Regulation of a Senescence Checkpoint Response by the E2F1 Transcription Factor and p14ARF Tumor Suppressor (original) (raw)
Abstract
Normal cells do not divide indefinitely due to a process known as replicative senescence. Human cells arrest growth with a senescent phenotype when they acquire one or more critically short telomeres as a consequence of cell division. Recent evidence suggests that certain types of DNA damage, chromatin remodeling, and oncogenic forms of Ras or Raf can also elicit a senescence response. We show here that E2F1, a multifunctional transcription factor that binds the retinoblastoma (pRb) tumor suppressor and that can either promote or suppress tumorigenesis, induces a senescent phenotype when overexpressed in normal human fibroblasts. Normal human cells stably arrested proliferation and expressed several markers of replicative senescence in response to E2F1. This activity of E2F1 was independent of its pRb binding activity but dependent on its ability to stimulate gene expression. The E2F1 target gene critical for the senescence response appeared to be the p14ARF tumor suppressor. Replicatively senescent human fibroblasts overexpressed p14ARF, and ectopic expression of p14ARF in presenescent cells induced a phenotype similar to that induced by E2F1. Consistent with a critical role for p14ARF, cells with compromised p53 function were immune to senescence induction by E2F1, as were cells deficient in p14ARF. Our findings support the idea that the senescence response is a critical tumor-suppressive mechanism, provide an explanation for the apparently paradoxical roles of E2F1 in oncogenesis, and identify p14ARF as a potentially important mediator of the senescent phenotype.
Normal somatic cells undergo replicative senescence (26; reviewed in reference 9), whereby they irreversibly lose the ability to proliferate after completing a finite number of divisions. Replicative senescence does not result in cell death. Rather, senescent cells survive for long periods of time and are more resistant to apoptotic death than presenescent cells. Senescent cells also express an altered spectrum of differentiated functions. Thus, replicative senescence induces at least three phenotypic changes (growth arrest, apoptosis resistance, and altered differentiation), which we refer to as the senescent phenotype (reviewed in references 7–9). Several lines of evidence suggest that the senescence response curtails tumorigenesis and may also contribute to certain age-related pathologies (7–9, 56, 65).
Replicative senescence is particularly stringent in human cells. Cells from rodents and several other species spontaneously escape senescence with low but measurable frequencies, whereas human cells rarely acquire an indefinite or immortal replicative life span (9, 56, 60, 65, 71). It is now clear that human cells undergo replicative senescence because they acquire one or more critically short telomeres, the repetitive DNA sequences that cap the ends of linear eukaryotic chromosomes. Telomeres shorten with each cell cycle because DNA polymerases are unidirectional, whereas DNA replication is bidirectional and initiated from labile primers, and most somatic cells do not express telomerase, the enzyme that can replenish telomeric DNA de novo (25, 60). Ectopic expression of telomerase prevents telomere shortening, extends replicative life span, and immortalizes at least some normal human cells, including fibroblasts (6).
Recent findings suggest that telomere shortening is not the only inducer of the senescent phenotype. Normal human cells respond to certain types of DNA damage (10, 13, 55), histone deacetylase inhibitors (which remodel chromatin) (45), and oncogenic forms of Ras or Raf (which transduce mitogenic signals) (61, 76) by adopting a phenotype that closely resembles replicative senescence. Immortal cells, by contrast, tend to respond to DNA damage or oncogenes by undergoing apoptosis or neoplastic transformation. Thus, normal human cells differ markedly from immortal cells in their response to at least some potentially oncogenic stimuli (repeated cell division, DNA damage, and inappropriate mitogenic signals). Because it entails an essentially irreversible growth arrest, the senescence response may be a fail-safe program for suppressing tumorigenesis (61). In vivo, cells that express a marker of the senescent phenotype accumulate with age (16, 40, 49). It is not known whether such cells accumulate primarily due to replicative senescence or the senescence response elicited by DNA damage or inappropriate mitogenic signals.
The senescence-associated growth arrest is almost certainly due to the downregulation of selected positive-acting cell cycle regulatory genes. In fibroblasts and other cell types, these include the c-fos proto-oncogene, genes for Cdc2 and cyclin A and E components of cyclin-dependent protein kinases (Cdks), genes for Id1 and Id2 inhibitors of basic helix-loop-helix transcription factors, and the multifunctional transcription factor E2F1 (reviewed in references 9 and 14). In addition, senescent cells express high levels of selected growth inhibitors, most notably the Cdk inhibitors p16INK4a and p21 (2, 24, 44, 66). Senescent cells also over- or underexpress genes that have no obvious direct role in cell proliferation (7–9), but rather are associated with differentiated functions. For example, senescent dermal fibroblasts overexpress extracellular matrix-remodeling genes such as the interstitial collagenase (MMP-1) and stromelysin genes (39, 70). The senescence-associated growth arrest is undoubtedly critical for suppressing tumorigenesis; the functional changes, on the other hand, may contribute to aging (7–9).
Viral oncoproteins such as the simian virus 40 T antigen and human papillomavirus E6 and E7 proteins enable cells to bypass the checkpoint engaged by telomere shortening and thus extend the replicative life span of human cells (9, 14, 60, 71). With the exception of E6 in certain human epithelial cells (33), most viral oncoproteins do not induce telomerase. Consequently, viral oncogene-expressing cells continue to divide, despite telomere lengths shorter than those found in senescent cells. As cell division proceeds, telomere erosion continues and, ultimately, the cells enter an unstable state termed crisis from which rare immortal cells may emerge. Viral oncogenes act primarily by inactivating the cellular tumor suppressor proteins pRb and p53. Thus, pRb and p53 are critical mediators of the senescence response to telomere shortening. Depending on the cell type and stimulus, they can also mediate the senescence response to DNA damage and oncogene overexpression (10, 13, 55, 61, 76). Consistent with critical roles for pRb and p53, overexpression of p16INK4a, which acts upstream of pRb, and p21, which acts downstream of p53, induces a premature senescence-like phenotype in normal human fibroblasts (35). Little is known about other cellular proteins that control or mediate the senescence response.
E2F1 is among the growth-regulatory genes that are repressed in senescent human cells (15, 58). E2F1 belongs to a family of heterodimeric transcription factors that regulate cell cycle progression, primarily by activating the transcription of several genes needed for DNA synthesis (reviewed in references 19, 27, and 43). Growth factors induce E2F1 expression a few hours before S phase in quiescent, but not senescent, human cells (15). In addition to binding DNA and transactivating target genes, E2F1 binds several growth-regulatory proteins, including the inhibitory (unphosphorylated) form of pRb, as well as cyclin A and Mdm2 (11, 12, 19, 27, 28, 43). Ectopic expression of E2F1 induces DNA synthesis in quiescent immortal rodent cells (31, 52, 62, 63), confers neoplastic properties to immortal rodent cells (12, 19, 27), and increases tumorigenesis in transgenic mice that lack p53 function (50). These findings suggest that E2F1 is an oncogene. On the other hand, loss of E2F1 by germ line inactivation increases tumorigenesis in mice, suggesting that E2F1 is a tumor suppressor (21, 74).
How might these contradictory activities of E2F1 be explained? One possibility stems from the finding that under some circumstances E2F1 induces apoptosis in rodent cells in culture and in vivo (12, 21, 52, 62, 63, 72–74). Thus, E2F1 may promote cell cycle progression and thus neoplasia in some cell contexts, whereas in other contexts it may promote apoptosis, which suppresses tumorigenesis. Here, we provide evidence for an additional mechanism by which E2F1 may suppress tumorigenesis. We show that E2F1 induces a senescence-like phenotype when overexpressed in normal human fibroblasts. E2F1 transactivation activity and the presence of wild-type p53 were essential for this senescence response. Moreover, E2F1 was recently shown to induce p19ARF (the human homologue of p14ARF) (4, 12, 77), a tumor suppressor gene that derives from an alternate reading frame in the p16INK4a locus (32, 64, 67). We show that p14ARF is very likely a critical mediator of the E2F1-induced senescence response.
MATERIALS AND METHODS
Vectors, viruses, and cell culture.
pLXSN (38) and cells producing LXSN-E6 (23) were from D. Miller (Fred Hutchinson Cancer Center, Seattle, Wash.) and V. Band (Tufts-New England Medical Center, Boston, Mass.; originally from D. Galloway). pBabe-puro (41) (B0) was from A. Gualberto (Case Western Reserve University, Cleveland, Ohio; originally from H. Land). Babe-E2F1 vectors were constructed by inserting E2F1 cDNAs into the _Eco_RI site. Wild-type E2F1 cDNA was from W.-H. Lee (University of Texas, San Antonio) (63). CterSt and D423G mutants were generated by site-directed mutagenesis with the pAlter kit from Promega (Madison, Wis.), and CterD1 was generated by _Xho_I deletion of the E2F1 cDNA 5′ terminus. The E132 mutant was from K. Vousden (National Cancer Institute, Frederick, Md.; originally from K. Helin). mRNA from senescent WI-38 cells was reverse transcribed, and the p14ARF coding region was amplified by PCR and cloned into pBabe-puro. The mdm2 cDNA, obtained from B. Vogelstein (Johns Hopkins University, Baltimore, Md.), was subcloned into the _Bam_HI site in pLXSN.
Infectious virus was produced by transfecting retroviral vectors and the pIK packaging vector into tsa54 cells, both from CellGenesys (48). Virus-containing medium was collected 24 h later, frozen, thawed, and assayed for reverse transcriptase (RT). Viral titers were expressed as units of RT per milliliter. Proliferating cells (30 to 50% confluent) were infected with 2 to 4 U of RT twice for 6 h each with a 16-h interval and medium change between infections. Two days later, infected cells were given puromycin (Babe viruses; 1 μg/ml; 3 to 5 days) or G-418 (LXSN viruses; 400 μg/ml; 6 to 7 days); mock-infected cells were cultured without antibiotics. Under these selection conditions, uninfected cells did not survive. Based on the number of infected cells after selection compared to the number of mock-infected cells without selection, this protocol typically yielded 80 to 90% infected (antibiotic-resistant) cells.
WI-38 human fibroblasts were obtained and cultured as described previously (15–18). When 70 to 80% confluent, cells were subcultured at 104 to 2 × 104/cm2 and plated at 3 × 103 to 5 × 103/cm2 to determine the percentage that incorporated [3H]thymidine over a 72-h interval (percent labeled nuclei) and/or stained positive for senescence-associated beta-galactosidase (SA-β-Gal), as described previously (16). U2OS, A375, and HT1080 cells from the American Type Culture Collection and NIH 3T3 cells, originally from A. Pardee (Dana-Farber Cancer Institute, Boston, Mass.), were cultured as described for WI-38 cells.
Western analyses and immunoprecipitation.
Denatured protein lysates (30 μg) in 2× Laemmli lysis buffer were analyzed by gradient (4 to 15%) sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting (18). Western blots were probed with E2F1 (KH95) or pRb (C-15) monoclonal antibody or QM (loading control) (18) polyclonal antibody from Santa Cruz Biotechnology Inc. (Santa Cruz, Calif.), p53 (Ab 6) or α-tubulin monoclonal antibody from Oncogene Sciences (Cambridge, Mass.), Mdm2 (Ab 1) monoclonal antibody from Calbiochem (San Diego, Calif.), or p14ARF (Ab 1) polyclonal antibody from Neo Markers (Union City, Calif.). The E2F1 antibody recognizes amino acids 342 to 386 and reacted with all the E2F1 proteins used in this study (Fig. 1B). Rb and E2F1 immunoprecipitations were performed with serum-deprived cells, in which endogenous E2F1 is barely detectable. Protein lysates in radioimmunoprecipitation assay (RIPA) buffer (500 μg) were incubated with 2 μg of E2F1 antibody for 3 h, followed by overnight incubation with protein A-Sepharose beads. The beads were washed with RIPA buffer and phosphate-buffered saline. Bound proteins were released with 2× Laemmli buffer and analyzed for pRb by Western blotting. Fifteen micrograms of RIPA lysate protein was mixed with 2× Laemmli buffer and analyzed for pRb and tubulin by Western blotting.
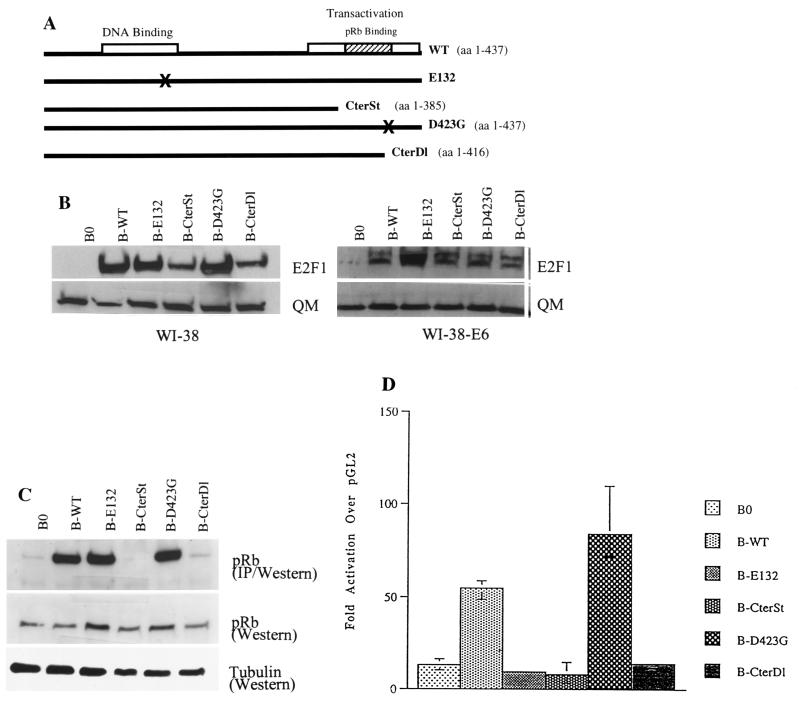
FIG. 1.
Analysis of E2F1 proteins used in this study. (A) Diagram of wild-type (WT) and mutant (E132, CterSt, D423G, CterDl) E2F1 proteins described in the text. (B) Expression levels. Normal (WI-38) or E6-expressing (WI-38-E6) human fibroblasts were infected with control (B0) or E2F1-expressing retroviruses (B-WT, B-E132, B-CterSt, B-D423G, B-CterDl). Protein lysates were prepared one passage after antibiotic selection and analyzed by Western blotting for the levels of E2F1 and QM (control) (18) proteins. The presence of two E2F1 protein species, migrating with slightly different mobilities (more prominent in E6-expressing cells), is consistent with phosphorylated and unphosphorylated forms. Under our SDS-PAGE conditions, wild-type and mutant E2F1 proteins migrated with approximately the same mobilities, despite slight differences in size. The estimated sizes of the E2F1 proteins are 52 kDa for WT, E132, and D423G; 46 kDa for CterSt; and 59 kDa for CterDl (due to the addition of 58 C-terminal amino acids from the vector). (C) pRb-binding activities. WI-38-E6 cells were infected with control or E2F1-expressing retroviruses, made quiescent by serum deprivation (to reduce endogenous E2F1), and lysed. Then, 500 μg of protein lysate was immunoprecipitated with anti-E2F1 antibody and analyzed for pRb by Western blotting (IP/Western). In parallel, 15 μg of protein was analyzed without immunoprecipitation (Western) for pRb and α-tubulin (control). (D) Transactivation activities. E6-expressing cells infected with control or E2F1-expressing viruses were cotransfected with a normalization vector and either pGL2, a promoterless luciferase vector, or E2F-luc, in which luciferase is driven by E2F binding sites (42); extracts were prepared and analyzed as described in Materials and Methods. The results shown are the amounts of normalized E2F-luc activity relative to the amount of normalized pGL2 activity.
Reporter assays.
Reporter assays were performed as described previously (17). E2F1-luc (pGL2-AN) (42) was obtained from W. Kaelin (Dana-Farber Cancer Institute) and contains the luciferase reporter gene driven by 275 bp (−211 to +64) of the E2F1 promoter, which includes four E2F binding sites. E2F-luc was cotransfected with the pCMV-β-gal normalization vector by using Lipofectamine Plus (Gibco BRL, Gaithersburg, Md.). Extracts were prepared 48 h after transfection, analyzed for luciferase activity, and normalized for bacterial β-galactosidase (pH 7.5) activity, as described previously (17).
DNA laddering and TUNEL assay.
Analysis of DNA fragmentation was done as described by Shan and Lee (63). Briefly, cells were collected and suspended in lysis buffer (10 mM Tris [pH 7.9], 5 mM EDTA, 0.5% SDS, 0.1 mg of proteinase K/ml). The lysate was incubated at 55°C for several hours and extracted with phenol-chloroform, and the DNA was precipitated, dissolved in TE (10 mM Tris [pH 8], 1 mM EDTA) containing 20 μg of RNase A/ml, separated on a 1.5% agarose gel, and stained with ethidium bromide. Terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling (TUNEL) was performed with a kit (S7160) from Intergen (Purchase, N.Y.). MG132 was purchased from Calbiochem.
Semiquantitative RT-PCR analyses.
Total RNA was isolated as described previously (15, 18), and 2 μg was reverse transcribed with Super-scriptase (Gibco BRL). Hot-start PCR was carried out for 20 (β-actin), 30 (c-myc, cyclin E, E2F1, p16INK4a, interstitial collagenase [MMP-1], plasminogen activator inhibitor-1 [PAI-1], and stromelysin-1), or 35 (p14ARF) cycles. The linear range of amplification was determined from PCRs run with serially diluted cDNA and β-actin primers. The results were verified by varying the number of PCR cycles for each cDNA and set of primers (not shown). The c-myc and β-actin primers were from Clontech; the other primers were as follows: cyclin E, 5′ CAGAGACAGCTTGGATTTGCTG and 3′ AGGCGCGCAACTGTCTTTGGTG; E2F1, 5′ GTCCCGGATGGGCAGCCTG and 3′ GTAGCCAGACCCCAGAGCTAG (endogenous) or TAACTGACACACATTCCACAGG (virally expressed); p16, 5′ CAGCATGGAGCCTTCGGCTGAC and 3′ CAGCCGCGCGCAGGTACCGTGCGA; p14, 5′ GAAGATGGTGCGCAGGTTCT and 3′ CCTCAGCCAGGTCCACGGG; PAI-1, 5′ GTCATAGTCTCAGCCCGCATG and 3′ TTTCCTTCAGAAAGAGTCATAAC; MMP-1, 5′ CCAGTGACTGCACATGAGGTTC and 3′ CCTCTAGAGTCACTGATACACA; stromelysin: 5′ GACACACACTTTGAAGAGTAACAG and 3′ GTCTGTTGCACACGAGTGCTTCC. PCR products were separated on agarose gels, visualized by ethidium bromide staining, analyzed with an Alpha Innotech imager, and quantified with ImageQuant software.
RESULTS
Analysis of wild-type and mutant E2F1 proteins.
The importance of E2F1 in G1 progression and its repression in senescent cells suggested that constitutive E2F1 expression might extend the replicative life span of normal cells. To test this idea, Babe retroviruses (41) were used to constitutively overexpress wild-type or mutant E2F1 proteins in human cells. Initially, E2F1 proteins were expressed in WI-38 normal human fibroblasts and WI-38 cells having an extended life span owing to inactivation of p53 by E6 (59) (WI-38-E6 cells). Four E2F1 protein mutants were studied (Fig. 1A): E132, a mutant with a two-amino-acid substitution that disrupts DNA binding (28); CterSt, a mutant in which a stop codon after the codon for amino acid 385 deletes the Mdm2-binding, pRb-binding, and transactivation domains; D423G, a mutant with Gly-for-Asp substitution that is reported to disrupt pRb binding but retain transactivation activity (63); and CterDl, which lacks 21 C-terminal amino acids and thus pRb-binding and transactivation activity but which retains Mdm2 binding.
Western analyses of infected cell lysates showed that all the retroviruses, except the B0 control, produced a 5- to 10-fold overexpression of E2F1 proteins (Fig. 1B). This was true in both normal WI-38 cells, which, as described below, arrested growth in response to E2F1, and WI-38-E6 cells, in which E2F1 did not arrest growth. To avoid potentially confounding effects of growth arrest on E2F activity, we characterized the retrovirally expressed proteins in WI-38-E6 cells. Coimmunoprecipitation confirmed that CterDl and CterSt proteins were essentially devoid of pRb binding, whereas wild-type and E132 proteins were fully capable of pRb binding (Fig. 1C). Although yeast two-hybrid analysis confirmed that D423G does not bind pRb (not shown) as reported previously (63), coimmunoprecipitation showed that D423G retained substantial pRb binding (Fig. 1C). Transient transfection experiments confirmed that only wild-type and D423G proteins transactivated an E2F-driven reporter gene (four- and sixfold, respectively, over the endogenous E2F activity seen in B0-infected cells) (Fig. 1D). D423G transactivation activity was somewhat greater than wild-type activity, consistent with published data (63). Thus, all the retrovirally produced E2F1 proteins were stably expressed, and, with the exception of the pRb-binding activity of D423G, all showed the expected deficits in activity. The E132, CterSt, and CterDl mutants did not appear to be strong dominant inhibitors of endogenous E2F activity (i.e., CterDl had no effect on endogenous E2F activity, whereas E132 and CterSt reduced it by <30%), although the transient transfection experiments do not rigorously exclude this possibility.
E2F1 arrests the proliferation of normal human fibroblasts.
Wild-type E2F1, when constitutively overexpressed, unexpectedly arrested cell proliferation within two population doublings (PD) after selection (Fig. 2A). Rapid growth arrest was also seen in response to D423G. In both cases, cell number showed little or no increase up to 10 days after selection (Fig. 2A and F), and the fraction of cells that synthesized DNA over a 3-day interval declined from >80 to <25% (Fig. 2B). By contrast, cells expressing the other E2F1 mutants (E132, CterD1, or CterSt) proliferated in a manner that was indistinguishable from that of control cells (Fig. 2A, B, and F). Thus, constitutive overexpression of E2F1 proteins lacking transactivation activity, but retaining DNA-, DP-, cyclin A-, Mdm2-, and/or pRb-binding activity, had no effect on cell proliferation. However, E2F1 proteins with both DNA-binding and transactivation activity (wild type and D423G) strongly inhibited the proliferation of normal human fibroblasts.
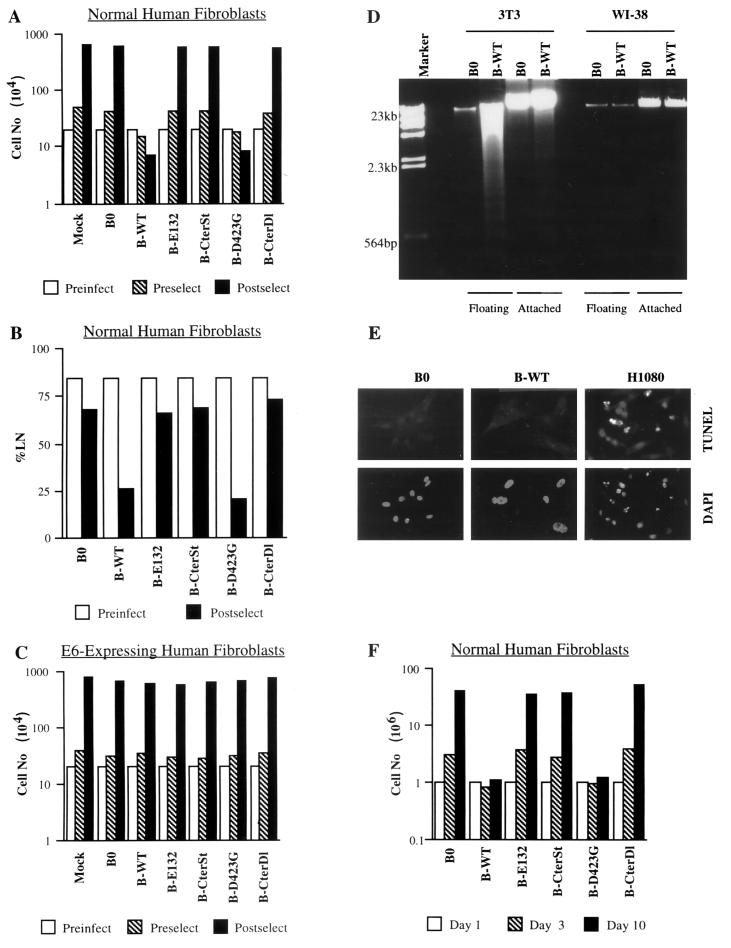
FIG. 2.
Effects of E2F1 proteins on cell proliferation, DNA synthesis, and apoptosis. (A) Proliferation of normal human cells. WI-38 normal human fibroblasts (Preinfect; 2 × 105 cells) were mock infected (Mock) or infected with control (B0) or E2F1-expressing (wild-type [WT], E132, CterSt, D423G, CterDl) retroviruses. Three days after infection, cell number was determined (Preselect). Infected cells were then selected with puromycin for 5 days and cultured in medium without antibiotic for an additional 3 days; mock-infected cells were cultured without antibiotic for 8 days. The cell number was then again determined (Postselect). Similar results were obtained in at least three independent experiments using different batches of virus. (B) DNA synthesis in normal human cells. WI-38 cells (Preinfect) were infected and selected as described for panel A. After selection, [3H]thymidine was added in antibiotic-free medium and the cells were processed for autoradiography 3 days later (Postselect). A minimum of 200 cells were counted to determine the percentages of radiolabeled nuclei (%LN). (C) Proliferation of E6-expressing human cells. WI-38 cells were infected with an LXSN retrovirus carrying E6 and selected in G-418, as described in Materials and Methods (WI-38-E6 cells). The resulting WI-38-E6 cultures were then infected with E2F1-expressing retroviruses, selected in puromycin, and counted as described for normal WI-38 cells (A). (D) Apoptosis in normal human and immortal mouse cells. WI-38 and mouse NIH 3T3 fibroblasts were infected with control (B0) or E2F1-expressing (B-WT) retroviruses, as described in Materials and Methods. Seventy-two hours later, detached (floating) cells were collected by gentle centrifugation. DNA was isolated from the floating cells and the remaining attached cells and analyzed as described in Materials and Methods. WI-38 cells also showed no DNA fragmentation if they were selected in puromycin prior to DNA isolation (not shown). Left lane, DNA size markers. (E) Apoptosis in normal human cells. WI-38 cells were infected with control (B0) or E2F1-expressing (B-WT) retroviruses, selected in puromycin for 3 days, and plated on chambered glass slides; 24 h later, they were assayed in situ for DNA fragmentation by TUNEL, and then stained with DAPI (4′,6′-diamidino-α-phenylindole) as described in Materials and Methods. HT1080 cells treated for 16 h with 10 μM MG132, a proteasome inhibitor that induces apoptosis in human tumor cells (37), served as a positive control for the reaction. No TUNEL-positive cells were observed among >1,000 B0- or B-WT-infected WI-38 cells (<0.1% apoptosis), whereas 52 of 611 TUNEL-positive cells were observed among MG132-treated HT1080 cells (8.5% apoptosis). (F) Stability of E2F1 growth arrest. WI-38 cells were infected and selected as described for panel A. After selection 106 cells were plated in antibiotic-free medium (day 1). The numbers of cells were determined 2 (day 3) and 9 days (day 10) later.
Three lines of evidence argue that the growth arrest induced by E2F1 was not due to nonspecific toxicity. First, all the E2F1 proteins were expressed at similar levels (Fig. 1) yet only two, the wild type and D423G, inhibited cell proliferation. Second, none of the E2F1 proteins arrested the proliferation of isogenic E6-expressing cells (Fig. 2C). Third, E2F1 overexpression did not induce apoptosis, as judged by the absence of detached cells, DNA laddering (Fig. 2D), TUNEL staining, and apoptotic nuclear morphology (Fig. 2E). By contrast, the same E2F1 retrovirus readily induced apoptosis in immortal mouse (3T3) fibroblasts, as judged by abundant cell detachment (not shown) and DNA laddering (Fig. 2D). The slight decline in the number of E2F1-expressing human cells seen after infection and selection was due to the slightly reduced replating efficiency of E2F1-arrested cells. Moreover, the E2F1-arrested cells remained viable for at least 10 days in culture (Fig. 2F), with no discernible loss in cell number. After 10 days or so, a few colonies of proliferating cells were often apparent in E2F1-arrested cultures. Upon expansion and assay for E2F1, these E2F1-resistant cells were found to express only very low levels of the retrovirally introduced E2F1 gene.
E2F1 induces a senescence-like phenotype in normal human cells.
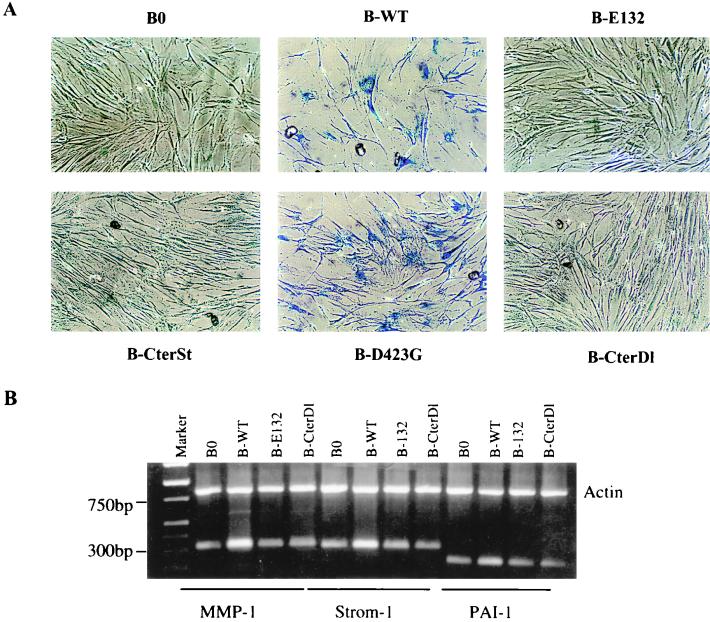
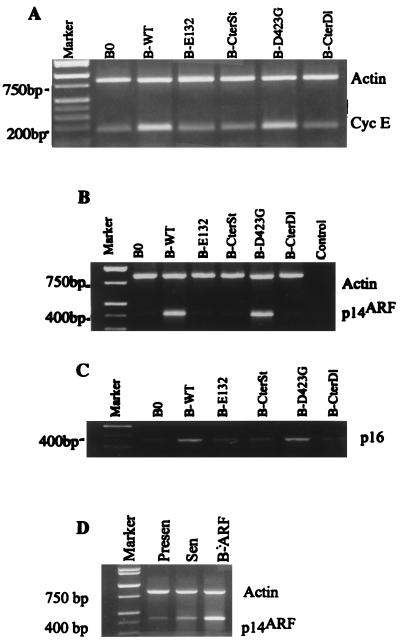
In addition to arresting cell proliferation, wild-type and D423G proteins, but not the other E2F1 proteins, also induced >50% of the cells to assume a flat morphology, typical of senescent cells (Fig. 3A). We therefore stained the cells for the pH 6.0 SA-β-Gal, a histochemically detectable marker that correlates well with the senescent phenotype (16). Approximately 80% of cells expressing wild-type (Fig. 3A; Table 1) or D423G (Fig. 3A) E2F1 proteins also expressed SA-β-Gal. Control cells (Table 1) and cells expressing the other E2F1 mutants were essentially devoid of SA-β-Gal staining (Fig. 3A). These findings suggest that E2F1 induces a senescence-like phenotype in normal human cells. Consistent with this idea, semiquantitative PCR showed that wild-type E2F1 induced the expression of three extracellular matrix-remodeling genes that are overexpressed by senescent human fibroblasts (reviewed in reference 9; 39, 70): genes for interstitial collagenase (MMP-1), stromelysin-1, and PAI-1 (Fig. 3B). The E2F1 protein and the D423G mutant (not shown) induced all three genes to levels that were similar to those expressed by replicatively senescent cells (Fig. 5D; Table 2) (39, 70). By contrast, the E132 and CterDl E2F1 mutants (and CterSt [not shown]), which did not arrest growth or induce SA-β-Gal, did not induce these senescence-associated genes (Fig. 3B).
FIG. 3.
E2F1 proteins induce a senescence-like phenotype. (A) Morphology and SA-β-Gal expression. WI-38 cells were infected with the indicated E2F1-expressing retroviruses, selected, and, 2 to 3 days thereafter, stained for SA-β-Gal, as described in Materials and Methods. The cells were photographed under phase-contrast optics. WT, wild type. (B) Expression of matrix-remodeling genes. RNA, isolated from WI-38 cells infected as described for panel A, was analyzed for interstitial collagenase (MMP-1), stromelysin-1 (Strom-1), PAI-1, and β-actin (control) mRNA by RT-PCR, as described in Materials and Methods. Signals were normalized to β-actin, and the MMP-1, stromelysin-1, and PAI-1 mRNA levels in control cells (B0) were each set at 1. Relative to that by B0, levels of induction by B-WT were 2.7, 2.2, and 1.8, respectively; levels of induction by B-E132 were 0.9, 1.0, and 1.1, respectively; and levels of induction by B-CterDl were 0.9, 1.2, and 1.0, respectively.
TABLE 1.
E6 and Mdm2 abrogate the growth arrest and SA-β-Gal expression induced by p14ARF and E2F1a
| Retroviruses | % SA-β-Gal positive | % Labeled nuclei |
|---|---|---|
| L0 + B0 | 8 | 65 |
| L0 + B-ARF | 85 | 15 |
| L0 + B-WT (E2F1) | 78 | 18 |
| L-E6 + B0 | 1 | 88 |
| L-E6 + B-ARF | 3 | 85 |
| L-E6 + B-WT (E2F1) | 1 | 89 |
| L-Mdm2 + B0 | 5 | 70 |
| L-Mdm2 + B-ARF | 19 | 62 |
| L-Mdm2 + B-WT (E2F1) | 14 | 68 |
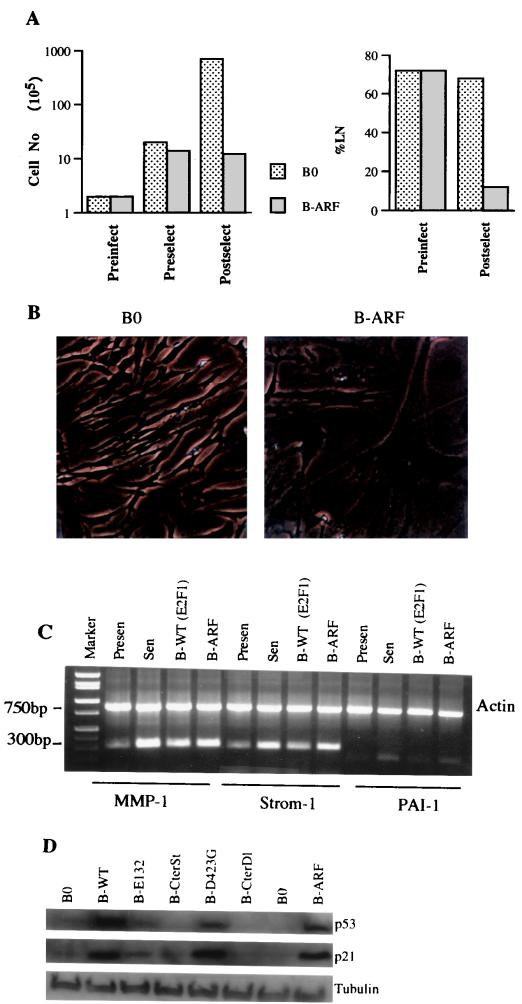
FIG. 5.
Senescent phenotype induced by p14ARF. (A) Growth inhibition. Cell number was determined prior to infection by control (B0) or p14ARF-expressing (B-ARF) retroviruses (Preinfect), prior to selection (Preselect), and after selection (Postselect), as described in the legend to Fig. 2A. Postselected cells were also given [3H]thymidine for 72 h to determine the fraction of cells that were capable of synthesizing DNA (%LN). (B) Morphology and SA-β-Gal expression. Postselected cells were stained for SA-β-Gal and photographed under phase-contrast optics. (C) Expression of matrix-remodeling genes. RNA was isolated from postselected WI-38 cells, as well as presenescent and senescent WI-38 cells and analyzed by semiquantitative RT-PCR for actin (control) mRNA and interstitial collagenase (MMP-1), stromelysin-1 (Strom-1), and PAI-1 mRNA. See Table 2 for quantitation. (D) Induction of p53 and p21 by 14ARF and E2F1. Protein extracts were prepared from postselected cells and analyzed by Western blotting for p53, p21, and tubulin (control).
TABLE 2.
Induction of MMP-1, stromelysin-1, and PAI-1 mRNA by replicative senescence, E2F1, and p14ARFa
| Protein | Cell or virus type | mRNA induction (fold) |
|---|---|---|
| MMP-1 | Presenescent | 1.0 |
| Senescent | 5.6 | |
| B-WT (E2F1) | 3.5 | |
| B-ARF | 3.7 | |
| Strom-1 | Presenescent | 1.0 |
| Senescent | 5.1 | |
| B-WT (E2F1) | 2.6 | |
| B-ARF | 3.6 | |
| PAI-1 | Presenescent | 1.0 |
| Senescent | 5.6 | |
| B-WT (E2F1) | 2.6 | |
| B-ARF | 3.4 |
Thus, in normal human cells, constitutive E2F1 overexpression rapidly induced a senescence-like phenotype, as judged by a stable growth arrest, morphology, SA-β-Gal expression, and elevated expression of MMP-1, stromelysin, and PAI-1. Because only wild-type and D423G proteins are capable of transactivation, this phenotype may be controlled by one or more E2F-inducible genes.
p14ARF, a potential senescence-inducing E2F1 target gene.
E2F1 has been shown to induce several genes needed for DNA metabolism, as well as positive-acting growth-regulatory genes such as the cyclin E and c-myc genes (12, 27, 36, 46). Although E2F1 induced the cyclin E (Fig. 4A) and c-myc (not shown) genes when overexpressed in normal human cells, growth stimulatory genes per se are unlikely to mediate the E2F1-induced senescence response. E2F1 was recently shown to induce expression of ARF (p19ARF in mice; p14ARF in humans) (4, 12, 77), a tumor suppressor gene that derives from an alternate reading frame in the 16INK4a locus (32, 67), and the human ARF promoter was shown to contain E2F binding sites (54). ARF inhibits cell proliferation and tumorigenesis (reviewed in reference 64) and thus is a good candidate for mediating the senescence-inducing activity of E2F1.
FIG. 4.
Induction of cyclin E, p14ARF, and p16INK4a by E2F1 proteins. (A) Cyclin E. RNA was isolated from postselected WI-38 cells that had been infected with the indicated E2F1-expressing retroviruses (WT, wild type) and analyzed by semiquantitative RT-PCR for β-actin (control) and cyclin E mRNA. The cyclin E signal was normalized to β-actin, and the expression level in control infected cells (B0) was set at 1. Relative to that for B0, levels of induction of cyclin E were as follows: B-WT, 3.2; B-E132, 1.2; B-CterSt, 1.3; B-D423G, 3.1; B-CterSt, 1.2. (B) p14ARF. RNA was isolated, analyzed for p14ARF and β-actin, and normalized as described for panel A. Relative to that for B0, the levels of induction of p14ARF were as follows: B-WT, 5.7; B-E132, 1.6; B-CterSt, 0.9; B-D423G, 5.5; B-CterSt, 1.0. (C) p16INK4a. RNA was isolated, analyzed for p16INK4a and β-actin, and normalized as described for panel A. p16INK4a was analyzed in the same experiment as p14ARF and has the same β-actin (B) control. (D) Expression of p14ARF in presenescent, senescent, and retrovirally infected cells. RNA was isolated from presenescent (Presen) and senescent (Sen) WI-38 cells, as well as WI-38 cells infected with the p14ARF retrovirus, and cells were cultured in serum-containing medium. RNA was analyzed and normalized as described for panel A, and the p14ARF expression in senescent cells was set at 1. Relative to that for senescent cells, p14ARF expression levels were 0.15 in presenescent cells and 3 in retrovirally infected cells.
To test this idea, normal cells expressing wild-type or mutant E2F1 proteins were assayed for p14ARF expression. E2F1 proteins that had transactivation activity (wild type and D423G), but not the other E2F1 proteins, strongly induced p14ARF mRNA (Fig. 4B). Transactivation-competent E2F1 proteins also induced p16INK4a expression (Fig. 4C), but this induction was substantially less vigorous than the induction of p14ARF (Fig. 4B). p14ARF was also highly expressed by replicatively senescent cells (six- to sevenfold over the level expressed by presenescent cells; Fig. 4D). This result suggests that p14ARF, like p21 and p16, may participate in establishing and/or maintaining the senescent phenotype. Together, these findings suggest that the E2F1 protein may induce the senescence-like phenotype by virtue of its ability to induce p14ARF and possibly p16INK4a.
Senescent phenotype induced by p14ARF.
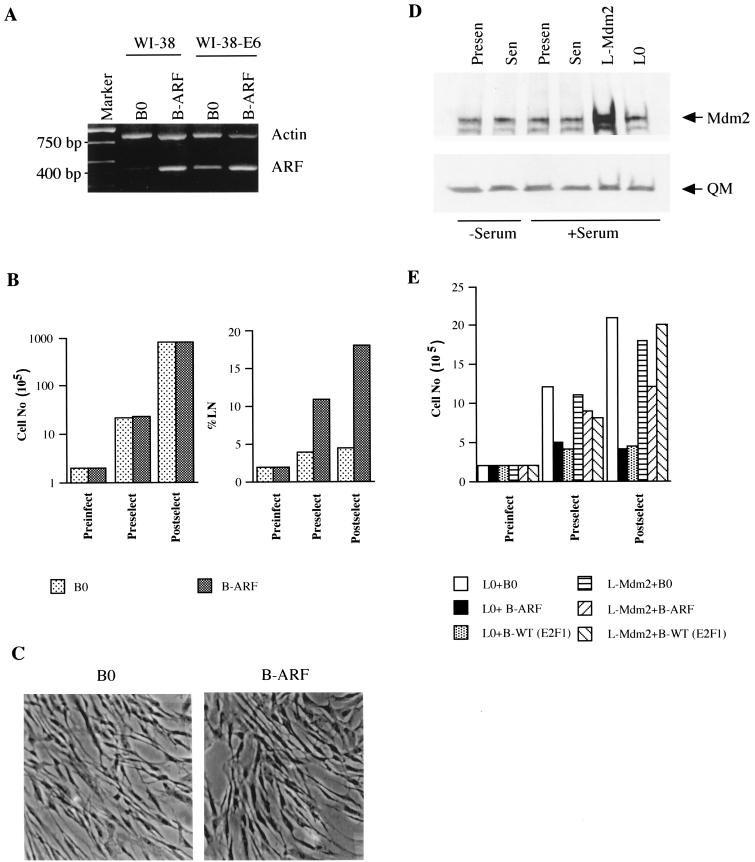
To test the idea that p14ARF mediates the senescent phenotype induced by E2F1, p14ARF was introduced by retroviral transfer into normal human fibroblasts. Retroviral transfer (B-ARF) increased p14ARF to a level that was threefold greater than that expressed by senescent cells (Fig. 4D). Retrovirally expressed p14ARF strongly inhibited cell proliferation (Fig. 5A). Cell division was suppressed within two PD after selection, and the fraction of cells that synthesized DNA over a 3-day interval declined from >70 to <10% (Fig. 5A). In addition, the majority (>80%) of p14ARF-expressing normal cells had an enlarged and flat morphology (Fig. 5B) and stained positive for SA-β-Gal (Table 1). p14ARF also induced expression of the matrix-remodeling genes for MMP-1, stromelysin-1, and PAI-1 to levels that were similar to those induced by E2F1 and replicative senescence (Fig. 5C; Table 2).
p14ARF attenuates Mdm2-mediated p53 degradation (64, 67, 68, 75), thereby elevating p53 protein levels, which can transcriptionally induce p21 (20). If the senescent phenotype induced by E2F is due to its ability to induce p14ARF, both p14ARF and transactivation-competent E2F1 proteins should induce p53 and p21. Indeed, wild-type E2F1 and D423G proteins, as well as the p14ARF protein, strongly induced p53 (Fig. 5D). p21 was also induced by these proteins (Fig. 5D), presumably as a consequence of the induction of p53. By contrast, E2F1 proteins that were incapable of transactivation (E132, CterSt, and CterDl) failed to induce either p53 or p21 (Fig. 5D).
These data support the idea that the senescence response to constitutive E2F1 overexpression can be accounted for by the induction of p14ARF by E2F1.
The E2F1- and p14ARF-induced phenotypes are p53 dependent.
p14ARF requires p53 for its growth-inhibitory and tumor-suppressive activities (51, 64, 67, 75). If p14ARF is a critical mediator of E2F1-induced senescence, p53 should be essential for the senescent phenotypes induced by both p14ARF and E2F1.
To test this prediction, we introduced E6, which accelerates p53 degradation (59), into normal WI-38 cells with an LXSN retrovirus (WI-38-E6 cells). WI-38-E6 cells have an extended replicative life span (five to eight PD longer than normal), but eventually senesce (66; our unpublished data). Western analysis confirmed that WI-38-E6 cells had substantially less p53 protein than normal cells (not shown) and that the levels of retrovirally expressed E2F1 and p14ARF in normal and E6-expressing cells were similar (Fig. 1B and 6A). E6 expression completely prevented the growth arrest induced by both E2F1 (Fig. 2B) and p14ARF (Fig. 6B). E6 also prevented the senescence-like morphology (Fig. 6C) and SA-β-Gal expression (Table 1) induced by E2F1 and p14ARF. Because E6 has multiple functions in addition to facilitating p53 degradation (53), we asked whether overexpression of Mdm2 also prevented E2F1- or p14ARF-induced senescence. Mdm2 functionally inactivates p53 (22), and p14ARF increases p53 levels by interfering with Mdm2 function (51, 64, 67, 68, 75).
FIG. 6.
Senescent phenotype induced by E2F1 and p14ARF is p53 dependent. (A) p14ARF expression in normal and E6-expressing cells. WI-38-E6 cells were infected with control (B0) or p14ARF-expressing (B-ARF) retroviruses and selected. RNA was isolated from postselected cells and analyzed by RT-PCR for actin (control) and p14ARF mRNA. The same analysis performed on normal WI-38 cells is shown for comparison. (B) Growth inhibition by p14ARF in E6-expressing cells. WI-38-E6 cells were infected and selected as described for panel A. Cell numbers were determined prior to infection (Preinfect) and selection (Preselect) and after selection (Postselect). Postselected cells were labeled with [3H]thymidine for 72 h. A minimum of 200 cells were counted to determine the percentage of radiolabeled nuclei (%LN). (C) Morphology and SA-β-Gal expression. Postselected WI-38-E6 cells were stained for SA-β-Gal and photographed under phase-contrast optics. (D) Mdm2 expression. Protein extracts were prepared from presenescent and senescent WI-38 cells maintained for 3 days in 0.2 (−Serum) or 10% (+Serum) serum, and proliferating cells were infected with control (B0) or Mdm2 (B-Mdm2)-expressing retroviruses. Extracts were analyzed by Western blotting for Mdm2 and QM (control) proteins. (E) Growth inhibition in Mdm2-expressing cells. WI-38 cells were infected with control (L0) or Mdm2-expressing (L-Mdm2) viruses, selected in G-418, and superinfected with control (B0), p14ARF (B-ARF), or wild-type E2F1 [WT (E2F1)] viruses, and selected in puromycin. Cells were counted prior to infection (Preinfect) and selection (Preselect) and 3 days after selection (Postselect).
Mdm2 was expressed at similar levels in presenescent and senescent cells (Fig. 6D). Presenescent cells that overexpress Mdm2 were generated with an LXSN retrovirus. Western analysis showed that the retrovirus produced a three- to fivefold elevation of Mdm2 protein relative to control (L0)-infected cells (Fig. 6D). After infection and selection, control and Mdm2-expressing cells were infected with Babe retroviruses carrying either no insert (control, B0), p14ARF, or wild-type E2F1. The doubly infected cells were then monitored for growth and SA-β-Gal expression. p14ARF and E2F1 did not arrest growth (Fig. 6E) or induce SA-β-Gal staining (Table 1) in cells that overexpress Mdm2.
Together, these results support the prediction that p53 is required for the senescence response induced by either E2F1 or p14ARF.
E2F1, but not p14ARF, fails to induce a senescence response in p14ARF-deficient cells.
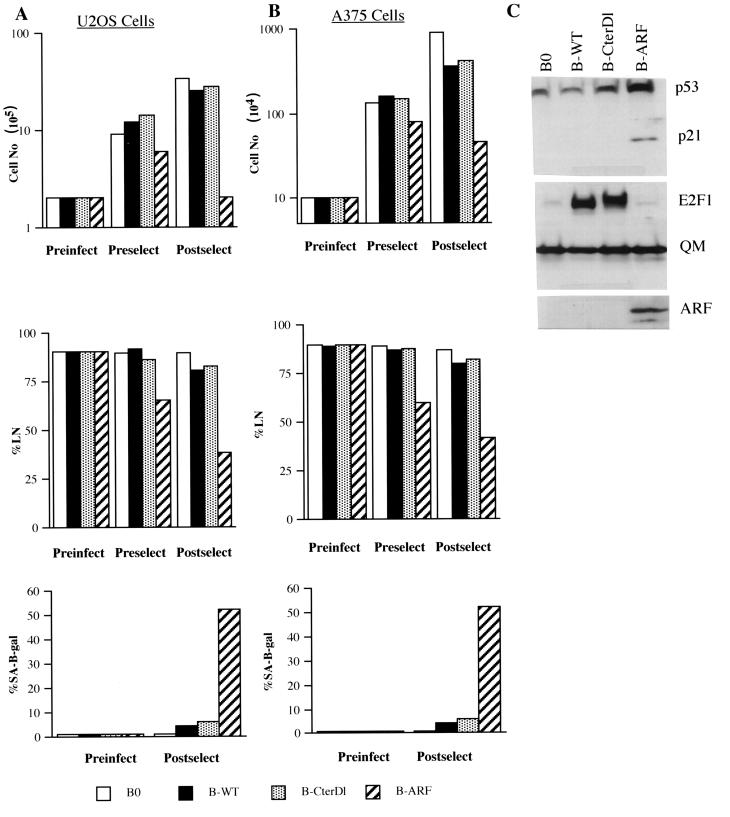
Additional support for a critical role for p14ARF in mediating the senescence response to E2F1 was obtained with human tumor cells deficient in p14ARF. U2OS and A375 are human tumor cell lines that have retained wild-type p53 activity but that have lost p14ARF expression owing to methylation of the INK4a locus (U2OS) or deletion of INK4a exon 1β (A375) (67). Retroviral transfer was used to express p14ARF protein in these cells (shown for U2OS in Fig. 7C). In both cell lines, p14ARF caused a rapid arrest of growth as well as expression of SA-β-Gal (Fig. 7A and B). In U2OS cells, p14ARF increased p53 and p21 protein expression (Fig. 7C) but not p16 expression (not shown). Thus, U2OS and A375 cells behaved similarly to normal cells in their response to ectopic p14ARF expression, consistent with their wild-type p53 status. By contrast, U2OS and A375 cells failed to arrest proliferation or express SA-β-Gal in response to retrovirally expressed E2F1 (Fig. 7A and B). For U2OS, E2F1 also failed to induce p53 or p21 (Fig. 7C). These results strongly suggest that p14ARF is a critical mediator of the senescence response to constitutive E2F1 overexpression.
FIG. 7.
Senescent phenotype induced by E2F1 is p14ARF dependent. (A) U2OS cells were infected with control (B0)-, wild-type E2F1 (B-WT)-, transactivation defective CterD1 E2F1 (B-CterD1)-, or p14ARF (B-ARF)-expressing retroviruses and selected in puromycin. The cells were counted or labeled with [3H]thymidine for 3 days to determine the percentage that synthesized DNA (%LN) prior to infection (Preinfect), prior to selection (Preselect), or 2 to 3 days after selection (Postselect). In parallel, the cells were stained for SA-β-Gal activity prior to infection (Preinfect) and after selection (Postselect). For each determination of percent labeled nuclei and SA-β-Gal activity, at least 400 cells were counted from two independent culture dishes. (B) A375 cells were infected and analyzed for cell number, percent labeled nuclei, and SA-β-Gal activity, as described for U2OS cells (A). (C) Protein lysates were prepared from postselected U2OS cells infected with control (B0)-, wild-type E2F1 (B-WT)-, CterDl E2F1 (B-CterDl)-, or p14ARF (B-ARF)-expressing retroviruses and analyzed by Western blotting for p53, p21, E2F1, p14ARF, and QM (control) protein.
DISCUSSION
Significance of E2F1-induced senescence response.
We describe a novel activity of E2F1: the ability to induce a premature senescence-like phenotype when constitutively overexpressed in normal human cells. Our results suggest that although E2F1, like activated Ras or Raf (61, 76), is potentially oncogenic when highly expressed, normal cells respond to highly expressed E2F1 by a stable arrest of cell proliferation resembling that of replicative senescence. As such, our findings add to the mounting evidence that senescence arrest is a checkpoint response that acts to prevent tumorigenesis. They also raise the possibility that the accumulation of senescent cells in vivo (16, 40, 49) may be due not solely to replicatively senescent cells but also to damaged or oncogenically stimulated cells.
Our findings also help explain the apparently conflicting biological activities of E2F1. The senescence response elicited by E2F1 depended on the status of the cell. Cells with compromised p53 or p14ARF function failed to senesce in response to constitutive E2F1 overexpression. Moreover, our unpublished data suggest that constitutively expressed E2F1 extends the replicative life span of E6-expressing cells, causing them to enter crisis (unpublished data). This finding complements published data showing that E2F1 stimulates DNA synthesis in quiescent immortal cells (12, 27, 31, 62, 63) but not quiescent normal cells (15, 36). Thus, E2F1 may either inhibit or stimulate cell proliferation, depending on the p53, p14ARF, and immortalization status of the cell.
E2F1 did not induce apoptosis in normal human fibroblasts, although it has been shown to induce apoptosis in immortal cells (12, 52, 62, 63, 69, 72) and certain tissues. E2F1 deficiency suppressed apoptosis in selected pRb −/− tissues (69) and a murine brain tumor model in which p53-mediated apoptosis suppresses tumorigenesis (47). The increase in lymphomas in mice with germ line inactivation of E2F1 is though to result from loss of E2F1-induced apoptosis, which was evident in the thymus of E2F1 −/− animals (21, 74). Our results suggest that the ability to induce a senescence response may be an additional, equally important mechanism by which E2F1 suppresses tumorigenesis. Whether E2F1 induces apoptosis or senescence may be cell type and genotype specific. Our preliminary data suggest that E2F1 stimulates cell death, possibly by apoptosis, in human fibroblasts that lack pRb function. Thus, at least in human fibroblasts, inappropriate E2F1 expression may result in senescence (normal cells), life span extension (p53-compromised cells), or possibly apoptosis (pRb-compromised cells). In vivo, inappropriate or excess E2F1 can result from a number of mechanisms, including gene amplification (57), upstream oncogene or mitogenic activation (19, 27, 42, 43), or DNA damage (5, 29). The high levels of E2F1 that can result from any of these potentially oncogenic stimuli can elicit a senescence response. Because the senescence growth arrest is irreversible, the senescence response would suppress neoplastic transformation.
Mechanism of the E2F1-induced senescence response.
The senescence response induced by E2F1 depended on its ability to transactivate, and the E2F1 target gene that appeared to be primarily responsible was p14ARF. p14ARF was identified as an E2F1 target gene in p53-deficient SAOS-2 osteosarcoma cells (4) and immortal rodent fibroblasts (12, 77). We show here that p14ARF is also an E2F1 target gene in normal human fibroblasts, which have an intact p53 response and stringent control of replicative senescence. Consistent with findings for immortal cells, p14ARF, whether induced by E2F1 or overexpressed directly, increased p53, and consequently p21, protein levels. Although the outcome of these direct and indirect effects of E2F1 in immortal cells was growth stimulation or apoptosis, in normal human fibroblasts the outcome was a senescence-like phenotype.
Consistent with a critical role in the E2F1 senescence response, p14ARF alone induced a senescence-like phenotype when introduced into presenescent cells. This result and our finding that p14ARF is highly expressed by replicatively senescent human fibroblasts suggest that p14ARF is an important regulator of senescence, whether induced by cell division or E2F1. We do not yet know how p14ARF expression is upregulated and maintained during replicative senescence. It is almost certainly not due to induction by E2F, since the level of E2F1 expression, as well as that of E2F DNA-binding activity, is very low in senescent cells (15). It is also unlikely that the high levels of p14ARF in senescent cells are due to DNA damage, since p14ARF does not appear to be induced by DNA damage (64, 67). One possibility is that it is due to reduced levels of BMI-1, an oncogene and transcriptional repressor that regulates the INK4a locus (30). Our preliminary data suggest that the level of BMI-1 expression is lower in senescent human fibroblasts than in presenescent proliferating cells. Whatever the mechanism, our results suggest that p14ARF can account for the senescence response elicited by ectopic E2F1 expression and that the presence of elevated levels of p14ARF is also a feature of replicatively senescent cells.
Nature of the E2F1-induced senescence response.
The E2F1- and p14ARF-induced phenotype shared several features with that of replicatively senescent cells. These features include a stable growth arrest that was not reversed by physiological mitogens (serum growth factors), an enlarged, flat morphology, expression of SA-β-Gal, and elevated expression of p21, p16INK4a, and p14ARF, as well as extracellular matrix-remodeling genes (MMP-1, stromelysin-1, and PAI-1 genes). On the other hand, p14ARF- and E2F1-overexpressing cells had high levels of p53 protein, which is not found in replicatively senescent cells (1, 3; our unpublished results). One possibility is that telomere shortening, overexpression of E2F1, and possibly other senescence-inducing stimuli induce similar, but not identical, phenotypes. It is also possible that p53 does increase during replicative senescence but that because p53 induces mdm2, which in turn reduces p53, the increase is transient. This possibility may explain why the p21 gene, a p53 target gene (20), increases as cultures reach replicative senescence but thereafter gradually declines (2, 66).
Normal human keratinocytes were reported to arrest growth in response to very high levels of E2F1 (36). The nature of this growth arrest was not characterized but differed from the senescence response we describe here in two major respects. First, neither E6 nor E7 overcame the growth arrest. Second, the growth arrest required E2F1 DNA binding, but not transactivation activity. By contrast, the E2F1-induced senescence response was abolished by E6 and was strictly dependent on E2F1 transactivation activity. Whether these dissimilarities are due to differences in the experimentally induced levels of E2F1, cell type-specific differences in the response to E2F1, or differences in the nature of the growth arrest (quiescence, differentiation, or senescence) remains to be determined.
Role of p53.
p53 played a critical role in determining how human cells responded to E2F1 overexpression. E2F1-induced senescence was p53 dependent, since it did not occur in cells that expressed E6 or high levels of Mdm2. This result provides a plausible molecular explanation for the finding that elevated E2F1 activity increases the incidence of tumors in mice that are deficient in p53 (50). p53 activity was also required for the senescence response to overexpression of activated Ras (61) and its downstream effector, MEK (34). In contrast, p53 was not required for the senescence response to overexpression of activated Raf (76). One possible difference between the response elicited by activated Raf and that elicited by E2F1 may be the upregulation of p14ARF. The E2F1-induced response was mediated by p14ARF, which requires p53 for its growth-inhibitory effects (reviewed in reference 64), and p14ARF required p53 for its ability to induce a senescence response. It is possible, then, that activated Raf induces senescence via a p14ARF-independent pathway.
Our preliminary data based on microarrayed cDNAs suggest there are both similarities and differences in the phenotypes induced by replicative senescence, E2F1, and p14ARF. Since E2F1 induces growth stimulators, in addition to p14ARF, this result is not surprising. It will be important to determine the molecular correlates of the phenotypes induced by telomere shortening, DNA damage, and mitogenic signals in order to understand how senescent cells influence tumorigenesis and aging.
ACKNOWLEDGMENTS
We thank D. Miller, V. Band, D. Galloway, A. Gualberto, H. Land, W.-H. Lee, K. Vousden, K. Helin, W. Kaelin, B. Vogelstein, and CellGenesys for generously providing reagents used in this study and P. Yaswen for helpful discussions.
This work was supported by grant AG09909 from the National Institute on Aging.
REFERENCES
- 1.Afshari C A, Vojta P J, Annab L A, Futreal P A, Willard T B, Barrett J C. Investigation of the role of G1/S cell cycle mediators in cellular senescence. Exp Cell Res. 1993;209:231–237. doi: 10.1006/excr.1993.1306. [DOI] [PubMed] [Google Scholar]
- 2.Alcorta D A, Xiong Y, Phelps D, Hannon G, Beach D, Barrett J C. Involvement of the cyclin-dependent kinase inhibitor p16 (INK4a) in replicative senescence of normal human fibroblasts. Proc Natl Acad Sci USA. 1996;93:13742–13747. doi: 10.1073/pnas.93.24.13742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atadja P, Wong H, Garkavstev I, Veillette C, Riabowol K. Increased activity of p53 in senescing fibroblasts. Proc Natl Acad Sci USA. 1995;92:8348–8352. doi: 10.1073/pnas.92.18.8348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bates S, Phillips A C, Clark P A, Stott F, Peters G, Ludwig R L, Voudsen K H. p14ARF links the tumor suppressors RB and p53. Nature. 1998;395:124–125. doi: 10.1038/25867. [DOI] [PubMed] [Google Scholar]
- 5.Blattner C, Sparks A, Lane D. Transcription factor E2F-1 is upregulated in response to DNA damage in a manner analogous to that of p53. Mol Cell Biol. 1999;19:3704–3713. doi: 10.1128/mcb.19.5.3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bodnar A G, Ouellette M, Frolkis M, Holt S E, Chiu C P, Morin G B, Harley C B, Shay J W, Lichtsteiner S, Wright W E. Extension of life span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 7.Campisi J. Aging and cancer: the double-edged sword of replicative senescence. J Am Geriatrics Soc. 1997;45:1–6. doi: 10.1111/j.1532-5415.1997.tb05175.x. [DOI] [PubMed] [Google Scholar]
- 8.Campisi J. Replicative senescence: an old lives tale? Cell. 1996;84:497–500. doi: 10.1016/s0092-8674(00)81023-5. [DOI] [PubMed] [Google Scholar]
- 9.Campisi J, Dimri G P, Hara E. Control of replicative senescence. In: Schneider E, Rowe J, editors. Handbook of the biology of aging. 4th ed. New York, N.Y: Academic Press; 1996. pp. 121–149. [Google Scholar]
- 10.Chen Q, Bartholomew J C, Campisi J, Acosta M, Reagen J D, Ames B. Molecular analysis of H2O2-induced senescent-like growth arrest in normal human fibroblasts: p53 and Rb control G(1) arrest but not cell replication. Biochem J. 1998;332:43–50. doi: 10.1042/bj3320043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeGregori J, Kowalik T, Nevins J R. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis and G1/S regulatory genes. Mol Cell Biol. 1995;15:4215–4224. doi: 10.1128/mcb.15.8.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeGregori J, Leone G, Miron A, Jakoi L, Nevins J R. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc Natl Acad Sci USA. 1997;94:7245–7250. doi: 10.1073/pnas.94.14.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DiLeonardo A, Linke S P, Clarkin K, Wahl G M. DNA damage triggers a prolonged p53-dependent G1 arrest and long-term induction of Cip1 in normal human fibroblasts. Genes Dev. 1994;8:2540–2551. doi: 10.1101/gad.8.21.2540. [DOI] [PubMed] [Google Scholar]
- 14.Dimri G P, Campisi J Cold Spring Harbor Press. Molecular and cell biology of replicative senescence. Mol Gen Cancer. 1994;59:67–73. doi: 10.1101/sqb.1994.059.01.010. [DOI] [PubMed] [Google Scholar]
- 15.Dimri G P, Hara E E, Campisi J. Regulation of two E2F-related genes in presenescent and senescent human fibroblasts. J Biol Chem. 1994;269:16180–16186. [PubMed] [Google Scholar]
- 16.Dimri G P, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano E E, Linskens M, Rubelj I, Pereira-Smith O M, Peacocke M, Campisi J. A novel biomarker identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dimri G P, Nakanishi M, Desprez P Y, Smith J R, Campisi J. Inhibition of E2F activity by the p21 inhibitor of cyclin dependent protein kinases in cells expressing or lacking a functional retinoblastoma protein. Mol Cell Biol. 1996;16:2987–2997. doi: 10.1128/mcb.16.6.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dimri G P, Testori A, Acosta M, Campisi J. Replicative senescence, aging and growth regulatory transcription factors. Biol Signals. 1996;5:154–162. doi: 10.1159/000109185. [DOI] [PubMed] [Google Scholar]
- 19.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 20.El-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 21.Field S J, Tsai F Y, Kuo F, Zubiaga A M, Kaelin W G, Livingston D M, Orkin S H, Greenberg M E. E2F1 functions in mice to promote apoptosis and suppress proliferation. Cell. 1996;85:549–561. doi: 10.1016/s0092-8674(00)81255-6. [DOI] [PubMed] [Google Scholar]
- 22.Freedman D A, Wu L, Levine A J. Functions of the MDM2 oncoprotein. Mol Life Sci. 1999;55:96–107. doi: 10.1007/s000180050273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Halbert C L, Demes G W, Galloway D A. The E7 gene of human papillomavirus type 16 is sufficient for immortalization of human epithelial cells. J Virol. 1992;66:2125–2134. doi: 10.1128/jvi.65.1.473-478.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hara E, Smith R, Parry D, Tahara H, Stone S, Peters G. Regulation of p16/CDKN2 expression and its implications for cell immortalization and senescence. Mol Cell Biol. 1996;16:859–867. doi: 10.1128/mcb.16.3.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harley C B. Telomere loss: mitotic clock or genetic time bomb? Mutat Res. 1991;256:271–282. doi: 10.1016/0921-8734(91)90018-7. [DOI] [PubMed] [Google Scholar]
- 26.Hayflick L. The limited in vitro lifetime of human diploid cell strains. Exp Cell Res. 1965;37:614–636. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- 27.Helin K. Regulation of cell proliferation by E2F transcription factors. Curr Opin Genet Dev. 1997;8:28–35. doi: 10.1016/s0959-437x(98)80058-0. [DOI] [PubMed] [Google Scholar]
- 28.Helin K, Lees J A, Vidal M, Dyson N, Harlow E, Fattaey A. A cDNA encoding a pRB binding protein with properties of the transcription factor E2F. Cell. 1992;70:337–350. doi: 10.1016/0092-8674(92)90107-n. [DOI] [PubMed] [Google Scholar]
- 29.Hofferer M, Wirbelauer C, Humar B, Kerk W. Increased levels of E2F1 dependent DNA binding activity after UV- or gamma-irradiation. Nucleic Acids Res. 1999;15:491–495. doi: 10.1093/nar/27.2.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacobs J J, Kieboom K, Marino S, DePinho R A, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 31.Johnson D G, Schwarz J K, Cress W D, Nevins J R. Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature. 1993;365:349–352. doi: 10.1038/365349a0. [DOI] [PubMed] [Google Scholar]
- 32.Kamijo T, Zindy F, Roussel M F, Quelle D E, Downing R A, Ashmun G, Grosveld G, Sherr C J. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 33.Klingelhutz A, Foster S A, McDougall J K. Telomerase activation by the E6 gene product of human papillomavirus type 16. Nature. 1996;380:79–83. doi: 10.1038/380079a0. [DOI] [PubMed] [Google Scholar]
- 34.Lin A W, Barradas M, Stone J C, van Aelst L, Serrano M, Lowe S W. Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling. Genes Dev. 1998;12:3008–3019. doi: 10.1101/gad.12.19.3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McConnell B B, Starborg M, Brookes S, Peters G. Inhibitors of cyclin-dependent kinases induce features of replicative senescence in early passage human diploid fibroblasts. Curr Biol. 1998;8:351–354. doi: 10.1016/s0960-9822(98)70137-x. [DOI] [PubMed] [Google Scholar]
- 36.Melillo R M, Helin K, Lowy D R, Schiller J T. Positive and negative regulation of cell proliferation by E2F1: influence of protein level and human papilloma virus oncoproteins. Mol Cell Biol. 1994;14:8241–8249. doi: 10.1128/mcb.14.12.8241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Merlin A B, Gabai V L, Yaglom J, Shifrin V I, Sherman M Y. Proteasome inhibitors activate stress kinases and induce Hsp72. J Biol Chem. 1998;273:6373–6379. doi: 10.1074/jbc.273.11.6373. [DOI] [PubMed] [Google Scholar]
- 38.Miller A D, Rosman G J. Improved retroviral vectors for gene transfer and expression. BioTechniques. 1989;7:980–988. [PMC free article] [PubMed] [Google Scholar]
- 39.Millis A J T, Hoyle M, McCue H M, Martini H. Differential expression of metalloproteinase and tissue inhibitor of metalloproteinase genes in diploid human fibroblasts. Exp Cell Res. 1992;201:373–379. doi: 10.1016/0014-4827(92)90286-h. [DOI] [PubMed] [Google Scholar]
- 40.Mishima K, Handa J T, Aotaki-Keen A, Lutty G A, Morse L S, Hjelmeland L M. Senescence-associated beta-galactosidase histochemistry for the primate eye. Invest Ophthalmol Vis Sci. 1999;40:1590–1593. [PubMed] [Google Scholar]
- 41.Morgenstern J P, Land H. Advanced mammalian gene transfer: high titer retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neuman E, Flemington E K, Sellers W R, Kaelin W G., Jr Transcription of the E2F-1 gene is rendered cell cycle dependent by E2F DNA-binding sites within its promoter. Mol Cell Biol. 1994;14:6607–6615. doi: 10.1128/mcb.14.10.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nevins J R. Toward an understanding of the functional complexity of the E2F and retinoblastoma families. Nat Genet. 1998;9:585–593. [PubMed] [Google Scholar]
- 44.Noda A, Ning Y, Venable S F, Pereira-Smith O M, Smith J. Cloning of senescent cell-derived inhibitors of DNA synthesis using an expression screen. Exp Cell Res. 1994;21:90–98. doi: 10.1006/excr.1994.1063. [DOI] [PubMed] [Google Scholar]
- 45.Ogryzko V V, Hirai T H, Russanova V R, Barbie D A, Howard B H. Human fibroblast commitment to a senescence-like state in response to histone deacetylase inhibitors is cell cycle dependent. Mol Cell Biol. 1996;16:5210–5218. doi: 10.1128/mcb.16.9.5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oswald F, Lovec H, Moroy T, Lipp M. E2F dependent regulation of human MYC: transactivation by cyclins D1 and A overrides tumor suppressor protein functions. Oncogene. 1994;9:2029–2036. [PubMed] [Google Scholar]
- 47.Pan H, Yin C, Dyson N J, Harlow E, Yamasaki L, van Dyke T. Key roles for E2F1 in signaling p53-dependent apoptosis and in cell division within developing tumors. Mol Cell. 1998;2:283–292. doi: 10.1016/s1097-2765(00)80273-7. [DOI] [PubMed] [Google Scholar]
- 48.Pear W S, Nolan G P, Scott M L, Baltimore D. Production of high titer helper free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pendergrass W R, Lane M A, Bodkin N L, Hansen B C, Ingram D K, Roth G S, Yi L, Bin H, Wolf N S. Cellular proliferation potential during aging and caloric restriction in rhesus monkeys (Macaca mulatta) J Cell Physiol. 1999;180:123–130. doi: 10.1002/(SICI)1097-4652(199907)180:1<123::AID-JCP14>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 50.Pierce A M, Gimenez-Conti I B, Schneider-Broussard R, Martinez L A, Conti C J, Johnson D G. Increased E2F1 activity induces skin tumors in mice heterozygous and nullizygous for p53. Proc Natl Acad Sci USA. 1998;95:8858–8863. doi: 10.1073/pnas.95.15.8858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pomerantz J, Schreiber-Agus N, Liegeois N J, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee H W, Cordon-Cardo C, DePinho R A. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 52.Qin X Q, Livingston D M, Kaelin W G, Adams P D. Deregulated transcription factor E2F-1 expression leads to S-phase entry and p53-mediated apoptosis. Proc Natl Acad Sci USA. 1994;91:10918–10922. doi: 10.1073/pnas.91.23.10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rapp L, Chen J J. The papillomavirus E6 proteins. Biochim Biophys Acta. 1998;1378:F1–F19. doi: 10.1016/s0304-419x(98)00009-2. [DOI] [PubMed] [Google Scholar]
- 54.Robertson K D, Jones P A. The human ARF cell cycle regulatory gene promoter is a CpG island which can be silenced by DNA methylation and down-regulated by wild-type p53. Mol Cell Biol. 1998;18:6457–6473. doi: 10.1128/mcb.18.11.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robles S J, Adami G R. Agents that cause DNA double strand breaks lead to p16INK4a enrichment and the premature senescence of normal fibroblasts. Oncogene. 1998;16:1113–1123. doi: 10.1038/sj.onc.1201862. [DOI] [PubMed] [Google Scholar]
- 56.Sager R. Senescence as a mode of tumor suppression. Environ Health Perspect. 1991;93:59–62. doi: 10.1289/ehp.919359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saito M, Helin K, Valentine M B, Griffith B B, Willman C L, Harlow E, Look A T. Amplification of the E2F1 transcription factor gene in the HEL erythroleukemia cell line. Genomics. 1995;25:130–138. doi: 10.1016/0888-7543(95)80118-6. [DOI] [PubMed] [Google Scholar]
- 58.Saunders N A, Smith R J, Jetten A M. Regulation of proliferation-specific and differentiation-specific genes during senescence of human epidermal keratinocytes and mammary epithelial cells. Biochem Biophys Res Commun. 1994;197:46–54. doi: 10.1006/bbrc.1993.2439. [DOI] [PubMed] [Google Scholar]
- 59.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 60.Sedivy J M. Can ends justify the means? Telomeres and the mechanisms of replicative senescence and immortalization in mammalian cells. Proc Natl Acad Sci USA. 1998;95:9078–9081. doi: 10.1073/pnas.95.16.9078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Serrano M, Lin A W, McCurrach M E, Beach D, Lowe S W. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 62.Shan B, Durfee T, Lee W H. Disruption of RB/E2F-1 interaction by single point mutations in E2F-1 enhances S-phase entry and apoptosis. Proc Natl Acad Sci USA. 1996;93:679–684. doi: 10.1073/pnas.93.2.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shan B, Lee W H. Deregulated expression of E2F-1 induces S-phase entry and leads to apoptosis. Mol Cell Biol. 1994;14:8166–8173. doi: 10.1128/mcb.14.12.8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sherr C J. Tumor surveillance via the ARF-p53 pathway. Genes Dev. 1998;12:2984–2981. doi: 10.1101/gad.12.19.2984. [DOI] [PubMed] [Google Scholar]
- 65.Smith J R, Pereira-Smith O M. Replicative senescence: implications for in vivo aging and tumor suppression. Science. 1996;273:63–67. doi: 10.1126/science.273.5271.63. [DOI] [PubMed] [Google Scholar]
- 66.Stein G H, Drullinger L F, Soulard A, Dulic V. Differential roles for cyclin-dependent kinase inhibitors p21 and p16 in the mechanisms of senescence and differentiation in human fibroblasts. Mol Cell Biol. 1999;19:2109–2117. doi: 10.1128/mcb.19.3.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stott F J, Bates S, James M C, McConnell B B, Starborg M, Brookes S, Palmero I, Ryan K, Hara E, Vousden K H, Peters G. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 1998;17:5001–5014. doi: 10.1093/emboj/17.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tao W, Levine A J. P19 (ARF) stabilizes p53 by blocking nucleo-cytoplasmic shuttling of Mdm2. Proc Natl Acad Sci USA. 1999;96:6937–6941. doi: 10.1073/pnas.96.12.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tsai K Y, Hu Y, Macleod K F, Crowley D, Yamasaki L, Jacks T. Mutation of E2F-1 suppresses apoptosis and inappropriate S phase entry and extends survival of Rb-deficient mouse embryos. Mol Cell. 1998;2:293–304. doi: 10.1016/s1097-2765(00)80274-9. [DOI] [PubMed] [Google Scholar]
- 70.West M D, Pereira-Smith O M, Smith J R. Replicative senescence of human skin fibroblasts correlates with a loss of regulation and overexpression of collagenase activity. Exp Cell Res. 1989;184:138–147. doi: 10.1016/0014-4827(89)90372-8. [DOI] [PubMed] [Google Scholar]
- 71.Wright W E, Shay J W. Mechanism of escaping senescence in human diploid cells. In: Holbrook N J, Martin G R, Lockshin R A, editors. Modern cell biology series. Cellular aging and cell death. New York, N.Y: Wiley & Sons, Inc.; 1996. pp. 153–167. [Google Scholar]
- 72.Wu X, Levine A J. P53 and E2F-1 cooperate to mediate apoptosis. Proc Natl Acad Sci USA. 1994;91:3601–3606. doi: 10.1073/pnas.91.9.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yamasaki L, Bronson R, Williams B O, Dyson N J, Harlow E, Jacks T. Loss of E2F-1 reduces tumorigenesis and extends the lifespan of Rb1(+/−) mice. Nat Genet. 1998;18:360–364. doi: 10.1038/ng0498-360. [DOI] [PubMed] [Google Scholar]
- 74.Yamasaki L, Jacks T, Bronson R, Goillot E, Harlow E, Dyson N J. Tumor induction and tissue atrophy in mice lacking E2F-1. Cell. 1996;85:537–548. doi: 10.1016/s0092-8674(00)81254-4. [DOI] [PubMed] [Google Scholar]
- 75.Zhang Y, Xiong Y, Yarbrough W G. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppressor pathways. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 76.Zhu J, Woods D, McMahon M, Bishop J M. Senescence of human fibroblasts induced by oncogenic raf. Genes Dev. 1998;12:2997–3007. doi: 10.1101/gad.12.19.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zindy F, Eischen C M, Randle D, Kamijo T, Cleveland J L, Sherr C J, Roussel M F. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998;12:2424–2434. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]