Translation of NRF mRNA Is Mediated by Highly Efficient Internal Ribosome Entry (original) (raw)
Abstract
The ubiquitous transcription factor NRF (NF-κB repressing factor) is a constitutive transcriptional silencer of the multifunctional cytokine interferon-β. NRF mRNA contains a long 5′ untranslated region (5′UTR) predicted to fold into a strong secondary structure. The presence of stable hairpins is known to be incompatible with efficient translation by ribosomal scanning. Using dicistronic reporter gene constructs, we show that the NRF 5′UTR acts as an internal ribosome entry site (IRES) which directs ribosomes to the downstream start codon by a cap-independent mechanism. The relative activity of this IRES in various cell lines is at least 30-fold higher than that of picornaviral IRESs. The NRF 5′UTR also functions as a translational enhancer in the context of monocistronic mRNAs. Our results indicate that the NRF 5′UTR contains a highly potent IRES, which may allow for an alternate mode of translation under physiological conditions in which cap-dependent translation is inhibited.
NRF (NF-κB repressing factor) was identified as a constitutively expressed silencer protein which binds to the beta interferon (IFN-β) promoter and represses the basal transcription of this gene (12). IFN-β belongs to a group of inducible cytokines which mediate antiviral defense, immune activation, and cell growth regulation. Based on these important biological activities, a sufficient level of NRF protein is essential for the complete silencing of the IFN-β gene and thus for cell viability. For example, reduction of NRF protein level through expression of NRF antisense RNA results in basal activation of IFN-β gene transcription (13).
Transcription of the NRF gene generates two mRNAs of 3.7 and 2.8 kb (13). Gene mapping analysis revealed that both mRNAs begin at a single transcription initiation site and differ only in their use of alternate polyadenylation sites in their 3′ untranslated regions (UTRs) (13). The 5′UTR of NRF is unusually long (653 nucleotides), contains 11 AUG codons, and is predicted to fold into a complex secondary structure (−126.3 kcal/mol) with several stable hairpins (Fig. 1). Some of the 5′-proximal AUG codons are in a favorable context for initiation and are followed by open reading frames as long as 30 codons.
FIG. 1.
Sequence and secondary structure of NRF 5′UTR (EMBL accession number AJO11812). Eleven open reading frames are shown as arrows; the initiating AUG is shown in boldface. The secondary structure was predicted by using Zuker's RNAFOLD (25).
In contrast to the NRF 5′UTR, the 5′UTRs of typical mammalian genes are relatively short (∼150 nucleotides is the average length), lack AUGs, and do not contain stable secondary structures. Eukaryotic mRNAs are generally translated by a mechanism known as cap-dependent translation initiation. This mechanism is characterized by ribosomes scanning from the capped mRNA 5′ end along the mRNA molecule in the 3′ direction until they recognize an AUG codon in a favorable context. The limiting step of this process is ribosome binding to the cap structure, since it depends on the cap-binding eukaryotic initiation factor 4E (eIF4E), which is present in only small amounts in cells (3). The selection of a particular mRNA from the pool of translatable mRNAs is determined by the relative efficiency by which eIF4E binds to its cap structure and by the efficiency of translation initiation by ribosome scanning, which is governed largely by the composition and structure of the 5′UTR of the mRNA (19). The presence of upstream AUG codons and stable secondary structures is known to interfere with ribosome scanning and to inhibit translation initiation at the authentic AUG start site (19).
At least two initiation mechanisms which are able to bypass extraordinarily long 5′UTRs have been described, shunting and internal ribosome entry (23). Shunting is also cap dependent but differs from scanning in that the 40S subunits bypass the majority of the 5′UTR by shunting or jumping to a region at or near the authentic site of translation initiation. Internal ribosome entry involves binding of the 40S ribosomal subunits to an internal ribosome entry site (IRES) at or near upstream of the authentic AUG. This mechanism does not require a 5′ cap structure or scanning through the greater part of the 5′UTR. Initially, translation initiation by internal ribosome entry was proposed for the mRNAs of picornaviruses (9). These viruses produce noncapped transcripts with long 5′UTRs containing multiple noninitiating AUG codons. The infective strategy of some picornaviruses involves inactivation of the essential cap-binding complex eIF4F and hence the capture of the cellular translation apparatus for their own use. The function of these IRESs does not depend on viral gene products but relies on interaction with cellular proteins (10). Indeed, a number of IRESs have also been identified in cellular mRNAs (e.g., fibroblast growth factor, vascular endothelial growth factor, immunoglobulin-binding, protein Bip, proto-oncoprotein c-Myc, and voltage-gated potassium channel), but they exert a lower activity than the viral IRESs (11, 20–23). While the advantages of internal initiation in viral mRNAs are clear, the rationales for maintaining cap-independent translation by cellular IRESs are not fully understood.
IRESs have been categorized into three groups (17). Type III IRESs are extremely poor candidates for dicistronic expression in vivo, whereas types I and II function efficiently. The main difference between type I and type II is the location of the IRES with respect to the initiating AUG. Type I IRESs, for example, poliovirus IRES, can be located as far as 50 to 100 nucleotides upstream of the initiation codon. Type II IRESs, for example, encephalomyocarditis virus (EMCV) IRES, include the initiation codon at their 3′ boundary.
In principle, mRNAs containing unusually long leader sequences with multiple short upstream reading frames are good candidates for translation initiating via a cap-independent internal ribosome-binding mechanism (10). This led us to examine whether the translation initiation of NRF may occur by a mechanism other than 5′ cap-dependent scanning. Here we report that the 5′UTR of NRF contains an IRES that can initiate efficient translation of a reporter protein in a cap-independent fashion. We propose that translation initiation by internal ribosome entry may represent a general and important mechanism for the regulation of NRF expression.
MATERIALS AND METHODS
Expression plasmids.
The reporter plasmids are derivatives of the expression plasmid pSBC-1, which contains the simian virus 40 (SV40) enhancer/promoter and the poliovirus IRES (2). Renilla reniformis luciferase and Photinus pyralis firefly luciferase coding sequences were PCR amplified from pRL (Promega) and pL (2) and inserted into the _Eco_RI and _Not_I sites of pSBC-1 to obtain the dicistronic pBSRPF. PCR amplification was carried out with a Taq DNA polymerase possessing a proofreading activity (PWO DNA polymerase; Boehringer) and the oligonucleotide primers Renilla luciferase 5′ primer (5′CGAATTCGAAATGACTTCGAAAG3′) and Renilla luciferase 3′ primer (5′CGAATTCTTATTGTTCATTTTTC3′) or firefly luciferase 5′ primer (5′GCGGCCGCATGGAAGACGCC3′) and firefly luciferase 3′ primer (5′GCGGCCGCTTACAATTTGGACTTTCC3′). The poliovirus IRES in pBSRPF was replaced by the EMCV IRES (pCREL; kindly provided by L. Creancier, Institut Federatif de Recherche Louis Bugnard, Toulouse, France) or the NRF 5′UTR, which were amplified by PCR as described above with the EMCV UTR 5′ primer (5′GCGGCCGCGAAATTAATACGACTCAC3′) and EMCV UTR 3′ primer (5′CCATGGTATCATCGTGTTTTTCAAAGG3′) or the NRF UTR 5′ primer (5′CGAATTCCACGAGCAGAGTAATGACATGG3′) and NRF UTR 3′ primer (5′GCGGCCGCCAAGCGTGGGCTGTACC3′) (corresponding to nucleotides 1 to 22 and 637 to 653, respectively, of the human NRF cDNA; GenBank accession number AJO11812) to obtain pBSREF and pBSRNF, respectively. pBSRN110F was designed by insertion of a 110-bp linker sequence of pSBC-1 5′ to the firefly luciferase AUG codon of pBSRNF. The Renilla luciferase coding sequences in pBSRPF and pBSRNF were deleted by _Eco_RI digestion to obtain the monocistronic plasmids pMSPF and pMSNF, respectively. pMSF was constructed by deletion of the poliovirus IRES from pMSPF via _Nco_I/_Eco_RI digestion and fill-in and ligation reactions. All plasmid constructs were partially sequenced to examine their structure and correct open reading frames within PCR fragments.
Cell lines and DNA transfection.
ES-CCE (7), C243 (14), HeLa (ATCC CCL2), NIH 3T3 (ATCC CRL 1658), BHK (ATCC CCL10), and CHO (ATCC CCL61) cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum (BioWhittaker), 2 mM glutamine, penicillin, and streptomycin and were transfected by calcium phosphate coprecipitation. Porcine EF cells were maintained as described by Kues et al. (7). Murine ES cells and porcine EF cells were transfected with the Fugene reagent (Boehringer). A total of 5 μg of DNA containing 1 μg of reporter plasmid, 1 μg of chloramphenicol acetyltransferase (CAT) control plasmid, and a suitable complementing amount of high-molecular-weight DNA were transfected per 1.5 × 105 ES and EF cells using the Fugene reagent or per 7.5 × 104 cells of the other cell lines by calcium phosphate coprecipitation (16). Cells were harvested after 72 h and pooled for reporter gene analysis.
Quantification.
Firefly and Renilla luciferase activities were determined by using the Dual-Luciferase reporter system (Promega) as described in the manufacturer's protocol. The amount of CAT enzyme was measured with the CAT-Elisa kit (Boehringer) according to the manufacturer's protocol. Luciferase activity was measured in a Lumat LB 9501 luminometer (Berthold) and is reported as relative light units and corrected for protein content and the amount of CAT enzyme as a transfection efficiency control.
Northern blot analysis.
The total RNA from C234 cells was isolated by using the Trizol reagent (Gibco/BRL, Life Technologies) according to the manufacturer's instructions. Polyadenylated RNA was isolated from total RNA by using Dynabeads oligo(dT)25 (Dynal) according to the manufacturer's instructions. Northern blot analysis was performed as described by Sambrook et al. (18).
Western blot analysis.
Cellular extracts were prepared as described in the Dual Luciferase Assay protocol. Total extracts (20 μg) were run on a 10% polyacrylamide gel. Running conditions were optimized to separate 40- to 70-kDa proteins by a range of 1 mm per kDa. The Western blot analysis was performed as described by Sambrook et al. (18). Rabbit polyclonal antibody against firefly luciferase was kindly provided by M. Grashoff (GBF-National Research Institute for Biotechnology, Braunschweig, Germany).
RESULTS
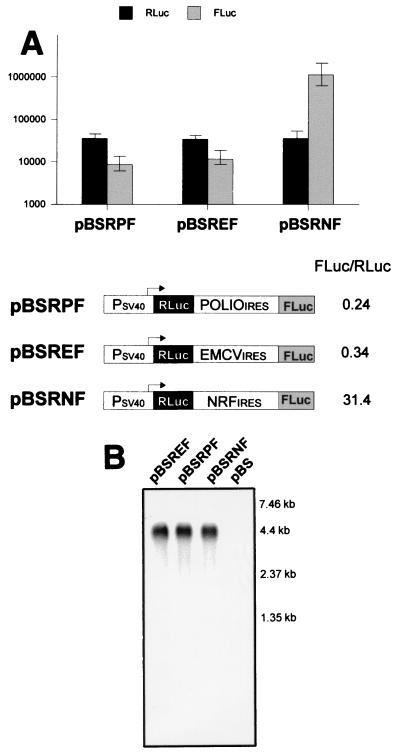
The 5′UTR of NRF mRNA has several features which are incompatible with efficient ribosomal scanning. This prompted us to examine whether NRF mRNA can be translated by internal ribosome entry. A eukaryotic expression vector (pBSRF) was constructed in which an SV40 promoter drives the expression of a dicistronic RNA. The first cistron, encoding Renilla luciferase, should be translated by a cap-dependent scanning mechanism. The second cistron, encoding firefly luciferase, requires translation by internal ribosome entry, as ribosomes will not be able to scan through an intercistronic region containing termination codons in all reading frames. The full-length 5′UTR of NRF, the EMCV IRES, or the poliovirus IRES was inserted between the Renilla luciferase stop codon and the firefly luciferase start codon. All constructs were transfected into murine C243 cells and tested for Renilla and firefly luciferase activities. In all transfectants, comparable levels of Renilla luciferase were detected (Fig. 2A). This indicates that the Renilla luciferase activity is directly proportional to the transfection efficiency and the steady-state level of dicistronic mRNA (Fig. 2B). Thus, we have normalized firefly luciferase activity to Renilla luciferase activity in order to estimate a relative factor for IRES activity. IRES-dependent expression of the second cistron varied widely, whereas expression of the first cistron was almost identical in all three reporter plasmids. The relative activities of the poliovirus and EMCV IRESs (<1) clearly demonstrate that the level of internal initiation at the second cistron is lower than that of cap-dependent initiation at the first cistron. In contrast, NRF IRES-mediated internal initiation at the second cistron is significantly higher than cap-dependent initiation (up to a factor of 31.4). A comparison of the relative IRES activities in C243 cells shows that the NRF IRES is 92-fold more active than the EMCV IRES and 130-fold more active than the poliovirus IRES.
FIG. 2.
(A) Comparison of poliovirus, EMCV, and NRF IRES activities in C243 cells. C243 cells were transfected with dicistronic constructs (pBSRNF contains the NRF IRES, pBSRPF contains the poliovirus IRES, and pBSREF contains the EMCV IRES) and a monocistronic CAT control plasmid. Luciferase activity was normalized to the CAT activity protein content. The relative firefly luciferase (FLuc) and Renilla luciferase (RLuc) activities are shown as mean relative light units ± standard errors of the means for at least five independent transfection experiments. The relative IRES activities are calculated as the ratio of firefly and Renilla luciferase activities for each transfected construct. (B) Integrity of dicistronic mRNAs. Northern blot analysis was performed with 1 μg of polyadenylated mRNA isolated from transfected C243 cells. The RNA blot was hybridized with full-length firefly luciferase cDNA. The transfected constructs are indicated above each lane. The sizes of the mRNA marker bands are indicated on the right.
RNA blot hybridization analysis was performed to ensure that translation of firefly luciferase occurred from a dicistronic mRNA. As shown in Fig. 2B, a single mRNA species, corresponding to the expected size of the respective dicistronic transcript (4.4 kb), hybridized with the firefly luciferase specific probe (second cistron). This single dicistronic mRNA was also detected by hybridization with the Renilla luciferase probe (data not shown). These results indicate that firefly and Renilla luciferase proteins were translated from a single dicistronic mRNA species. Importantly, the measured activity of Renilla luciferase in all transfections can be directly compared, as they expressed equal concentrations of dicistronic mRNA.
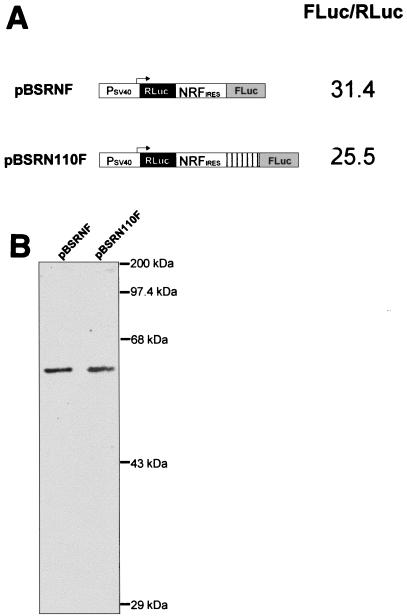
We have examined whether the NRF IRES acts in a distance-independent manner. Our results show that the NRF IRES behaves like a type I IRES, since its relative activity is not reduced by insertion of an AUG-free 110-bp spacer between its 3′ end and the initiation codon of firefly luciferase (Fig. 3A). We also ruled out expression of functional firefly luciferase enzyme from the next in-frame AUG following the authentic AUG. Initiation at this downstream AUG would result in a firefly luciferase lacking its N-terminal 18 amino acid residues (∼2 kDa). To demonstrate that translation initiation in our dicistronic mRNAs occurs at the authentic AUG of firefly luciferase, we determined the sizes of the resulting proteins by Western immunoblotting. As shown in Fig. 3B, the size of the firefly luciferase protein (60.67 kDa) does not change after insertion of the 110-bp spacer.
FIG. 3.
NRF IRES activity in constructs with different placements of the initiating AUG. (A) IRES activity was determined as described in the legend to Fig. 2A. The 3′ end of the NRF IRES and the AUG start codon of the firefly luciferase gene are separated by a single _Not_I site (8 bp) in pBSRNF and by an additional 110-bp spacer sequence in pBSRN110F. (B) Sizes of resulting firefly luciferase. Cellular extracts (20 μg) were subjected to sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis and Western blot analysis with a rabbit polyclonal antibody directed against firefly luciferase. The sizes of protein marker bands are indicated on the right.
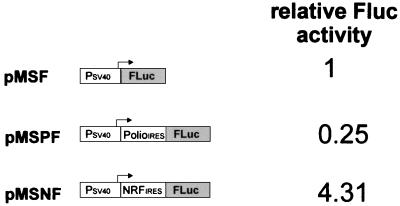
Previous studies with the EMCV IRES as well as a functional analysis of the cellular fibroblast growth factor IRES have shown that both act as translational enhancers in the context of a monocistronic mRNA (4, 21). We have examined whether the 5′UTR of NRF may also act as a translational enhancer. Monocistronic constructs (pMSF, pMSPF, and pMSNF) were designed in which either no IRES, the NRF 5′UTR, or the poliovirus IRES is positioned between an SV40 promoter and the coding region of firefly luciferase. As shown in Fig. 4, insertion of the poliovirus IRES inhibits the level of firefly luciferase expression compared with that in the control construct without the IRES. In contrast, the presence of the NRF IRES element leads to a more than 4-fold-higher expression level. Analysis of RNA extracted from these transfectants showed similar levels of firefly luciferase mRNA (data not shown).
FIG. 4.
Effect of the NRF and poliovirus IRESs on monocistronic expression. The Renilla luciferase coding regions in the dicistronic expression plasmids (see Fig. 2A) were removed to obtain monocistronic expression plasmids. The firefly luciferase (Fluc) activity of the monocistronic construct without the IRES (pMSF) was set at 1.
Type I IRESs are known to exhibit the greatest cell type-specific variations in internal translation initiation (1, 5). To determine the variation of NRF IRES activity in various cell lines, we separately transfected two dicistronic constructs (pBSRNF and pBSRPF) into murine embryonic stem cells, porcine embryonic fibroblasts, NIH 3T3, HeLa, BHK, and CHO cells. The results show that NRF IRES activity varies in different cell lines (Fig. 5). However, in all cell lines tested, the efficiency of the NRF IRES is at least 30-fold higher than that of the poliovirus IRES. These observations, together with the distance-independent action of the NRF IRES, indicate that it belongs to the group of type I IRES elements.
FIG. 5.
Comparison of NRF and poliovirus IRES activities in murine embryonic stem cells (ES), embryonic fibroblasts (EF), and NIH 3T3, HeLa, BHK, and CHO cells. (Top) Expression plasmids used. (Bottom) IRES activities were determined as described in the legend to Fig. 2A. Data are shown for pBSRPF (top row) and pBSRNF (bottom row).
DISCUSSION
Most vertebrate mRNAs have a 5′UTR 20 to 100 nucleotides long. Those mRNAs that are translated at a high rate generally have a short and unstructured 5′UTR. The mRNA of NRF has an unusually long and highly structured 5′UTR, but it is very efficiently translated in mammalian cells (13). This observation led us to examine whether NRF mRNA can be translated through a mechanism of cap-independent internal initiation. Various experimental methods have been applied to examine internal initiation, including the use of cap analogs, analysis of poliovirus-infected cells, and the dicistronic mRNA expression assay (6). The dicistronic assay is considered the most valid test if complemented by proof that the downstream cistron is translated from a dicistronic mRNA (6). The experiments whose results are shown in Fig. 2 provide compelling evidence that the NRF 5′UTR acts as a functional IRES in dicistronic reporter constructs.
The most common use of IRES elements is in artificial dicistronic constructs, where expression of the first cistron is cap dependent and expression of the second cistron, located on the same mRNA, is dependent on internal ribosome entry. Naturally, however, IRES elements are located 5′ to the cistron of monocistronic mRNAs. It has been suggested that cellular IRESs may improve the competition of an mRNA with other highly translated mRNAs and may allow translation under conditions in which cap-dependent translation is not functional (17). The results shown in Fig. 4 show that the level of translation initiation directed by the NRF 5′UTR is significantly higher than that of cap-dependent translation. Although it has to be confirmed experimentally, addition of the NRF IRES 5′ to the coding sequence of a monocistronic mRNA may maximize the efficiency of translation initiation in physiological situations when cap-dependent translation is inhibited, e.g., during viral infection or hypoxia.
The most important feature of the NRF IRES is its remarkably high efficiency of translation initiation compared with picornaviral IRESs. In the past, viral IRESs were the elements of choice for construction of vectors in which efficient expression of one or more cistrons is directed by internal initiation (1). A number of cellular IRESs have also been compared, but they are at best as efficient as viral IRESs (10). Several reports support the hypothesis that the cell line-dependent efficiency of IRES elements results from interaction with the specific mix of protein factors present in different cell lines or under different physiological conditions (11, 20–23). Consistent with this hypothesis, we found that NRF IRES activity varies in different cell lines. However, compared with the efficiency of the poliovirus IRES, the activity of the NRF IRES is remarkably high in all cell lines tested.
IRESs have been classified into three distinct groups (17). Type III (e.g., hepatitis A virus) IRESs are extremely poor candidates for dicistronic expression in vivo, whereas type I (poliovirus) and type II (EMCV) IRESs function efficiently in different cell lines. The use of type II IRESs for artificial constructs is difficult because the initiation codon of the downstream gene has to be fused precisely to the 3′ end of the IRES. In contrast, type I IRESs act in a distance-independent manner, allowing more flexible placement of the initiating AUG of the downstream gene. The results shown in Fig. 3 demonstrate that the NRF IRES is a type I IRES. Thus, the NRF IRES described here is a highly potent IRES element suitable for easy and flexible construction of mono- or dicistronic vectors directing cap-independent expression of any gene of interest.
Cellular and viral IRESs do not have any significant homology in their primary sequences. Among the known viral IRESs, a homologous pyrimidine-rich tract was identified which is located about 25 nucleotides upstream of the initiation site (4). None of the cellular IRESs, including the NRF IRES, contain this polypyrimidine tract. A search for sequence homology between some of the cellular 5′UTRs revealed only an overall high GC content (64 to 80%) (20). However, the GC content of the NRF 5′UTR is only about 34%. The lack of significant sequence homology among cellular IRESs and the fact that IRES activity is likely to depend on the recognition of specific secondary structure elements by protein factors led to the identification of a putative conserved common RNA structure, termed Y-type (8). Interestingly, the region between nucleotides 450 and 600 of the NRF 5′UTR can fold into a stable (−33.4 kcal/mol) Y-shaped structure. However, the functional significance of this structure is not proven and will require detailed mutational analysis.
ACKNOWLEDGMENTS
We thank M. G. Gruber (Promega, Madison, Wis.) for discussions and critical reading of the manuscript. We also thank R. Kaempfer (The Hebrew University of Jerusalem, Jerusalem, Israel) for helpful suggestions.
REFERENCES
- 1.Borman A M, LeMercier P, Girard M, Kean K M. Comparison of picornaviral IRES-driven internal initiation of translation in cultured cells of different origins. Nucleic Acids Res. 1997;25:925–932. doi: 10.1093/nar/25.5.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dirks W, Wirth M, Hauser H. Dicistronic transcription units for gene expression in mammalian cells. Gene. 1993;128:247–249. doi: 10.1016/0378-1119(93)90569-o. [DOI] [PubMed] [Google Scholar]
- 3.Duncan R, Milburn S C, Hershey J W B. Regulated phosphorylation and low abundance of HeLa cells eIF-4E suggest a role in translational control. J Biol Chem. 1987;262:380–388. [PubMed] [Google Scholar]
- 4.Ehrenfeld E. Initiation of translation by picornavirus RNAs. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translation control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 549–574. [Google Scholar]
- 5.Hershey J W B, editor. Translation control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- 6.Jackson R J A. A comparative view of initiation site selection mechanisms. In: Hershey J W B, editor. Translation control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 71–112. [Google Scholar]
- 7.Kues W A, Anger M, Carnwath J W, Motlik J, Niemann H, Paul D. Cell cycle synchronization of porcin primary fibroblasts: effects of serum withdrawal and reversible cell cycle inhibitors. Theriogenology. 1999;51:205. doi: 10.1095/biolreprod62.2.412. [DOI] [PubMed] [Google Scholar]
- 8.Le S Y, Maizel J V., Jr A common RNA structural motif involved in the internal initiation of translation of cellular mRNAs. Nucleic Acids Res. 1997;25:362–369. doi: 10.1093/nar/25.2.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mountford P, Zevnik B, Duwel A, Nicols J, Li M, Dani C, Robertson M, Chamber E, Smith A. Dicistronic targeting constructs: reporters and modifiers of mammalian gene expression. Proc Natl Acad Sci USA. 1994;91:4303–4307. doi: 10.1073/pnas.91.10.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mountford P S, Smith A G. Internal ribosome entry sites and dicistronic RNAs in mammalian transgenesis. Trends Genet. 1995;11:179–184. doi: 10.1016/S0168-9525(00)89040-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Negulescu D, Leong L E C, Chandy K G, Semler B M. Translation initiation of a cardiac voltage-gated potassium channel by internal ribosome entry. J Biol Chem. 1998;273:20109–20113. doi: 10.1074/jbc.273.32.20109. [DOI] [PubMed] [Google Scholar]
- 12.Nourbakhsh M, Hauser H. The transcriptional silencer protein NRF: a repressor of NF-kappaB enhancers. Immunobiology. 1997;198:65–72. doi: 10.1016/s0171-2985(97)80027-7. [DOI] [PubMed] [Google Scholar]
- 13.Nourbakhsh M, Hauser H. Constitutive silencing of IFN-β promoter is mediated by NRF (NF-κB repressing factor), a nuclear inhibitor of NF-κB. EMBO J. 1999;18:6415–6425. doi: 10.1093/emboj/18.22.6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oie H K, Gazdar A F, Buckler C E, Baron S. High interferon producing line of transformed murine cells. J Gen Virol. 1977;17:107–109. doi: 10.1099/0022-1317-17-1-107. [DOI] [PubMed] [Google Scholar]
- 15.Pelletier J, Sonenberg N. Insertion mutagenesis to increase secondary structure within the 5′ noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell. 1985;40:515–526. doi: 10.1016/0092-8674(85)90200-4. [DOI] [PubMed] [Google Scholar]
- 16.Sabe H, Kondo S, Shimuzu A, Tagaya Y, Yodoi J, Kobayashi N, Hatanaka M, Matsunamim N, Maeda M, Noma T, Honjo T. Properties of human interleukin-2 receptors expressed on non-lymphoid cells by cDNA transfection. Mol Biol Med. 1984;2:379–396. [PubMed] [Google Scholar]
- 17.Sachs A B, Sarnow P, Hentze P M. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 19.Sonenberg N, Gingras A. The mRNA 5′ cap-binding protein eiF4E and control of cell growth. Curr Opin Cell Biol. 1998;10:268–275. doi: 10.1016/s0955-0674(98)80150-6. [DOI] [PubMed] [Google Scholar]
- 20.Stein I, Itin A, Einat P, Skaliter R, Grossman Z, Keshet E. Translation of vascular endothelial growth factor mRNA by internal ribosome entry: implications for translation under hypoxia. Mol Cell Biol. 1998;18:3112–3119. doi: 10.1128/mcb.18.6.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stoneley M, Paulin F E M, Le Quense J P C, Chappell S A, Willis A E. C-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene. 1998;16:423–428. doi: 10.1038/sj.onc.1201763. [DOI] [PubMed] [Google Scholar]
- 22.Vagner S. Alternative translation of human fibroblast growth factor 2 mRNA occurs by internal entry of ribosomes. Mol Cell Biol. 1995;15:35–44. doi: 10.1128/mcb.15.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang Q, Sarnow P. Location of the internal ribosome entry site in the 5′ non-coding region of the immunoglobulin heavy-chain binding protein (BiP) mRNA: evidence for specific RNA-protein interactions. Nucleic Acids Res. 1997;25:2800–2807. doi: 10.1093/nar/25.14.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yueh A, Schneider R J. Selective translation initiation by ribosome jumping in adenovirus-infected and heat-shocked cells. Genes Dev. 1996;10:1557–1567. doi: 10.1101/gad.10.12.1557. [DOI] [PubMed] [Google Scholar]
- 25.Zuker M. Computer prediction of RNA structure. Methods Enzymol. 1989;180:262–288. doi: 10.1016/0076-6879(89)80106-5. [DOI] [PubMed] [Google Scholar]