PKR Stimulates NF-κB Irrespective of Its Kinase Function by Interacting with the IκB Kinase Complex (original) (raw)
Abstract
The interferon (IFN)-induced double-stranded RNA-activated protein kinase PKR mediates inhibition of protein synthesis through phosphorylation of the α subunit of eukaryotic initiation factor 2 (eIF2α) and is also involved in the induction of the IFN gene through the activation of the transcription factor NF-κB. NF-κB is retained in the cytoplasm through binding to its inhibitor IκBα. The critical step in NF-κB activation is the phosphorylation of IκBα by the IκB kinase (IKK) complex. This activity releases NF-κB from IκBα and allows its translocation to the nucleus. Here, we have studied the ability of PKR to activate NF-κB in a reporter assay and have shown for the first time that two catalytically inactive PKR mutants, PKR/KR296 and a deletion mutant (PKR/Del42) which lacks the potential eIF2α-binding domain, can also activate NF-κB. This result indicated that NF-κB activation by PKR does not require its kinase activity and that it is independent of the PKR-eIF2α relationship. Transfection of either wild-type PKR or catalytically inactive PKR in PKR0/0 mouse embryo fibroblasts resulted in the activation of the IKK complex. By using a glutathione _S_-transferase pull-down assay, we showed that PKR interacts with the IKKβ subunit of the IKK complex. This interaction apparently does not require the integrity of the IKK complex, as it was found to occur with extracts from cells deficient in the NF-κB essential modulator, one of the components of the IKK complex. Therefore, our results reveal a novel pathway by which PKR can modulate the NF-κB signaling pathway without using its kinase activity.
The double-stranded RNA (dsRNA)-activated protein kinase PKR is a serine/threonine protein kinase which is present in most cells at basal levels and which can be induced upon interferon (IFN) treatment (22, 42, 50). Its best-characterized substrate is the α subunit of eukaryotic initiation factor 2 (eIF2α), the phosphorylation of which leads to inhibition of protein synthesis (21). PKR was shown to phosphorylate eIF2α in yeast (8) or during a viral infection in murine clones stably expressing PKR (44); for a review, see reference 11. The use of these two in vivo systems firmly established that PKR-mediated phosphorylation of eIF2α is directly responsible for inhibition of cellular growth and inhibition of viral growth, two properties related to the antiviral and antiproliferative mechanisms of action of IFN.
PKR was also reported by different but convergent experiments to be involved in transcriptional stimulation through activation of the NF-κB pathway (see below). This activity was initially suggested by the observation that the PKR inhibitor 2-aminopurine could inhibit the induction of the immediate-early genes c-myc and c-fos as well as the induction of the beta IFN (IFNβ) gene and IFNβ-induced genes (65); for a review, see reference 58. More direct evidence was provided by in vivo experiments where selective ablation of PKR mRNAs led to inhibition of NF-κB activation in response to dsRNA (38). Moreover, mouse embryo fibroblasts (MEFs) from PKR knockout mice (PKR0/0) showed a much lower response than the corresponding PKR+/+ MEFs for the induction of IFNβ in response to dsRNA (30, 60). These data collectively implicate PKR as playing a role in the induction of genes, in addition to regulating other metabolic events, such as protein translation, through eIF2α phosphorylation. PKR has now also been shown to be involved in some of the mechanisms leading to apoptosis, in particular, in the response of cells to viral infection or to dsRNA treatment (for a review, see reference 17). This property could be due, at least in part, to the ability of PKR to activate NF-κB (18).
NF-κB, first identified as a transcription factor required for B-cell-specific gene expression, is essential in the cellular response to inflammatory and stress signals (3, 28). NF-κB is negatively regulated in the cytoplasm of unstimulated cells through interaction at its nuclear localization sites with the IκB proteins. This activity prevents its translocation to the nucleus and therefore its ability to activate gene transcription (20). The NF-κB transcription pathway is activated by proinflammatory cytokines, such as tumor necrosis factor alpha and interleukin 1 (IL-1); by bacterial or viral products, such as lipopolysaccharide (LPS), dsRNA, or the human T-cell leukemia virus type 1 Tax protein; and by oxidative stress molecules (2). All these stimuli trigger the phosphorylation of IκB and its subsequent ubiquitination and degradation by the 26S proteasome (1, 7, 61). As a consequence, NF-κB is liberated and migrates to the nucleus.
IκB phosphorylation is achieved by a 700- to 900-kDa multimeric complex, referred to as the IκB kinase (IKK) complex (15, 41, 48, 54, 63). IKK contains two catalytic subunits, IKKα and IKKβ, which can form homo- or heterodimers. Both kinases can be activated upon phosphorylation by the NF-κB inducing kinase (36) and by the MAP kinase kinase kinase 1 (32). Recent data show that IKKβ is the major effector of IκB phosphorylation in response to cytokines (24, 33, 55). Another component of the multimeric IKK complex is the NF-κB essential modulator (NEMO), which interacts with IKKβ and regulates the kinase activity of IKK (49). Mutant cell lines which do not express NEMO cannot activate NF-κB in response to multiple stimuli, such as the ones cited above (59).
In order to study the mechanism by which PKR stimulates gene expression through NF-κB activation, we have used a functional microassay for PKR with luciferase as a reporter gene under the control of NF-κB response elements. In this assay, both wild-type PKR (PKRwt) and inactive PKR mutants were used in cells either expressing the PKR gene (PKR+/+ MEFs) or not expressing it (PKR0/0 MEFs). This strategy allowed us to demonstrate that the ability of PKR to stimulate NF-κB-dependent gene expression is a property of PKR independent of its kinase activity. Transfection of PKRwt and PKR mutants in PKR0/0 cells allowed the activation of NF-κB and of IKK, thus demonstrating that PKR does not require its kinase function to activate IKK. Accordingly, a recent report has also presented evidence that an inactive PKR mutant can activate IKKβ (9). Finally, PKR was found to interact with the IKKβ subunit of the complex in a glutathione _S_-transferase (GST) pull-down assay. This finding indicates that PKR can act upstream of the IKK signaling pathway and emphasizes its role in signaling for IFN production.
MATERIALS AND METHODS
Plasmids.
Plasmid pcDNA1/Amp expressing PKRwt or its catalytically inactive form PKR/KR296 has been previously described (44). The pHIV-1 LTR-luc plasmid, corresponding to the full-length human immunodeficiency virus type 1 (HIV-1) long terminal repeat (LTR), and the pHIV-1 LTRΔNF-κB-luc plasmid, corresponding to the same region but with a deletion in the 26-bp segment spanning the two NF-κB binding sites (16), were provided by N. Israel (Institut Pasteur, Paris, France). The plasmids IgκCona-luc and Cona-luc (45), as well as pcDNA3 (hemagglutinin epitope-tagged NEMO), were provided by A. Israel (Institut Pasteur). The pGL2 HIV-1 LTR-luc plasmid was a gift from Anne Gatignol (Jewish General Hospital, Montreal, Quebec, Canada). For the construction of pCMV-Tax, the Tax sequence was copied by PCR from the vector HTLoligophX (obtained from M. Nerenberg, Scripps Research Institute, La Jolla, Calif.) with the primers AGATCCAAGCTTCCACCATG (5′ end) and TTAAGCTTCCTTTTCAGACT (3′ end) tailed with _Hin_dIII restriction sites. The PCR-amplified Tax DNA was first subcloned in PcRII (Invitrogen), cut with _Hin_dIII, and inserted in a pCMV vector using a pCMV-Tat plasmid previously cut with _Hin_dIII to remove the Tat sequence (pCMV-Tat was a gift from M. Emerman, Hutchinson Cancer Research, Seattle, Wash.). The pGEX-murine PKRwt plasmid was provided by S. Kadereit and B. R. G. Williams (Cleveland Clinic Foundation, Cleveland, Ohio). The pcDNA3-IKKα and pcDNA3-Flag epitope-tagged IKKβ plasmids were gifts from M. Kroll and F. Arenzana-Seisdedos (Institut Pasteur).
Construction of a PKR deletion mutant.
The plasmid BluescriptSK (PKRwt) (42) was cut with _Bcl_I and _Afl_II to remove the PKR region located between subdomains IV and VI. The resulting PKR truncation plasmid was religated between the _Bcl_I and _Afl_II sites with the use of a 25-bp adapter obtained by annealing two complementary oligonucleotides of sense strand sequence: 5′-AACTTGATCATCGCGAGATCTTAAG-3′. This adapter was designed to contain an internal _Nru_I digestion site (TCG/CGA) which will give a blunt end in addition to restoring an arginine residue (CGA codon) in the PKR sequence at its original position (residue 412). The religated plasmid, designated BluescriptSK(PKR-Nru), was cut with _Bcl_I and _Nru_I. This construct contains all of the PKRwt sequence except for a 100-amino-acid gap between residues 312 and 412.
A PKR fragment located between the PKR unique sites _Bcl_I and _Afl_II was amplified by PCR using the Vent polymerase (New England Biolabs) which, since it has no 5′-adenosyltransferase activity, allowed us to insert the product directly in BluescriptSK(PKR-Nru) and create the PKR deletion mutant PKR/Del42. For the PCR, we used a 5′ primer (5′-TGGCAAAACTTGATCATGTA-3′) corresponding to the _Bcl_I site and a 3′ primer (5′-ATCACAGAATTCCATTTGGA-3′) corresponding to the region between PKR subdomains V and VI (corresponding to the motif LFIQMEFCD). A PCR fragment of 175 bp was cut with _Bcl_I and inserted in BluescriptSK(PKR-Nru) previously cut with _Bcl_I and _Nru_I. This procedure restored the PKR sequence up to position 369 and left the desired 42-amino-acid deletion. The PKR/Del42 deletion mutant was excised from the BluescriptSK plasmid with _Hin_dIII and _Bam_HI and transferred to the pcDNA1/Amp vector.
RT-PCR of luciferase transcripts.
PKR0/0 cells cultured in 10-cm petri dishes were transfected with 10 μg of pGL2 HIV-1 LTR-luc in the presence of various amounts of PKRwt or mutant PKR using the calcium phosphate precipitation-glycerol shock technique. Total RNA was extracted 24 h posttransfection with RNA-plus extraction solution (Quantum-Bioprobe). After 30 min of DNase I treatment, 10 μg of total RNA was submitted to reverse transcription (RT) with an oligo(dT) primer. Five microliters of the RT reaction mixture was used as a matrix to perform a PCR with the following primers: 5′ luc 1600 (5′-CCGCGAAAAAGTTGCGCGGAGGA-3′) and 3′ UTR SV40 (5′-GGAGGAGTAGAATGTTGAGAGTCA-3′). The 3′ UTR SV40 primer was designed to hybridize to a sequence located after the simian virus 40 intron on the pGL2 plasmid in order to discriminate between the DNA from the plasmid and the DNA obtained by PCR after RT of Luc mRNA. An internal control RT-PCR was performed with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) primers (gapdh+, 5′ TGA AGG TCG GAG TCA ACG GAT TTG GT; gapdh−, 5′ CAT GTG GGC CAT GAG GTC CAC CAC).
In vitro transcription and translation of PKR.
The pcDNA1/Amp (PKR) plasmids (wt, KR296, and Del42) were linearized with _Bam_HI. Transcription from the T7 promoter in the presence of the cap analog 7-mGpppG was carried out using an mRNA capping kit (Stratagene). The in vitro-transcribed mRNAs were translated in a rabbit reticulocyte lysate (Stratagene) using 10 μg of template RNA in the presence of 20 μCi of [35S]methionine/cysteine (ProMix; Amersham) and 6 mM 2-aminopurine (Sigma) in a 25-μl reaction volume. After 60 min of incubation at 30°C, the samples were analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) either directly or after subsequent analysis [immunoprecipitation or binding to poly(I)-poly(C)–agarose].
Immunoprecipitation of PKR.
In vitro-translated PKR was incubated with anti-PKR monoclonal antibody 71/10 (31) in 200 μl of BI buffer (20 mM Tris-HCl [pH 7.6], 50 mM KCl, 400 mM NaCl, 1 mM EDTA, 1% Triton X-100, 5 mM 2-mercaptoethanol, 1% aprotinin [Sigma], 0.2 mM phenylmethylsulfonyl fluoride [PMSF], 20% glycerol). After 60 min of incubation at 4°C, 50 μl of protein A/G-agarose (Protein AG PLUS-agarose; Santa Cruz Biotechnology) was added, and the samples were further incubated overnight at 4°C. After three cycles of centrifugation (4°C, 3,000 × g) and resuspension in BI buffer, the samples were either directly analyzed after separation of the proteins by SDS-PAGE or used for in vitro phosphorylation of PKR.
Immunoprecipitation of IKK.
For immunoprecipitation with anti-IKKα and anti-IKKβ polyclonal antibodies (sc-7218 and sc-7607, respectively; Santa Cruz) or with anti-NEMO antibody (59), cell extracts were prepared in buffer L (20 mM Tris-HCl [pH 7.6], 50 mM KCl, 40 mM NaCl, 1 mM EDTA, 1% Triton X-100, 3 mM β-mercaptoethanol, 1% aprotinin, 0.2 mM PMSF, 25% glycerol) containing sodium orthovanadate (Na3VO4) (1 mM), _para_-nitrophenyl phosphate (PNPP) (10 mM), β-glycerophosphate (10 mM), and sodium fluoride (NaF) (5 mM) as phosphatase inhibitors. Cell extracts were incubated overnight at 4°C with the desired antibodies previously bound to protein A/G-agarose (1 μl of antibody for 30 μl of agarose). After three cycles of centrifugation (4°C, 3,000 × g) and resuspension in buffer L-phosphatase inhibitors, the samples were used in in vitro phosphorylation assays.
In vitro phosphorylation of PKR.
The immunoprecipitated in vitro-translated PKR proteins (recovered by 50 μl of protein A/G-agarose beads) were washed with BI buffer, further washed three times with buffer II (20 mM Tris-HCl [pH 7.6], 100 mM KCl, 0.1 mM EDTA, 1% aprotinin, 20% glycerol), and resuspended in buffer III (buffer II supplemented with 2 mM MgCl2). For the phosphorylation reaction, each sample was incubated with 40 μl of buffer III supplemented with 2 mM MnCl2 and 10 μl of [γ-32P]ATP solution (1.25 mCi of [γ-32P]ATP per ml [3,000 Ci/mmol; ICN], 10 mM ATP, and 1 mM MgCl2 in buffer II). The reaction was performed in the absence or in the presence of 1 μg of poly(I)-poly(C) (Pharmacia) per ml. After incubation for 15 min at 30°C, 2 μl (165 ng) of a pure preparation of rabbit eIF2 complex (a gift from C. Proud, University of Dundee, Dundee, United Kingdom) was added, and the reaction was continued for another 15 min. An equal volume of 2× SDS electrophoresis sample buffer was added, and the products were analyzed by SDS-PAGE.
Cell cultures and transfections.
Human HeLa cells were grown in Glutamax-1 Dulbecco's modified Eagle's medium (Gibco) supplemented with 5 μg of penicillin-streptomycin per ml and containing 10% fetal calf serum. PKR0/0 and PKR+/+ MEFs were provided by B. R. G. Williams. They were cultured in the same medium as HeLa cells. The 70Z/3 murine pre-B-cell line and the NF-κB-unresponsive mutant 1.3E2 (NEMO deficient) were provided by G. Courtois and A. Israel. They were cultured in Glutamax-1 RPMI medium (Gibco) supplemented with 5 μg of penicillin-streptomycin per ml and 50 μM β-mercaptoethanol and containing 10% fetal calf serum. For microtransfection, 18 to 24 h before transfection, the cells were seeded at 20,000 cells/well in 96-well microplates (Costar) with 200 μl of complete culture medium. At 3 h before transfection, the medium was aspirated and replaced with fresh medium. The desired amounts of plasmids were adjusted to the same final concentration with the addition of pcDNA1/Amp and were transfected into the cells using the calcium phosphate precipitation-glycerol shock technique as previously described (43). At 48 h after transfection, the culture medium was aspirated, and the cells were washed twice with phosphate-buffered saline (PBS). Cells were then scraped and lysed in 140 μl of luc lysis buffer (25 mM Tris-phosphate [pH 7.6], 8 mM MgCl2, 1 mM dithiothreitol [DTT], 0.1% Triton X-100, 15% glycerol) per well. For each sample, 100 μl was analyzed for luciferase activity in a luminometer by automatic mixing with luc lysis buffer solution containing 0.25 mM Luciferine (Sigma), 1 mM ATP, and 1% bovine serum albumin (BSA). The protein content of each sample was measured in 96-well microplates using a Bio-Rad Bradford kit and the absorbance at 590 nm was determined in an LP400 spectrophotometer. Each result corresponds to the average of four independent transfections.
Nuclear extracts.
PKR0/0 cells cultured in 10-cm dishes were transfected with the different PKR constructs using the calcium phosphate precipitation-glycerol shock technique. pCMV-Tax was used as a positive control for the induction of NF-κB after transfection. PKR0/0 cells were transfected with 250 ng or 1 μg of PKRwt or PKR/KR296 per plate in the presence of 10 μg of pcDNA1/Amp. Four hours after transfection, the cells were submitted to glycerol shock and then further incubated for 4 h in 2% serum medium, after which they were scraped and washed with PBS. The pellet was resuspended in 200 μl of electrophoretic mobility shift assay (EMSA) I buffer (50 mM Tris-HCl [pH 7.9], 10 mM KCl, 1 mM EDTA, 0.2% Nonidet P-40, 1 mM DTT, 1 mM PMSF, 1% aprotinin, 10% glycerol), incubated for 1 min on ice, and centrifuged for 3 min at 6,000 × g and 4°C. The nuclear pellet was then resuspended in 20 μl of EMSA II buffer (20 mM HEPES [pH 7.9], 400 mM NaCl, 10 mM KCl, 1 mM EDTA, 1 mM DTT, 1 mM PMSF, 1% aprotinin, 20% glycerol), incubated for 20 min on ice, and centrifuged for 10 min at 12,000 × g and 4°C. The nuclear extracts were collected and stored at −80°C. The protein concentration was estimated using the Bio-Rad Bradford kit.
EMSA.
Nuclear extracts (5 μg) were preincubated with 1 μg of poly(dI-dC) · poly(dI-dC) in 20 μl of binding buffer (20 mM HEPES [pH 7.5], 70 mM NaCl, 2 mM DTT, 100 μg of BSA per ml, 0.01% Nonidet P-40, 4% Ficoll) for 5 min at room temperature. Then, 0.2 pmol of 32P-labeled NF-κB probe (30,000 cpm) was added, and the extracts were further incubated for 20 min. Complexes were resolved by electrophoresis at 180 V on a prerun 5% native polyacrylamide gel in 0.5× Tris-borate-EDTA. The gel was dried and exposed overnight for autoradiography in a PhosphorImager. For the supershift assay, nuclear extracts were incubated for 10 min at room temperature in binding buffer in the presence of 1 μl of anti-p50 (1157; a kind gift from N. Israel) or anti-p65 (sc-109; Santa Cruz) polyclonal antibody prior to the addition of the probe.
IKK phosphorylation.
Cell extracts corresponding to 106 cells were immunoprecipitated with anti-NEMO antibody previously used to coat 50 μl of protein A/G-agarose beads in low-salt buffer (20 mM Tris [pH 7.6], 40 mM NaCl, 50 mM KCl, 1 mM EDTA, 1% Triton X-100, 25% glycerol, 0.2 mM PMSF, 3 mM 2-mercaptoethanol, 10 μg of aprotinin per ml) containing 5 mM NaF, 10 mM PNPP, 1 mM Na3VO4, and 10 mM β-glycerophosphate. After overnight incubation at 4°C, the beads were washed with low-salt buffer two times, washed once with kinase buffer (see below) without ATP, and incubated with 40 μl of kinase buffer (20 mM Tris-HCl [pH 7.6], 2 mM MgCl2, 2 mM MnCl2, 10 μM ATP, 3 μCi of [γ-32P]ATP (3,000 Ci/mmol), 10 mM β-glycerophosphate, 10 mM NaF, 10 mM PNPP, 300 μM Na3VO4, 2 μM PMSF, 10 μg of aprotinin per ml, 1 mM DTT) at 30°C for 30 min in the presence of 200 ng of IκBα per reaction (IκBα was provided by M. Kroll and F. Arenzana-Seisdedos). An equal volume of 2× SDS electrophoresis sample buffer was added, and the products were analyzed by SDS-PAGE.
Protein expression and purification.
Escherichia coli BL21(DE3) cells containing the pGEX-murine PKR expression vector were grown overnight at 37°C in 100 ml of Terrific Broth containing 100 μg of ampicillin per ml. After 1:50 dilution in 100 ml of fresh medium, cells were grown at 30°C to an optical density of approximately 0.5 (6 h) and induced with 0.5 mM isopropyl-1-thio-β-d-galactopyranoside (IPTG) for an additional 2 h. Bacteria were pelleted at 4,000 × g for 10 min at 4°C, resuspended in 5 ml of PBS containing 0.2 mM PMSF and 1 mM DTT, and frozen at −20°C overnight. After being thawed on ice, the bacterial suspensions were frozen again in a dry ice-methanol bath, thawed on ice, sonicated three times for 30 s each time on ice, adjusted to 1% (wt/vol) with Triton X-100, and centrifuged at 9,000 × g for 20 min at 4°C.
The supernatant (5 ml) was incubated with 500 μl of glutathione-Sepharose 4B beads (Amersham) in 1% Triton X-100 buffer for 60 min at 4°C with gentle rotation. After centrifugation at 1,500 × g, the bound protein was washed four times with 1% Triton X-100 buffer and resuspended in 2.5 ml of 30% glycerol–PBS. The concentration of bound GST-PKR was estimated to be 1.25 μg/30 μl of beads in comparison with known concentrations of BSA by SDS-PAGE with Coomassie blue staining (Bio-Safe Coomassie; Bio-Rad). For protein interaction, 30 μl of glutathione-Sepharose-bound GST-PKR or of glutathione-Sepharose-bound GST-HP1α (HP1α is a nuclear protein [52] [provided by J. Seeler]) was incubated overnight at 4°C with in vitro-translated proteins in 200 μl of HNGT buffer (20 mM HEPES [pH 7.9], 150 mM NaCl, 10% glycerol, 0.1% Triton X-100). After three washes with 500 μl of HNGT buffer, 20 μl of SDS sample buffer was added, and the proteins were analyzed by SDS-PAGE.
RESULTS
NF-κB-dependent stimulation of gene expression by PKR is independent of its protein kinase activity.
PKR is best known as a strong inhibitor of protein synthesis; however, it can also stimulate gene expression through NF-κB activation. In order to discriminate between these two opposite properties, we have analyzed the effect of different concentrations of PKR on the expression of reporter-expressing plasmids dependent or not dependent on the presence of NF-κB response elements. For this analysis, we chose a natural NF-κB-responsive promoter, the HIV-1 LTR, and an artificial construct, IgκCona, containing three NF-κB response elements cloned upstream of a minimal promoter. The assay was performed with PKR+/+ MEFs, and the effect of PKRwt was compared with that of the catalytically inactive PKR/KR296 mutant (26, 44), allowing us to determine whether the observed effect is related to the kinase activity of PKR.
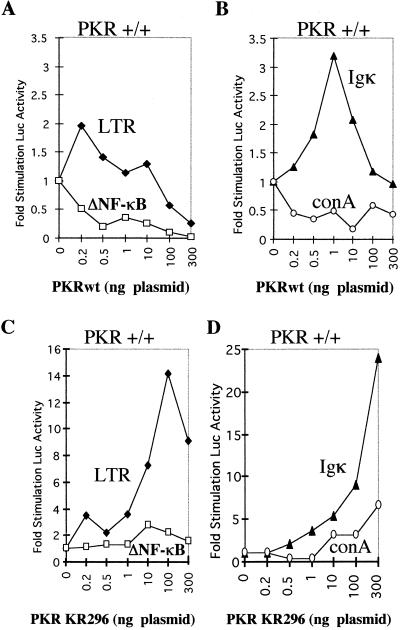
The data presented in Fig. 1 show the effect of PKRwt or PKR/KR296 on the pair pHIV-1 LTR-luc–pHIV-1 LTRΔNF-κB-luc (Fig. 1A and C) and on the pair IgκCona-luc–Cona-luc (Fig. 1B and D). PKRwt was found to stimulate gene expression when transfected at low concentrations (0.2 to 1 ng of the PKR plasmid for 100 ng of the reporter; transfection in 20,000 cells). This stimulatory effect, although not strong (two- to threefold), was specific, since it could not stimulate pHIV-1 LTRΔNF-κB-luc and Cona-luc, used as controls (Fig. 1A and B). Thus, our assay can be used to monitor the NF-κB stimulation of gene expression by PKR. Surprisingly, the PKR/KR296 mutant was also found to stimulate the expression of pHIV-1 LTR-luc and IgκCona-luc, since this stimulation was dependent on the presence of the NF-κB response elements (Fig. 1C and D). These data illustrate that NF-κB-mediated stimulation of gene expression by PKR does not require its kinase activity. The lower stimulatory effect of PKRwt than of PKR/KR296 is probably due to the ability of PKRwt to also inhibit the translation of the reporter gene (see below).
FIG. 1.
NF-κB-dependent stimulation of gene expression by PKRwt and PKR/KR296 in a reporter assay. Different concentrations (0 to 300 ng) of pcDNA1/Amp expressing either PKRwt (A and B) or PKR/KR296 (C and D) were cotransfected in PKR+/+ MEFs with 100 ng of the reporter plasmid expressing luciferase under the control of different promoters: pHIV-1 LTR-luc (LTR; closed diamonds) or pHIV-1 LTRΔNF-κB-luc (ΔNF-κB; open squares) (A and C) and IgκCona-luc (Igκ; closed triangles) or Cona-luc (conA; open circles) (B and D). Each DNA mixture was adjusted to the same final DNA content with the addition of the empty pcDNA1/Amp vector. The luciferase activity per microgram of protein content per well was calculated as the mean of the transfections in four different wells (data not shown). Each value (fold stimulation of luciferase [Luc] activity) corresponds to the ratio of luciferase activity obtained by transfecting PKR with the reporter gene to luciferase activity obtained by transfecting the reporter gene alone.
The PKR-eIF2α relationship is not required for PKR-mediated stimulation of NF-κB-dependent gene expression.
Figure 1 shows that when PKRwt was transfected at concentrations higher than 1 ng/well, it could inhibit gene expression. This result can be attributed to its ability as a kinase to phosphorylate eIF2α, prevent its normal recycling, and provoke arrest in protein synthesis. On the other hand, when PKR/KR296 was transfected at concentrations higher than 1 ng/well, the stimulation of the reporter continued to rise. The stimulation appeared to be specific with regard to the presence or absence of the NF-κB response elements in the promoters. However, the possibility that PKR/KR296 upregulates the NF-κB-responsive reporter gene by binding eIF2α and preventing access of eIF2α to PKR or other eIF2α kinases could not be excluded. We therefore generated a new PKR mutant, PKR/Del42, by removing 42 amino acids between residues 369 and 412 in the catalytic subdomains V and VI (Fig. 2A). The rationale for this deletion mutant construct was based on a sequence comparison which revealed that this 42-amino-acid PKR motif could present an α-helix structure similar to that of protein kinase A, known to bind its pseudosubstrate inhibitor (27). Furthermore, this PKR domain was shown to bind K3L, a vaccinia virus protein analog of the natural PKR substrate eIF2α which acts as a pseudosubstrate inhibitor for PKR (13). Consequently, this domain is the most susceptible binding domain for eIF2α, and its deletion allows study of the function of PKR independently of its interaction with its substrate.
FIG. 2.
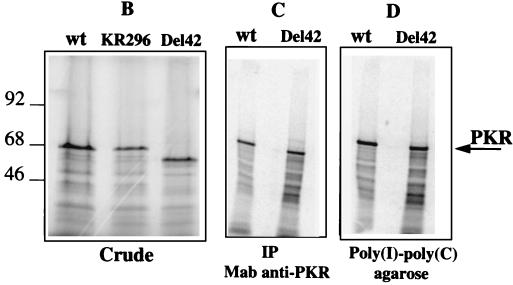
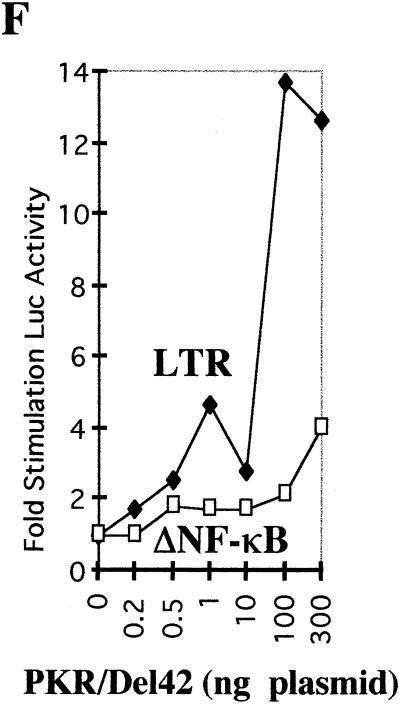
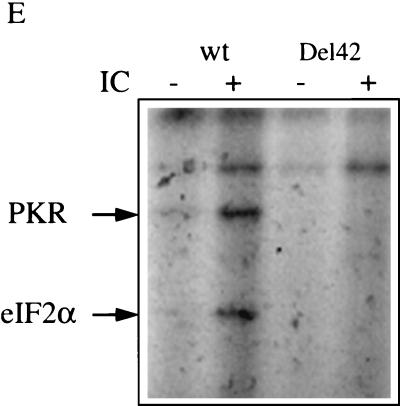
Construction and characterization of the PKR/Del42 mutant. (A) The PKR deletion mutant PKR/Del42 was generated by removing the region between subdomains V and VI (see Materials and Methods). The deletion starts after residue 369 and stops before residue 412. The length of the deletion is 42 amino acids. A schematic presentation of the PKR/Del42 mutant is shown, along with the amino acid sequence of wild-type (wt) PKR residues 357 to 422. The approximate positions of the _Bcl_I and _Afl_II restriction sites used for the construction are indicated. DRDB, dsRNA binding domain. (B) RNA preparations were in vitro transcribed from plasmid pcDNA1/Amp expressing PKRwt, PKR/KR296, and PKR/Del42. They were then translated in rabbit reticulocyte lysates in the presence of ProMix and 6 mM 2-aminopurine. After 60 min of incubation at 30°C, a 1/10 volume of the translation mixture was analyzed by SDS-PAGE on 12.5% acrylamide gels. The molecular weights (103) of marker proteins are indicated on the left. (C) Equivalent amounts of in vitro-translated PKRwt and PKR/Del42 (calculated by integrating areas from the different PKR bands shown in panel B) were immunoprecipitated (IP) with 0.2 μl of anti-PKR monoclonal antibody (Mab) 71/10 and 50 μl of protein A/G-agarose. After 18 h of incubation at 4°C and extensive washing with BI buffer, the immunoprecipitated 35S-labeled PKR preparations were analyzed by SDS-PAGE on 12.5% acrylamide gels. (D) Equivalent amounts of in vitro-translated PKRwt and PKR/Del42 (as in panel C) were analyzed for binding to poly(I)-poly(C)–agarose (Pharmacia). After 18 h of incubation at 4°C and extensive washing with BI, the poly(I)-poly(C)-bound 35S-labeled PKR preparations were analyzed by SDS-PAGE on 12.5% acrylamide gels. (E) Preparations corresponding to PKRwt and PKR/Del42 RNAs were in vitro translated in rabbit reticulocyte lysates as described for panel B but in the absence of 35S-labeled amino acids. The proteins were then immunoprecipitated with anti-PKR monoclonal antibody 71/10 as described for panel C. The immunoprecipitated PKR samples were incubated at 30°C in the absence or presence of 1 μg of poly(I)-poly(C) (IC) per ml and in the presence of 0.25 μCi of [γ-32P]ATP per ml. After 15 min of incubation, 2 μl (165 ng) of a purified eIF2 complex preparation was added, and incubation was continued for another 15 min. After the addition of an equal volume of 2× SDS buffer, samples were heated (5 min at 95°C) and analyzed by SDS-PAGE on 12.5% acrylamide gels. Arrows show the positions of PKR (68 kDa) and eIF2α (35 kDa). (F) PKR/Del42 was cotransfected with pHIV-1 LTR-luc (LTR; closed diamonds) or pHIV-1 LTRΔNF-κB-luc (ΔNF-κB; open squares), and the fold stimulation of luciferase (Luc) was measured as described in the legend to Fig. 1.
The PKR/Del42 mutant migrated in acrylamide gels slightly faster than PKRwt or the PKR/KR296 mutant (Fig. 2B). It retained its ability to be immunoprecipitated with a monoclonal antibody directed against the N terminus of PKR (Fig. 2C) as well as its ability to bind dsRNA (Fig. 2D; purification on poly(I)-poly(C)–agarose). In contrast, the PKR/Del42 mutant did not demonstrate any kinase activity when analyzed for its ability to autophosphorylate in the presence of poly(I)-poly(C) and to phosphorylate eIF2α, in contrast to PKRwt (Fig. 2E). The PKR/Del42 mutant was assayed for its effect on the expression of the pHIV-1 LTR-luc reporter plasmid using the same procedure as that described for Fig. 1. The results show that, like PKRwt and PKR/KR296, PKR/Del42 activated NF-κB (Fig. 2F).
NF-κB-dependent stimulation of gene expression in PKR0/0 MEFs by PKRwt and the mutants PKR/KR296 and PKR/Del42.
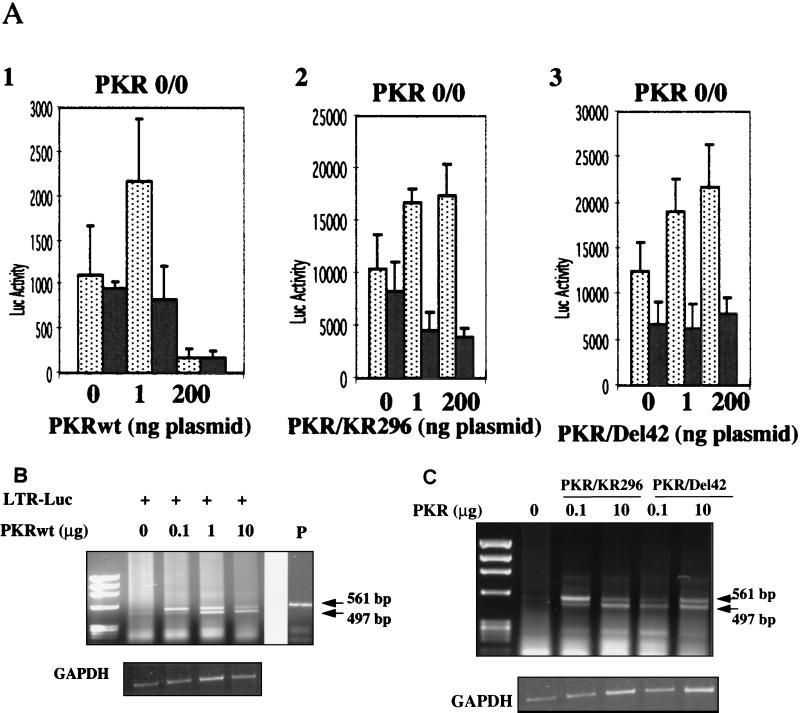
We next assayed the effects of PKRwt, PKR/KR296, and PKR/Del42 on NF-κB activation after transfection in a cellular PKR-free environment, i.e., in PKR0/0 MEFs. For each PKR construct, we observed that the stimulation of the reporter was dependent on the presence of the NF-κB response elements (Fig. 3A). This observation confirmed that PKR does not need its kinase function to stimulate gene expression. The fact that stimulation could take place in PKR0/0 cells demonstrates that the stimulatory effect of mutant PKR is direct and cannot be explained by sequestration of endogenous active PKR. It was striking to note that, at high concentrations, PKRwt inhibited strongly reporter expression, contrary to the PKR mutants (Fig. 1 and 3).
FIG. 3.
PKR/wt and mutants PKR/KR296 and PKR/Del42 stimulate NF-κB-dependent reporter gene expression in PKR0/0 MEFs. (A) The assay was performed as described in the legend to Fig. 1, except that PKR0/0 MEFs were used instead of PKR+/+ MEFs. The reporter plasmids pHIV-1 LTR-luc (dotted bars) and pHIV-1 LTRΔNF-κB-luc (grey bars) (100 ng each) were transfected either as such (0) or in the presence of 1 or 200 ng of PKRwt, PKR/KR296, or PKR/Del42. Data (luciferase [Luc] activity) are expressed as the mean of four independent transfections ± the standard error. (B) RT-PCR analysis of total RNA from PKR0/0 MEFs (in 10-cm petri dishes) transfected with pGL2 HIV-1 LTR-luc (LTR-Luc) alone or in the presence of increasing concentrations of PKRwt (0.1, 1, and 10 μg of plasmid). The sizes of the PCR products were estimated with a φX174 DNA _Hae_III digest as a marker. The upper band (561 bp) corresponds to amplification from the transfected DNA as compared to the size of the PCR product from the pHIV-1 LTR-luc plasmid (P). The lower band (497 bp) is specific for DNA amplified from the reverse-transcribed luciferase mRNA. Control GAPDH RT-PCR is shown below. (C) RT-PCR analysis of total RNA from PKR0/0 MEFs transfected as described for panel B with the vector (LTR-Luc) alone or in the presence of PKR/KR296 or PKR/Del42 (0.1 and 10 μg of plasmid).
In order to determine whether this PKRwt-mediated inhibition was at the transcriptional or the translational level, we assayed RNA levels by RT-PCR. We found that the levels of the luciferase mRNAs transcribed from the reporter plasmid increased with the concentration of PKR, either wild type (Fig. 3B) or mutated (Fig. 3C), whereas the corresponding luciferase activity was either inhibited (70% for PKRwt) or stimulated (2.5- to 3-fold for PKR mutants) at the highest concentrations of PKR (data not shown). These findings are in agreement with the known property of PKR to control gene expression at the translational level when it is functional as a kinase, in contrast to the PKR/KR296 and PKR/Del42 mutants.
Our data confirmed the ability of PKRwt to stimulate NF-κB-dependent gene expression. Moreover, the use of different concentrations of PKRwt allowed us to discriminate between the abilities of PKR to stimulate and to inhibit gene expression. Finally, the use of different PKR mutants revealed the intrinsic ability of PKR to stimulate NF-κB-dependent gene expression in the absence of its kinase function.
NF-κB activation by PKRwt and catalytically inactive PKR in EMSAs.
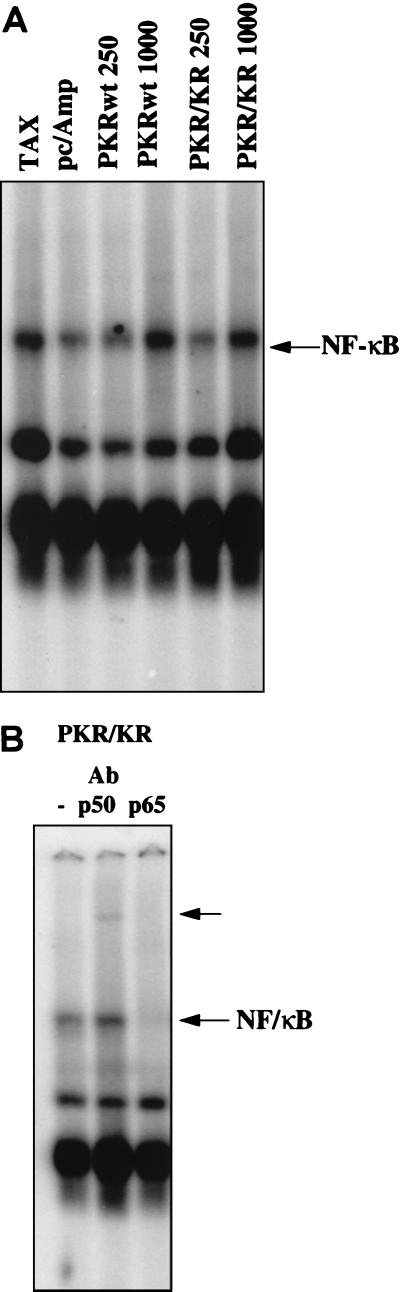
In order to confirm in an independent experiment that catalytically inactive forms of PKR could activate NF-κB, we performed a gel shift assay using cell extracts from PKR0/0 MEFs transfected with plasmids coding for PKRwt or PKR/KR296 (Fig. 4). These experiments were performed with PKR0/0 cells in order to evaluate our data in response to the expression of ectopically expressed PKR in a PKR-free environment. Basal NF-κB activity was determined by transfection with the control pcDNA1/Amp vector. As expected, transfection of a plasmid encoding Tax, a known NF-κB activator (57), led to NF-κB activation. Transfection of the PKRwt- or PKR/KR296-expressing plasmids revealed their ability to stimulate NF-κB. The identity of the complex retained on the probe as NF-κB was confirmed by a supershift assay using nuclear extracts from PKR/KR296-transfected cells (Fig. 4B).
FIG. 4.
PKRwt and mutant PKR/KR296 can activate NF-κB in EMSAs. (A) PKR0/0 cells were transfected with 250 ng or 1 μg of plasmid pcDNA1/Amp expressing PKRwt or PKR/KR296 as described in Materials and Methods. The total amounts of plasmids were adjusted to 10 μg with the vector pcDNA1/Amp. All plasmids were prepared with an Endo-free Plasmid kit (Qiagen). Positive controls for NF-κB activation were obtained by transfecting 10 μg of pCMV-Tax (TAX). Background NF-κB activity is shown for cells transfected with pcDNA1/Amp alone (pc/Amp). Five micrograms of nuclear extracts was incubated with 0.2 pmol of 32P-labeled NF-κB probe for 20 min in the presence of 1 μg of poly(dI-dC) · poly(dI-dC) at 20°C and analyzed by an EMSA on 5% native acrylamide gels. Inducible NF-κB complex is indicated by the arrow. (B) Five micrograms of nuclear extracts from PKR/KR296-transfected cells was incubated for 10 min either as such (−) or in the presence of anti-p50 antibody (Ab) (shift indicated by upper arrow) or anti-p65 antibody (disappearance of the complex) prior to the addition of the probe and processed as described above.
The capacity of PKR to activate NF-κB is in agreement with previous reports (18, 29, 60). Moreover, our results reveal that catalytically inactive PKR also brings about such activation.
PKRwt and catalytically inactive PKR activate the IKK complex.
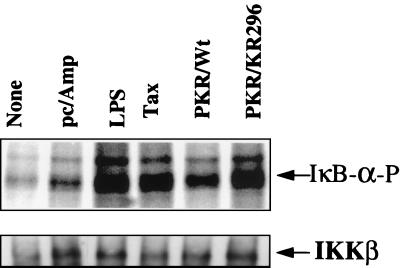
In order to further investigate the mechanism by which PKR, either wild type or mutant, activates NF-κB, we have assayed its effect on the activation of the IKK complex. PKR0/0 cells were transfected with plasmids expressing PKRwt or PKR/KR296, and the IKK complex was immunoprecipitated from cytoplasmic extracts using antibodies against NEMO, a component of the IKK complex necessary for its assembly and activity (59). IKK activity was then assayed in vitro by phosphorylation of IκBα as described previously (41). Compared to LPS treatment or transfection with Tax, transfection of both PKRwt and catalytically inactive PKR/KR296 efficiently activated IKK, whereas the transfection of a control vector resulted in basal-level activation similar to that in untreated cells (Fig. 5).
FIG. 5.
PKRwt and the PKR mutants activate IKK. PKR0/0 cells were either untreated (None) or transfected with 5 μg of pcDNA1/Amp, alone (pc/Amp) or in the presence of 250 ng of pcDNA1/Amp expressing PKRwt or PKR/KR296. As a positive control, they were also treated for 30 min with 7 μg of LPS (Sigma) per ml or transfected with pCMV-Tax (Tax). Cell extracts (300 μg of protein/sample) were immunoprecipitated with anti-NEMO antibodies and analyzed for IKK activity in an in vitro phosphorylation assay with IκBα as a substrate (see Materials and Methods). The positions of phosphorylated IκBα (IκB-α-P) and IKKβ immunoprecipitated from the different cell extracts and revealed by immunoblotting are indicated.
Interaction of PKR with the IKK complex.
Since PKRwt and catalytically inactive PKR/KR296 both can activate NF-κB through activation of the IKK complex, it is plausible to suggest that PKR could activate NF-κB simply by interacting with the IKK complex.
In order to test for a potential physical interaction between PKR and the IKK complex, we submitted HeLa cell extracts from control or IFN-treated cells to immunoprecipitation with anti-IKKα antibodies. The proteins, retained on the immunosorbent, were then analyzed for the presence of PKR in immunoblots. The presence of IKKα and PKR in crude extracts from HeLa cells and the increase in PKR levels after IFN treatment are shown in Fig. 6A (left side). Immunoblot analysis of the immunoprecipitates revealed that PKR could be recovered from the cell extracts by coimmunoprecipitation with anti-IKKα antibodies. This coprecipitation was specific, since PKR was not retained by the blank antibodies.
FIG. 6.
Interaction of PKR with IKKβ. (A) Extracts from HeLa cells treated or not treated with 500 U of IFNα per ml were incubated with protein A/G-agarose previously coated with anti-IKKα or anti-IKKβ antibodies. Although PKR specifically interacts with IKKβ (see text), the antibodies directed against IKKβ (Santa Cruz) that we have used are unable to immunoprecipitate IKKβ. Therefore, they were conveniently used here as a control for nonspecific binding (Blank). After extensive washes, the proteins which were retained on the beads were separated by SDS-PAGE on 12.5% acrylamide gels and analyzed by immunoblotting with anti-PKR antibodies. Each lane represents the analysis of proteins immunoprecipitated (IP) from extracts equivalent to 5 × 106 cells. In parallel, crude extracts were analyzed by immunoblotting for the presence of IKKα or PKR. (B) The proteins IKKβ, IKKα, NEMO, PKR/KR296, and luciferase (Luc) were translated in vitro from 2 μg of their respective plasmids in 25 μl of reticulocyte lysate in the presence of ProMix using a TNT-coupled Reticulocyte Lysate System (Promega). The translation products were analyzed by SDS-PAGE either as such (5 μl; 10% input) or after incubation for 18 h at 4°C with 1.25 μg of GST-PKR or an identical amount of GST-HP1α, a nuclear protein (52). Beads were adjusted to 50 μl in all samples with glutathione-Sepharose 4B (Amersham). (C) Cell extracts from 75 × 106 70Z/3 and 1.3E2 (NEMO-deficient) cells were incubated as described above with GST-PKR or GST-HP1α. Crude extracts corresponding to 7.5 × 106 cells (10% input) and purified extracts were analyzed by immunoblotting with antibodies directed against murine IKKβ after SDS-PAGE.
To determine whether PKR binds to one of the known components of the IKK complex, we analyzed the ability of GST-PKR to retain in vitro-translated IKKα, IKKβ, and NEMO. In addition, PKR/KR296 and luciferase were included as controls. Nonspecific binding was determined using GST-HP1α, a nuclear protein (52). The results (Fig. 6B) revealed that, within the IKK complex, only the IKKβ subunit has the ability to bind specifically to GST-PKR. IKKα was found to bind similarly to GST-PKR and to the irrelevant GST protein. As a negative control, luciferase showed no binding to either type of GST protein, while as a positive control, PKR/KR296 bound to GST-PKR, in accord with its capacity to dimerize (for a review, see reference 10). A small amount of PKR/KR296 could be recovered from the irrelevant GST protein. This latter result was most probably due to a nonspecific interaction, as was the case for IKKα.
In order to confirm the association between PKR and IKKβ, we performed another GST-PKR pull-down assay using cell extracts from the murine NEMO-deficient cell line 1.3E2 and the corresponding parental cell line 70Z/3 (12, 59). Yamaoka and colleagues have shown that IKKs are not in a complex when the NEMO protein is absent, as in the 1.3E2 cell line (59). The results (Fig. 6C) showed that despite the difference in its localization, IKKβ from both cell types could bind to PKR and not to the irrelevant protein. It should be noted that IKKβ was detected as two bands with anti-IKKβ polyclonal antibodies in crude extracts from both NEMO-deficient cells (1.3E2) and the corresponding parental cells (70Z/3) (10% input), whereas only one of the bands could be detected after purification using GST-PKR. The reason for this difference is not known and requires further analysis. Whatever the case, the fact that PKR bound to similar amounts of IKKβ in NEMO-deficient and parental cells suggests that this binding is direct and does not require the integrity of the IKK structure.
DISCUSSION
The ability of PKR to activate NF-κB under various experimental conditions has been reported by others (18, 38, 46, 60). In this work, we have confirmed this finding and further demonstrated that the PKR-mediated activation of NF-κB does not require its kinase function.
We first showed that defective kinase mutants of PKR activate the expression of NF-κB-responsive reporter genes. The PKR constructs were found to stimulate the expression of luciferase placed under the control of the HIV-1 LTR, whereas they had no effect on its expression when placed under the control of HIV-1 ΔNF-κB. Alternative possibilities to explain this increase in luciferase expression, such as sequestration of endogenous PKR or competition for the substrate eIF2α with other eIF2α kinases (19, 51), could be ruled out. Indeed, activation of NF-κB was also observed in a PKR-free environment (PKR0/0 MEFs) and even by the PKR/Del42 mutant, which has a deletion of the region thought to bind eIF2α. The ability of a catalytically inactive PKR to activate NF-κB was next confirmed by EMSAs and by activation of the IKK complex. Finally, in vitro protein-protein interaction experiments allowed us to show that PKR binds specifically to the IKKβ subunit of the IKK complex.
In a recent report, Chu et al. have shown that PKR/KR296 can activate IKKβ, either after cotransfection in 3T3 cells with an epitope-tagged IKKβ-expressing plasmid or by direct interaction with purified proteins (9). The experiments that we have performed using PKR0/0 cells clearly show the activation of the natural endogenous IKK complex by both PKRwt and mutants and indicate that this activation is independent of the kinase function of PKR.
The fact that inactive PKR mutants have the ability to activate NF-κB through the activation of the IKK complex and the observation that PKR can physically interact with this complex indicate that PKR can be used as an adapter protein in this signaling pathway. Such a situation has also been reported for two other kinases. (i) One is the RIP kinase family, the members of which associate with the tumor necrosis factor alpha signaling complex (23, 40, 62). RIP contains three major domains: a kinase domain at its N terminus, an intermediate domain, and a death domain at its C terminus. Use of different RIP mutants and, in particular, their introduction into RIP-deficient cell lines showed that NF-κB activation did not require the kinase domain of RIP but did require the integrity of a charged domain located in its intermediate domain (23, 56). In contrast, the death domain of RIP was both necessary and sufficient for the induction of apoptosis (23). (ii) Another example of the participation of a kinase as an adapter protein is the IL-1 receptor-associated kinase IRAK. It has recently been reported that catalytically inactive IRAK mutants can restore IL-1 signaling in IL-1-unresponsive cell lines (34).
PKR is, however, best known as an inhibitor of protein synthesis through its ability to phosphorylate eIF2α and, in this respect, belongs to the growing family of stress-activated eIF2α kinases, such as GCN2, HRI, and PERK (5, 6, 14, 19, 53). Through this ability to inhibit protein translation, PKR participates in the antiviral action of IFN (44). Indeed, overexpression of PKR after insertion at the nef gene site of an infectious HIV-1 genome proved to severely inhibit the growth of HIV-1 (4). Our results suggest that PKR may activate IKK through protein-protein interactions with the IKKβ subunit. This notion is surprising, since some signaling pathways can be activated through the inhibition of the synthesis of repressors, as we have shown recently for c-Jun NH2-terminal kinase (JNK) (25) (see below). Therefore, activation of NF-κB through binding of PKR to IKK instead of inhibition of the synthesis of an inhibitor for IKK is a novel mechanism.
NF-κB is one of the transcription factors necessary for IFN induction, together with the IFN regulatory factors and the factor AP-1, itself dependent on the activation of JNK and the p38 MAP kinase (37). The exact mechanism by which a viral infection triggers PKR to bind to IKK and activate NF-κB is not known; however, dsRNA could be involved in this process, since PKR-deficient cells have been reported to be unresponsive to dsRNA for NF-κB activation (9, 30, 60).
Except for our reporter experiments performed with pHIV-1 LTR-luc, which led to the transcription of dsRNA-containing transcripts (the HIV-1 TAR region at the 5′ end of luc mRNAs), we have not addressed here the importance of dsRNA for PKR-mediated NF-κB or IKK activation. All of our activation data were obtained after transfecting the PKR plasmids alone in PKR0/0 MEFs. Similarly, the PKR/KR296-mediated activation of IKK analyzed by Chu et al. was also studied in the absence of added or transfected dsRNA (9). Therefore, the exact mechanism by which viral infection or dsRNA treatment leads to PKR docking to IKK remains an open question. It is possible, however, that the binding of PKR to dsRNA is sufficient to change the conformation of the protein (47) in a way that favors its interaction with IKKβ. It should be noted that the binding of PKR to IKKβ may not be sufficient to trigger IKK activation. We have shown that PKR can bind IKKβ from NEMO-deficient cells. However, NEMO-deficient cells are unresponsive to dsRNA for NF-κB activation (12, 59), although they do contain normal amounts of PKR (M. C. Bonnet and E. F. Meurs, unpublished data). Therefore, this result suggests that the PKR-mediated activation of NF-κB through IKKβ requires the presence of a complete IKK complex.
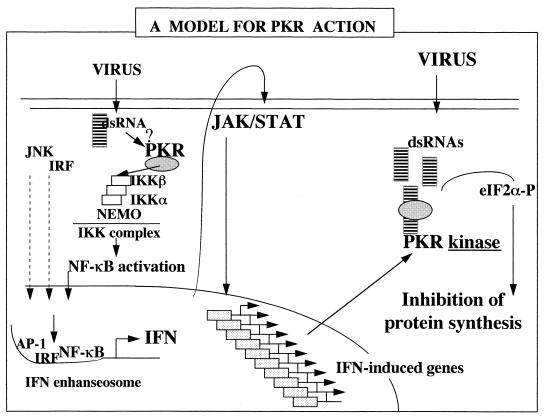
dsRNA has long been known to activate in IFN-treated cells, independently of PKR, a pathway leading to the activation of a cytosolic RNase (RNase L) which can degrade diverse RNA substrates, including 18S and 28S rRNAs, thus inhibiting cellular protein synthesis (35, 39, 64). We have recently shown, with the use of PKR- and RNase L-deficient murine cells, that those cells were unresponsive to JNK activation in response to dsRNA or virus infection, an effect which could be linked to the defect in the regulation of translation by PKR and RNase L (25). This result indicates that PKR and RNase L are involved in the activation of JNK, probably through the inhibition of a negative regulator for JNK, leading to the induction of the IFN gene (25). Therefore, the participation of PKR in the IFN system is dual: (i) as a kinase, it can regulate protein synthesis, which both limits viral propagation and reinforces the induction of IFN, through JNK activation; and (ii) as a protein, it activates IKK by protein-protein interactions, leading to NF-κB activation and the induction of the IFN gene, together with AP-1 and the IFN regulatory factors (Fig. 7). In this respect, the nature of the PKR motifs involved in its interaction with IKK and the potential role of dsRNA in this interaction remain to be investigated.
FIG. 7.
Model for PKR action. The presence of small amounts of dsRNA in cells, such as at the onset of viral infection, may provoke the binding of endogenous PKR to IKK, leading to NF-κB activation and potentiating the induction of IFN genes. For this function, PKR would not need its kinase function. Once IFN is synthesized, it provokes the induction of several genes, through JAK/STAT signaling, including the PKR gene. As the viral infection develops, there is an accumulation in the cytoplasm of dsRNA structures which activate PKR, the levels of which are now increased after IFN induction. As a kinase, PKR phosphorylates eIF2α and provokes the inhibition of protein synthesis, which can limit the propagation of the virus. eIF2α-P, phosphorylated eIF2α; IRF, IFN regulatory factor.
ACKNOWLEDGMENTS
We thank M. Kroll, F. Arenzana-Seisdedos, A. Gatignol, S. Ozden, G. Courtois, and A. Israel for reagents and helpful discussions.
This work was supported in part by a grant from the Agence Nationale de la Recherche sur le SIDA (ANRS 61852), Paris, France. M. Bonnet was supported by a grant from the Ministère de l'Enseignement, de la Recherche, et de la Technologie, Paris, France (MENRT).
REFERENCES
- 1.Alkalay I, Yaron A, Hatzubai A, Orian A, Ciechanover A, Ben-Neriah Y. Stimulation-dependent IκB phosphorylation marks the NF-κB inhibitor for degradation via the ubiquitin-proteasome pathway. Proc Natl Acad Sci USA. 1995;92:10599–10603. doi: 10.1073/pnas.92.23.10599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baeuerle P, Baichwal V. NF-κB as a frequent target for immunosuppressive and anti-inflammatory molecules. Adv Immunol. 1997;65:111–136. [PubMed] [Google Scholar]
- 3.Baldwin A. The NF-κB and IκB proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–681. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 4.Benkirane M, Neuveut C, Chun R F, Smith S M, Samuel C E, Gatignol A, Jeang K-T. Oncogenic potential of TAR RNA-binding TRBP and its regulatory interaction with protein kinase R. EMBO J. 1997;16:611–624. doi: 10.1093/emboj/16.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berlanga J J, Santoyo J, De Haro C. Characterization of a mammalian homolog of the GCN2 eukaryotic initiation factor 2α kinase. Eur J Biochem. 1999;265:754–762. doi: 10.1046/j.1432-1327.1999.00780.x. [DOI] [PubMed] [Google Scholar]
- 6.Berlanga J J, Herrero S, De Haro C. Characterization of the hemin-sensitive eukaryotic initiation factor 2α kinase from mouse nonerythroid cells. J Biol Chem. 1998;273:32340–32346. doi: 10.1074/jbc.273.48.32340. [DOI] [PubMed] [Google Scholar]
- 7.Chen Z, Hagler J, Palombella V, Melandri F, Scherer D, Ballard D, Maniatis T. Signal-induced site-specific phosphorylation targets IκBα to the ubiquitin-proteasome pathway. Genes Dev. 1995;9:1586–1597. doi: 10.1101/gad.9.13.1586. [DOI] [PubMed] [Google Scholar]
- 8.Chong K, Feng L, Donahue T F, Friesen J D, Meurs E, Hovanessian A G, Williams B R G. Human p68 kinase exhibits growth suppression in yeast and homology to the translational regulator GCN2. EMBO J. 1992;11:1553–1562. doi: 10.1002/j.1460-2075.1992.tb05200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chu W M, Ostertag D, Li Z, Chang L, Chen Y, Hu Y, Williams B R G, Perrault J, Karin M. JNK2 and IKKβ are required for activating the innate response to viral infection. Immunity. 1999;11:721–731. doi: 10.1016/s1074-7613(00)80146-6. [DOI] [PubMed] [Google Scholar]
- 10.Clemens M J, Androulla E. The double-stranded RNA-dependent protein kinase PKR: structure and function. J Interferon Res. 1997;17:503–524. doi: 10.1089/jir.1997.17.503. [DOI] [PubMed] [Google Scholar]
- 11.Clemens M J, Bommer U A. Translational control: the cancer connection. Int J Biochem Cell Biol. 1999;31:1–23. doi: 10.1016/s1357-2725(98)00127-7. [DOI] [PubMed] [Google Scholar]
- 12.Courtois G, Whiteside S T, Sibley C H, Israel A. Characterization of a mutant cell line that does not activate NF-κB in response to multiple stimuli. Mol Cell Biol. 1997;17:1441–1449. doi: 10.1128/mcb.17.3.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Craig A, Cosentino G, Donze O, Sonenberg N. The kinase insert domain of interferon-induced protein kinase PKR is required for activity but not for interaction with the pseudosubstrate K3L. J Biol Chem. 1996;271:24526–24533. doi: 10.1074/jbc.271.40.24526. [DOI] [PubMed] [Google Scholar]
- 14.Dever T E, Chen J J, Barber G N, Cigan A M, Feng L, Donahue T F, London I M, Katze M G, Hinnebusch A G. Mammalian eIF-2α kinases functionally substitute for GCN2 and stimulate GCN4 translation in yeast. Proc Natl Acad Sci USA. 1993;90:4616–4620. doi: 10.1073/pnas.90.10.4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DiDonato J A, Hayakawa M, Rothwarf D M, Zandi E, Karin M. A cytokine-responsive IκB kinase that activates the transcription factor NF-κB. Nature. 1997;388:548–554. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- 16.Duh E J, Maury W J, Folks T M, Fauci A S, Rabson A B. Tumor necrosis factor α activates human immunodeficiency virus type 1 through induction of nuclear factor binding to the NF-κB sites in the long terminal repeat. Proc Natl Acad Sci USA. 1989;86:5974–5978. [Google Scholar]
- 17.Everett H, McFadden G. Apoptosis: an innate immune response to virus infection. Trends Microbiol. 1999;7:160–165. doi: 10.1016/s0966-842x(99)01487-0. [DOI] [PubMed] [Google Scholar]
- 18.Gil J, Alcami J, Esteban M. Induction of apoptosis by double-stranded-RNA-dependent protein kinase (PKR) involves the α subunit of eukaryotic translation initiation factor 2 and NF-κB. Mol Cell Biol. 1999;19:4653–4663. doi: 10.1128/mcb.19.7.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harding H P, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 20.Henkel T, Zabel U, Van Zee K, Muller J, Fanning E, Baeuerle P A. Intramolecular masking of the nuclear location signal and dimerization domain in the precursor for the p50 NF-κB subunit. Cell. 1992;68:1121–1133. doi: 10.1016/0092-8674(92)90083-o. [DOI] [PubMed] [Google Scholar]
- 21.Hershey J W. Translational control in mammalian cells. Annu Rev Biochem. 1991;60:717–755. doi: 10.1146/annurev.bi.60.070191.003441. [DOI] [PubMed] [Google Scholar]
- 22.Hovanessian A G. The double-stranded RNA-activated protein kinase induced by interferon: dsRNA-PK. J Interferon Res. 1989;9:641–647. doi: 10.1089/jir.1989.9.641. [DOI] [PubMed] [Google Scholar]
- 23.Hsu H, Huang J, Shu H-B, Baichwal V, Goeddel D V. TNF-dependent recruitment of the protein kinase RIP to the TNF receptor-1 signaling complex. Immunity. 1996;4:387–396. doi: 10.1016/s1074-7613(00)80252-6. [DOI] [PubMed] [Google Scholar]
- 24.Hu Y, Baud V, Delhase M, Zhang P, Deerinck T, Ellisman M, Johnson R, Karin M. Abnormal morphogenesis but intact IKK activation in mice lacking the IKK α subunit of the IκB kinase. Science. 1999;284:316–320. doi: 10.1126/science.284.5412.316. [DOI] [PubMed] [Google Scholar]
- 25.Iordanov M S, Paranjape J M, Zhou A, Wong J, Williams B R G, Meurs E F, Silverman R H, Magun B E. Activation of p38 mitogen-activated protein kinase and c-jun NH2-terminal kinase by double-stranded RNA and encephalomyocarditis virus: involvement of RNase L, protein kinase R, and alternative pathways. Mol Cell Biol. 2000;20:617–627. doi: 10.1128/mcb.20.2.617-627.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katze M G, Wambach M, Wong M L, Garfinkel M, Meurs E, Chong K, Williams B R G, Hovanessian A G, Barber G N. Functional expression and RNA binding analysis of the interferon-induced, double-stranded RNA-activated 68,000-Mr protein kinase in a cell-free system. Mol Cell Biol. 1991;11:5497–5505. doi: 10.1128/mcb.11.11.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knighton D R, Zheng J H, Ten Eyck L F, Xuong N H, Taylor S S, Sowadski J M. Structure of a peptide inhibitor bound to the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991;253:414–420. doi: 10.1126/science.1862343. [DOI] [PubMed] [Google Scholar]
- 28.Kopp E, Gosh S. NF-κB: ten years after. Adv Immunol. 1995;58:1–27. doi: 10.1016/s0065-2776(08)60618-5. [DOI] [PubMed] [Google Scholar]
- 29.Kumar A, Haque J, Lacoste J, Hiscott J, Williams B R G. The dsRNA-dependent protein kinase, PKR, activates transcription factor NF-κB by phosphorylating IκB. Proc Natl Acad Sci USA. 1994;91:6288–6292. doi: 10.1073/pnas.91.14.6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar A, Yang Y L, Flati V, Der S, Kadereit S, Deb A, Hapque J, Reis L, Weissmann C, Williams B R. Deficient cytokine signaling in mouse embryo fibroblasts with a targeted deletion in the PKR gene: role of IRF-1 and NF-κB. EMBO J. 1996;16:406–416. doi: 10.1093/emboj/16.2.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laurent A G, Krust B, Galabru J, Svab J, Hovanessian A G. Monoclonal antibodies to interferon induced 68,000 Mr protein and their use for the detection of double-stranded RNA dependent protein kinase in human cells. Proc Natl Acad Sci USA. 1985;82:4341–4345. doi: 10.1073/pnas.82.13.4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee F, Hagler J, Chen Z, Maniatis T. Activation of the IκB α complex by MEKK1, a kinase of the JNK pathway. Cell. 1997;88:213–222. doi: 10.1016/s0092-8674(00)81842-5. [DOI] [PubMed] [Google Scholar]
- 33.Li Q, Van Antwerp D, Mercurio F, Lee K, Verma I. Severe liver degeneration in mice lacking the IκB kinase 2 gene. Science. 1999;284:321–325. doi: 10.1126/science.284.5412.321. [DOI] [PubMed] [Google Scholar]
- 34.Li X, Commane M, Burns C, Vithalani K, Cao Z, Stark G R. Mutant cells that do not respond to interleukin-1 (IL-1) reveal a novel role for IL-1 receptor-associated kinase. Mol Cell Biol. 1999;19:4643–4652. doi: 10.1128/mcb.19.7.4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li X L, Blackford J A, Hassel B A. RNase L mediates the antiviral effect of interferon through a selective reduction in viral RNA during encephalomyocarditis virus infection. J Virol. 1998;72:2752–2759. doi: 10.1128/jvi.72.4.2752-2759.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malinin N, Boldin M, Kovalenko A, Wallach D. MAP3K-related kinase involved in NF-κB induction by TNF, CD95 and IL-1. Nature. 1997;385:540–544. doi: 10.1038/385540a0. [DOI] [PubMed] [Google Scholar]
- 37.Mamane Y, Heylbroeck C, Génin P, Algarté M, Servant M J, LePage C, DeLuca C, Kwon H, Lin R, Hiscott J. Interferon regulatory factors: the next generation. Gene. 1999;237:1–14. doi: 10.1016/s0378-1119(99)00262-0. [DOI] [PubMed] [Google Scholar]
- 38.Maran A, Maitra R K, Kumar A, Dong B, Xiao W, Li G, Williams B R, Torrence P F, Silverman R H. Blockage of NF-kappa B signaling by selective ablation of an mRNA target by 2-5A antisense chimeras. Science. 1994;265:789–792. doi: 10.1126/science.7914032. [DOI] [PubMed] [Google Scholar]
- 39.Maran A, Waller C F, Paranjape J M, Li G, Xiao W, Zhang K, et al. 2′,5′-Oligoadenylate-antisense chimeras cause RNAse L to selectively degrade bcr/abl mRNA in chronic myelogenous leukemia cells. Blood. 1998;92:4336–4343. [PubMed] [Google Scholar]
- 40.McCarthy J V, Ni J, Dixit V M. RIP2 is a novel NF-κB-activating and cell death-inducing kinase. J Biol Chem. 1998;273:16968–16975. doi: 10.1074/jbc.273.27.16968. [DOI] [PubMed] [Google Scholar]
- 41.Mercurio F, Zhu H, Murray B, Shevchenko A, Bennett B, Li J, Young D, Barbosa M, Mann M, Manning A, Rao A. IKK-1 and IKK-2: cytokine-activated IκB kinases essential for NF-κB activation. Science. 1997;278:860–866. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- 42.Meurs E, Chong K, Galabru J, Thomas S B, Kerr I M, Williams B R G, Hovanessian A G. Molecular cloning and characterization of cDNA encoding human double-stranded RNA activated protein kinase induced by interferon. Cell. 1990;62:379–390. doi: 10.1016/0092-8674(90)90374-n. [DOI] [PubMed] [Google Scholar]
- 43.Meurs E F, McMillan N, Williams B R, Hovanessian A G, Southern P J. Human PKR transfected into murine cells stimulates expression of genes under control of the HIV1 or HTLV-1 LTR. Virology. 1995;214:653–659. doi: 10.1006/viro.1995.0080. [DOI] [PubMed] [Google Scholar]
- 44.Meurs E F, Watanabe Y, Kadereit S, Barber G N, Katze M G, Chong K, Williams B R G, Hovanessian A G. Constitutive expression of human double-stranded RNA-activated p68 kinase in murine cells mediates phosphorylation of eucaryotic initiation factor 2 and partial resistance to encephalomyocarditis virus growth. J Virol. 1992;66:5805–5814. doi: 10.1128/jvi.66.10.5805-5814.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Munoz E, Courtois G, Veschambre P, Jalinot P, Israel A. Tax induces nuclear translocation of NF-κB through dissociation of cytoplasmic complexes containing p105 or p100 but does not induce degradation of IκBα/MAD3. J Virol. 1994;68:8035–8044. doi: 10.1128/jvi.68.12.8035-8044.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nagai K, Hoi-Tao Wong A, Li S, Wai Ning Tam N, Cuddihy A R, Sonenberg N, Mathews M B, Hiscott J, Wainberg M A, Koromilas A E. Induction of CD4 expression and human immunodeficiency virus type 1 replication by mutants of the interferon-inducible protein kinase PKR. J Virol. 1997;71:1718–1725. doi: 10.1128/jvi.71.2.1718-1725.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patel R C, Stanton P, Sen G C. Specific mutations near the amino terminus of double-stranded RNA-dependent protein kinase (PKR) differentially affect its double-stranded RNA binding and dimerization properties. J Biol Chem. 1996;271:25657–25663. doi: 10.1074/jbc.271.41.25657. [DOI] [PubMed] [Google Scholar]
- 48.Regnier C H, Song H Y, Gao X, Goeddel D V, Cao Z, Rothe M. Identification and characterization of an IkappaB kinase. Cell. 1997;90:373–383. doi: 10.1016/s0092-8674(00)80344-x. [DOI] [PubMed] [Google Scholar]
- 49.Rothwarf D, Zandi E, Natoli G, Karin M. IKK-γ is an essential regulatory subunit of the IκB kinase complex. Nature. 1998;395:297–300. doi: 10.1038/26261. [DOI] [PubMed] [Google Scholar]
- 50.Samuel C E. Antiviral actions of interferon. Interferon-regulated cellular proteins and their surprisingly selective antiviral activities. Virology. 1991;183:1–11. doi: 10.1016/0042-6822(91)90112-o. [DOI] [PubMed] [Google Scholar]
- 51.Santoyo J, Alcalde J, Mendez R, Pulido D, De Haro C. Cloning and characterization of a cDNA encoding a protein synthesis initiation factor-2α (eIF-2α) kinase from Drosophila melanogaster. J Biol Chem. 1997;272:12544–12550. doi: 10.1074/jbc.272.19.12544. [DOI] [PubMed] [Google Scholar]
- 52.Seeler J S, Marchio A, Sitterlin D, Transy C, Dejean A. Interaction of SP100 with HP1 proteins: a link between the promyelocytic leukemia-associated nuclear bodies and the chromatin compartment. Proc Natl Acad Sci USA. 1998;95:7316–7321. doi: 10.1073/pnas.95.13.7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shi Y, Vattem K M, Sood R, An J, Liang J, Stramm L, Wek R C. Identification and characterization of pancreatic eukaryotic initiation factor 2 α-subunit, PEK, involved in translational control. Mol Cell Biol. 1998;18:7499–7509. doi: 10.1128/mcb.18.12.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stancovski I, Baltimore D. NF-κB activation: the IκB kinase revealed? Cell. 1997;91:299–302. doi: 10.1016/s0092-8674(00)80413-4. [DOI] [PubMed] [Google Scholar]
- 55.Takeda K, Takeuchi O, Tsujimura T, Itami S, Adachi O, Kawai T, Sanjo H, Yoshikawa K, Terada N, Akira S. Limb and skin abnormalities in mice lacking IKKα. Science. 1999;284:313–316. doi: 10.1126/science.284.5412.313. [DOI] [PubMed] [Google Scholar]
- 56.Ting A T, Pimentel-Muinos F X, Seed B. RIP mediates tumor necrosis factor receptor 1 activation of NF-κB but not Fas/APO-1-initiated apoptosis. EMBO J. 1996;15:6189–6196. [PMC free article] [PubMed] [Google Scholar]
- 57.Watanabe M, Muramatsu M, Hirai H, Suzuki T, Fujisawa J, Yoshida M, Arai K, Arai N. HTLV-1 encoded Tax in association with NF-κB precursor p105 enhances nuclear localization of NF-κB p50 and p65 in transfected cells. Oncogene. 1993;8:2949–2958. [PubMed] [Google Scholar]
- 58.Williams B R G. The role of the dsRNA-activated protein kinase, PKR, in signal transduction. Semin Virol. 1995;6:191–202. [Google Scholar]
- 59.Yamaoka S, Courtois G, Bessia C, Whiteside S T, Weil R, Agout F, Kirk H E, Kay R J, Israel A. Complementation cloning of NEMO, a component of the IκB kinase complex essential for NF-κB activation. Cell. 1998;93:1231–1240. doi: 10.1016/s0092-8674(00)81466-x. [DOI] [PubMed] [Google Scholar]
- 60.Yang Y L, Reis L F, Pavlovic J, Aguzzi A, Schafer R, Kumar A, Williams B R, Aguet M, Weissmann C. Deficient signaling in mice devoid of double-stranded RNA-dependent protein kinase. EMBO J. 1995;14:6095–6106. doi: 10.1002/j.1460-2075.1995.tb00300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yaron A, Gonen H, Alkalay I, Hatzubai A, Jung S, Beyth S, Mercurio F, Manning A, Ciechanover A, Ben-Neriah Y. Inhibition of NF-κB cellular function via specific targeting of the IκB ubiquitin ligase. EMBO J. 1997;16:101–107. doi: 10.1093/emboj/16.21.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu P W, Huang B C B, Shen M, Quast J, Chan E, Xu X, Nolan G P, Payan D G, Luo Y. Identification of RIP3, a RIP-like kinase that activates apoptosis and NF-κB. Curr Biol. 1999;9:539–542. doi: 10.1016/s0960-9822(99)80239-5. [DOI] [PubMed] [Google Scholar]
- 63.Zandi E, Rothwarf D M, Delhasse M, Hayakawa M, Karin M. The IκB kinase complex (IKK) contains two kinase subunits, IKKα and IKKβ, necessary for IκB phosphorylation and NF-κB activation. Cell. 1997;91:243–252. doi: 10.1016/s0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]
- 64.Zhou A, Hassel B A, Silverman R H. Expression cloning of 2-5A-dependent RNAse: a uniquely regulatory mediator of interferon action. Cell. 1993;72:753–765. doi: 10.1016/0092-8674(93)90403-d. [DOI] [PubMed] [Google Scholar]
- 65.Zinn K, Keller A, Whittemore L A, Maniatis T. 2-Aminopurine selectively inhibits the induction of β-interferon, c-fos, and c-myc gene expression. Science. 1988;240:210–213. doi: 10.1126/science.3281258. [DOI] [PubMed] [Google Scholar]