases significantly. These c-kit+ CD44++ CD24− CD25− progenitor cells in the thymus resemble the fraction A cells in the bone marrow not only in their ability to differentiate into several lineages, as mentioned above, but also in their high levels of endogenous Id1 expression. The fraction A cells, particularly fraction A0 cells, also express Id1 at a high level (32). Significantly, when Id1 is ectopically expressed either in the bone marrow (57) or the thymus, B- or T-cell development is arrested at the stages when the Id1 gene is normally expressed, implying that down-regulation of Id1 gene expression is essential for B- and T-cell development to proceed.
Although the major developmental block in the transgenic mice occurs in the DN stage, abnormalities have also been found in the CD4+ or CD8+ single-positive thymocytes. The total number of single-positive thymocytes in the transgenic mice is estimated to be less than 10% of the wild-type count in animals 5 weeks of age or older. The identities of these single-positive cells are rather intriguing. In the transgenic thymus, DP cells are scarce, so where are these single-positive cells derived from? One possibility is that these small populations of cells, which have escaped the initial block, pass through the DP stage extremely fast due to the lack of competition in the DP compartment. Supporting this hypothesis is that the number of single-positive cells correlates inversely with the level of Id1 transgene expression; i.e., homozygous Id1 transgenic mice have fewer single-positive cells than the heterozygotes do. Furthermore, the CD8+ cells carry both the α and β chains of the CD8 coreceptor, which would suggest a thymic origin of these single-positive cells (data not shown). The alternative possibility for the source of these single-positive cells in the transgenic thymus is that they have reentered the thymus from peripheral organs. The lack of CD24 on the surface of most CD8+ cells and some CD4+ cells would identify these cells as peripheral T cells. In addition, γδ T cells contribute to the CD8+ single-positive population. To determine the origin of these T cells, further investigation is necessary.
Possible mechanisms by which E proteins control lymphocyte differentiation.
T-cell development in the Id1 transgenic mice seems to be blocked primarily during the commitment of progenitor cells to the T-cell lineage, since the predominant population of cells accumulated in these mice was made up of DN cells carrying the cell surface markers c-kit+ CD44+ CD24− CD25− or c-kit− CD44+ CD24+ CD25−, depending on the age of the animals. This phenotype of Id1 transgenic mice is in sharp contrast to the phenotypes of knockout mice, in which genes encoding various proteins involved in pre-T or TCR signaling are disrupted (20, 35, 38, 53). In these mice, T-cell development is arrested at the CD44− CD25+ DN stage. Therefore, it is unlikely that inhibition of the expression of the genes encoding TCR pre-Tα or subunits of CD3 and impairment of TCR gene rearrangement and expression are the crucial factors involved. Our data from analyses of gene expression and TCR gene rearrangement would support this notion. Indeed, we have found that expression of functionally rearranged TCR genes in the Id1 transgenic mice could not rescue the developmental defect (data not shown). The phenotype of the Id1 transgenic mice is also distinct from that of the IL-7 or IL-7 receptor-deficient mice, in which T-cell development is blocked primarily at the CD44+ CD25+ DN stage (39, 47, 61). Our study has shown that expression of the IL-7 receptor α gene is not repressed by Id1. Furthermore, the phenotype of Id1 transgenic mice is different from those of the PU.1-deficient mice (37, 52) and the mice expressing the dominant negative form of the Ikaros protein that inhibits the function of the Ikaros family of proteins (18). In these mice, the lymphoid lineage is lacking, suggesting that the E proteins act at different points from the PU.1 and Ikaros transcription factors during hematopoiesis.
The apparent developmental block at the progenitor stage in the Id1 transgenic mice may be explained either by the inability of these progenitor cells to differentiate or by the failure of differentiating T cells to survive or both. It has been shown that tumor necrosis factor alpha and IL-1α signals can stimulate the differentiation of CD44+ CD25− cells along the T-cell lineage (67). Whether this signaling pathway is defective in the Id1 transgenic mice remains to be determined. On the other hand, we have indeed found massive apoptosis in the Id1 transgenic thymus. The apoptotic cells have undergone rearrangement of their TCR α and β loci, suggesting that apoptosis occurs after the initiation of T-cell differentiation. Several possible molecular mechanisms involved in triggering apoptosis in Id1 transgenic mice may be proposed. First, Id1 might inhibit E-protein-activated expression of genes involved in cellular antiapoptotic functions, rendering the differentiating T cells highly susceptible to apoptosis. However, since apoptosis occurs in at least 50% of the thymocytes, there would have to be an active mechanism that causes apoptosis in the differentiating T cells by default and that is normally suppressed by cellular antiapoptotic functions. Second, developmental blocks at various stages may lead to apoptosis of immature T cells.
Alternatively, Id1 expression might indirectly trigger apoptosis in the transgenic mice. How would Id1 cause apoptosis? Id1 has been known to stimulate cell growth in nonlymphoid cells (6, 21, 49). We have attempted to test if Id1 transgenic thymocytes are more proliferative than their wild-type counterparts but have found no significant difference in their abilities to incorporate bromodeoxyuridine in vivo (data not shown). However, if excessive proliferation leads to cell death, we may not have been able to detect any changes in the live population of cells. Obviously, it is extremely difficult to examine the proliferative status of dead cells, even though proliferation might have taken place before the cells died. If Id1 overexpression indeed alters the proliferative state of differentiating T cells, it might be incompatible with the differentiation process occurring in the cells. For example, one of the major events taking place during T-cell development is the rearrangement of TCR genes. V(D)J recombination reactions are likely to be carried out in cells in the resting state (34, 46, 59). If Id1 overexpression places cells in the wrong phase of the cell cycle or prevents the necessary cell cycle arrest for TCR rearrangement, rearrangement of the TCR genes which involves the cutting and joining of DNA strands might appear to cells as DNA damage, thus triggering apoptosis. In support of this hypothesis, we have found that the majority of apoptotic cells have undergone gene rearrangement whereas the surviving DN cells have not rearranged their TCR genes. This hypothesis is currently being tested experimentally by eliminating the V(D)J recombination reactions in the Id1 transgenic mice.
The proliferative effect of Id1 could also lead to the overstimulation of T cells and create a situation analogous to that of negative selection in the thymus, which results in apoptosis of the thymocytes (26). Supporting this hypothesis, the Id1-expressing single-positive T cells in the thymus and spleen also carry high levels of the CD44 surface marker, which serves as an indication of T-cell activation.
Mechanisms of T-cell lymphomagenesis.
The Id proteins are essential for the G1-to-S-phase transition in the cell cycle of fibroblasts, as shown by experiments with antisense oligonucleotides (6, 21). Conversely, E2A proteins have been found to block cell cycle progression at the G1-to-S-phase transition (48), perhaps by activating the transcription of the gene encoding the cyclin-dependent kinase inhibitor p21CIP/WAF1 (49). Furthermore, we have shown that in a human T-cell lymphoblastic leukemia cell line, Jurkat, restoration of E2A activity, which is inhibited by the aberrant expression of the Tal1 oncogene, leads to cell growth arrest and apoptosis (45). It is therefore not surprising that overexpression of Id1 or disruption of the E2A gene leads to T-cell lymphomagenesis (5, 63). The molecular mechanisms leading to tumor formation, however, remain to be determined. Lack of p21 gene expression is unlikely to be solely responsible for lymphomagenesis in the Id1 transgenic mice, because p21 is normally not expressed at a high level in T cells and the p21-deficient mice do not develop lymphomas (14).
The proliferative effect of Id1 is probably a major contributor to lymphomagenesis. However, tumor formation might require a second hit. The second hit may result from spontaneous genetic mutations causing the repression of tumor suppressor genes or the activation of oncogenes. It may simply be due to growth stimulation by lymphokines that are present during normal T-cell development. Furthermore, the second hit could also be created by Id1 overexpression, which may lead to alteration of the expression of growth-regulating genes or to genetic instability. The heterogeneous phenotypes of the Id1 transgenic lymphomas with regard to their developmental stages would suggest that the second hit may occur at various stages in T-cell development and may have diverse tumorigenic potentials.
It will also be interesting to determine whether the tumorigenic effect and the ability of Id1 to block T-cell development are mediated through similar mechanisms. Perhaps it is because of the proliferative effect of Id1 that massive apoptosis of developing thymocytes occurs and T-cell lymphoma develops. An excellent example of proteins with such dual functions is the c-myc oncogene, which is oncogenic in certain situations (58) but apoptotic under other circumstances (16). Overexpression of c-myc in T cells does cause T-cell lymphoma (8, 28, 54). In fact, Bain et al. (5) have suggested that c-myc expression is elevated in the lymphomas found in E2A-deficient mice. High levels of c-myc expression have also been detected in the lymphomas from the Id1 transgenic mice (data not shown). However, whether c-myc expression is a causal factor in lymphomagenesis induced by Id1 overexpression remains to be determined.
ACKNOWLEDGMENTS
We are grateful to Sankar Ghosh for discussion and critical reading of the manuscript. We thank Yang Liu, Stanislav Vukmanovic, and Alan Frey for advice and John Hirst of the cell-sorting facility of Kaplan Cancer Center for excellent assistance in cell-sorting analyses. The Id1 transgenic mice were produced by the Transgenic Facility of Rockefeller University and the facility of the Kaplan Cancer Center at NYU School of Medicine.
This work was supported by grants from NIH (1R01AI33597 and 1R01CA77553), the American Cancer Society (RPG-98-247-01-LBC), and the Life and Health Insurance Fund. X.H.S. is an Irma T. Hirschl Trust Scholar.
REFERENCES
- 1.Amsen D, Kruisbeek A M. Thymocyte selection: not by TCR alone. Immunol Rev. 1998;165:209–229. doi: 10.1111/j.1600-065x.1998.tb01241.x. [DOI] [PubMed] [Google Scholar]
- 2.Anderson S J, Abraham K M, Nakayama T, Singer A, Perlmutter R M. Inhibition of T-cell receptor β-chain gene rearrangement by overexpression of the non-receptor protein tyrosine kinase p56lck. EMBO J. 1992;11:4877–4886. doi: 10.1002/j.1460-2075.1992.tb05594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bain G, Robanus Maandag E C, Izon D J, Amsen D, Kruisbeek A M, Weintraub B, Krop I, Schlissel M S, Feeney A J, van Roon M, van der Valk M, te Riele H P J, Berns A, Murre C. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell. 1994;79:885–892. doi: 10.1016/0092-8674(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 4.Bain G, Robanus Maandag E C, te Riele H P, Feeney A J, Sheehy A, Schlissel M, Shinton S A, Hardy R R, Murre C. Both E12 and E47 allow commitment to the B cell lineage. Immunity. 1997;6:145–154. doi: 10.1016/s1074-7613(00)80421-5. [DOI] [PubMed] [Google Scholar]
- 5.Bain G, Engel I, Robanus Maandag E C, te Riele H P, Voland J R, Sharp L L, Chun J, Huey B, Pinkel D, Murre C. E2A deficiency leads to abnormalities in alphabeta T-cell development and to rapid development of T-cell lymphomas. Mol Cell Biol. 1997;17:4782–4791. doi: 10.1128/mcb.17.8.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barone V M, Pepperkok R, Peverali F A, Philipson L. Id proteins control growth induction in mammalian cells. Proc Natl Acad Sci USA. 1994;91:4985–4988. doi: 10.1073/pnas.91.11.4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benezra R, Davis R L, Lockshon D, Turner D L, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 8.Benvenisty N, Ornitz D M, Bennett G L, Sahagan B G, Kuo A, Cardiff R D, Leder P. Brain tumours and lymphomas in transgenic mice that carry HTLV-I LTR/c-myc and Ig/tax genes. Oncogene. 1992;7:2399–2405. [PubMed] [Google Scholar]
- 9.Blom B, Heemskerk M H, Verschuren M C, van Dongen J J, Stegmann A P, Bakker A Q, Couwenberg F, Res P C, Spits H. Disruption of alphabeta but not of gammadelta T cell development by overexpression of the helix-loop-helix protein Id3 in committed T cell progenitors. EMBO J. 1999;18:2793–2802. doi: 10.1093/emboj/18.10.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burtrum D B, Kim S, Dudley E C, Hayday A C, Petrie H T. TCR gene recombination and alpha beta-gamma delta lineage divergence: productive TCR-beta rearrangement is neither exclusive nor preclusive of gamma delta cell development. J Immunol. 1996;157:4293–4296. [PubMed] [Google Scholar]
- 11.Carroll A M, Bosma M J. T-lymphocyte development in scid mice is arrested shortly after the initiation of T-cell receptor δ gene recombination. Genes Dev. 1991;5:1357–1366. doi: 10.1101/gad.5.8.1357. [DOI] [PubMed] [Google Scholar]
- 12.Casanova J L, Romero P, Widmann C, Kourilsky P, Maryansky J L. T cell receptor genes in a series of class I major histocompatibility complex-restricted cytotoxic T lymphocyte clones specific for a Plasmodium berghei nonapeptide: implications for T cell allelic exclusion and antigen-specific repertoire. J Exp Med. 1991;174:1371–1383. doi: 10.1084/jem.174.6.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chun J M, Schatz D G, Oettinger M A, Jaenisch R, Baltimore D. The recombination activating gene-1 (RAG-1) transcript is present in the murine central nervous system. Cell. 1991;64:189–200. doi: 10.1016/0092-8674(91)90220-s. [DOI] [PubMed] [Google Scholar]
- 14.Deng C, Zhang P, Harper J W, Elledge S J, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 15.Diamond R A, Ward Owada-Makabe S B K, Wang H, Rothenberg E V. Different developmental arrest points in RAG-2−/− and SCID thymocytes on two genetic backgrounds: developmental choices and cell death mechanisms before TCR gene rearrangement. J Immunol. 1997;158:4052–4064. [PubMed] [Google Scholar]
- 16.Evan G I, Wyllie A H, Gilbert C S, Littlewood T D, Land H, Brooks M, Waters C M, Penn L Z, Hancock D C. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- 17.Garvin A M, Abraham K M, Forbush K A, Farr A G, Davison B L, Perlmutter R M. Disruption of thymocyte development and lymphomagenesis induced by SV40 T-antigen. Int Immunol. 1990;2:173–180. doi: 10.1093/intimm/2.2.173. [DOI] [PubMed] [Google Scholar]
- 18.Georgopoulos K, Bigby M, Wang J H, Molnar A, Wu P, Winandy S, Sharpe A. The Ikaros gene is required for the development of all lymphoid lineages. Cell. 1994;79:143–156. doi: 10.1016/0092-8674(94)90407-3. [DOI] [PubMed] [Google Scholar]
- 19.Hahm K, Ernst P, Lo K, Kim G S, Turck C, Smale S T. The lymphoid transcription factor LyF-1 is encoded by specific, alternatively spliced mRNAs derived from the Ikaros gene. Mol Cell Biol. 1994;14:7111–7123. doi: 10.1128/mcb.14.11.7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haks M C, Krimpenfort P, Borst J, Kruisbeek A M. The CD3γ chain is essential for development of both the TCRαβ and TCRγδ lineages. EMBO J. 1998;17:1871–1882. doi: 10.1093/emboj/17.7.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hara E, Yamaguchi T, Nojima H, Ide T, Campisi J, Okayama H, Oda K. Id-related genes encoding helix-loop-helix proteins are required for G1 progression and are repressed in senescent human fibroblasts. J Bio Chem. 1994;269:2139–2145. [PubMed] [Google Scholar]
- 22.Hardy R R, Carmack C E, Shinton S A, Kemp J D, Hayakawa K. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J Exp Med. 1991;173:1213–1225. doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heemskerk M H, Blom B, Nolan G, Stegmann A P, Bakker A Q, Weijer K, Res P C, Spits H. Inhibition of T cell and promotion of natural killer cell development by the dominant negative helix loop helix factor Id3. J Exp Med. 1997;186:1597–1602. doi: 10.1084/jem.186.9.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henthorn P, Kiledjian M, Kadesch T. Two distinct transcription factors that bind the immunoglobulin enhancer μE5/κE2 motif. Science. 1990;247:467–470. doi: 10.1126/science.2105528. [DOI] [PubMed] [Google Scholar]
- 25.Hu J-S, Olson E N, Kingston R E. HEB, a helix-loop-helix protein related to E2A and ITF2 that can modulate the DNA-binding activity of myogenic regulatory factors. Mol Cell Biol. 1992;12:1031–1042. doi: 10.1128/mcb.12.3.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janeway C A, Travers P. Immunobiology: the immune system in health and disease. 3rd ed. New York, N.Y: Current Biology Ltd./Garland Publishing, Inc.; 1997. [Google Scholar]
- 27.Koyasu S, Clayton L K, Lerner A, Heiken H, Parkes A, Reinherz E L. Pre-TCR signaling components trigger transcriptional activation of a rearranged TCRα gene locus and silencing of the pre-TCRα locus: implications for intrathymic differentiation. Int Immunol. 1997;9:1475–1480. doi: 10.1093/intimm/9.10.1475. [DOI] [PubMed] [Google Scholar]
- 28.Leder A, Pattengale P K, Kuo A, Stewart T A, Leder P. Consequences of widespread deregulation of the c-myc gene in transgenic mice: multiple neoplasms and normal development. Cell. 1986;45:485–495. doi: 10.1016/0092-8674(86)90280-1. [DOI] [PubMed] [Google Scholar]
- 29.Lee J E, Hollenberg S M, Snider L, Turner D L, Lipnick N, Weintraub H. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop-helix protein. Science. 1995;268:836–844. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- 30.Li Y S, Wasserman R, Hayakawa K, Hardy R R. Identification of the earliest B lineage stage in mouse bone marrow. Immunity. 1996;5:527–535. doi: 10.1016/s1074-7613(00)80268-x. [DOI] [PubMed] [Google Scholar]
- 31.Levin S D, Anderson S J, Forbush K A, Perlmutter R M. A dominant-negative transgene defines a role for p56lck in thymopoieses. EMBO J. 1993;12:1671–1680. doi: 10.1002/j.1460-2075.1993.tb05812.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y S, Hayakawa K, Hardy R R. The regulated expression of B lineage associated genes during B cell differentiation in bone marrow and fetal liver. J Exp Med. 1993;178:951–960. doi: 10.1084/jem.178.3.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin H, Grosschedl R. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature. 1995;376:263–267. doi: 10.1038/376263a0. [DOI] [PubMed] [Google Scholar]
- 34.Lin W C, Desiderio S. Cell cycle regulation of V(D)J recombination-activating protein RAG-2. Proc Natl Acad Sci USA. 1994;91:2733–2737. doi: 10.1073/pnas.91.7.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malissen M, Gillet A, Ardouin L, Bouvier G, Trucy J, Ferrier P, Vivier E, Malissen B. Altered T cell development in mice with a targeted mutation of the CD3-epsilon gene. EMBO J. 1995;14:4641–4653. doi: 10.1002/j.1460-2075.1995.tb00146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Massari M E, Jennings P A, Murre C. The AD1 transactivation domain of E2A contains a highly conserved helix which is required for its activity in both Saccharomyces cerevisiae and mammalian cells. Mol Cell Biol. 1996;16:121–129. doi: 10.1128/mcb.16.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKercher S R, Torbett B E, Anderson K L, Henkel G W, Vestal D J, Baribault H, Klemsz M, Feeney A J, Wu G E, Paige C J, Maki R A. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 1996;15:5647–5658. [PMC free article] [PubMed] [Google Scholar]
- 38.Mombaerts P, Iacomini J, Johnson R S, Herrup K, Tonegawa S, Papaioannou V E. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 39.Moore T A, von Freeden-Jeffry U, Murray R, Zlotnik A. Inhibition of gamma delta T cell development and early thymocyte maturation in IL-7 −/− mice. J Immunol. 1996;157:2366–2373. [PubMed] [Google Scholar]
- 40.Murre C, McCaw P S, Baltimore D. A new DNA binding and dimerization motif in immunoglobin enhancer binding, daughterless, MyoD and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- 41.Murre C, McCaw P S, Vaessin H, Caudy M, Jan L Y, Jan Y N, Cabrera C V, Buskin J N, Hauschka S D, Lassar A B, Weintraub H, Baltimore D. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- 42.Naya F J, Stellrecht C M, Tsai M J. Tissue-specific regulation of the insulin gene by a novel basic helix-loop-helix transcription factor. Genes Dev. 1995;9:1009–1019. doi: 10.1101/gad.9.8.1009. [DOI] [PubMed] [Google Scholar]
- 43.Okamura R M, Sigvardsson M, Galceran J, Verbeek S, Clevers H, Grosschedl R. Redundant regulation of T cell differentiation and TCRα gene expression by the transcription factors LEF-1 and TCF-1. Immunity. 1998;8:11–20. doi: 10.1016/s1074-7613(00)80454-9. [DOI] [PubMed] [Google Scholar]
- 44.Pardoll D M, Fowlkes B J, Bluestone J A, Kruisbeek A, Maloy W L, Coligan J E, Schwartz R H. Differential expression of two distinct T-cell receptors during thymocyte development. Nature. 1987;326:79–81. doi: 10.1038/326079a0. [DOI] [PubMed] [Google Scholar]
- 45.Park S T, Nolan G P, Sun X-H. Growth inhibition and apoptosis due to restoration of E2A activity in T cell acute lymphoblastic leukemia cells. J Exp Med. 1999;189:501–508. doi: 10.1084/jem.189.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Penit C, Lucas B, Vasseur F. Cell expansion and growth arrest phases during the transition from precursor (CD4−8−) to immature (CD4+8+) thymocytes in normal and genetically modified mice. J Immunol. 1995;154:5103–5113. [PubMed] [Google Scholar]
- 47.Peschon J J, Morrissey P J, Grabstein K H, Ramsdell F J, Maraskovsky E, Gliniak B C, Park L S, Ziegler S F, Williams D E, Ware C B, Meyer J D, Davison B L. Early lymphocyte expansion is severely impaired in interleukin 7 receptor-deficient mice. J Exp Med. 1994;180:1955–1960. doi: 10.1084/jem.180.5.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peverali F A, Ramqvist T, Saffrich R, Pepperkik R, Barone M V, Philipson L. Regulation of G1 progression by E2A and Id helix-loop-helix proteins. EMBO J. 1994;13:4291–4301. doi: 10.1002/j.1460-2075.1994.tb06749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prabhu S, Ignatova A, Park S T, Sun X-H. Regulation of the expression of cyclin-dependent kinase inhibitor, p21, by E2A and Id proteins. Mol Cell Biol. 1997;17:5888–5896. doi: 10.1128/mcb.17.10.5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quong M W, Massari M E, Zwart R, Murre C. A new transcriptional-activation motif restricted to a class of helix-loop-helix proteins is functionally conserved in both yeast and mammalian cells. Mol Cell Biol. 1993;13:792–800. doi: 10.1128/mcb.13.2.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rothenberg E V. Gene regulation in T-cell lineage conpartment. In: Monroe J G, Rothenberg E V, editors. Molecular biology of B-cell and T-cell development. Totowa, N.J: Humana Press Inc.; 1998. pp. 337–365. [Google Scholar]
- 52.Scott E W, Simon M C, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 53.Shinkai Y, Rathbun G, Lam K P, Oltz E M, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall A M, Alt F W. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 54.Stewart M, Cameron E, Campbell M, McFarlane R, Toth S, Lang K, Onions D, Neil J C. Conditional expression and oncogenicity of c-myc linked to a CD2 gene dominant control region. Int J Cancer. 1993;53:1023–1030. doi: 10.1002/ijc.2910530628. [DOI] [PubMed] [Google Scholar]
- 55.Sun X-H, Baltimore D. An inhibitory domain of E12 prevents DNA binding in E12 homodimers but not in E12 heterodimers. Cell. 1991;64:459–470. doi: 10.1016/0092-8674(91)90653-g. [DOI] [PubMed] [Google Scholar]
- 56.Sun X-H, Copeland N G, Jenkins N A, Baltimore D. Id proteins, Id1 and Id2, selectively inhibit DNA binding by one class of helix-loop-helix proteins. Mol Cell Biol. 1991;11:5603–5611. doi: 10.1128/mcb.11.11.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun X-H. Constitutive expression of the Id1 gene impairs mouse B cell development. Cell. 1994;79:893–900. doi: 10.1016/0092-8674(94)90078-7. [DOI] [PubMed] [Google Scholar]
- 58.Taub R, Kirsch I, Morton C, Lenoir G, Swan D, Tronick S, Aaronson S, Leder P. Translocation of the c-myc gene into the immunoglobulin heavy chain locus in human Burkitt lymphoma and murine plasmacytoma cells. Proc Natl Acad Sci USA. 1982;79:7837–7841. doi: 10.1073/pnas.79.24.7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tourigny M R, Mazel S, Burtrum D B, Petrie H T. T cell receptor (TCR)-beta gene recombination: dissociation from cell cycle regulation and developmental progression during T cell ontogeny. J Exp Med. 1997;185:1549–1556. doi: 10.1084/jem.185.9.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vacchio M S, Ashwell J D, King L B. A positive role for thymus-derived steroids in formation of the T-cell repertoire. Ann N Y Acad Sci. 1998;840:317–327. doi: 10.1111/j.1749-6632.1998.tb09571.x. [DOI] [PubMed] [Google Scholar]
- 61.von Freeden-Jeffry U, Vieira P, Lucian L A, McNeil T, Burdach S E G, Murray R. Lymphopenia in interleukin (IL)-7 gene-deleted mice identifies IL-7 as a nonredundant cytokine. J Exp Med. 1995;181:1519–1526. doi: 10.1084/jem.181.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu L, Livak F, Petrie H T. TCR-independent development of pluripotent T-cell precursors. In: Monroe J G, Rothenberg E V, editors. Molecular biology of B-cell and T-cell development. Totowa, N.J: Humana Press Inc.; 1998. pp. 285–303. [Google Scholar]
- 63.Yan W, Young A Z, Soares V C, Kelley R, Benezra R, Zhuang Y. High incidence of T-cell tumors in E2A-null mice and E2A/Id1 double-knockout mice. Mol Cell Biol. 1997;17:7317–7327. doi: 10.1128/mcb.17.12.7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhuang Y, Soriano P, Weintraub H. The helix-loop-helix gene E2A is required for B cell differentiation. Cell. 1994;79:875–884. doi: 10.1016/0092-8674(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 65.Zhuang Y, Cheng P, Weintraub H. B-lymphocyte development is regulated by the combined dosage of three basic helix-loop-helix genes, E2A, E2-2 and HEB. Mol Cell Biol. 1996;16:2898–2905. doi: 10.1128/mcb.16.6.2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhuang Y, Barndt R J, Pan L, Kelley R, Dai M. Functional replacement of the mouse E2A gene with a human HEB cDNA. Mol Cell Biol. 1998;18:3340–3349. doi: 10.1128/mcb.18.6.3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zuniga-Pflucker J C, Di J, Lenardo M J. Requirement for TNF-alpha and IL-1 alpha in fetal thymocyte commitment and differentiation. Science. 1995;268:1906–1909. doi: 10.1126/science.7541554. [DOI] [PubMed] [Google Scholar]