Prevalence of Corynebacterial 16S rRNA Sequences in Patients with Bacterial and “Nonbacterial” Prostatitis (original) (raw)
Abstract
The etiology of chronic prostatitis syndromes in men is controversial, particularly when positive cultures for established uropathogens are lacking. Although identification of bacteria in prostatic fluid has relied on cultivation and microscopy, most microorganisms in the environment, including some human pathogens, are resistant to cultivation. We report here on an rRNA-based molecular phylogenetic approach to the identification of bacteria in prostate fluid from prostatitis patients. Positive bacterial signals were seen for 65% of patients with chronic prostatitis overall. Seven of 11 patients with bacterial signals but none of 6 patients without bacterial signals were cured with antibiotic-based therapy. Results indicate the occurrence in the prostate fluid of a wide spectrum of bacterial species representing several genera. Most rRNA genes were closely related to those of species belonging to the genera Corynebacterium, Staphylococcus, Peptostreptococcus, Streptococcus, and Escherichia. Unexpectedly, a wide diversity of Corynebacterium species was found in high proportion compared to the proportions of other bacterial species found. A subset of these 16S rRNA sequences represent those of undescribed species on the basis of their positions in phylogenetic trees. These uncharacterized organisms were not detected in control samples, suggesting that the organisms have a role in the disease or are the consequence of the disease. These studies show that microorganisms associated with prostatitis generally occur as complex microbial communities that differ between patients. The results also indicate that microbial communities distinct from those associated with prostatitis may occur at low levels in normal prostatic fluid.
Chronic prostatitis is a common disorder in adult men of all ages. It is estimated to affect 10% of all men at any time, and 50% of all men are treated for it at least once in their lives (25, 37). The most common symptom is pain, which is felt in the perineum, scrotum, lower abdomen, pelvis, or back. Urinary symptoms include hesitancy, weak stream, painful urination, frequency, and nocturia. Up to a quarter of men complain of associated impotence or ejaculatory dysfunction. Acute prostatitis is clearly a bacterial urinary tract infection, with associated symptoms typical of an acute septic illness and rapid response to appropriate antibiotics. The etiology of chronic prostatitis is far less clear (27). On the basis of routine cultures of expressed prostatic secretions (EPSs), EPSs from fewer than 10% of patients show significant growth of known uropathogens (Escherichia coli, Enterobacter spp., Klebsiella spp., Pseudomonas spp., etc.), and the diagnosis is chronic bacterial prostatitis (CBP). The disease in men with negative cultures but leukocytosis in the EPSs (usually considered >10 leukocytes per high-power field) is classified as nonbacterial prostatitis (NBP), and the disease in those with negative cultures and no leukocytosis is classified as prostatodynia.
The etiology of these syndromes remains unclear. Central to the controversy is whether the underlying cause is bacterial, autoimmune, or a primary neuromuscular pain syndrome. Clinical response to antibiotic therapy is variable and often disappointing, regardless of the culture results. The role of the frequently cultured gram-positive organisms (Staphylococcus epidermidis, viridans group streptococci) as commensal organisms or pathogens is controversial, as are the roles of Chlamydia spp., Ureaplasma spp., and anaerobes. Nevertheless, some patients with NBP have antibodies to uropathogenic bacterial antigens (23, 36), and molecular methods have indicated bacterial DNA in prostate biopsy specimens (22, 34).
Studies with samples from diverse environments have established that most microbial organisms (>99%) are not cultivated by standard methods (2, 18, 21, 29). The application of PCR to amplification of the 16S rRNA gene (rDNA) from mixed communities, however, has been successful for the identification of organisms without cultivation. These molecular methods have been applied to samples from diverse environments, including the clinical arena.
Establishment of the etiology of human infections has traditionally relied on cultivation of microorganisms and a demonstration of infection in animal models, i.e., Koch’s postulates. However, it is apparent that many human clinical syndromes traditionally thought to be nonmicrobial may in fact be of a microbial etiology (6, 10, 24, 30). There are well-known microbial pathogens resistant to culture, including Mycobacterium leprae, Treponema pallidum, and Tropheryma whippelii, the causative agents of leprosy, syphilis, and Whipple’s disease, respectively. Molecular methods were used to determine the 16S rRNA sequence of the uncultured organism from patients with Whipple’s disease, and an evolutionary tree was generated to identify the nearest relatives (31). Some estimation of the physiology of the otherwise unknown organism can be made on the basis of the properties of cultivated relatives (7, 20, 29).
In the present study, we determined the 16S rDNA sequences of bacteria in EPSs from patients with chronic prostatitis and controls and correlate the sequences with those of the rDNAs of cultured organisms. We uncovered a number of previously unidentified organisms in the chronic prostatitis patients. The sequence collection is a resource for further study of this important affliction.
MATERIALS AND METHODS
Patients.
EPSs were collected by prostatic massage following cleansing of the glans penis with an alcohol swab and retraction of the prepuce if present. One drop was examined microscopically for leukocytes, several drops were sent for standard aerobic culture on agar and incubated for 5 to 7 days, and the remainder was stored at −70°C for use in the molecular studies. First-void urine (voided bladder; VB1 samples) was collected prior to prostatic massage and was treated in an identical fashion.
The study included 17 patients with chronic prostatitis. These patients had suffered from symptoms for a median of 3.5 years (range, 1 to 9 years). All patients had previously failed multiple courses of treatment with antibiotics. This is the common presentation history for patients with long-standing chronic prostatitis. None of the control patients had recently received antimicrobial therapy. The diagnosis was CBP in six patients (although only one patient was infected with a typical uropathogen), NBP in seven patients, and prostatodynia in four patients. Two patients with prostatodynia had equal counts of gram-positive bacteria in both the urethra and prostate.
All patients were treated with a combination of regular prostate massage and oral antimicrobial therapy. On initial examination all patients had been off of antimicrobial agents for at least 2 weeks. Antimicrobial agents were chosen on the basis of culture sensitivity testing or, in the case of negative cultures, empirically (usually trovafloxacin). Common choices were azithromycin (Biaxin) at 500 mg twice a day, amoxicillin-clavulanate (Augmentin) at 875 mg twice a day, minocycline at 100 mg twice a day, or cephalexin (Keflex) at 500 mg three times a day (all antimicrobial agents were given for at least 4 weeks).
Eight samples were collected from control patients, none of whom had pelvic pain. Two patients had symptomatic benign prostatic hyperplasia (BPH), one patient had prostate cancer, and five patients had no urologic disorders (prevasectomy patients).
DNA extraction.
A total of 50 to 100 μl of the EPS or VB1 sample was mixed with 500 μl of TEN buffer (200 mM Tris HCl [pH 8.0], 20 mM EDTA, 200 mM NaCl) in a sterile 2-ml screw-cap tube, and the mixture was transferred to ice. After the addition of 20 μl of poly(A) (10 mg/ml) and 30 μl of lysozyme (100 mg/ml), the samples were incubated for 30 min at 37°C. This was followed by the addition of 10 μl of 20% sodium dodecyl sulfate and 60 μl of proteinase K (20 mg/ml) and incubation for 30 min at 50°C. An additional 200 μl of 20% sodium dodecyl sulfate was added, followed by the addition of 500 μl of phenol-chloroform-isoamyl alcohol (25:24:1; vol/vol) and approximately 0.5 g of untreated zirconia-silica beads (diameter, 0.1 mm; Biospec Products, Bartlesville, Okla.) according to the manufacturer’s recommendations. All solutions were prepared with pure sterile water (Fluka Chemical Corporation, Milwaukee, Wis.). Samples were subjected to bead beating with a Mini-Beadbeater (Biospec Products) on low for 2 min and on high for 0.5 min. After collection of the aqueous phase and reextraction, the DNA was precipitated, rinsed in 70% ethanol, dried, and resuspended in 100 μl of water.
PCR.
The bacterium-specific and universal primers designed to amplify the 16S rDNA were 27F (AGAGTTTGATCMTGGCTCAG), 515F (GTGCCAGCMGCCGCGGTAA), 805R (GACTACCAGGGTATCTAATCC), 1391R (GACGGGCGGTGWGTRCA), and 1492R (GGYTACCTTGTTACGACTT) (where M is A or C, Y is C or T, W is A or T, and R is A or G). The following primer pairs were selected for amplification of the genomic DNA: 27F-805R (bacteria only), 27F-1492R (bacteria only), and 515F-1391R (universal primer). The primers that were designed for this study and that are specific to the genus Corynebacterium and close relatives are Cory52F (GAACGCTGSCGGCGTGCTTAAC) and Cory1479R (TTGTTACRRCTTCGTCCCAATCGCC) (where S is G or C and R is A or G). These primers amplify the region between nucleotides 52 and 1479 (E. coli numbering).
PCRs were carried out with 100-μl PCR mixtures consisting of 1 μl of template DNA, 200 μM deoxynucleoside triphosphates, 1.5 mM MgCl2, and 40 cycles of amplification, consisting of 20 cycles of 1 min at 94°C, 40 s starting at 67°C and decreasing by 1°C/cycle, and 1 min at 72°C and 20 cycles of 1 min at 94°C, 1 min at 47°C, and 1.5 min plus 1 s/cycle at 72°C. The PCR had a 5-min preincubation at 94°C to activate the Ampli_Taq_ Gold (Perkin-Elmer, Norwalk, Conn.) and a final step at 72°C for 8 min to ensure complete extension for efficient cloning. Five units of Ampli_Taq_ Gold was added per 100-μl reaction mixture. The primer concentration was 0.4 μM.
Cloning and RFLP analysis.
PCR products were gel purified and cloned with the TOPO TA cloning kit pCR2.1-TOPO vector (Invitrogen Corporation, Carlsbad, Calif.) by following the manufacturer’s recommendations. Ninety-six-well minipreps (26) were carried out to isolate plasmids from individual clones for restriction fragment length polymorphism (RFLP) analysis and sequencing.
rDNA inserts from pCR2.1 vector clones were amplified by PCR with vector primers equidistant from the 5′ and 3′ ends of the insert. We used RFLP analysis to estimate the diversity of bacteria in the samples from patients with prostatitis and to identify unique clones for sequence analysis. For RFLP analysis, DNA was digested with _Msp_I and _Hin_P1I in NEBuffer 2 (New England Biolabs, Beverly, Mass.) and 0.01% Triton X-100. The digested DNA was separated on a 3% MetaPhor gel (FMC Bioproducts, Rockland, Maine) in 1× TBE (Tris-borate-EDTA) for about 2.5 h at 50 V. Ethidium bromide-stained gels were visualized with a NucleoVision digital imaging system (NucleoTech Corporation, San Carlos, Calif.).
Sequencing and phylogenetic analysis.
Clones determined to be unique by RFLP analysis were sequenced on an ABI 373 stretch DNA sequencer (Dye-Terminator Cycle Sequencing Ready Reaction FS kit; PE Applied Biosystems, Foster City, Calif.) according to the manufacturer’s instructions. The 16S rDNA sequences were compared to known sequences in GenBank with the advanced gapped BLAST (basic local alignment search tool) algorithm (1, 4). The 16S rDNA sequences were compiled in Sequence Navigator (PE Applied Biosystems), aligned with the genetic database environment (GDE) alignment editor, and placed into a phylogenetic tree containing approximately 8,000 rDNA sequences. The neighbor-joining, distance matrix, and maximum-likelihood methods were carried out on the ARB platform (38). Statistical evaluation was performed by ARB bootstrap analysis.
Nucleotide sequence accession numbers.
The rDNA sequences of the 28 corynebacterium-like prostatitis-associated clones have the GenBank accession no. AF115927 to AF115954, respectively.
RESULTS
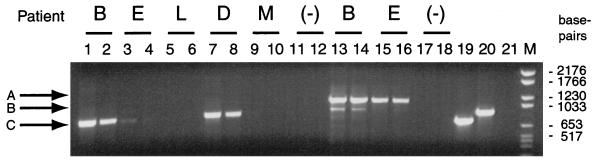
DNAs isolated directly from EPSs from 17 patients and 8 controls were subjected to PCR with universal and, alternatively, bacterium-specific 16S rRNA primers. First-void urine (VB1) samples, collected prior to the collection of EPSs, served as controls for the bacteria present in the urine or urethra of the patients. PCR products were analyzed by agarose gel electrophoresis. Products were detected in DNA from EPSs from 11 patients and 6 controls, ranging from a qualitatively strong band of the expected size to a faint band (Fig. 1). Patients whose samples produced a bacterial signal by PCR and the histories of some of the patients are described in Table 1. As seen from Table 1, positive cultures were found for only 7 of 11 patients from whom bacterial DNA was detected by PCR. These seven patients (64%) with positive bacterial signals responded successfully to antibiotic and prostate message therapy. Of particular note, all three patients with NBP and positive bacterial signals responded to treatment, while none of the six patients without bacterial signals responded.
FIG. 1.
PCR of genomic DNA from EPSs. The 16S rDNA gene segments were amplified with primers 27F-805R (lanes 1 to 12) or primers 515F-1391R (lanes 13 to 18). Genomic DNA from E. coli served as a positive control and was amplified with primers 27F-805R (lane 19) and 515F-1391R (lane 20). Lane (−), negative extraction control; lane 21, PCR in the absence of template DNA (515F-1391R); lane M, DNA size markers. Arrows A, B, and C denote 16S rDNA (27F-805R), 16S rDNA (515F-1391R), and 18S rDNA (515F-1391R), respectively.
TABLE 1.
Patient diagnosis, cultures, and response to antibiotics
| Patient | Age (yr) | Diagnosis | Culture findings | Response to antibiotic and prostate massage therapya |
|---|---|---|---|---|
| A | 37 | NBP | Multiple negative culturesb | Yes |
| B | 60 | CBP | Viridans group streptococci (35,000 CFU/ml) and yeast | No |
| C | 50 | CBP | Escherichia coli | Yes |
| D | 54 | CBP | Viridans group streptococci (1,000 CFU/ml) and Corynebacterium afermentans (3,000 CFU/ml) | Yes |
| E | 25 | CBP | Corynebacterium group ANF (35,000 CFU/ml) | No |
| F | 34 | CBP | Enterococcus, viridans group streptococci, coagulase-negative Staphylococcus and Corynebacterium group ANF | Yes |
| G | 50 | NBP | Negative | Yes |
| H | 59 | NBP | Negative | Yes |
| I | 35 | CBP | Staphylococcus epidermidis | Yes |
| J | 55 | Prostatodynia | Corynebacterium sp.c | No |
| K | 31 | Prostatodynia | Corynebacterium xerosis and Corynebacterium afermentans (2,000 CFU/ml)c | No |
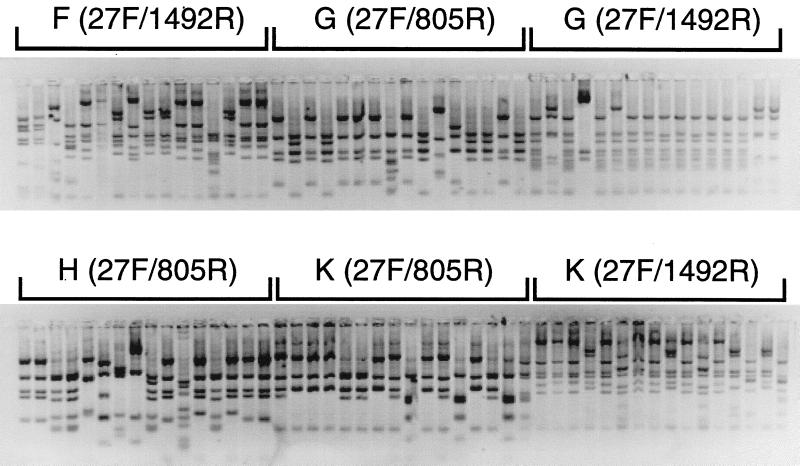
A representative RFLP gel electrophoresis profile for four patient samples is shown in Fig. 2. Instances of identical RFLP patterns were observed in all patients, and the sequences of the unique types were determined. About 45 RFLP types were encountered overall in this study. Approximately 200 partial or nearly complete 16S rRNA sequences were determined. A detailed account of the results and analysis of the sequences is presented on the web site supplement (39).
FIG. 2.
RFLP analysis of 16S rDNA clones from the EPSs from four patients. Agarose gel electrophoresis of partial 16S rDNA digested with _Msp_I and _Hin_P1I from one 96-well plate was performed. The RFLP types were counted, and unique banding patterns were analyzed further by sequencing.
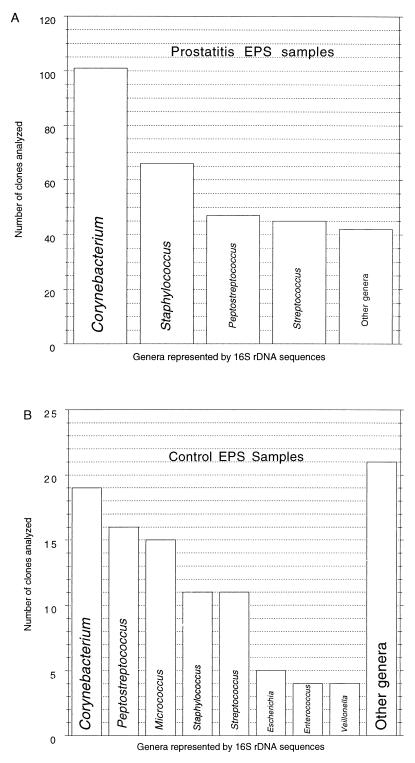
Each of the 200 sequences determined was >90% identical to the 16S rDNAs of known organisms; most were closely related (>98% identity) but not identical to 16S rDNA sequences in the database. Phylogenetic analysis of the 16S rDNAs allowed the corresponding organisms to be resolved to the species level in some cases. All sequences were clearly from organisms representing described genera. The distribution of genera represented by the 16S rDNA sequences isolated from samples from the prostatitis patients and the controls is summarized in Fig. 3. Table 2 details the occurrences of the most conspicuous prostatitis-associated organisms, Corynebacterium spp., and the noncorynebacterial spp. The species distribution differed significantly between disease-related and control samples, as discussed below (see the Discussion). The most prominent species detected in the VB1 samples, S. epidermidis, Propionibacterium acnes, Enterococcus faecium, and Pseudomonas sp., also were detected in EPSs, but they were not conspicuous. The rDNA sequences encountered in this survey of EPSs represent overall a limited extent of bacterial diversity, only 3 of the 38 known divisions (21) of Bacteria: Actinobacteria, gram-positive bacteria with low G+C contents, and Proteobacteria. The majority of sequences detected were from representatives of the Actinobacteria and the gram-positive bacteria with low G+C contents.
FIG. 3.
(A) Frequency of detection of bacterial genera in EPSs from prostatitis patients. The bar graph is a compilation of the total number of clones (n = 301) from the 11 prostatitis patients examined in this study. (B) Representative genera from the EPSs from control subjects (106 total clones). Other genera of lesser abundance are listed on the web site (39).
TABLE 2.
Distribution of corynebacterial 16S rDNA sequences in prostatitis patient and control samplesa
| Patient | A | B | C | D | E | F | G | H | I | J | K | C1 | C2 | C3 | C4 | C5 | C6 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detection of bacteria by cultivation | − | + | + | +b | + | + | − | − | + | + | +c | ||||||
| Total no. of clones analyzed | 70 | 43 | 18 | 29 | 19 | 16 | 32 | 16 | 23 | 20 | 31 | 14 | 16 | 15 | 15 | 26 | 19 |
| Corynebacterium sp. strain CDC B8037 | 15 | ||||||||||||||||
| “Corynebacterium genitalium” | 2 | 4 | 2 | ||||||||||||||
| Corynebacterium accolens | 3 | ||||||||||||||||
| Corynebacterium mucifaciens | • | ||||||||||||||||
| “Corynebacterium tuberculostearicum” | 3 | • | 1 | 1 | 10 | 1 | 12 | 6 | 5 | ||||||||
| Corynebacterium seminale | 11 | 8 | |||||||||||||||
| Corynebacterium jeikeium | • | • | 1 | ||||||||||||||
| Corynebacterium amycolatum | • | ||||||||||||||||
| Corynebacterium xerosis | • | ||||||||||||||||
| Corynebacterium glucuronolyticum | • | ||||||||||||||||
| Corynebacterium thomssenii | • | ||||||||||||||||
| Clone MTcory19R | 3 | • | • | ||||||||||||||
| Clone MTcory21R | • | • | 5 | ||||||||||||||
| Clone MTcory16R | 1 | 7 | |||||||||||||||
| Clone MTcory11W | 10 | 1 | 1 | • | |||||||||||||
| Staphylococcus aureus | 20 | ||||||||||||||||
| Staphylococcus epidermidis | 11 | 4 | 2 | ||||||||||||||
| Staphylococcus haemolyticus | 7 | 11 | 7 | 6 | 8 | 1 | |||||||||||
| Streptococcus oralis | 16 | ||||||||||||||||
| Streptococcus mitis | 10 | 12 | 1 | 7 | 1 | ||||||||||||
| Streptococcus pneumoniae | 6 | ||||||||||||||||
| Streptococcus parasanguis | 1 | ||||||||||||||||
| Streptococcus gordonii | 2 | ||||||||||||||||
| Streptococcus milleri | 1 | ||||||||||||||||
| Escherichia coli | 10 | 3 | 2 | ||||||||||||||
| Gemella haemolysans | 1 | 6 | 1 | ||||||||||||||
| Peptostreptococcus magnus | 6 | 8 | 2 | ||||||||||||||
| Peptostreptococcus asaccharolyticus | 9 | ||||||||||||||||
| Micrococcus luteus | 15 | ||||||||||||||||
| Propionibacterium acnes | 1 | 18 | 1 | ||||||||||||||
| Actinomyces neuii | 4 |
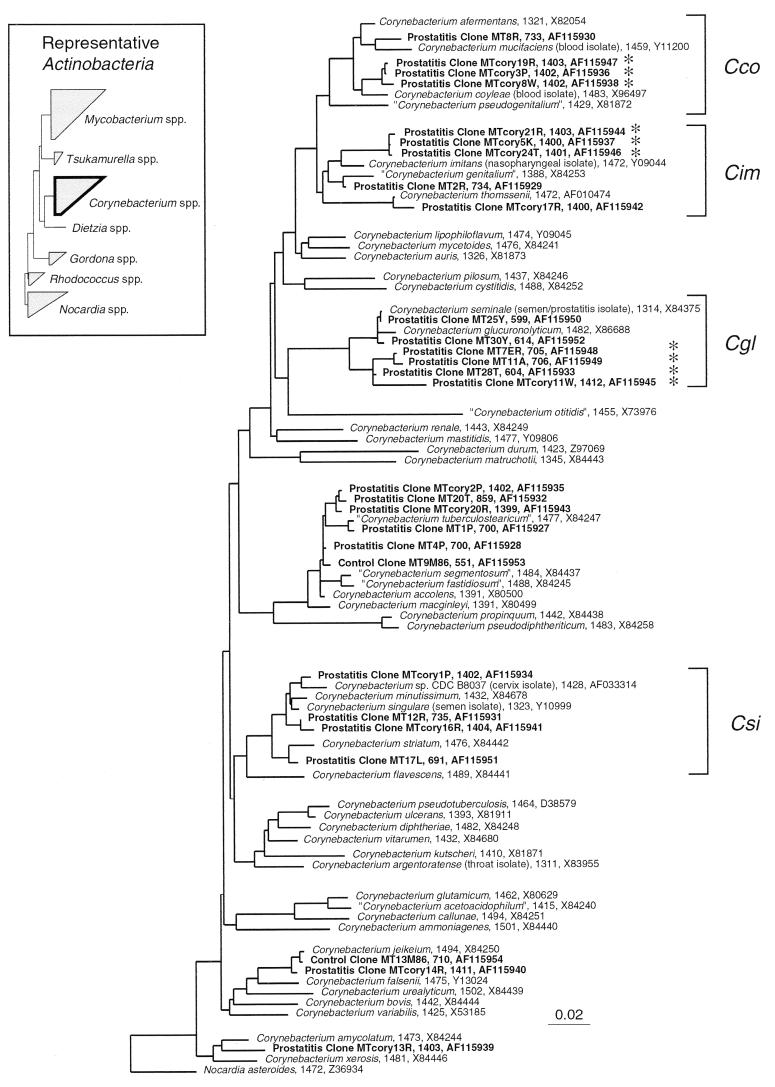
The most diverse and, for samples from patients, the most abundant sequences are characteristic of corynebacteria (Table 2; Fig. 4). The 16S rDNA sequences of 15 distinct Corynebacterium spp. were identified, with each patient having at least one species. One patient, patient D, yielded 9 of the 15 different Corynebacterium species detected (corynebacterial rDNAs were cleared from the EPSs from patient D after treatment with trovafloxacin and prostate massage). Ten of the corynebacterial sequences detected are nearly identical (>98%) to those of known Corynebacterium spp. Control samples had sequences indicating only three corynebacterial species (see the Discussion), and most of the control samples lacked detectable corynebacterial sequences.
FIG. 4.
Distance dendrogram of the genus Corynebacterium. This genus is affiliated with the Actinobacteria division, with relatives diagrammed in the inset. Sequences are identified with the number of nucleotides analyzed and the corresponding GenBank entry number. The Corynebacteria 16S rDNA sequences from EPSs cluster into distinct groups. The four main groups discussed in the text are indicated with brackets. Abbreviations: Cco, C. coyleae group; Cim, C. imitans group; Cgl, C. glucuronolyticum group; and Csi, C. singulare group. Clones indicated by an asterisk occurred in EPSs from at least three patients. Tree topology was determined by neighbor joining. Nocardia asteroides was used as the outgroup. The bar at the bottom indicates nucleotide changes per site.
The genus Corynebacterium is an assemblage of at least 40 described species, several of which have been identified in diverse clinical samples over the last 5 years (13). A phylogenetic analysis of available rRNA sequences of Corynebacterium spp., including those determined in this study, is shown in Fig. 4. Although most of the corynebacterial sequences recovered from the samples from patients with prostatitis were nearly identical to those of characterized organisms, several of the sequences were less than 98% identical to known corynebacterial sequences and so represent those from organisms that differ from known organisms at the species level. These novel sequences fall into four relatedness groups (bracketed in Fig. 4). Representatives of three of these groups, indicated with asterisks in Fig. 4, were identified in at least three different patients and were not detected in the controls. The closest described relatives of these three clone types are Corynebacterium coyleae (98%), Corynebacterium imitans (97.5%), and Corynebacterium seminale (96%).
In analyses of microbes by PCR-based techniques it is crucial to avoid contamination. The sizes of the DNAs obtained with primers 27F-805R and 27F-1492R were consistent with those of an amplified portion of the 16S rDNA. Negative controls were included to account for potential contamination from the extraction procedure or the PCR mixture, and significant products from neither the extraction procedure nor the PCR mixture were detected (Fig. 1). Nonetheless, contaminating DNA from materials used in the processing of the samples is a threat. Although negative extraction controls did not produce a visible PCR product, cloning of the resulting small amounts of DNA yields some colonies when the control samples are plated in the same manner as the positive samples. We have analyzed rDNAs cloned from trace levels of PCR products in no-sample controls in order to determine the bacterial types that are potentially present as background contaminants (40). The organisms detected in these control samples were characteristic of contaminant organisms identified in a previous study (40) and presumably represent intrinsic contamination in the system: reagents, buffers, solvents, etc. Patient samples with weak signals were more likely to have small numbers of contaminating organisms, but still, these represented fewer than 5% of the total clones in libraries derived from low-yield samples. Contaminating clones, representing organisms such as Leptothrix sp., Flavobacterium sp., and Cytophaga sp., are typical in control, no-sample libraries.
DISCUSSION
Molecular phylogenetic methods based on PCR and sequence analysis were used to determine the bacterial species present in clinical samples from prostatitis patients and to compare the results with those from culture studies. Patient samples that had positive culture results generally yielded a strong PCR signal, whereas the three samples from patients with nonbacterial prostatitis generated weaker signals, albeit well above the background levels. Results of culture, when it was successful, generally agreed with the rDNA sequence data (compare Table 1 with Table 2). For example, the 16S rDNAs predominately obtained from patients B and C corresponded to those of Streptococcus spp. and E. coli, respectively, in agreement with the culture results. Additionally, patient E had only 16S rDNAs indicative of Corynebacterium spp., which correlated with the culture results. However, it is often difficult to distinguish bacteria to the species level on the basis of routine culture studies. One possible example of misidentification in culture analysis is the case of patient D. Culture analysis indicated the presence of Corynebacterium afermentans, but the molecular analysis showed that the EPSs from patient D had nine different types of rRNA sequences indicative of Corynebacterium spp., but none corresponded to C. fermentans, for which the 16S rRNA sequence is available.
The extent of diversity of the bacteria in the EPSs was assessed initially with restriction digests of the cloned portions of the 16S rDNAs. An average of 5 to 10 RFLP types were found in samples from a typical patient. The quantitative distribution of the sequences in any particular sample probably approximates the distribution of organisms in the sample, but the potential for differential PCR or cloning, various gene dosages in different organisms, etc., must be acknowledged (43). This extent of diversity, although not high, still exceeds the usual expectation that one type of organism is associated with a particular syndrome. Single organisms are seldom seen in the natural environment, however. The establishment of consortia, often syntrophic, is the generality (29). Complex microbial communities may be typical in some pathological states as well but are traditionally not detected because of cultivation difficulties.
Bacteria thought to be important causative agents of prostatitis include Klebsiella spp., Proteus spp., Pseudomonas spp., Neisseria gonorrhoeae, Serratia spp., Enterobacter spp., Enterococcus faecalis, E. coli, and Chlamydia trachomatis (5, 41). Except for E. coli in one patient, these bacteria were not detected in this study in samples from patients with prostatitis. The primer pairs 27F-805R and 27F-1492R would be expected to amplify the rDNA of the organisms mentioned above but not that of C. trachomatis, which has been implicated in a small number of prostatitis cases and which would possibly be missed due to three mismatches between the 27F primer and the 16S rDNA of C. trachomatis (this type of organism is phylogenetically distant from those of all other bacterial divisions [44]).
rDNAs characteristic of Staphylococcus spp. were detected by PCR in samples from five patients and two controls. Because of the potential for contamination from commensal organisms, it is difficult to assess the significance in prostatitis of these Staphylococcus spp. However, an association of Staphylococcus spp. with prostatitis has been documented (28), and Staphylococcus haemolyticus was the most prevalent staphylococcal species detected in the samples from patients with prostatitis. rDNA characteristic of this organism also was obtained from two controls, both BPH patients. It has been reported that 90% of BPH patients have prostatitis, despite a diagnosis of BPH, and Staphylococcus spp. have been isolated from tissue from patients with BPH (3). S. haemolyticus has been associated with urinary tract infections and was isolated from a patient previously diagnosed with and treated for prostatitis (17). Significant numbers of _Staphylococcus aureus_-like 16S rDNA clones were identified in the sample from patient A but were absent from samples from all controls. S. aureus is a well-known pathogen and commensal organism, and it has been found in chronic prostatitis patients (16). The absence of this organism from the controls and samples from other patients suggests that the high concentration of S. aureus may contribute to or result from the disease in patient A. Perhaps Staphylococcus spp. are commonly present in healthy individuals and contribute to prostatitis when their numbers are elevated.
Streptococcus spp., most commonly, close relatives of Streptococcus mitis, were found in both patients and controls and so probably play only a passive role in infection. Representatives of Streptococcus pneumoniae and Streptococcus parasanguis, which were absent from the controls, were found in patients B and F. Patient B had a high proportion (about 75%) of Streptococcus spp., indicating a possible contribution of these species to the disease. Other streptococcal species, S. gordonii and S. milleri, were found in control sample C3 but not in any patient samples.
Detection of difficult-to-culture bacteria in expressed prostatic secretions is a recognized diagnostic problem for urologists (9). Three known types of such organisms that were not found in multiple patients by culture were detected in this molecular analysis. Two of these, Propionibacterium acnes and Peptostreptococcus magnus, may go undetected in clinical laboratories because some strains have complex growth requirements and grow slowly under anaerobic conditions (19). The other organism is a close relative of Gemella haemolysans, which is normally found in the pharyngeal mucosa and which has been isolated from patients with endocarditis, meningitis, and knee arthroplasty (35). The 16S rDNA sequences with close matches to G. haemolysans were found in three patients but in none of the controls.
The most abundant and diverse group of EPS-associated organisms identified in this study was the corynebacteria. Although coryneform bacteria were detected by culture in some patients, the PCR and sequencing methods used in the present study indicate that the number of species detected (species diversity) is much greater than the one or two species detected by culture. In a recent study by Domingue et al. (8), difficult-to-culture corynebacteria that would have been missed by routine culture were found in EPSs. Although Corynebacterium group ANF and Corynebacterium minutissimum were identified in that study, our results indicate that more species are typically undetected than are detected even by aggressive culture attempts. Indeed, it appears that diverse corynebacteria thrive in abundance in the inflamed prostate.
Four main relatedness groups of corynebacteria were detected in the samples from patients with prostatitis and are indicated in Fig. 4. The first group (Cco) is represented by _C. coyleae_-like organisms, and organisms of this group were detected in three patients, patients A, B, and D. The sequences of these three clones are about 98% identical to that of C. coyleae, a species isolated from patients with blood infections (14). The corresponding organisms may represent undescribed species of Corynebacterium or subspecies of C. coyleae. The members of the second group of clonal types (Cim) are essentially identical to one another, and their rRNAs are closely related to the rRNA of C. imitans (97.5% identity). Members of this group were found in three patients, patients C, D, and E. C. imitans was first identified in nasopharyngeal samples from a 5-month old boy (12). Other organisms in this group of closely related organisms include Corynebacterium genitalium, isolated from a urethritis patient (15), and Corynebacterium thomssenii, isolated from the pleural fluid of a patient with renal failure (45). _Corynebacterium glucuronolyticum_-like clones represent the third group, Cgl in Fig. 4, which contains organisms previously identified from patients with prostatitis and urinary infections (11). The rRNA sequences of the clones differed only slightly from each other but were distinct (3 to 4% differences in rRNA sequences) from those of the nearest relative, C. seminale, an organism isolated from a prostatitis patient (32). The fourth group (Csi) is represented by an unusual organism, Corynebacterium sp. strain B8037, that was identified in patient A and that was independently cultured from the cervix of a 36-year-old woman at the Centers for Disease Control and Prevention (Atlanta, Ga.) (42), Corynebacterium singulare (a species found in human semen [33]), and C. minutissimum (recently cultured from EPSs [8]).
A previous study of 16S rDNA sequences from samples from prostatitis patients indicated the presence of gram-negative bacteria related to the genera Aeromonas, Proteus, Escherichia, and Vibrio (34). Samples were obtained from biopsy tissues by a double-needle method and differed from the EPSs examined in the present study. Organisms residing in the tissues and potential contaminants may indeed be different from those identified in the prostate fluid. Additionally, rDNA sequences related to Flavobacterium spp. and Pseudomonas testosteroni were reported by Riley et al. (34). We have identified similar clones of Flavobacterium spp. and Pseudomonas testosteroni in no-sample control libraries in this study and in a previous study (40). We therefore consider that the occurrence of rDNAs indicative of these organisms in prostatic samples is likely to have resulted from contamination.
Detection of bacteria in EPSs by sequence-based technology has diagnostic and therapeutic implications for chronic prostatitis. The bacterial signal in the PCR step correlated far better with the response to antimicrobial therapy than did the presence of leukocytes, particularly in patients with negative conventional cultures. While the pathogenic significance of these novel bacterial species requires study with a larger number of patients with chronic pelvic pain syndromes and controls, this type of approach is a concrete first step in sorting out the etiology and selection of treatment in this highly prevalent and challenging disorder.
Establishment of the presence of organisms in diseased tissue does not establish that the organisms are the causative agents of the disease state. The prostate or other tissues may harbor a specific, low-level suite of microorganisms that have adapted to a unique hostile environment both in the healthy state and in the diseased state. In the diseased state, the abnormal environment of the tissue may enrich a specific suite of such commensal microbes. The general association of particular corynebacterial rRNA sequences with diseased EPSs and their uncommon occurrence in fluids from controls is a strong correlation that indicates further study. The organisms detected in secretions likely are washed-off biofilms associated with prostate tissues. The rRNA sequences reported here are a basis for tools, for instance, hybridization probes, that can be used to continue to explore the association of the corresponding organisms and this disease state. Diagnostic tests based on the rRNA sequences may also be useful for clinical applications, such as the detection and classification of the pervasive but nebulous condition of prostatitis.
ACKNOWLEDGMENTS
We thank Dan Frank for comments on the manuscript and members of the Pace laboratory for stimulating discussions.
This work was supported by grants from the National Institutes of Health and the U.S. Department of Energy.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bedalov G, Vuckovic I, Fridrih S, Bruk M, Puskar D, Bartolin Z. Prostatitis in benign prostatic hyperplasia: a histological, bacteriological and clinical study. Acta Med Croat. 1994;48:105–109. [PubMed] [Google Scholar]
- 4.Benson D A, Boguski M S, Lipman D J, Ostell J, Ouellette B F F. GenBank. Nucleic Acids Res. 1998;26:1–7. doi: 10.1093/nar/26.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruce A W, Reid G. Prostatitis associated with Chlamydia trachomatis in 6 patients. J Urol. 1989;142:1006–1007. doi: 10.1016/s0022-5347(17)38970-x. [DOI] [PubMed] [Google Scholar]
- 6.Carson D A. An infectious origin of extraskeletal calcification. Proc Natl Acad Sci USA. 1998;95:7846–7847. doi: 10.1073/pnas.95.14.7846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dojka M A, Hugenholtz P, Haack S K, Pace N R. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl Environ Microbiol. 1998;64:3869–3877. doi: 10.1128/aem.64.10.3869-3877.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Domingue G J, Human L G, Hellstrom W J G. Hidden microorganisms in “abacterial” prostatitis/prostatodynia. J Urol. 1997;157:243. [Google Scholar]
- 9.Domingue G J, Hellstrom W J G. Prostatitis. Clin Microbiol Rev. 1998;11:604–613. doi: 10.1128/cmr.11.4.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fredricks D N, Relman D A. Sequence-based identification of microbial pathogens: a reconsideration of Koch’s postulates. Clin Microbiol Rev. 1996;9:18–33. doi: 10.1128/cmr.9.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Funke G, Bernard K A, Bucher C, Pfyffer G E, Collins M D. Corynebacterium glucuronolyticum sp. nov. isolated from male patients with genitourinary infections. Med Microbiol Lett. 1995;4:204–215. [Google Scholar]
- 12.Funke G, Efstratiou A, Kuklinska D, Hutson R A, de Zoysa A, Engler K H, Collins M D. Corynebacterium imitans sp. nov. isolated from patients with suspected diphtheria. J Clin Microbiol. 1997;35:1978–1983. doi: 10.1128/jcm.35.8.1978-1983.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Funke G, von Graevenitz A, Clarridge J E, Bernard K A. Clinical microbiology of coryneform bacteria. Clin Microbiol Rev. 1997;10:125–159. doi: 10.1128/cmr.10.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Funke G, Ramos C P, Collins M D. Corynebacterium coyleae sp. nov., isolated from human clinical specimens. Int J Syst Bacteriol. 1997;47:92–96. doi: 10.1099/00207713-47-1-92. [DOI] [PubMed] [Google Scholar]
- 15.Furness G, Evangelista A T. Infection of a nonspecific urethritis patient and his consort with a pathogenic species of nonspecific urethritis Corynebacteria, Corynebacterium genitalium n. sp. Invest Urol. 1976;14:202–205. [PubMed] [Google Scholar]
- 16.Giamarellou H, Kosmidis J, Leonidas M, Papadakis M, Daikos G K. A study of the effectiveness of rifaprim in chronic prostatitis caused mainly by Staphylococcus aureus. J Urol. 1982;128:321–324. doi: 10.1016/s0022-5347(17)52906-7. [DOI] [PubMed] [Google Scholar]
- 17.Gunn B A, Davis C E. Staphylococcus haemolyticus urinary tract infection in a male patient. J Clin Microbiol. 1988;26:1055–1057. doi: 10.1128/jcm.26.5.1055-1057.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Head I M, Saunders J R, Pickup R W. Microbial evolution, diversity, and ecology: a decade of ribosomal RNA analysis of uncultivated microorganisms. Microbiol Ecol. 1998;35:1–21. doi: 10.1007/s002489900056. [DOI] [PubMed] [Google Scholar]
- 19.Hillier S L, Moncla B J. Peptostreptococcus, Propionibacterium, Eubacterium, and other nonsporeforming anaerobic gram-positive bacteria. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C: American Society for Microbiology; 1995. pp. 587–602. [Google Scholar]
- 20.Hugenholtz P, Pitulle C, Hershberger K L, Pace N R. Novel division level bacterial diversity in a Yellowstone hot spring. J Bacteriol. 1998;180:366–376. doi: 10.1128/jb.180.2.366-376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hugenholtz P, Goebel B M, Pace N R. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol. 1998;180:4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krieger J N, Riley D E, Roberts M C, Berger R E. Prokaryotic DNA sequences in patients with chronic idiopathic prostatitis. J Clin Microbiol. 1996;34:3120–3128. doi: 10.1128/jcm.34.12.3120-3128.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumon H. Detection of a local prostatic immune response to bacterial prostatitis. Infection. 1992;20:236–238. doi: 10.1007/BF01704389. [DOI] [PubMed] [Google Scholar]
- 24.Lorber B. Are all diseases infectious? Ann Intern Med. 1996;125:844–851. doi: 10.7326/0003-4819-125-10-199611150-00010. [DOI] [PubMed] [Google Scholar]
- 25.National Center for Health Statistics. Advance data from vital and health statistics. Vital Health Stat. 1993;16:61–70. [Google Scholar]
- 26.Ng W-L, Schummer M, Cirisano F D, Baldwin R L, Karlan B Y, Hood L. High-throughput plasmid mini preparations facilitated by micro-mixing. Nucleic Acids Res. 1996;24:5045–5047. doi: 10.1093/nar/24.24.5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nickel J C. Prostatitis: myths and realities. Urology. 1998;51:362–366. doi: 10.1016/s0090-4295(97)00643-2. [DOI] [PubMed] [Google Scholar]
- 28.Nickel J C, Costerton J W. Coagulase-negative Staphylococcus in chronic prostatitis. J Urol. 1992;147:398–400. doi: 10.1016/s0022-5347(17)37247-6. [DOI] [PubMed] [Google Scholar]
- 29.Pace N R. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 30.Relman D A. Detection and identification of previously unrecognized microbial pathogens. Emerg Infect Dis. 1998;4:382–389. doi: 10.3201/eid0403.980310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Relman D A, Schmidt T M, MacDermott R P, Falkow S. Identification of the uncultured bacillus of Whipple’s disease. N Engl J Med. 1992;327:293–301. doi: 10.1056/NEJM199207303270501. [DOI] [PubMed] [Google Scholar]
- 32.Riegel P, Ruimy R, de Briel D, Prévost G, Jehl F, Bimet F, Christen R, Monteil H. Corynebacterium seminale sp. nov., a new species associated with genital infections in male patients. J Clin Microbiol. 1995;33:2244–2249. doi: 10.1128/jcm.33.9.2244-2249.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riegel P, Ruimy R, Renaud F N R, Freney J, Prevost G, Jehl F, Christen R, Monteil H. Corynebacterium singulare sp. nov., a new species for urease-positive strains related to Corynebacterium minutissimum. Int J Syst Bacteriol. 1997;47:1092–1096. doi: 10.1099/00207713-47-4-1092. [DOI] [PubMed] [Google Scholar]
- 34.Riley D E, Berger R E, Miner D C, Krieger J N. Diverse and related 16S rRNA-encoding DNA sequences in prostate tissues of men with chronic prostatitis. J Clin Microbiol. 1998;36:1646–1652. doi: 10.1128/jcm.36.6.1646-1652.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruoff K L. Leuconostoc, Pediococcus, Stomatococcus, and miscellaneous gram-positive cocci that grow aerobically. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C: American Society for Microbiology; 1995. pp. 315–323. [Google Scholar]
- 36.Shortliffe L, Elliot J K, Sellers R G. Measurement of urinary antibodies to crude bacterial antigen in patients with chronic bacterial prostatitis. J Urol. 1989;141:632–636. doi: 10.1016/s0022-5347(17)40921-9. [DOI] [PubMed] [Google Scholar]
- 37.Stamey T A. Pathogenesis and treatment of urinary tract infections. Baltimore, Md: The Williams & Wilkins Co.; 1980. Urinary infections in males; pp. 342–429. [Google Scholar]
- 38.Strunk, O., O. Gross, B. Reichel, M. Max, S. Hermann, N. Struckmann, B. Nonhoff, M. Lenke, A. Vilbig, T. Ludwig, A. Bode, K. H. Schleifer, and W. Ludwig. 1996. ARB: a software environment for sequence data. [Online.] http://www.mikro.biologie.tu-muenchen.de/pub/ARB/documentation/arb.ps. [1 March 1999, last date accessed.]
- 39.Tanner, M. A. 1999. A supplement to “Prevalence of Corynebacterial 16S rRNA Sequences in Patients with Bacterial and ‘Nonbacterial’ Prostatitis.” [Online.] http://pacelab.berkeley.edu/182.htm. [1 April 1999, last date accessed.] [DOI] [PMC free article] [PubMed]
- 40.Tanner M A, Goebel B M, Dojka M A, Pace N R. Specific ribosomal DNA sequences from diverse environmental settings correlate with experimental contaminants. Appl Environ Microbiol. 1998;64:3110–3113. doi: 10.1128/aem.64.8.3110-3113.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vetrosky D, White G L. Prostatitis. Lippincott’s Primary Care Pract. 1997;1:437–441. [PubMed] [Google Scholar]
- 42.Weyant, R. S. Personal communication.
- 43.Wintzingerode F, Göbel U B, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 44.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zimmermann O, Spröer C, Kroppenstedt R M, Fuchs E, Köchel H G, Funke G. Corynebacterium thomssenii sp. nov., a Corynebacterium with N-acetyl-β-glucosaminidase activity from human clinical specimens. Int J Syst Bacteriol. 1998;48:489–494. doi: 10.1099/00207713-48-2-489. [DOI] [PubMed] [Google Scholar]