A Nuclear 3′-5′ Exonuclease Involved in mRNA Degradation Interacts with Poly(A) Polymerase and the hnRNA Protein Npl3p (original) (raw)
Abstract
Inactivation of poly(A) polymerase (encoded by PAP1) in Saccharomyces cerevisiae cells carrying the temperature-sensitive, lethal pap1-1 mutation results in reduced levels of poly(A)+ mRNAs. Genetic selection for suppressors of pap1-1 yielded two recessive, cold-sensitive alleles of the gene RRP6. These suppressors, rrp6-1 and rrp6-2, as well as a deletion of RRP6, allow growth of pap1-1 strains at high temperature and partially restore the levels of poly(A)+ mRNA in a manner distinct from the cytoplasmic mRNA turnover pathway and without slowing a rate-limiting step in mRNA decay. Subcellular localization of an Rrp6p-green fluorescent protein fusion shows that the enzyme residues in the nucleus. Phylogenetic analysis and the nature of the rrp6-1 mutation suggest the existence of a highly conserved 3′-5′ exonuclease core domain within Rrp6p. As predicted, recombinant Rrp6p catalyzes the hydrolysis of a synthetic radiolabeled RNA in a manner consistent with a 3′-5′ exonucleolytic mechanism. Genetic and biochemical experiments indicate that Rrp6p interacts with poly(A) polymerase and with Npl3p, a poly(A)+ mRNA binding protein implicated in pre-mRNA processing and mRNA nuclear export. These findings suggest that Rrp6p may interact with the mRNA polyadenylation system and thereby play a role in a nuclear pathway for the degradation of aberrantly processed precursor mRNAs.
A large body of evidence indicates that nucleotide sequences in the 3′ untranslated regions (UTRs) of mRNAs specify the regulation of poly(A) tail length, which in turn can have significant effects on the stability and translational activity of individual mRNAs (35). Many mRNAs, such as those encoding lymphokines and proto-oncogenes, carry 3′ UTR sequences that regulate transcript stability in cis, in some cases by altering the rate of poly(A) tail removal (6). The dependence of translation on polyadenylation plays a central role during oogenesis and embryogenesis, where maternal mRNAs undergo poly(A) tail lengthening and shortening that correlate with changes in translation activity (67). Thus, processes occurring posttranscriptionally at the 3′ ends of mRNAs may have a profound impact on the extent, timing, and spatial regulation of the expression of certain genes (19). The regulation and function of poly(A) tail lengths depend critically on proteins such as poly(A) polymerase that synthesize the poly(A) tail, as well as on those that interact with the poly(A) tail and with other parts of the mRNA. For example, the 5′-terminal cap structure and the poly(A) tail contact one another via an eIF4G-mediated interaction between the cap-binding protein eIF4E and poly(A)-binding protein (Pab1p) (66). Interactions between the cap and poly(A) tails manifest themselves as (i) synergistic enhancement of translation (26), (ii) regulation of the order of deadenylation and decapping during mRNA turnover (50), (iii) premature cap removal and mRNA degradation caused by disruption of the poly(A)-Pab1p complex (16), and (iv) dependence of cap methylation upon poly(A) tail extension in developing oocytes (28, 40).
Conditional lethal mutations in yeast mRNA 3′-end processing genes provided the first demonstration that inactivation of polyadenylation causes the inability of the cell to accumulate many mRNAs, thus leading to cell death (54, 55). The continued efficient translation of unadenylated mRNAs after polyadenylation shutoff implicated mRNA stabilization, rather than translational enhancement, as the essential function for poly(A)+ tails (54, 58). Although the disappearance of mRNAs after polyadenylation shutoff supports a role for poly(A) tails in conferring mRNA stability, it remained unclear why these mRNAs fail to accumulate (58). Evidence presented in this report indicates that loss of poly(A)+ mRNAs does not result from the action of the major cytoplasmic mRNA turnover system featuring Xrn1p/Ski1p (35). Instead, suppressor mutations which increase the levels of poly(A)+ mRNAs in cells with a poly(A) polymerase defect arise in genes encoding nuclear functions involved in the biogenesis of tRNA and rRNA (10, 11). These findings raise the possibility that the polyadenylation apparatus may communicate in a novel way with factors synthesizing and processing other types of RNA so as to ensure a balance of RNA molecules required for efficient protein synthesis. Proteins that play a role in such communication would likely constitute important targets for signal transduction pathways, as well as for pathogens seeking to subvert the host's gene expression system. Indeed, expression of the unadenylated mRNAs of yeast double-stranded RNA (dsRNA) killer viruses is controlled by host-encoded gene products involved in ribosome biogenesis and mRNA turnover (2, 37, 53).
We recently showed that a putative 3′-5′ riboexonuclease encoded by the RRP6 gene plays a role in 5.8S rRNA 3′-end processing and that defects in this gene suppress the growth defect associated with an mRNA polyadenylation defect (10). Here we report the results of experiments designed to determine the mechanism by which loss of Rrp6p function results in the growth of cells that have lost much of their poly(A)+ mRNA due to a temperature-sensitive defect in poly(A) polymerase. We found that deletion of RRP6 increases the level of poly(A)+ mRNA under these conditions without altering the rate of mRNA decay and in a manner distinct from the major cytoplasmic mRNA decay pathway. Consistent with this difference, subcellular localization of Rrp6p shows that the protein resides in the nucleus. Purified Rrp6p demonstrates the RNase activity expected of a 3′-5′ riboexonucleolytic mode of hydrolysis. Finally, we provide evidence that Rrp6p interacts with poly(A) polymerase and with the hnRNA protein Npl3p. These findings suggest that Rrp6p may interact with the mRNA 3′ processing system and thereby participate in a novel nuclear RNA degradation pathway that destroys slowly or incompletely processed mRNAs.
MATERIALS AND METHODS
Yeast strains, media, and genetic techniques.
Table 1 lists the strains used in these experiments. Yeast strains were grown in yeast extract-peptone-dextrose (YEPD) or synthetic complete medium lacking uracil and/or methionine. Transformation of yeast with plasmids was performed as described by Schiestl and Geitz (62). Escherichia coli DH5α and XL-1 were used for recombinant DNA manipulations.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| A364A | MATα ade1 ade2 lys2 gal1 ura1 his7 tyr1 | 54 |
| BP02 | MATα ade1 ade2 lys2 gal1 ura3-52 | 11 |
| BP02-12F | MATα ade1 ade2 lys2 gal1 ura3-52 rrp6::URA3 | 10 |
| BPKAN | MATα ade1 ade2 lys2 gal1 ura3-52 rrp6::KAN | This study |
| UR3148-1B | MATα ade1 ade2 lys2 gal1 ura3-52 pap1-1 | 54 |
| UR3148-1B-12F | MATα ade1 ade2 lys2 gal1 ura3-52 pap1-1 rrp6::URA3 | This study |
| UR3148-1BC1 | MATα ade1 ade2 lys2 gal1 ura3-52 pap1-1 rrp6-1 | 10 |
| UR3148-1BC12 | MATα ade1 ade2 lys2 gal1 ura3-52 pap1-1 rrp6-2 | 10 |
| UR3148-1B ΔX | MATα ade1 ade2 lys2 gal1 ura3-52 pap1-1 xrn1::URA3 | This study |
| UR3148-1B Δ | MATα ade1 ade2 lys2 gal1 ura3-52 pap1-1 upf1::URA3 | This study |
| YAP201 | MATα ade1 ade2 lys2 gal1 ura3-52 pap1-1 spb2(rpl46)::URA3 | 56 |
| ABC1-2D | MATα ade1 ade2 lys2 gal1 ura3-52 rrp6-1 | 10 |
| ABC1-4D | MATα ade1 ade2 lys2 gal1 ura3-52 rrp6-1 | This study |
| PSY1 | MATa ade2-1 trp1-1 ura3-1 leu2-3,112 his3-11,15 can1-100 | P. Silver |
| PSY1.RT | MATa ade2-1 trp1-1 ura3-1 leu2-3,112 his3-11,15 can1-100 rrp6::TRP1 | This study |
| PSY773 | MATa his3-11 ade2-1 leu2-1 ura3-1 trp1-1 npl3-1 | 31 |
| PSY773.RT | MATa his3-11 ade2-1 leu2-1 ura3-1 trp1-1 npl3-1 rrp6::TRP1 (YCpRRP6) | This study |
| EJ758 | MATa his3Δ200 leu2-3,112 ura3-52 pep4::HIS3 | E. Grayhack |
| Y190 | MATa gal4 gal80 his3 trp1-901 ade2-101 ura3-52 leu2-3,112 URA3::GAL-lacZ LYS2::GAL(UAS)-HIS3 cyhr | 4 |
| Y187 | MATα gal4 gal80 his3 trp1-901 ade2-101 ura3-52 leu2-3,112 URA3::GAL-lacZ | 4 |
Chromosomal deletions of UPF1 and XRN1 were carried out by transforming UR3148-1B with either _Eco_RI/_Bam_HI-cleaved pPL51 (44) or _Xho_I/_Sal_I-cleaved pFL306 (42), respectively. PCR analysis verified proper disruption of UPF1, while Southern blot analysis verified that of XRN1. Deletion of SPB2(RPL46) to create strain YAP201 was described previously (57).
Chromosomal deletion of RRP6 by using the loxP-kanMX-loxP disruption cassette was performed as described by Guldener et al. (29). BPO2 was transformed with the Kanr marker which had been PCR amplified with primers oSB87 and oSB88 (Oligos, Etc., Inc.), which carry at their 3′ ends segments homologous to the sequences flanking the loxP-kanMX-loxP module and at their 5′ ends segments homologous to RRP6. Gene disruptants were selected for on YPD plus geneticin plates (200 μg/ml; Gibco-BRL) and screened for temperature sensitivity at 37°C. Proper disruption of RRP6 was verified by PCR analysis with primers oSB41 and oSB42 (10).
Plasmids and oligonucleotides.
Table 2 lists the plasmids and deoxyoligonucleotides utilized in these experiments. Restriction enzymes were purchased from Gibco-BRL, Promega, or New England Biolabs, and digestions were performed according to the manufacturers' instructions. Double-stranded DNA probe templates were prepared by electroelution, using dialysis bags (Spectrum), from 1% agarose gels and labeled by random hexamer priming with a 5′-[α-32P]dCTP (NEN Life Science Products; 3,000 Ci/mmol) and the Klenow fragment of DNA polymerase I (Boehringer Mannheim) according to the manufacturer's instructions. Deoxyoligonucleotide probes (Oligos, Etc., Inc.) were radiolabeled with 5′-[γ-32P]ATP (NEN Life Science Products; 6,000 Ci/mmol) by using T4 polynucleotide kinase (Gibco-BRL) according to the manufacturer's instructions. Unincorporated nucleotides were removed from probes by chromatography on Sephadex G-25 (Amersham-Pharmacia Biotech).
TABLE 2.
Plasmids and oligonucleotides used in this study
| Plasmid or oligonucleotide | Relevant characteristics or sequence | Source or reference |
|---|---|---|
| Oligonucleotides | ||
| o5.8S | 5′-TGCGTTCTTCATCGATGCGAGAACC-3′ | 64 |
| oRP100 | 5′-GTCTAGCCGCGAGGAAGG-3′ | 51 |
| oSB7 | 5′-ATAGGAATAAGAATGCTCAA-3′ | This study |
| oSB86 | 5′-CATACGCGCACAAAAGCAGAG-3′ | This study |
| oSB41/oRRP6-2 | 5′-AGAATTTAGACAGGG-3′ | 10 |
| oSB42/oRRP6-3 | 5′-CATCGTCTCTTCTTGC-3′ | 10 |
| oSB43 | 5′-CGGAATTCCGCAAAATAAGTTCACGTG-3′ | This study |
| oSB44 | 5′-CGGAATTCCTAACATGAGTCAAATCCC-3′ | This study |
| RP29 | Random-primed probe from 1-kb _Eco_RI-_Hin_dIII fragment in A13 | 32 |
| oSB87 | 5′-GATTTAGTAAAGATCTGAAATCAAAAGCAGACAAAC-3′ | This study |
| oSB88 | 5′-GCAATAAGATCTCTCCATTGGTATAGCTCTCTTACT | This study |
| oSB89 | 5′-CATACCACCTGAGAGGGAAG-3′ | This study |
| oSB100 | 5′-CTATCTATTCGATGATGAAG-3′ | 4 |
| oSB101 | 5′-ACAGTTGAAGTGAACTTGCG-3′ | 4 |
| oSB116 | 5′-GAATTTAGACAGGGGATTTAGTAAAGATCTGAAATCAAAAGCAGACA AACTTGCGGATAGGCGTATCACGAGGCC-3′ | This study |
| oSB117 | 5′-CGAATAAGATCTCTCCATTGGTATAGCTCTCTTACTAATACTTCCCTCT CACCGAAACGCGCGAGACGAAAGGG-3′ | This study |
| Plasmids | ||
| YCplac33 | CEN4, URA3 library cloning vector | 27 |
| YCpRRP6 | CEN4, URA3 vector containing RRP6 | 10 |
| pGFP-FOR11 | RRP6 cloned in frame into pGFP-N-FUS in forward orientation | This study |
| pGFP-REV2 | RRP6 cloned into pGFP-N-FUS in the reverse orientation | This study |
| pRST66 | RRP6 cloned in frame into pGEX2T | This study |
| pAS2-RRP6 | RRP6 cloned in frame to the GAL4 DNA binding domain of pAS2 | This study |
| pACT-RRP43 | RRP43 cloned to the GAL4 activation domain of pACT2 | 71 |
| pGAD424 | 2μ, GAL4 activation domain vector | 5 |
| pACT-NPL3 | NPL3 fused to the GAL4 activation domain of pACT2 | This study |
| pAS2 | 2μ, GAL4 DNA binding domain | 4 |
| pAS2-CDK2 | CDK2 fused to the GAL4 DNA binding domain of pAS2 | 4 |
| pAS2-SNF1 | SNF1 fused to the GAL4 DNA binding domain of pAS2 | 4 |
| pAS2-p53 | p53 fused to the GAL4 DNA binding domain of pAS2 | 4 |
| pEG(KT) | 2μ, URA3 plasmid containing GST under control of the GAL1-10 UAS | 47 |
| pEGP65 | RRP6 cloned in frame to GST of pEGKT | This study |
Plasmids pGFP-FOR11 and pGFP-REV2 were constructed by filling in the ends of a 2.3-kb _Bsp_EI fragment containing RRP6 from pUN9D4 (10) with deoxynucleoside triphosphates, using the Klenow fragment of DNA polymerase I, followed by ligation of the fragment in either the forward or the reverse orientation, respectively, into the _Sma_I site of pGFP-N-FUS (52). DNA sequence analysis of the junction between pGFP-N-FUS and RRP6 verified that green fluorescent protein (GFP) was cloned in frame with the sixth codon of RRP6 in pGFP-FOR11. Plasmids pRST66, pAS2-RRP6, and pEGP65 were constructed by inserting a 2.3-kb _Bsp_EI fragment containing RRP6 from pUN9D4 into the _Xma_I site in frame with glutathione _S_-transferase (GST), the GAL4 DNA binding domain, or GST in pGEX2T (Amersham-Pharmacia Biotech), pAS2, or pEG(KT), respectively. These plasmids express fusions of the respective plasmid-encoded proteins to codon 6 of RRP6.
RNA analyses.
Total and poly(A)+ RNAs were prepared and Northern analysis was carried out as described by Patel and Butler (54). mRNA levels were quantified by storage PhosphorImager analysis (Molecular Dynamics) and normalized to the levels of an RNA polymerase III transcript, SCR1.
Total poly(A) content in vivo was determined as described by Butler et al. (12), except that RNAs were separated by electrophoresis on a 15% polyacrylamide–7.5 M urea gel. mRNA decay rates were measured by using 4 mg of thiolutin (a kind gift of Saul B. Kadin, Pfizer) per ml as described by Briggs and Butler (11).
Sequencing of rrp6-1.
Chromosomal DNAs from UR3148-1B and UR3148-1BC1 were isolated, and RRP6 and rrp6-1 were PCR amplified by using two sets of primers: (i) oSB41 and oSB42 and (ii) oSB43 and oSB44. The DNA templates were independently sequenced by using primer oSB41, oSB42, or oSB89 and the BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems) according to the manufacturer's instructions; the products were then analyzed by the University of Rochester Core Nucleic Acid Laboratory.
Subcellular localization of Rrp6p.
Subcellular localization of Rrp6p was analyzed by visualizing the fluorescent signal produced by GFP fusions to the amino terminus of Rrp6p (GFP-FOR11). As a control, Rrp6p was also cloned in reverse (GFP-REV2). BPKAN was transformed with either pGFP-FOR11 or pGFP-REV2 and grown to the exponential phase in liquid SC medium without uracil (to maintain the plasmid) and without methionine (to induce expression of GFP fusion proteins from the MET25 promoter). Hoechst dye (10 μM; Sigma Chemical Co.) was added to stain the nuclei in living cells, and the cells were visualized by fluorescent microscopy.
Preparation and assay of recombinant Rrp6p.
Escherichia coli (XL-1) containing plasmid pRST66 or pGEX2T was grown in Luria-Bertani broth containing ampicillin (0.2 mg/ml) at 30°C to an _A_650 of 2.0, and expression of the plasmid-encoded proteins was induced by the addition of IPTG (isopropyl-β-d-thiogalactopyranoside) to a final concentration of 0.4 mM. Cells were shaken at the same temperature for an additional 4 h, collected by centrifugation at 4°C, washed in 100 ml of phosphate-buffered saline (4°C), and resuspended in 4 ml of buffer A (4°C; 50 mM Tris-Cl [pH 7.9], 5% glycerol, 2 mM Na2EDTA, 0.1 mM dithiothreitol [DTT], 1 mM 2-mercaptoethanol, 0.3 M KCl, and 2 μg of aprotinin, 0.5 μg of leupeptin, and 1 mg of Pefabloc SC per [Boehringer-Mannheim] per ml). The cells were lysed by two passages through a French pressure cell (15,000 lb/in2), and the cell debris was removed by centrifugation at 8,000 × g for 20 min. The supernatant was mixed with 0.2 ml of glutathione-Sepharose 4B (Amersham-Pharmacia) equilibrated with buffer A (50% [wt/vol]) and incubated with gentle rocking at 25°C for 30 min. The resin was collected by low-speed centrifugation and then washed three times with 2 ml of buffer A and then three times with 2 ml of buffer B (100 mM Tris-Cl, pH 7.5; 25% glycerol; 0.1 mM DTT; 1 mg of Pefabloc SC per ml) at 4°C. GST or GST-Rrp6p was eluted from the resin by incubation with 0.2 ml of buffer B supplemented with 25 mM glutathione, with gentle rocking, for 30 min at 25°C and stored at −20°C.
RNA substrates for exonuclease assays were prepared by SP6 RNA polymerase transcription of pGYC1 cut with _Bsr_GI or pSP65t′ cut with _Pvu_II (13). Selective labeling of the first four adenosines of the transcript was achieved by creating stalled complexes and chasing the complexes with an excess of all four unlabeled nucleotide triphosphates. One microgram of plasmid template was incubated in a 50-μl reaction mixture containing 0.2 M Tris-Cl (pH 7.9), 30 mM MgCl2, 10 mM spermidine, 0.5 mM GTP, 0.5 mM UTP, 0.01 mM ATP, 50 μCi of 5′-[α-32P]ATP (3,000 Ci/mmol; New England Nuclear), 100 U of RNasin (Promega), and 30 U of SP6 RNA polymerase at 37°C. After 15 min of incubation, the stalled complexes were chased by incubation for 60 min in the presence of all four nucleotide triphosphates at a final concentration of 0.25 mM. The 5′-end-labeled RNA product was separated from unincorporated nucleotides by chromatography on Sephadex G-25 and stored at −20°C. The 3′ end of unlabeled trpt′ RNA transcribed from pSP65t′ in the absence of radiolabeled nucleotides was phosphorylated by incubation with 5′-[α-32P]pCP as described elsewhere (12).
RNase assays were carried out at 30°C in 70-μl reaction mixtures containing 10 mM Tris-Cl (pH 7.5), 2.5% glycerol, 2% polyethylene glycol, 5 mM Mg acetate, 1 mM DTT, 10 nM 5′-α-32P-labeled CYC1 RNA, and 0.1 nM GST-Rrp6 or 0.1 nM GST. Then, 10-μl samples were removed at various time points, and the reaction was terminated by the addition of 2 μl of stop solution (15 mM Na2EDTA, 2% sodium dodecyl sulfate [SDS], 1 μg of proteinase K per μl) and incubation at 37°C for 15 min. The products were precipitated by the addition of 5 μl of carrier solution (0.3 M potassium acetate [pH 5], 15 μg of tRNA per μl) and incubation at −20°C for at least 20 min. The RNA precipitate was collected by centrifugation at 13,000 × g for 10 min, dried in a Speed-Vac (Savant), resuspended in 80% formamide–0.01% bromophenol blue–0.01% xylene cyanol, heated at 90°C for 5 min, and separated by electrophoresis on a 6% acrylamide-bisacrylamide (39:1)–8 M urea gel. The gel was dried and subjected to storage PhosphorImager analysis. Thin-layer chromatography analysis was carried out on a Brinkman Cel 300 UV254 plate developed with saturated NH4SO4-H2O-isopropanol (40:9:1) at pH 3.5.
Two-hybrid screen.
The two-hybrid screen was performed as described by Bai and Elledge (4). Bait plasmid pAS2-RRP6 was transformed into Y190 and was tested for the ability to activate lacZ transcription. The plasmid alone or in combination with either pACT-RRP43 (72) or pGAD424 cannot activate lacZ or HIS3 expression. A yeast genomic library fused to the GAL4 activating domain in pACT2 (E. Phizicky, University of Rochester) was then transformed into Y190 (pAS2-RRP6). Transformants were selected on synthetic medium lacking histidine, leucine, and tryptophan and supplemented with 3-aminotriazole (3-AT; 50 mM; Sigma) and then screened for lacZ expression by using an X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) colony filter lift assay (4). To determine whether lacZ expression in these colonies was dependent on both plasmids, the clones were plated on synthetic medium lacking leucine and supplemented with cycloheximide (2.5 μg/ml; Sigma) to select for loss of pAS2-RRP6 plasmid. Total nucleic acid was isolated from each strain and electroporated into E. coli DH5α to isolate the activation domain plasmids, which were subjected to DNA sequence analyses by using oSB100 and oSB101 (Oligos, Etc., Inc.). Yeast strains containing only the activation domain plasmids were then crossed with Y187 strains expressing the GAL4 DNA binding domain fused to CDK2, SNF1, and p53 and then retested for β-galactosidase activity. Clones positive for interaction with pAS2-RRP6 but negative for interactions with the other plasmids were chosen for further study.
Synthetic lethality of npl3-1 allele and rrp6::TRP1.
To test for synthetic lethality between RRP6 and the temperature-sensitive npl3-1 allele, an rrp6::TRP1 PCR product was transformed into PSY1 and PSY773, each containing YCpRRP6. The rrp6::TRP1 fragment was constructed by PCR amplification of TRP1 from YIplac204 (27) by using primers oSB116 and oSB117 (Oligos, Etc., Inc.). Transformants were selected on synthetic medium lacking uracil and tryptophan, and the TRP1 disruption of RRP6 was confirmed by PCR analysis by using oSB41 and oSB42 (Oligos, Etc., Inc.). Synthetic lethality was monitored on plates containing 5-fluoroorotic acid (1 g/liter; Toronto Research Chemicals).
Affinity purification of GST and GST-Rrp6p.
EJ758 (E. Grayhack, University of Rochester) was transformed with either pEGKT or pEGP65. Cells were grown in 250 ml of synthetic medium with 2% raffinose lacking uracil at 30°C to an _A_600 of 1.0. Expression of GST or GST-Rrp6p was induced by addition of galactose to a final concentration of 4% (wt/vol). Cells were harvested after 3 h by centrifugation, washed in 10 ml of water (4°C), and resuspended in 3 ml of buffer C (50 mM Tris-Cl [pH 7.9], 5% glycerol, 2 mM Na2EDTA, 0.1 mM DTT, 1 mM 2-mercaptoethanol, 300 mM KCl, 0.5 μg of leupeptin [Roche Biochemicals] per ml, 1 mg of Pefabloc SC [Roche Biochemicals] per ml, 5 mM Mg2(CH3COO)2] at 4°C. Approximately 1.5 ml of acid-washed glass beads (Sigma) was added, followed by 15 cycles of vortexing for 30 s and then 30 s of incubation on ice. The sample was centrifuged for 2 min at 720 × g, and the supernatant was transferred to a clean tube. Approximately 100 μl of the supernatant was set aside as the “input” fraction. The remainder was mixed with 350 μl of glutathione-Sepharose 4B (Amersham-Pharmacia) equilibrated with buffer C (50% [wt/vol]) and incubated with gentle rocking overnight at 4°C. The resin was collected by centrifugation for 30 s at 181 × g, and the supernatant was transferred to a clean tube. The resin was washed four times in 3 ml of buffer C for 20 min at 4°C, washed five times in 5 ml of buffer C plus 0.1% NP-40 for 15 min at room temperature, and finally resuspended in 350 μl of buffer C and labeled the “bound” fraction.
Western blot analysis.
SDS-polyacrylamide gel electrophoresis (PAGE) was performed as described by Laemmli (41). Proteins were transferred to nitrocellulose (Schleicher & Schuell) at 40 mA of constant current overnight in transfer buffer (25 mM Tris [pH 8.3], 192 mM glycine, 20% methanol). Following transfer, the membrane was blocked for 1 h with 1% gelatin (Bio-Rad) in T-TBS (0.1% Tween 20, 100 mM Tris-Cl [pH 7.5], 0.9% NaCl) at room temperature. The membrane was then incubated with primary antibodies diluted in 1% gelatin–T-TBS for 1 h, washed twice for 20 min in T-TBS, incubated for 1 h with the secondary antibody (goat anti-mouse or goat anti-rabbit immunoglobulin horseradish peroxidase; 1:2,000 dilution; Santa Cruz Biotechnology), washed three times for 20 min in T-TBS, and washed twice for 1 min in TBS (100 mM Tris-Cl [pH 7.5], 0.9% NaCl). Antibodies were detected by enhanced chemiluminescence (NEN Life Science) according to the manufacturer's instructions. Monoclonal anti-Npl3 (1E4; 1:500), anti-Pap1p (1:500), and anti-Tcm1p (1:500) antibodies were generously provided by M. Swanson (University of Florida), Claire Moore (Tufts University), and J. Warner (Albert Einstein College of Medicine), respectively. GST-Rrp6p was detected with rabbit anti-GST antibody (1:600; Molecular Probes).
RESULTS
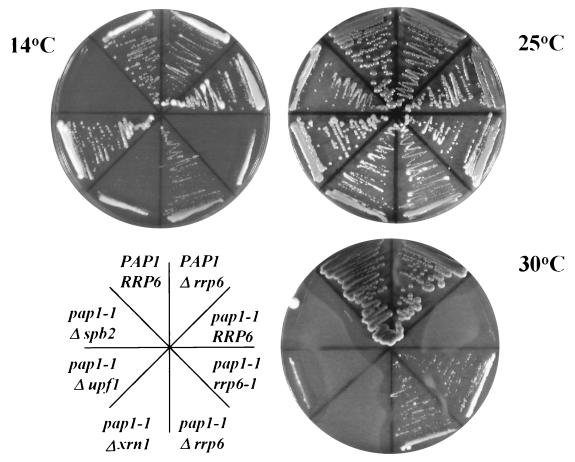
We reported previously the isolation of spontaneous, extragenic suppressors of a temperature-sensitive, lethal poly(A) polymerase mutation (pap1-1) that restore growth at the nonpermissive temperature 30°C (10). Characterization of two of these cold-sensitive suppressors, rrp6-1 and rrp6-2 (previously called pds1-1 and pds1-2 [11]), revealed that the normal, nonessential allele, RRP6, encodes a putative 3′-5′ riboexonuclease required for efficient 5.8S rRNA 3′-end processing. Since poly(A) tails appear to play a role in enhancing mRNA stability and translation initiation, we compared the ability of a rrp6::URA3 knockout to suppress pap1-1 with knockouts of several genes known to play a role in these processes. The XRN1/SKI1 and UPF1 gene products function in the degradation of mRNAs after deadenylation and during the translation of mRNAs with nonsense codons, respectively (44, 50). Loss of Xrn1p activity stabilizes mRNAs and leads to the accumulation of uncapped, deadenylated transcripts (21, 34). Loss of Upf1p leads to the stabilization of mRNAs bearing nonsense codons and suppresses _cis_-acting polyadenylation defects in Saccharomyces cerevisiae and Caenorhabditis elegans (44, 59; B. Das, Z. Guo, P. Chartrand, P. Russo, R. Singer, and F. Sherman, submitted for publication). Knockout mutations in either of these genes do not suppress the temperature sensitivity caused by the pap1-1 mutation, while the recessive rrp6-1 mutation, or a knockout of RRP6, allows pap1-1 cells to grow at 30°C (Fig. 1).
FIG. 1.
Suppression of pap1-1 temperature sensitivity by mutations in RRP6. Strains BPO2 (PAP1 RRP6), BPO2-12F (PAP1 rrp6::URA3), UR3148-1B (pap1-1), UR3148-1BC-12 (pap1-1 rrp6-1), UR3148-1B-12F (pap1-1 rrp6::URA3), UR3148-1B ΔX (pap1-1 xrn1::URA3), UR3148-1B ΔU (pap1-1 upf1::URA3), and YA201 (pap1-1 spb2::URA3) were grown on YPD plates at the indicated temperatures.
Deletion of the SPB2(RPL46) gene encoding the large subunit ribosomal protein L46 causes a decrease in 60S ribosomal subunit levels and bypasses the requirement for the otherwise essential poly(A)-binding protein (Pab1p), presumably by increasing the relative concentration of 40S ribosomes, thereby enhancing the rate of translation initiation of Pab1p-deficient mRNAs at the 40S binding step (16, 61, 65). RRP6 mutations also cause a decrease in 60S ribosomal subunit levels due to inefficient 5.8S rRNA 3′-end formation (11). Based on the proposed mechanism for the suppression of pab1 mutations by spb2, we initially thought that rrp6 mutations might belong to a general class of mutations that bypass the requirement for the poly(A) tail by enhancing the binding of poly(A)− mRNAs to the 40S subunit. Accordingly, we expected loss of SPB2 to suppress pap1-1 under conditions in which suppression by loss of RRP6 occurs. However, deletion of SPB2 does not suppress the growth defect caused by pap1-1 (Fig. 1).
RRP6 mutations enhance the accumulation of poly(A)+ mRNAs in pap1-1 cells.
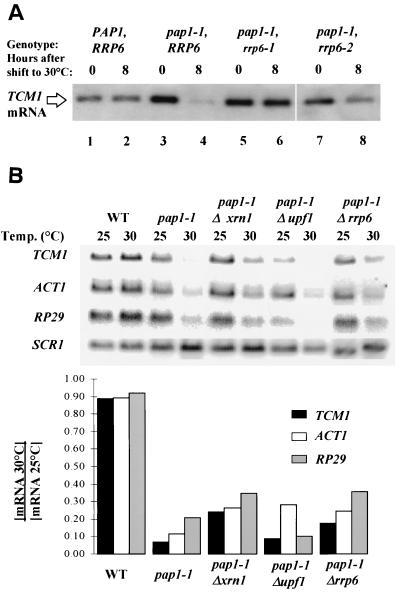
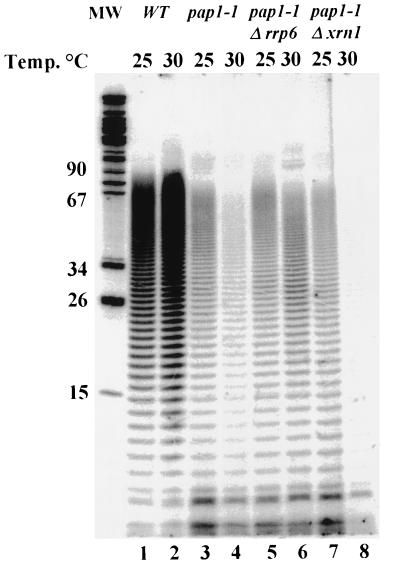
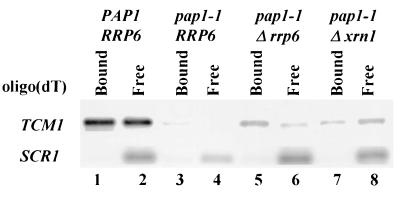
Our previous characterization of the effects of polyadenylation shutoff in a pap1-1 strain indicated that the cells die because they fail to accumulate many mRNAs and that inhibition of translation is a secondary consequence of mRNA loss (58). Thus, suppression of the pap1-1 growth defect under these conditions should result in an increase in steady-state mRNA levels. We quantitated by Northern blot analysis mRNA levels in pap1-1 cells after a shift to 30°C and normalized them to the levels of the stable RNA polymerase III transcript SCR1. In an effort to avoid secondary effects of Pap1p inactivation, we prepared RNA from cells harvested 2 h prior to growth cessation. Results representative of several experiments show suppression of the mRNA loss phenotype by rrp6-1, rrp6-2, and rrp6::URA3 (Fig. 2). Two experiments show that the mRNAs produced in the _rrp6_Δ pap1-1 strains carry poly(A) tails. First, visualization of total poly(A) after [5′-32P]pCp 3′-end labeling and PAGE shows a two- to threefold increase in the amount of poly(A) in an _rrp6_Δ pap1-1 strain compared to a pap1-1 strain at 30°C (Fig. 3). Second, oligo(dT)-mediated separation of total RNA samples into those with long poly(A) tails (Fig. 4, lanes 1, 3, 5, and 7) and those with short or no poly(A) tails (Fig. 4, lanes 2, 4, 6, and 8) reveals that the RRP6 deletion increase two- to threefold the levels of poly(A)+ TCM1 mRNA in pap1-1 cells at 30°C. Both of these experiments detect relatively low levels (ca. 10% of wild type) of poly(A)+ mRNA in pap1-1 cells at 30°C, suggesting that the mutant Pap1p remains partially active at the nonpermissive temperature. This finding contrasts with our previous studies of the effects of the pap1-1 mutation at 37°C, where we detected only unadenylated transcripts (58).
FIG. 2.
Restoration of mRNA levels in pap1-1 strains caused by mutations in RRP6. (A) Steady-state levels of TCM1 mRNA in rrp6 mutants. Total RNA was isolated from strains with the indicated genotypes before and 6 h after a shift to 30°C. TCM1 mRNA was revealed by Northern blot analysis as described in Materials and Methods. (B) Steady-state levels of TCM1, ACT1, and RP29 mRNA and the stable RNA polymerase III transcript SCR1 in pap1-1 mutants. Total RNA was isolated from strains with the indicated genotypes before and 6 h after a shift to 30°C. mRNA levels were revealed by Northern blot analysis as described in Materials and Methods. The bar graphs represent the ratios of mRNA levels after and before a shift to 30°C and were calculated after normalization of the level of each transcript to the level of SCR1 RNA.
FIG. 3.
RRP6 mutation allows the accumulation of poly(A)+ RNA in pap1-1 strains. Total RNA isolated as in Fig. 2 was 3′-end labeled with 5′-[32P]pCp and T4 RNA ligase. After hydrolysis of non-poly(A) tracts, the RNA was separated by PAGE, and the labeled poly(A) was visualized by storage PhosphorImager analysis.
FIG. 4.
RRP6 mutation allows the accumulation of poly(A)+ RNA in pap1-1 strains. Northern blot analysis of total RNA isolated as in Fig. 2 after fractionation on oligo(dT)-cellulose (54). Lanes 1, 3, 5, and 7 contain RNA that was bound to oligo(dT)-cellulose, and lanes 2, 4, 6, and 8 contain RNA that does not bind. Note that RNAs with poly(A) tails of fewer than ca. 20 nucleotides do not bind to oligo(dT) under these conditions; thus, 40 to 50% of yeast mRNAs are not retained on the resin (54).
Deletion of UPF1 does not increase the levels of TCM1 or RP29 transcripts, but it does reproducibly increase the level of ACT1 mRNA (Fig. 2B). Incomplete resolution of the unspliced and mature forms of the transcript in this experiment leaves open the issue of whether loss of Upf1p preferentially stabilizes unspliced ACT1 mRNA, as it does with other intron-containing mRNAs (30).
Deletion of XRN1/SKI1 results in an increase in the levels of all three test mRNAs (Fig. 2B) but does not increase the levels of poly(A) in a pap1-1 background at 30°C (Fig. 3, lane 8). Moreover, the XRN1/SKI1 deletion results in a relative increase in TCM1 mRNA that does not bind to oligo(dT)-cellulose, suggesting that this mutation results in the accumulation of unadenylated or deadenylated mRNAs (Fig. 4, lane 8). Since loss of Xrn1p/Ski1p activity does not suppress the growth defect of pap1-1 cells under these conditions, and based on the accepted role of Xrn1p/Ski1p as the major 5′-3′ riboexonuclease involved in mRNA degradation (37, 50), we propose that these transcripts accumulate as uncapped, deadenylated intermediates of the normal mRNA decay pathway and that the lack of a cap or poly(A) tail on these transcripts likely results in their inefficient translation.
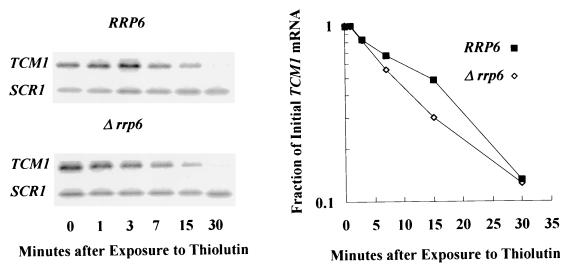
Rrp6p does not play a role in the rate-limiting step of mRNA decay.
The increase in poly(A)+ mRNA levels in the absence of Rrp6p and the homology of the enzyme to E. coli 3′-5′ RNase D (see below) suggest that loss of Rrp6p activity could suppress the pap1-1 defect if the enzyme played a role in the rate-limiting step of poly(A)+ mRNA decay. We tested this hypothesis by measuring the decay rates of several mRNAs after inhibition of RNA polymerase with the drug thiolutin. Northern blot and graphical analyses of TCM1 mRNA decay rates show that the loss of Rrp6p function does not decrease mRNA decay rates (Fig. 5; Table 3). These experiments were carried out with several mRNAs in a PAP1 background and in a pap1-1 background after a shift to 30°C (Table 3); in neither case did the rrp6::URA3 knockout slow the mRNA decay rates. Differential sensitivity of distinct strains to thiolutin could conceivably result in incomplete inhibition of transcription in some strains, resulting in incorrect decay rate values. We do not believe that this problem affects our measurements of the effects of deletion of RRP6 on decay rates since continued transcription would cause an apparent decrease in the measured rate of decay, which we did not observe. We conclude that Rrp6p does not play a role in the rate-limiting step of mRNA degradation and that suppression of the pap1-1 mRNA accumulation defect by rrp6 mutations may occur at a step prior to the major mRNA decay pathway.
FIG. 5.
TCM1 mRNA decay rates in RRP6 and Δ_rrp6_ cells. Shown are the results of a Northern blot analysis of TCM1 mRNA levels in total RNA samples from cells as a function of time after treatment with the transcriptional inhibitor thiolutin. The graph on the right illustrates the decay rates of mRNAs from the two strains, plotted after normalization to the levels of the stable SCR1 RNA.
TABLE 3.
Effect of RRP6 mutation on mRNA half-lives
| Strain (genotype) | mRNA half-life (min)a with: | ||
|---|---|---|---|
| TCM1 | ACT1 | RP29 | |
| PAP1 RRP6 | 14 | 42 | 11 |
| PAP1 rrp6::URA3 | 15 | 38 | 11 |
| pap1-1 RRP6 | 32 | 51 | 39 |
| pap1-1 rrp6::URA3 | 30 | 46 | 40 |
Nuclear localization of Rrp6p.
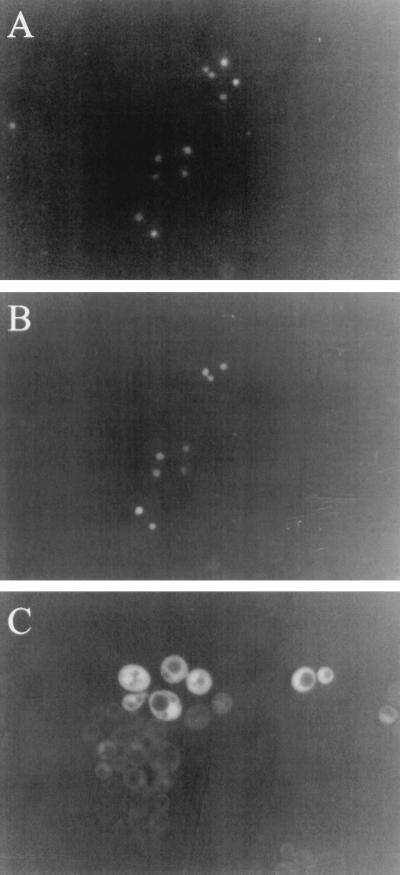
Since Rrp6p defects increase poly(A)+ mRNA levels without slowing mRNA decay, we surmised that the enzyme may play a role in limiting the concentration of unadenylated mRNAs at an early step in mRNA biogenesis. Since mRNA maturation, including polyadenylation, occurs in the nucleus, we determined the subcellular localization of Rrp6p. We fused GFP to the amino terminus of Rrp6p and expressed this fusion from a low-copy plasmid in a strain carrying an rrp6::KAN knockout. Expression of the fusion protein in these cells suppresses both the temperature sensitivity and the 5.8S rRNA processing defect caused by the rrp6::KAN mutation, indicating that the fusion protein functions as Rrp6p (data not shown). Fluorescence microscopy of logarithmically growing cells expressing GFP-Rrp6p and comparison with nuclear DNA staining of the same cells shows that the majority of the protein resides in the nucleus (compare Fig. 6A and B). GFP alone distributes itself evenly between the nucleus and the cytoplasm, but it is excluded from the vacuoles, as expected (Fig. 6C). Although this experiment does not exclude the possibility that some small fraction of Rrp6p resides in the cytoplasm, it does suggest, along with the protein's role in rRNA processing, that its major activity likely takes place within the nucleus.
FIG. 6.
Subcellular localization of GFP-Rrp6p in logarithmically growing yeast cells. Strain BPKAN carrying plasmid pGFP-RRP6-FOR11 (A and B) or pGFP-RRP6-REV2 (C) were grown in synthetic complete medium lacking uracil at 30°C to a density of approximately 106/ml. Green fluorescence (B and C) or Hoechst fluorescence (A) was visualized as described in Materials and Methods.
RRP6 mutation causes a decrease in LA RNA levels.
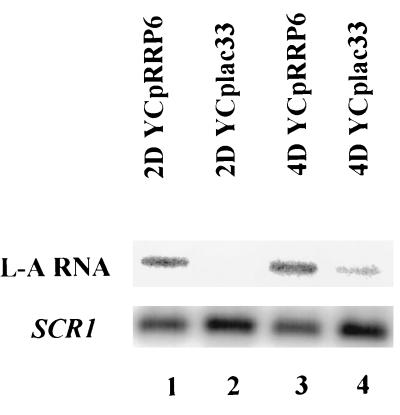
Many strains of S. cerevisiae harbor a dsRNA virus (LA virus) whose life cycle takes place in the cytoplasm (68). LA virus produces uncapped, unadenylated mRNAs that compete with cellular mRNAs during translation initiation. Like cellular mRNAs, the concentration of LA mRNAs is regulated by the cytoplasmic 5′-3′ riboexonuclease encoded by XRN1/SKI1 (33, 36, 37). Mutation of host SKI genes such as XRN1/SKI1, as well as SKI6/RRP41 encoding the exosomal 3′-5′ riboexonuclease Rrp41p, causes an increase in the expression and the amount of LA RNA (8, 60). In contrast, mutations in host MAK genes, many of which affect ribosomal subunit biogenesis, cause a decrease in LA RNA levels (24, 53). We reasoned that if Rrp6p plays a role in degrading unadenylated mRNAs in the cytoplasm, then rrp6 mutants should display an SKI phenotype. We tested this by Northern blot analysis of the levels of LA RNA in rrp6-1 strains and in the same strains complemented by a plasmid-borne copy of RRP6. Instead of an SKI phenotype, loss of Rrp6p activity causes a decrease in LA RNA levels reminiscent of an MAK phenotype (Fig. 7). We suggest that the decrease in LA RNA levels, like that associated with many MAK mutations, results from the decrease in the 60S/40S subunit ratio caused by rrp6-1 (10). These findings, along with the nuclear localization of Rrp6p demonstrated above, support a nuclear role for Rrp6p in RNA processing.
FIG. 7.
RRP6 mutations cause a decrease in LA RNA levels. Northern blot analysis of LA RNA levels from two different rrp6-1 strains carrying the indicated plasmids is shown. LA RNA levels were normalized to SCR1 RNA levels as indicated below.
Rrp6p is a 3′-5′ riboexonuclease.
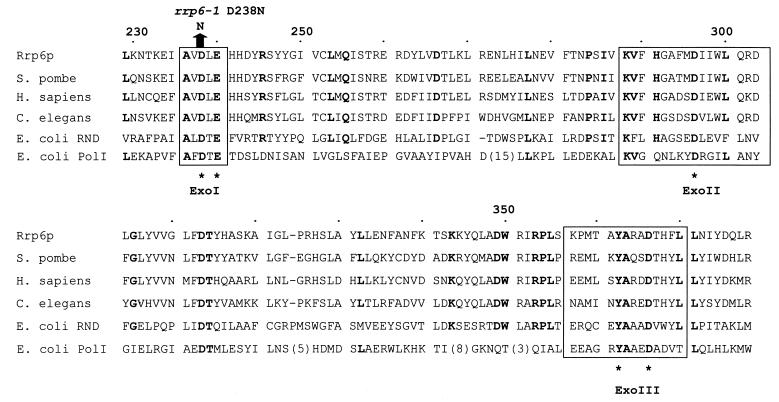
The homology of Rrp6p to the E. coli 3′-5′ riboexonuclease RNase D and the fact that rrp6 mutations result in the accumulation of a 3′-extended form of 5.8S rRNA led us to suggest a 3′-5′ riboexonuclease activity for Rrp6p (10). Figure 8 shows the homology of the core region of Rrp6p with that of a representative group of related proteins, including E. coli RNase D and the 3′-5′ deoxyriboexonuclease domain of E. coli DNA polymerase I (7, 72). Mian and colleagues have pointed out that enzymes of the RNase D class have domains whose sequence and spatial conservation resemble the catalytic domain required for the 3′-5′ deoxyriboexonuclease activity of DNA polymerase I (46, 49). Based on enzymatic analysis of specific amino acid changes in these domains and comparison with the enzyme's crystal structure, Steitz and colleagues proposed a two-metal ligand mechanism for phosphodiester bond cleavage that features the coordination of nucleophilic metal ions by specific amino acid side chains in these domains (7, 23). Nucleotide sequence analysis of rrp6-1 shows that it contains an aspartate-to-asparagine mutation at position 238 which, based on the two-metal ligand mechanism, would inactivate the exonuclease activity of the enzyme. Indeed, the rrp6-1 mutation leads to the accumulation of 3′-extended 5.8S rRNA molecules, a finding consistent with loss of the enzyme's 3′-5′ riboexonuclease activity (10).
FIG. 8.
Comparison of the predicted catalytic core of Rrp6p with homologues from Homo sapiens (PM-Scl 100 kDa; Q01780), Schizosaccharomyces pombe (Q10146), C. elegans (P34607), and E. coli (RNase D, P09155; POL, P00582). Amino acid identities occurring in five of the six homologues are highlighted in boldface. ExoI, ExoII, and ExoIII indicate the portions of the sequence homologous to the exonuclease domains of DNA polymerase I and are set off by boxes. The asterisks below the sequences indicate the positions of amino acids essential for the two-metal ligand mechanism of exonuclease activity of DNA polymerase I. The comparison is adapted from reference 46, which compares a larger set of sequences.
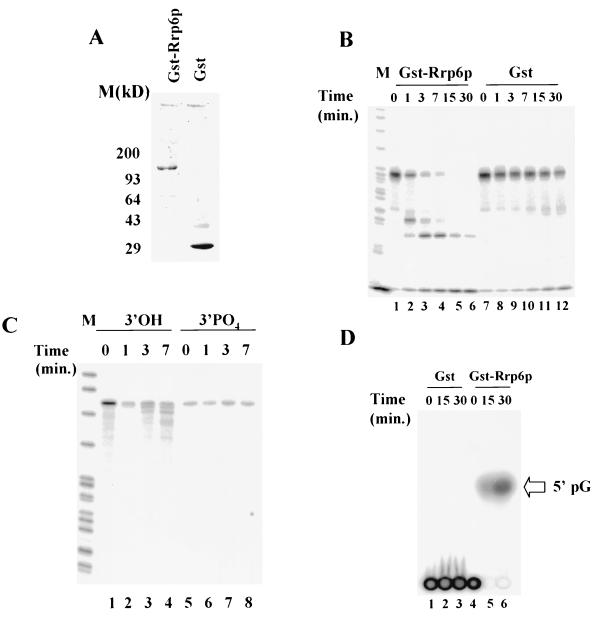
We tested Rrp6p for exonuclease activity by purifying a GST-Rrp6p fusion after expression in E. coli and by incubating it with different 5′-32P-labeled RNAs (Fig. 9). Expression of this GST-Rrp6p fusion in yeast complements the growth defects of an rrp6::KAN mutant, indicating normal function of the fusion protein in vivo (data not shown). Incubation of GST-Rrp6p with a 5′-32P-labeled CYC1 pre-mRNA substrate results in the production of specific degradation products, while GST alone shows little degradation of the substrate (Fig. 9B). The production of increasingly shorter 5′-32P-labeled CYC1 pre-mRNA products as a function of time and the fact that we observe this same pattern of intermediates when the substrate carries 5′-terminal cap (data not shown) imply a 3′-5′ directionality for the enzyme. A different 5′-32P-labeled RNA substrate yields a similar pattern of hydrolysis (Fig. 9C; lanes 1 to 4). However, placement of a 3′ PO4 at the RNA's 3′ end by ligation of 5′-[α-32P]pCp inhibits hydrolysis by GST-Rrp6p (Fig. 9C, lanes 5 to 8). Finally, we incubated GST or GST-Rrp6p with an internally labeled RNA substrate synthesized with SP6 RNA polymerase, unlabeled nucleotide triphosphates, and 5′-[α-32P]GTP (Fig. 9D). Analysis of the reaction products by thin-layer chromatography and comparison to nucleotide monophosphate standards show that Gst-Rrp6p produces a single radiolabeled product that comigrates with 5′pG, the product expected of a hydrolytic exonuclease (Fig. 9D, lanes 5 and 6). Taken together, these findings indicate that Rrp6p hydrolyzes RNA substrates by a 3′-5′ exonucleolytic mechanism. Unfortunately, we were unable to test the activity of the rrp6-1 mutant protein since it is unstable when expressed in E. coli.
FIG. 9.
Exonuclease activity of recombinant Rrp6p. (A) SDS-PAGE analysis of GST-Rrp6p and GST. After gel electrophoretic separation, the gel was stained with SYBRO RED (Molecular Probes) and analyzed by fluorimager analysis. (B) Storage phosphorimager analysis of PAGE separation of the products of incubation of 5′-32P-labeled CYC1 RNA (10 nM) with GST-Rrp6p (0.1 nM; lanes 1 to 6) or GST (0.1 nM; lanes 7 to 12) for the indicated amounts of time at 30°C. (C) Same as in panel B except that the substrate is 5′-32P-labeled E. coli trpt′ RNA (lanes 3 and 4) or trpt′ RNA 3′-end labeled with 5′-[α-32P]pCp (lanes 5 to 8). (D) Thin-layer chromatographic analysis of the products of incubation of GST (0.5 nM; lanes 1 to 3) or GST-Rrp6p (0.5 nM; lanes 4 to 6) with 5′-[α-32P]GTP-labeled CYC1 RNA (10 nM). Reactions were carried out and analyzed as described in Materials and Methods. The arrow at the right of the figure indicates the position of the 5′pG standard included on the thin-layer chromatography plate.
Rrp6p interacts with poly(A) polymerase and the hnRNA protein Npl3p.
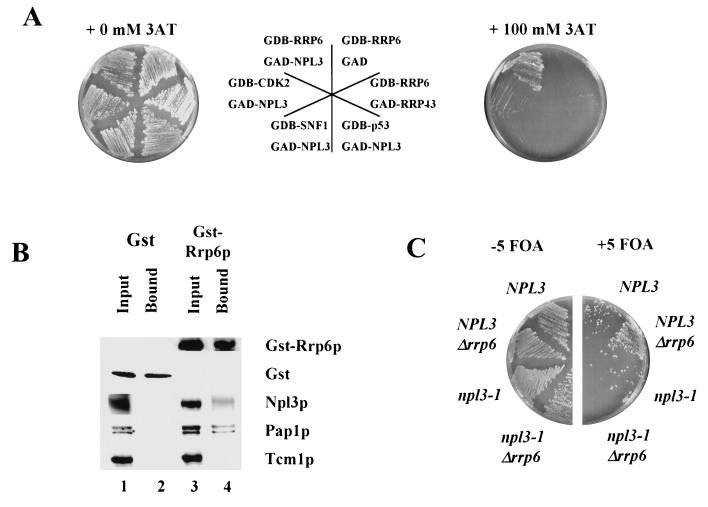
In an effort to extend our understanding of the role of Rrp6p in mRNA processing, we searched for proteins that interact with Rrp6p by using the two-hybrid screen (4). We fused Rrp6p to the DNA binding domain of Gal4p (GDB-RRP6) and screened a library of Gal4p-activation domain fusions (GAD) for their ability to activate transcription of GAL1-lacZ and GAL1-HIS3 reporters. Of 58,000 transformants screened, 20 grew in the presence of 3-AT and produced blue colonies in the presence of X-Gal, indicating the expression of GAL1-HIS3 and GAL1-lacZ, respectively. DNA sequence analysis of the inserts in the candidate plasmids revealed 14 different genes, including one example of NPL3. We chose NPL3 for further study because it is thought to function as an hnRNA protein involved in mRNA processing and transport of mRNA out of the nucleus (43, 63). Moreover, NPL3 interacts directly with mRNA and genetically with RNA15 and HRP1, which encode proteins directly involved in mRNA 3′-end processing (31, 38, 70). Neither GDB-RRP6 nor GAD-NPL3 activates transcription of GAL1-HIS3 on its own, as evidenced by the inability of reporter strains carrying either of these plasmids to grow in the presence of 100 mM 3-AT (Fig. 10A and data not shown). Each of these fusions also fails to promote growth under these conditions in the presence of other fusions, including, in the case of GDB-RRP6, the exosomal riboexonuclease GAD-RRP43 (Fig. 10A) (48). These findings suggest a specific interaction between Rrp6p and Npl3p in vivo (Fig. 10A).
FIG. 10.
Interaction of Rrp6p with Npl3p and Pap1p. (A) Growth at 30°C of strains carrying various GAL4 DNA binding domain (GBD) fusions and GAL4 activation domain (GAD) fusions on plates with or without the addition of 100 mM 3-AT. The diagram in the center indicates the position of each strain on the adjacent plates. (B) Western blot analysis of proteins bound to glutathione-Sepharose 4B beads from lysates of cells expressing GST (lanes 1 and 2) or GST-Rrp6p (lanes 3 and 4). Input (lanes 1 and 3) represents protein extracts prior to incubation with glutathione-Sepharose 4B beads, and Bound (lanes 2 and 4) represents proteins bound after incubation with glutathione-Sepharose 4B beads (see Materials and Methods). Each of the proteins listed to the right of the figure was detected with specific antisera described in Materials and Methods. (C) Synthetic lethality of the combination of npl3-1 and Δ_rrp6_. PSY1 (NPL3), PSY1.RT (NPL3 Δrrp6), PSY773 (npl3-1), and PSY773.RT (npl3-1 Δrrp6) containing YCpRRP6 were grown at 25°C in the absence (−5 FOA) or the presence (+5 FOA) of 1 g of 5-fluoroorotic acid per liter, which selects for the loss of YCpRRP6, thereby revealing the phenotypes of the chromosomal alleles indicated in the figure.
We sought further evidence for an interaction between Rrp6p and Npl3p by assaying for copurification of the two proteins. We constructed a plasmid capable of expressing in yeast cells the same GST-Rrp6p fusion protein used in the experiments illustrated in Fig. 9, and we showed that this plasmid complements the growth defect caused by an RRP6 knockout (data not shown). Affinity purification of GST-Rrp6p from yeast cells resulted in copurification of Npl3p, as judged by Western blot analysis (Fig. 10B, lane 4). GST was efficiently purified by this procedure, but Npl3p did not copurify with it (Fig. 10B, lane 2). Moreover, a negative control large subunit ribosomal protein, Tcm1p, does not copurify with GST or GST-Rrp6p (Fig. 10B, lane 2). These results confirm that Rrp6p and Npl3p interact specifically in vivo.
Next, we investigated the functional consequences of disrupting the interaction between Rrp6p and Npl3p. We knocked out the chromosomal allele of RRP6 in a strain carrying a chromosomal npl3-1 mutation and a plasmid-borne RRP6 allele. Plating such cells on 5-fluoroorotic acid (5 FOA) selects for those that have lost the plasmid and reveals that the combination of the npl3-1 mutation and the RRP6 deletion is lethal (Fig. 10C). Since cells carrying either of these mutations alone survive this test, we conclude that the mutations are synthetically lethal and that Rrp6p and Npl3p interact functionally in vivo.
Because our findings indicated that Rrp6p may function to degrade unadenylated mRNAs and that it exists as part of a complex containing Npl3p, we asked whether Rrp6p also interacts with Pap1p. The affinity purification of Rrp6p illustrated in Fig. 10B shows that Pap1p also copurifies with Rrp6p. We did not identify PAP1 as a result of our two-hybrid screen with GDB-Rrp6p, and others have also failed to identify interaction of Rrp6p with Pap1p by similar methods (22, 55). Thus, Pap1p and Rrp6p may interact together indirectly. Nevertheless, copurification of Rrp6p and Pap1p supports our model, based on the findings presented above, that Rrp6p may function as part of a complex of proteins that monitors the polyadenylation step of mRNA 3′-end processing such that it degrades mRNAs that fail to be polyadenylated.
DISCUSSION
The experimental results reported here provide evidence that the S. cerevisiae gene RRP6 encodes a nuclear 3′-5′ riboexonuclease that interacts with the mRNA polyadenylation apparatus and degrades unadenylated mRNAs. This conclusion is based on four major findings. First, RRP6 mutations, including a deletion of RRP6, partially restore the steady-state levels of poly(A)+ mRNAs, which decrease after a shift of pap1-1 strains to the nonpermissive temperature, and this increase occurs without slowing a rate-limiting step in mRNA decay. Second, subcellular localization shows that the majority of Rrp6p resides in the nucleus. Third, recombinant Rrp6p demonstrates 3′-5′ riboexonucleolytic activity, and the rrp6-1 mutation, which allows accumulation of poly(A)+ mRNA in pap1-1 cells, alters an amino acid predicted to be essential for exonuclease activity. Fourth, biochemical and genetic experiments provide evidence that Rrp6p interacts with Pap1p and with Npl3p, a poly(A)+ RNA binding protein that plays a role in mRNA export. Thus, our results suggest the existence of a novel nuclear degradation pathway that appears to monitor the integrity of pre-mRNAs prior to export to the cytoplasm.
Evidence for a nuclear mRNA degradation pathway.
Loss of Rrp6p function leads to increased levels of poly(A)+ mRNA after inhibition of poly(A) polymerase activity, suggesting that Rrp6p may normally act to limit the concentration of unadenylated mRNA in the nucleus. Several of our findings suggest that Rrp6p carries out this function by a mechanism distinct from the major mRNA degradation pathway in yeast. First, the majority of Rrp6p resides in the nucleus, while the mRNA turnover systems featuring Ufp1p and Xrn1p/Ski1p function in the cytoplasm (3, 33, 36). Second, loss of Rrp6p function results in the accumulation of poly(A)+ mRNA in a pap1-1 background, while loss of Upf1p function does not restore mRNA levels, and loss of Xrn1p/Ski1p enhances the accumulation of unadenylated or poly(A)-deficient mRNAs. Third, loss of Rrp6p function causes a decrease in cytoplasmic LA RNA levels, while mutations affecting the exonucleases Xrn1p/Ski1p and Ski6p/Rrp41p result in increased levels of LA RNA (8, 60). Xrn1p/Ski1p and the exosomal Ski6p/Rrp41p appear to limit LA RNA levels by virtue of their respective 5′-3′ and 3′-5′ riboexonuclease activities. In contrast, many MAK mutations, like RRP6 mutations, limit 60S ribosome levels and cause decreased LA RNA levels (53). We suggest that the negative effect of RRP6 mutations on LA RNA levels arises as an indirect consequence of the imbalance in ribosome subunit levels caused by the requirement for Rrp6p in efficient 5.8S rRNA 3′-end processing (10). Xrn1p/Ski1p and Ski6p/Rrp41p play essential roles, respectively, in the 5′-3′ and 3′-5′ degradation pathways common to many, if not all, yeast mRNAs (2, 21). Mutations inactivating either of these enzymes cause an increase in the half-lives of cellular mRNAs. In contrast, we found that mutations inactivating Rrp6p do not increase mRNA half-lives, suggesting that Rrp6p does not play a role in cytoplasmic mRNA decay.
An alternative to our model arises from the fact that loss of Rrp6p activity inhibits 60S ribosome subunit biogenesis, resulting in a decrease in the 60S/40S subunit ratio (10). Previous studies showed that some mutations that alter ribosomal subunit ratios suppress poly(A)-binding (Pab1p) protein defects, implying that such subunit deficiencies might bypass the loss of poly(A) tail function in vivo (61, 65). Several lines of evidence argue against this notion. First, MAK mutations, many of which decrease 60S ribosomal subunit levels, result in decreased levels of unadenylated LA virus RNAs (53). Second, mutation of LCP1, which leads to decreased levels of 40S ribosomal subunits, causes synthetic lethality in the presence of PAP1 mutations (69). Third, mutations that decrease 60S or 40S subunit levels inhibit translation of unadenylated mRNAs produced in pap1-1 strains, and one of these mutations (spb2) fails to suppress the growth defect caused by this mutation (Fig. 1) (57). These observations suggest that alterations in ribosomal subunit levels inhibit the translation of unadenylated mRNAs and therefore cannot bypass the requirement for poly(A) tails in mRNA function.
In S. cerevisiae, mRNA 3′-end processing produces poly(A) tails of between 60 and 90 adenosines in length (14). Cleavage of pre-mRNAs occurs at the nonpermissive temperature of 37°C in pap1-1 strains but results in undetectable steady-state levels of many mRNAs and low levels of other unadenylated mRNAs (54, 58). Evidence presented here suggests that Pap1p remains partially active at the nonpermissive temperature of 30°C since we detected low levels of poly(A)+ mRNAs and because deletion of RRP6 results in the accumulation of mRNA with normal poly(A) tails. The fact that our suppressors allow growth of pap1-1 cells at 30°C but not at 37°C (11) suggests that these mutations do not bypass the requirement for poly(A) polymerase but instead somehow enhance the rate of mRNA polyadenylation.
Alternatively, the increase in poly(A)+ mRNA in rrp6 mutants could conceivably result from the loss of deadenylase activity. Our data indicate that Rrp6p is a hydrolytic 3′-5′ riboexonuclease, like its bacterial homologue RNase D (17). Indeed, Rrp6p belongs to a small group of RNase D homologues, including the poly(A) nuclease subunit Pan2p and the vertebrate deadenylase PARN, that play roles in mRNA processing in eukaryotes (9, 39). After transport to the cytoplasm, an uncharacterized deadenylation system removes adenosines at mRNA-specific rates, resulting in populations of transcripts with tail lengths between 10 and 90 nucleotides (see Fig. 3 and reference 50). Since deadenylation comprises the rate-limiting step in the decay of some mRNAs, loss of poly(A) nuclease activity could increase the steady-state levels of a polyadenylated population of mRNAs and, thereby, suppress a partial polyadenylation defect. Several observations argue against a role for Rrp6p in deadenylation. First, Rrp6p resides largely in the nucleus, while deadenylation of mRNAs occurs, most likely, in the cytoplasm. Second, deadenylation continues in strains carrying a deletion of RRP6, as evidenced by the fact that poly(A) profiles from such strains appear normal (Fig. 3). Third, recombinant Rrp6p shows no preference for hydrolyzing poly(A)+ RNAs (data not shown). Finally, the fact that deletion of RRP6 does not increase the steady-state levels of mRNAs in the absence of a polyadenylation defect argues against a role for Rrp6p as a general inhibitor of transcription. These considerations and our results discussed above suggest that Rrp6p limits poly(A)− mRNA levels by degrading such transcripts in the nucleus.
Rrp6p as component of the nuclear exosome.
The recent discovery of a complex of riboexonucleases called the exosome and the demonstration that members of this complex play a role in rRNA and mRNA processing provides evidence for the potential coregulation of the processing of these RNAs (2, 20, 48). The exosome appears to contain 10 3′-5′ riboexonucleases, including Rrp6p, which participate in 5.8S rRNA 3′-end processing (1). One of these riboexonucleases, Rrp41p/Ski6p, along with Ski2p, Ski3p, and Ski8p, also plays a role in the 3′-5′ mRNA turnover pathway (2). The SKI gene products were originally discovered by virtue of their role in controlling the expression of yeast cytoplasmic dsRNA viruses by limiting the levels and translation of uncapped, unadenylated viral mRNAs (8, 45). Interestingly, efficient 5.8S rRNA 3′-end processing requires Rrp41p/Ski6p, but not Ski2p, Ski3p, or Ski8p. This observation led to the hypothesis that Ski2p, Ski3p, and Ski8p serve as adapters that facilitate degradation of cytoplasmic mRNA by a core exosomal complex containing Rrp41p/Ski6p (2). Consistent with the exosome's ability to process pre-rRNA and mRNA substrates, the complex copurifies with nuclear and cytoplasmic fractions from human cells (48). Rrp6p differs from other known exosomal components in that (i) its localization appears to be exclusively nuclear, (ii) it is not essential for viability, and (iii) loss of its activity yields a single aberrant 5.8S rRNA precursor (1, 10). Moreover, our findings indicate that Rrp6p degrades unadenylated mRNAs and interacts with Pap1p and Npl3p, neither of which appears to be associated with the exosome. These considerations raise the possibility that Rrp6p functions independently of the exosome in addition to its exosomal role in rRNA processing. Thus, Rrp6p may process different types of RNA molecules depending on its interaction with an adapter such as Npl3p or Pap1p in the case of mRNA or as a core component of the nuclear exosome in the case of rRNA.
Rrp6p as a monitor of mRNA 3′-end processing.
The interaction of Rrp6p with Pap1p and Npl3p provides further evidence for a role for Rrp6p in a nuclear pathway of mRNA decay. Npl3p is a predominantly nuclear, mRNA binding protein implicated in the transport of mRNA to the cytoplasm (25, 43, 63). Genetic interactions between Npl3p and the mRNA 3′-end formation factors Rna15p and Hrp1p suggest that Npl3p may function as part of an mRNP substrate during the polyadenylation and export phases of mRNA biogenesis (31, 38). Perhaps Rrp6p and Npl3p interact together to monitor this complex prior to export in a way that allows Rrp6p to degrade incorrectly or incompletely processed mRNAs. Surveillance of this complex by Rrp6p and the subsequent export of the normal mRNP would require Npl3p and, presumably, other nuclear export factors. Thus, the synthetic lethality observed between RRP6 and NPL3 mutations may reflect the failure to form such a complex, which, in turn, slows nuclear mRNA export below the threshold of viability.
Our findings suggest that Rrp6p degrades mRNAs that fail to be efficiently polyadenylated and may, therefore, constitute part of a system that limits the accumulation of unadenylated mRNAs in the nucleus. The copurification of Pap1p and Rrp6p supports the idea that Rrp6p interacts with the polyadenylation apparatus. This interaction could provide the cell with a mechanism for recognizing and destroying prematurely terminated or damaged mRNAs. mRNAs lacking a bona fide 3′ end could prove toxic to the cell if truncation of the transcript occurred within the coding sequence or within an internal intron. Translation of such incomplete messages could potentially produce incomplete polypeptides that have dominant-negative effects on cell metabolism. In an evolutionary sense, the benefits to a cell of a system that degrades truncated mRNAs would parallel those ascribed to the nonsense-mediated decay pathway, which limits the levels of mRNAs bearing stop codons early in their translational reading frames (15, 18). The evidence presented thus far in favor of such a role for Rrp6p limits this hypothesis to the realm of speculation. However, we believe that this working model best explains the evidence, and our present efforts are focused on further characterization of the role of Rrp6p in RNA processing.
ACKNOWLEDGMENTS
We thank Michael Briggs, Mark Dumont, Beth Grayhack, Eric Phizicky, Fred Sherman, and Terry Platt for helpful discussions and Terry Platt, Biswadip Das, Roy Parker, and the members of our laboratory for comments on the manuscript. We are grateful to Geoff Dance, Dave Goldfarb, Elizabeth Grayhack, Mark Martzen, Pam Silver, Maurice Swanson, Jon Warner, and Nilsen Zanchin for providing antibodies, plasmids, and strains, to Thomas “Trey” Westbrook for constructing pAS2-RRP6, and to Mark Burkard and Sherry Spinelli for help with thin-layer chromatography analysis.
This work was supported by grants from the National Science Foundation (MCB 9603893) and the National Institutes of Health (GM 59898) to J.S.B.
REFERENCES
- 1.Allmang C, Petfalski E, Podtelejnikov A, Mann M, Tollervey D, Mitchell P. The yeast exosome and human PM-Scl are related complexes of 3′→5′ exonucleases. Genes Dev. 1999;13:2148–2158. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson J S J, Parker R P. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atkin A L, Altamura N, Leeds P, Culbertson M R. The majority of yeast UPF1 co-localizes with polyribosomes in the cytoplasm. Mol Biol Cell. 1995;6:611–625. doi: 10.1091/mbc.6.5.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bai C, Elledge S J. Gene identification using the yeast two-hybrid system. Methods Enzymol. 1997;283:141–156. doi: 10.1016/s0076-6879(97)83013-3. [DOI] [PubMed] [Google Scholar]
- 5.Bartel P L, Fields S. Analyzing protein-protein interactions using two-hybrid system. Methods Enzymol. 1995;254:241–263. doi: 10.1016/0076-6879(95)54018-0. [DOI] [PubMed] [Google Scholar]
- 6.Beelman C A, Stevens A, Caponigro G, LaGrandeur T E, Hatfield L, Fortner D M, Parker R. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature. 1996;382:642–646. doi: 10.1038/382642a0. [DOI] [PubMed] [Google Scholar]
- 7.Beese L S, Steitz T A. Structural basis for the 3′-5′ exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J. 1991;10:25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benard L, Carroll K, Valle R C, Wickner R B. Ski6p is a homolog of RNA-processing enzymes that affects translation of non-poly(A) mRNAs and 60S ribosomal subunit biogenesis. Mol Cell Biol. 1998;18:2688–2696. doi: 10.1128/mcb.18.5.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boeck R, Tarun S, Jr, Rieger M, Deardorff J A, Muller-Auer S, Sachs A B. The yeast Pan2 protein is required for poly(A)-binding protein-stimulated poly(A)-nuclease activity. J Biol Chem. 1996;271:432–438. doi: 10.1074/jbc.271.1.432. [DOI] [PubMed] [Google Scholar]
- 10.Briggs M W, Burkard K T, Butler J S. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation. J Biol Chem. 1998;273:13255–13263. doi: 10.1074/jbc.273.21.13255. [DOI] [PubMed] [Google Scholar]
- 11.Briggs M W, Butler J S. RNA polymerase III defects suppress a conditional-lethal poly(A) polymerase mutation in Saccharomyces cerevisiae. Genetics. 1996;143:1149–1161. doi: 10.1093/genetics/143.3.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butler J S, Briggs M W, Proweller A. Analysis of polyadenylation phenotypes in Saccharomyces cerevisiae. In: Richter J, editor. mRNA formation and function. New York, N.Y: Academic Press; 1997. pp. 111–124. [Google Scholar]
- 13.Butler J S, Platt T. RNA processing generates the mature 3′ end of yeast CYC1 messenger RNA in vitro. Science. 1988;242:1270–1274. doi: 10.1126/science.2848317. [DOI] [PubMed] [Google Scholar]
- 14.Butler J S, Sadhale P P, Platt T. RNA processing in vitro produces mature 3′ ends of a variety of Saccharomyces cerevisiae mRNAs. Mol Cell Biol. 1990;10:2599–2605. doi: 10.1128/mcb.10.6.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cali B M, Anderson P. mRNA surveillance mitigates genetic dominance in Caenorhabditis elegans. Mol Gen Genet. 1998;260:176–184. doi: 10.1007/s004380050883. [DOI] [PubMed] [Google Scholar]
- 16.Caponigro G, Parker R. Multiple functions for the poly(A)-binding protein in mRNA decapping and deadenylation in yeast. Genes Dev. 1995;9:2421–2432. doi: 10.1101/gad.9.19.2421. [DOI] [PubMed] [Google Scholar]
- 17.Cudny H, Zaniewski R, Deutscher M P. Escherichia coli RNase D. Catalytic properties and substrate specificity of Escherichia coli RNase D. Purification and structural characterization of a putative processing nuclease. J Biol Chem. 1981;256:5633–5637. [PubMed] [Google Scholar]
- 18.Culbertson M R. RNA surveillance. Unforeseen consequences for gene expression, inherited genetic disorders and cancer. Trends Genet. 1999;15:74–80. doi: 10.1016/s0168-9525(98)01658-8. [DOI] [PubMed] [Google Scholar]
- 19.Curtis D, Lehman R, Zamore P D. Translational regulation in development. Cell. 1995;81:171–178. doi: 10.1016/0092-8674(95)90325-9. [DOI] [PubMed] [Google Scholar]
- 20.Decker C J. The exosome: a versatile RNA processing machine. Curr Biol. 1998;8:R238–R240. doi: 10.1016/s0960-9822(98)70149-6. [DOI] [PubMed] [Google Scholar]
- 21.Decker C J, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 22.del Olmo M, Mizrahi N, Gross S, Moore C L. The Uba2 and Ufd1 proteins of Saccharomyces cerevisiae interact with poly(A) polymerase and affect the polyadenylation activity of cell extracts. Mol Gen Genet. 1997;255:209–218. doi: 10.1007/s004380050491. [DOI] [PubMed] [Google Scholar]
- 23.Derbyshire V, Freemont P S, Sanderson M R, Beese L, Friedman J M, Joyce C M, Steitz T A. Genetic and crystallographic studies of the 3′,5′-exonucleolytic site of DNA polymerase I. Science. 1988;240:199–201. doi: 10.1126/science.2832946. [DOI] [PubMed] [Google Scholar]
- 24.Edskes H K, Ohtake Y, Wickner R B. Mak21p of Saccharomyces cerevisiae, a homolog of human CAATT-binding protein, is essential for 60S ribosomal subunit biogenesis. J Biol Chem. 1998;273:28912–28920. doi: 10.1074/jbc.273.44.28912. [DOI] [PubMed] [Google Scholar]
- 25.Flach J, Bossie M, Vogel J, Corbett A, Jinks T, Willins D A, Silver P A. A yeast RNA-binding protein shuttles between the nucleus and the cytoplasm. Mol Cell Biol. 1994;14:8399–8407. doi: 10.1128/mcb.14.12.8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gallie D R. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 27.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 28.Gillian-Daniel D L, Gray N K, Astrom J, Barkoff A, Wickens M. Modifications of the 5′ cap of mRNAs during Xenopus oocyte maturation: independence from changes in poly(A) length and impact on translation. Mol Cell Biol. 1998;18:6152–6163. doi: 10.1128/mcb.18.10.6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guldener U, Heck S, Fielder T, Beinhauer J, Hegemann J H. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He F, Peltz S W, Donahue J L, Rosbash M, Jacobson A. Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1− mutant. Proc Natl Acad Sci USA. 1993;90:7034–7038. doi: 10.1073/pnas.90.15.7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henry M, Borland C Z, Bossie M, Silver P A. Potential RNA binding proteins in Saccharomyces cerevisiae identified as suppressors of temperature-sensitive mutations in NPL3. Genetics. 1996;142:103–115. doi: 10.1093/genetics/142.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herrick D, Parker R, Jacobson A. Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:2269–2284. doi: 10.1128/mcb.10.5.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heyer W D, Johnson A W, Reinhart U, Kolodner R D. Regulation and intracellular localization of Saccharomyces cerevisiae strand exchange protein 1 (Sep1/Xrn1/Kem1), a multifunctional exonuclease. Mol Cell Biol. 1995;15:2728–2736. doi: 10.1128/mcb.15.5.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsu C L, Stevens A. Yeast cells lacking 5′→3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol Cell Biol. 1993;13:4826–4835. doi: 10.1128/mcb.13.8.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jacobson A, Peltz S W. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu Rev Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- 36.Johnson A W. Rat1p and Xrn1p are functionally interchangeable exoribonucleases that are restricted to and required in the nucleus and cytoplasm, respectively. Mol Cell Biol. 1997;17:6122–6130. doi: 10.1128/mcb.17.10.6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson A W, Kolodner R D. Synthetic lethality of sep1 (xrn1) ski2 and sep1 (xrn1) ski3 mutants of Saccharomyces cerevisiae is independent of killer virus and suggests a general role for these genes in translation control. Mol Cell Biol. 1995;15:2719–2727. doi: 10.1128/mcb.15.5.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kessler M M, Henry M F, Shen E, Zhao J, Gross S, Silver P A, Moore C L. Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′-end formation in yeast. Genes Dev. 1997;11:2545–2556. doi: 10.1101/gad.11.19.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Korner C G, Wormington M, Muckenthaler M, Schneider S, Dehlin E, Wahle E. The deadenylating nuclease (DAN) is involved in poly(A) tail removal during the meiotic maturation of Xenopus oocytes. EMBO J. 1998;17:5427–5437. doi: 10.1093/emboj/17.18.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuge H, Richter J D. Cytoplasmic 3′ poly(A) addition induces 5′ cap ribose methylation: implications for translational control of maternal mRNA. EMBO J. 1995;14:6301–6310. doi: 10.1002/j.1460-2075.1995.tb00320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 42.Larimer F W, Stevens A. Disruption of the gene XRN1, coding for a 5′----3′ exoribonuclease, restricts yeast cell growth. Gene. 1990;95:85–90. doi: 10.1016/0378-1119(90)90417-p. [DOI] [PubMed] [Google Scholar]
- 43.Lee M S, Henry M, Silver P A. A protein that shuttles between the nucleus and the cytoplasm is an important mediator of RNA export. Genes Dev. 1996;10:1233–1246. doi: 10.1101/gad.10.10.1233. [DOI] [PubMed] [Google Scholar]
- 44.Leeds P, Peltz S W, Jacobson A, Culbertson M R. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 1991;5:2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- 45.Masison D C, Blanc A, Ribas J C, Carroll K, Sonenberg N, Wickner R B. Decoying the cap-mRNA degradation system by a double-stranded RNA virus and poly(A)-mRNA surveillance by a yeast antiviral system. Mol Cell Biol. 1995;15:2763–2771. doi: 10.1128/mcb.15.5.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mian I S. Comparative sequence analysis of ribonucleases HII, III, II PH and D. Nucleic Acids Res. 1997;25:3187–3195. doi: 10.1093/nar/25.16.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mitchell D A, Marshall T K, Deschenes R J. Vectors for the inducible overexpression of glutathione S-transferase fusion proteins in yeast. Yeast. 1993;9:715–722. doi: 10.1002/yea.320090705. [DOI] [PubMed] [Google Scholar]
- 48.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 49.Moser M J, Holley W R, Chatterjee A, Mian I S. The proofreading domain of Escherichia coli DNA polymerase I and other DNA and/or RNA exonuclease domains. Nucleic Acids Res. 1997;25:5110–5118. doi: 10.1093/nar/25.24.5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muhlrad D, Decker C J, Parker R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′→3′ digestion of the transcript. Genes Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- 51.Muhlrad D, Parker R. Premature translational termination triggers mRNA decapping. Nature. 1994;370:578–581. doi: 10.1038/370578a0. [DOI] [PubMed] [Google Scholar]
- 52.Niedenthal R K, Riles L, Johnston M, Hegemann J H. Green fluorescent protein as a marker for gene expression and subcellular localization in budding yeast. Yeast. 1996;12:773–786. doi: 10.1002/(SICI)1097-0061(19960630)12:8%3C773::AID-YEA972%3E3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 53.Ohtake Y, Wickner R B. Yeast virus propagation depends critically on free 60S ribosomal subunit concentration. Mol Cell Biol. 1995;15:2772–2781. doi: 10.1128/mcb.15.5.2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Patel D, Butler J S. Conditional defect in mRNA 3′ end processing caused by a mutation in the gene for poly(A) polymerase. Mol Cell Biol. 1992;12:3297–3304. doi: 10.1128/mcb.12.7.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Preker P J, Lingner J, Minvielle-Sebastia L, Keller W. The FIP1 gene encodes a component of a yeast pre-mRNA polyadenylation factor that directly interacts with poly(A) polymerase. Cell. 1995;81:379–389. doi: 10.1016/0092-8674(95)90391-7. [DOI] [PubMed] [Google Scholar]
- 56.Proweller A, Butler J S. Ribosomal association of poly(A)-binding protein in poly(A)-deficient Saccharomyces cerevisiae. J Biol Chem. 1996;271:10859–10865. doi: 10.1074/jbc.271.18.10859. [DOI] [PubMed] [Google Scholar]
- 57.Proweller A, Butler J S. Ribosome concentration contributes to discrimination against poly(A)− mRNA during translation initiation in Saccharomyces cerevisiae. J Biol Chem. 1997;272:6004–6010. doi: 10.1074/jbc.272.9.6004. [DOI] [PubMed] [Google Scholar]
- 58.Proweller A, Butler S. Efficient translation of poly(A)-deficient mRNAs in Saccharomyces cerevisiae. Genes Dev. 1994;8:2629–2640. doi: 10.1101/gad.8.21.2629. [DOI] [PubMed] [Google Scholar]
- 59.Pulak R, Anderson P. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 1993;7:1885–1897. doi: 10.1101/gad.7.10.1885. [DOI] [PubMed] [Google Scholar]
- 60.Ridley S P, Sommer S S, Wickner R B. Superkiller mutations in Saccharomyces cerevisiae suppress exclusion of M2 double-stranded RNA by L-A-HN and confer cold sensitivity in the presence of M and L-A-HN. Mol Cell Biol. 1984;4:761–770. doi: 10.1128/mcb.4.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sachs A B, Davis R W. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989;58:857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- 62.Schiestl R H, Gietz R D. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 63.Singleton D R, Chen S, Hitomi M, Kumagai C, Tartakoff A M. A yeast protein that bidirectionally affects nucleocytoplasmic transport. J Cell Sci. 1995;108:265–272. doi: 10.1242/jcs.108.1.265. [DOI] [PubMed] [Google Scholar]
- 64.Skryabin K G, Eldarov M A, Larionov V L, Bayev A A, Klootwijk J, de Regt V C, Veldman G M, Planta R J, Georgiev O I, Hadjiolov A A. Structure and function of the nontranscribed spacer regions of yeast rDNA. Nucleic Acids Res. 1984;12:2955–2968. doi: 10.1093/nar/12.6.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tarun S Z, Jr, Sachs A B. A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes Dev. 1995;9:2997–3007. doi: 10.1101/gad.9.23.2997. [DOI] [PubMed] [Google Scholar]
- 66.Tarun S Z, Jr, Sachs A B. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 67.Wickens M, Anderson P, Jackson R J. Life and death in the cytoplasm: messages from the 3′ end. Curr Opin Genet Dev. 1997;7:220–232. doi: 10.1016/s0959-437x(97)80132-3. [DOI] [PubMed] [Google Scholar]
- 68.Wickner R B. Double-stranded RNA viruses of Saccharomyces cerevisiae. Microbiol Rev. 1996;60:250–265. doi: 10.1128/mr.60.1.250-265.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wiederkehr T, Pretot R F, Minvielle-Sebastia L. Synthetic lethal interactions with conditional poly(A) polymerase alleles identify LCP5, a gene involved in 18S rRNA maturation. RNA. 1998;4:1357–1372. doi: 10.1017/s1355838298980955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wilson S M, Datar K V, Paddy M R, Swedlow J R, Swanson M S. Characterization of nuclear polyadenylated RNA-binding proteins in Saccharomyces cerevisiae. J Cell Biol. 1994;127:1173–1184. doi: 10.1083/jcb.127.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zanchin N I T, Goldfarb D S. Nip7p interacts with Nop8p, an essential nucleolar protein required for 60S ribosome biogenesis, and the exosome subunit Rrp43p. Mol Cell Biol. 1999;19:1518–1525. doi: 10.1128/mcb.19.2.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang J R, Deutscher M P. Escherichia coli RNase D: sequencing of the rnd structural gene and purification of the overexpressed protein. Nucleic Acids Res. 1988;16:6265–6278. doi: 10.1093/nar/16.14.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]