Class II Transactivator: Mastering the Art of Major Histocompatibility Complex Expression (original) (raw)
Major histocompatibility complex (MHC) class II molecules are the predominant presenters of exogenous antigens to T helper cells (reviewed in references 21, 77, and 99). These key molecules are critical for numerous aspects of immune function, including T-cell selection, tolerance induction, antibody production, T-cell-mediated immunity, and the inflammatory response. As principal mediators of transplant rejection, these molecules are often common targets for immune therapies to prevent the rejection of grafted tissues. Class II MHC is implicated as a contributing factor in a host of diseases ranging from rheumatoid arthritis and diabetes to Alzheimer's disease and multiple sclerosis.
Constitutive expression of class II MHC is restricted to “professional” antigen-presenting cells but can be induced on various tissues by gamma interferon (IFN-γ). In humans, a congenital lack of both constitutive and inducible class II results in a profound and generally fatal immunodeficiency (type II bare lymphocyte syndrome [BLS]) (7, 29, 45, 61, 67, 75, 112) marked by a significant reduction of CD4+ T cells. Early molecular forays addressing BLS revealed that the genes encoding class II MHC were not defective. Instead, the defect lay in transcription factors controlling class II MHC gene expression. BLS thus became the first disease known to be caused by defective or absent transcription factors. The availability of patient-derived cell lines with class II MHC transcription defects provided a unique tool of nature to identify the requisite transcription factors.
Transcriptional regulation of class II MHC expression is complex. Class II MHC and related promoters are characterized by the presence of conserved W (or S), X, and Y boxes (Fig. 1) (reviewed in references 10, 67, and 75). The X element is bipartite. The upstream X1 region is recognized by RFX, a trimeric complex of RFX family members including RFX5, RFXANK (RFX-B), and RFXAP (32, 74, 87, 117). The downstream X2 box is bound by X2BP (NF-X2), a complex comprising CREB, and an unidentified 120-kDa protein (83, 84). Another trimeric complex, NF-Y (CBF), which is highly conserved in eukaryotes, binds the Y box (69, 71, 146; reviewed in reference 70). A number of factors interacting with the W box have been described, including the RFX complex (26, 48, 120). The factors involved in X and Y box binding are ubiquitous and expressed constitutively yet fail to account for either constitutive or IFN-γ-inducible class II MHC expression. Somatic cell fusions using BLS patient-derived cells allowed the definition of complementation groups, with each group containing a defect in a single genetic locus. This type of analysis revealed a crucial locus, aIr-1, which in all likelihood encodes the class II transactivator (CIITA), which explained the lack of class II transcription in BLS complementation group A (1, 118). Group A cells express the requisite X and Y binding proteins but fail to transcribe class II. CIITA expression appears to be a nearly absolute requisite for expression of class II MHC, whether constitutive or inducible (17, 19, 23, 47, 85, 103, 114, 118, 119). A number of class II MHC-related genes including genes encoding HLA-DM (H-2M in mice) and invariant chain (Ii), with promoters similar to those for classical class II genes, are also regulated by CIITA (17, 18, 22, 23, 50, 137). CIITA can also upregulate expression of class I MHC genes and beta-2-microglobulin (β2m) through effects at site α in addition to X- and Y-like sequences in the promoters for these genes (40, 72, 104). These initial observations have led to the view that CIITA is a master, or global, regulator for expression of class II MHC and related genes.
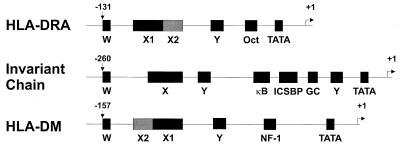
FIG. 1.
Organization of W, X, Y, and other motifs in the promoters of class II MHC and related genes. Genes coding for class II MHC and related proteins contain well conserved W, X, and Y boxes, the presence of which correlates with transcriptional regulation by CIITA.
Since the discovery of CIITA, numerous primary articles and several reviews on its role in regulating the class II MHC have been published. In this review, we will discuss the molecular structure of this novel protein, its mechanism of function, and its biologic and clinical relevance, which is broad.
A MASTER REGULATOR?
The father of all master regulators is MyoD, which when placed into 10T1/2 cells can cause these cells to acquire characteristics of muscle cells and the accompanying changes in gene transcription (reviewed in references 135 and 136). By this definition, CIITA is clearly a master regulator of class II MHC genes. Many reports have found that placing CIITA in an array of cell types can result in not only the induction of class II MHC promoters but also the expression of cell surface class II MHC proteins (8, 11, 18, 19, 23, 110, 118). Further, expression of class II MHC is controlled quantitatively by CIITA (96). In CIITA−/− mice, class II MHC is missing in almost all tissues and cells with one exception (see below). Invariant-chain expression is limited, but not absent, in CIITA−/− mice (17, 57). This is likely due to the presence of additional regulatory elements in the invariant-chain promoter (e.g., SP1 and NF-κB) (12, 13, 142). In addition, the expression of H-2O is not affected by the lack of CIITA (17). Most cytokines which alter class II MHC expression, such as IFN-γ, tumor necrosis factor α (TNF-α), transforming growth factor β, interleukin 1 (IL-1), IL-4, and IL-10, either up- or downregulate CIITA and class II MHC accordingly (18, 23, 42, 57, 88, 89, 92, 107, 119). In addition to cytokine-regulated class II MHC expression, the in situ expression of CIITA is also tightly linked to class II MHC gene expression (115, 116).
Developmentally, class II MHC is tightly associated with CIITA. Cell-type-specific and species-specific differences in class II MHC expression can also be explained by differences in CIITA. For example, CIITA is expressed in B cells but not in plasmacytomas where class II is extinguished (109, 114). CIITA is expressed in activated human T cells, which express class II MHC, but not in resting or activated mouse T cells, which lack class II MHC (20). However, one study has found some class II and CIITA message by reverse transcription-PCR (RT-PCR) in activated mouse T cells treated with IL-12 (41). Finally, CIITA regulates not only class II MHC but also Ii and DM molecules. Promoters of genes coding for all the aforementioned proteins have the W, X, and Y motifs, and CIITA works through these motifs (reviewed in references 67 and 126). Nonclassical class II genes are not obligatorily regulated by CIITA (17). Differential expression analyses show that the DNα gene is regulated by CIITA but that DOβ is not (125).
On the other hand, there are a few exceptions where CIITA is not required or associated with classical class II MHC gene expression. For example, it has been shown that in CIITA−/− mice, class II MHC+ dendritic cells are detected in lymph nodes and thymus, although the level is significantly lower than that for control mice (17, 139). These studies relied heavily on immunohistochemical staining and RT-PCR. The small amount of class II MHC found in these tissues is apparently not sufficient to permit development of a normal CD4+ T-cell population. We and others have found that TNF-α alteration of class II MHC may not be associated with changes in CIITA (30, 91). Indeed, it has been shown that negative modulation of class II MHC can occur via posttranscriptional mechanisms as has been observed with IFN-β (62), and posttranslational mechanisms affecting CIITA also occur with TNF-α (42). Additionally, NK cells have been reported to upregulate HLA-DR in fibrosarcoma cell lines in an IFN-γ- and CIITA-independent fashion dependent on cell contact (28). Finally, some invariant-chain and class II MHC isotype-specific expression in in vitro-generated mutant cell lines has been shown to occur in the absence of functional CIITA, which suggests the existence of isotype-specific transacting factors (31, 122, 149). However, in general, CIITA-independent class II expression appears to be the exception rather than the rule. This may be similar to gene regulation in muscle development, where the addition of MyoD cannot convert all cells to muscle cells and the lack of MyoD does not ablate all muscle development (4, 113). Hence, in a parallel comparison, CIITA compares favorably with MyoD to qualify as a master regulator.
GENETICS
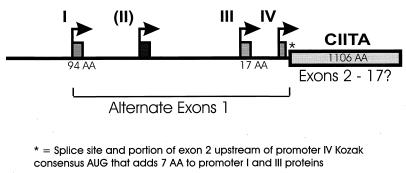
The coactivator function required for class II MHC expression was initially shown to reside on mouse chromosome 16 (the AIR-1 locus) (1). Human CIITA is encoded on chromosome 16 (119). The initial CIITA cDNA, cloned from the RJ2.2.5 B-cell lymphoma, encodes an 1,130-amino-acid protein. Genomic DNA from mice shows that CIITA is encoded by 19 exons (75), although the precise exon organization for human CIITA has not been reported. Expression of human CIITA is controlled by four distinct promoters, each with a distinct product, with three of the forms predominating. In mice only three promoters were identified (86) (Fig. 2). Promoter I is constitutively active in dendritic cells and has a promoter-specific first exon coding for 94 amino acids. In cDNA preparations, promoter II gave rise to a separate product, but this product is not present in significant amounts in any of the individual cell lines tested to date. Promoter III is constitutively active in B cells, responds to IFN-γ via a distal upstream sequence, and has a first exon encoding 17 amino acids (59, 86, 100, 101). The first 300 bp of promoter III is sufficient for B-cell function and contains two important sequences including a transcription elongation factor 2-like element (38). Promoter IV is IFN-γ responsive, active in the monocyte/macrophage lineage, endothelial cells, and fibroblasts, and drives expression of the shortest CIITA transcript (86, 101). Thus constitutive expression of CIITA generally results from promoters I and III. Promoter IV is likely responsible for the majority of IFN-γ-inducible expression, but sequences upstream of promoter III allow IFN-γ-mediated modulation of constitutive CIITA expression. The significance of the various isoforms of CIITA that differ only in the N terminus is presently unknown.
FIG. 2.
Promoter organization for CIITA. The CIITA upstream region in humans contains four promoters (I to IV) with independent start sites and alternate first exons. Significant mRNA populations have only been observed for I, III, and IV in vivo. The 3′ splice site for the first exons of promoters I and III is upstream of the translation start site for promoter IV mRNA and thus adds seven amino acid residues (∗). The core 1,106 amino acids of CIITA may be encoded by as many as 17 exons.
STRUCTURE AND FUNCTION
CIITA has predicted and apparent molecular masses of 123.5 and 135 to 140 kDa, respectively, suggesting some posttranslational modification (118). In those tissues and cell lines tested to date detection of the endogenous CIITA protein has been difficult at best, although some polyclonal-antibody preparations seem able to detect endogenous CIITA in whole-cell lysates of some cell types (14, 24). Intracellular concentrations are insufficient for immunohistochemical staining, and Western blotting requires larger numbers of cells for detection. Structure/function studies to date have been mostly driven by sequence comparison and have focused on the ability of transfected CIITA to activate transcription and subsequent expression from endogenous or engineered class II promoters, its compartmentalization, and its capacity to interact with other proteins.
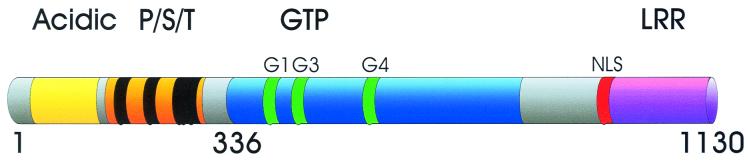
Sequence analyses of CIITA have revealed a complex domain structure (Fig. 3) composed of an amino-terminal acidic domain, proline-, serine-, and threonine-rich (PST) regions, a GTP-binding site, at least one nuclear localization sequence (NLS), and a series of leucine-rich repeats (LRR).
FIG. 3.
Schematic representation of protein domain organization for CIITA. Numbering corresponds to the B-cell form of CIITA. The first 336 amino acids of CIITA contain both an acidic domain and the PST domain. Residues 336 to 1130 contain the GTP-binding, NLS, and LRR motifs. G1, G3, and G4 mark the positions of specific motifs required for GTP binding (see text).
Acidic domain.
The N-terminal end of CIITA contains an acidic domain (residues 1 to 125) which through fusion to GAL4 DNA-binding sequences has been demonstrated to act as a transcription activation domain (54, 106, 148). The acidic activation domains of herpes simplex virus type 1α transinducing factor and VP16 (54, 148) can only partially substitute for that of CIITA (∼20 to 25% of wild type) using chimeric CIITA. Deletion of the acidic domain of CIITA results in a dominant-negative form of the protein (9, 14, 145) and has been touted as a possible vehicle for suppressing class II gene expression in transgenic animals. Much interest in proteins associating with CIITA has focused on this domain. A large number of proteins shown to interact with CIITA interact with residues in or near this domain. TFIIB, TAFIIs (30, 32, and 70), and CREB-binding protein (CBP) have all been demonstrated to interact with the acidic domain of CIITA in vitro, in vivo, and in some cases both (35, 36, 54, 68). In cotransfection experiments CBP cooperated with CIITA to increase DRA transcription, suggesting that CBP may provide a requisite histone acetyltransferase activity (36, 54).
PST domain.
CIITA contains a functionally necessary region with abundant proline, serine, and threonine (Fig. 3; residues 133 to 322) (24, 118). Domains rich in proline are common in a number of transcription factors, with either DNA-binding (16, 80) ability or activation domain (78, 123) properties independent of acidic sequences. In some instances, these proline-rich activation domains also have increased frequencies of serine, threonine, and glutamine as in the case of CTD-1, a prototypical transcription factor not utilizing an acidic activation domain (78). Limited functional data regarding the role of CIITA's PST domain exist. Deletion of the N-terminal or C-terminal half of this domain has no obvious impact on transactivation by CIITA, whereas complete deletion is highly detrimental to function and results in a dominant-negative protein (24). The nonessential carboxy-terminal half of this domain is absent in a CIITA mutant (106) and the published cDNA clone for mouse CIITA (115). Examination of the mouse genomic sequence reveals that the coding sequence for this region is present in a discrete exon of the mouse CIITA gene (J. F. Piskurich and J. P. Y. Ting, unpublished data), suggesting the possibility of splice variants. Deletion of 151 to 160 N-terminal residues from CIITA results in a potent dominant-negative form of the protein (9, 14, 145), indicating that the remaining, seemingly functional portion of the PST domain of CIITA likely fails to mediate activation events independent of the upstream acidic sequences. However, a GAL4-CIITA fusion protein containing residues 104 to 402 can bind CBP and enhance transcription from a GAL4 binding site containing a promoter (54). As some proline-rich sequences bind DNA (16), it is possible that this domain in CIITA may contact and/or bind DNA. The fact that CIITA has failed to bind DNA thus far makes this possibility seem unlikely.
Residues C-terminal to the acidic and PST domains (residues 317 to 1130) are necessary to promote transcription from a promoter comprising the W, X, and Y boxes (148). Relatively small perturbations of spacing and sequence in the DRA promoter render it unresponsive to IFN-γ (131), suggesting that CIITA is constrained by the promoter arrangement. An intact X box seems crucial to CIITA's ability to transactivate, suggesting cooperation between CIITA and X box binding proteins (83, 106). These observations suggest that a number of interactions, presumably with X box binding factors and NF-Y, occur with domains other than the acidic domain (see MODE OF ACTION). Amino acids 317 to 1130 of CIITA can be broken down further into a GTP-binding site, nuclear localization signal sequence, and a series of leucine-rich regions discussed below.
GTP-binding site.
The presence of a Walker A motif (also known as a P loop or G1 motif) known to be involved in ATP and GTP binding was noted with the initial description of CIITA (118). Further examination revealed GTP-binding motifs similar to those in other GTP-binding proteins, including a magnesium binding site (G3) and a guanine coordination site (G4). CIITA can bind GTP both in vitro and in vivo (43). Deletion or substantial mutation of individual sites (G1, G3, or G4) has a significant impact on both GTP binding and transactivation (14, 24, 43). In contrast, substitution with related sequences from Ras rescued CIITA function (43). Deletions in this region also have an impact on the ability of CIITA to “open” a closed promoter (157). GTP binding regulates a variety of cellular functions by acting as a molecular switch, where GTP binding results in one conformation (“on”) and GDP binding results in another (“off”). GTPase activity can be intrinsic or extrinsically provided by GTPase-activating proteins (GAPs) and can promote the off conformation. Exchange activity can be modified by guanine exchange factors (GEFs), which exchange GDP for GTP thus turning the protein on. In vitro, CIITA exhibits an apparent lack of intrinsic GTPase activity, and CIITA mutants that have intrinsic GTPase activity are functionally impaired (43). It is of great interest to determine if GAPs or GEFs regulate CIITA.
Recent experiments demonstrate that residues close to, if not within, the GTP-binding region (residues 336 to 702) can interact with themselves and with residues between 939 and 1130 (see M. W. Linhoff, J. A. Harton, B. J. Conti, D. E. Cressman, and J. P.-Y. Ting, submitted for publication; T. J. Sisk, S. Roys, and C.-H. Chang, submitted for publication). This finding suggests that cooperation within the GTP-binding domain or between the GTP-binding domain and C-terminal LRR, or both, may be important for nuclear localization or transcriptional activation.
NLSs.
CIITA is present in both the cytoplasm and the nuclei of transfected cells (28a), and this localization is critical for normal CIITA function. The simian virus 40 (SV40) NLS (KKRKK) recognized by the importins (karyopherins) has become the classic model for nuclear import of protein, with a plethora of nuclear proteins possessing some functionally essential iteration of the motif. A naturally occurring 24-amino-acid deletion in the C terminus of CIITA, identified in a BLS patient, lacks a 5-amino-acid motif very similar to the SV40 NLS. We have shown that this five-amino-acid deletion as well as the original patient-derived CIITA forms are present in the cytoplasm and not the nucleus. Functional studies of this region indicate that this motif in CIITA is essential for nuclear localization of CIITA and can function as an NLS independent of other CIITA-derived sequences (28a). GTP binding by CIITA is also required for nuclear import (43). We have postulated a relationship between the availability of GTP and the ability of CIITA to translocate to the nucleus as a means of regulating CIITA's activity. Elucidating such regulation will require further study. The relationship between NLS-dependent and GTP-dependent nuclear translocation of CIITA is unknown.
Cycling of CIITA between cytoplasm and the nucleus is suggested by cytoplasmic and nuclear expression (28a). CIITA exit from the nucleus, if occurring, should be mediated by nuclear export sequences (NES). Conforming roughly to the consensus LXXXLXXLXL, putative NES abound in CIITA, but functional studies have yet to elucidate which, if any, of these are relevant.
Leucine-rich regions.
Through sequence analysis of portions of CIITA it has become apparent that CIITA contains a number of leucine-rich sequences. These include leucine-charged domains (LCDs) (14), a series of leucine-rich repeats (LRR) (reviewed in reference 15) in the C terminus with similarity to those of Nod1, a nucleotide-dependent activator of caspases (46). Presumably these motifs mediate protein-protein interactions. Alanine substitution mutations in the LCD motifs of CIITA diminish class II MHC transcription (14). Some C-terminal deletions in CIITA (which happen to remove one or more LRR sequences) both abrogate transactivation function and confer a dominant-negative effect (9, 14, 22). While CIITA has fourLRR which conform precisely to the published consensus (15), CIITA also contains a number of LRR-like sequences (some of which only vary from the consensus by a single residue). Surprisingly, a point mutation changing F at 961 to S (within an LRR-like sequence adjacent to the identified NLS mentioned above) is responsible for an unusual BLS case. This particular patient was not diagnosed until his late twenties and died early in his thirties (102). In recent experiments examining CIITA self-association, specific point mutations within the LRR of CIITA diminished both transactivation and the ability of this region to self-associate with residues 336 to 702 (Linhoff et al., submitted); it remains unclear if these associations are direct or indirect. Mutations at various points in the LRR of CIITA affect the ability of CIITA to activate transcription, largely due to a defect in nuclear translocation (S. B. Hake, K. Masternak, C. Kammerbauer, C. Janzen, W. Reith, and V. Steimle, submitted for publication; J. Harton and J. P.-Y. Ting, unpublished data). Together these observations support an important role for CIITA's LRR.
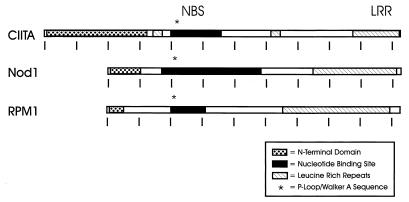
The LRR of CIITA have homology to a number of LRR-containing proteins. An example is the RNase inhibitor family, which includes human and pig RNase inhibitor and Schizosaccharomyces pombe rna1p (44, 52, 98). Surprisingly, Nod1 (see above) is similar to CIITA and contains a nucleotide binding site (NBS) motif upstream of its LRR (46). More intriguing still is the apparent conservation of NBS-LRR proteins, which function as disease resistance products in numerous species of plants (reviewed in reference 33). Remarkably, these proteins have NBS motifs and LRR with spacing and sizes similar to those of CIITA (Fig. 4), suggesting a divergent family of genes with a similar domain structure. It is tempting to speculate that these motifs are crucial for proteins which protect against infectious agents in both mammals and plants.
FIG. 4.
The domain organization of CIITA is similar to those of other NBS- and LRR-containing proteins. CIITA is compared to two NBS- and LRR-containing proteins, mammalian Nod1 and the plant disease resistance protein RPM1 (see text). The N-terminal domains of these proteins are dissimilar. Tick marks, intervals of approximately 100 amino acids.
MODE OF ACTION
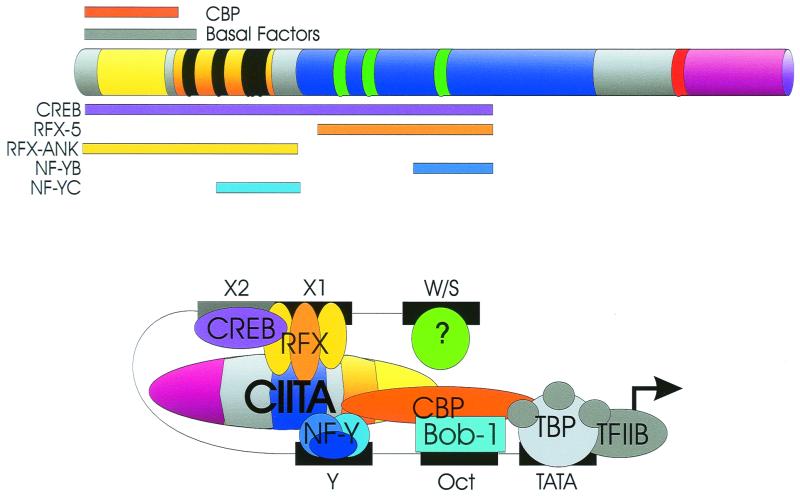
The mode of action of CIITA has been an enigma since its discovery, due to the lack of consensus DNA-binding motifs and its inability to bind W, X, and Y elements. However, CIITA requires intact W, X, and Y elements and requires their stereospecific alignment. Thus changes in X and Y by half a helical turn destroy the ability of CIITA to upregulate these genes, while insertion of a whole helical turn does not (150). This parallels early data demonstrating that IFN-γ induction of class II MHC promoters also requires an aligned promoter (130, 131). These data suggest that proteins binding to W, X, and Y may interact with CIITA in a highly specific three-dimensional structure allowing proper binding and intermolecular interactions. Indeed two lines of evidence now indicate that this is occurring. The first is indirect, showing that exogenously expressed CIITA can result in the in vivo protein binding of W, X, and Y of class II MHC, Ii, and DM promoters, as shown by in vivo footprinting (105, 132, 141). Second and more directly, in vitro and in vivo analyses have shown that CIITA interacts with RFX5, RFXANK, CREB, NF-YB, and NF-YC (75, 111, 150; Hake et al., submitted). Furthermore, recruitment of CIITA into the transcription complex requires the multiple, synergistic interactions provided by these transcription factors (76). Interactions between these factors and CIITA have been observed within the following residues of CIITA: RFX5, 335 to 612; RFXANK, 1 to 335; CREB, 1 to 612; NF-YB, 518 to 612; NF-YC, 218 to 335 (150). The recent observation that deleterious mutations in the LRR fail to disrupt RFX5, RFXANK, NF-YB, and NF-YC (Hake et al., submitted) is consistent with the mapping result above. Interactions between CIITA, RFX, CREB, and NF-Y are likely to be important as suggested by the observation that a C-terminal domain of RFX is required for cooperative binding between RFX and NF-Y (138). CIITA interactions with the B-cell-specific protein Bob-1 (OcaB) (34) have been described, but the domain involved is unknown. Thus many of these proteins could be contacting independent domains of CIITA (acidic, PST, or GTP binding), the exact sites of which will hopefully be informative. Considering the interactions between CIITA and components of the basal transcription machinery (see Structure and Function, “Acidic domain”), these observations have led to a firm view of CIITA as a scaffold which acts as an integrator or enhanceosome for class II transcription (37, 76, 127, 150; S. J. P. Gobin, M. van Zutphen, S. D. Westerheide, J. M. Boss, and P. van den Elsen, submitted for publication). Thus a composite picture would show all these proteins binding to CIITA and localized to the promoter (Fig. 5). A question of whether this binding is direct or indirect still has not been completely resolved and awaits the isolation of these proteins in pure form.
FIG. 5.
Multiple transcription factor contacts allow CIITA to function as a transcriptional scaffold and integrator. CIITA contacts components of the basal transcription machinery via its acidic domain. Interactions with RFX, RFXANK, CREB (X2BP), NF-YB, and NF-YC have been mapped. The region of CIITA required for each interaction is shown (see text). Domain markings for CIITA are the same as in Fig. 3. TBP, TATA-binding protein.
BIOLOGICAL SIGNIFICANCE
The implications of a master regulator controlling expression of class II MHC genes are immense. Aside from the obvious importance of CIITA to BLS, the regulation of CIITA largely determines the presence or absence of class II MHC and its degree of expression and thus the nature of the immune response. Consequently, CIITA is exceedingly important whenever class II MHC is required and will impact infectious diseases, neurologic disorders, autoimmune diseases, tumor rejection, graft acceptance, and perhaps even normal development. Furthermore, as CIITA is a single protein that affects a large family of genes, it represents an ideal target for pharmacological intervention in the class II antigen presentation pathway, whether for immune suppression in transplantation and autoimmunity or for enhancement for BLS and tumor rejection.
An emerging pathway by which pathogens escape the immune system involves modulating CIITA expression. Cytomegalovirus, for example, downregulates expression of CIITA by altering the IFN-γ signal pathway, thus diminishing the immune stimulatory function of class II MHC (60, 79). During Mycobacterium bovis BCG infection of murine macrophages, a mycobacterial N gene product diminishes IFN-γ-mediated phosphorylation of STAT-1α, thus reducing CIITA and class II MHC expression (140). Chlamydia similarly interferes with CIITA transcription through the degradation of upstream stimulatory factor 1 (147).
In contrast to examples where CIITA is downregulated during infection, the human immunodeficiency virus (HIV) utilizes CIITA to upregulate HIV long terminal repeat (LTR) function (108). There is a correlation between expression of class II MHC and higher HIV expression and replication. The introduction of CIITA into T cells leads to increased viral replication and transcription from the HIV LTR. CIITA is expressed by activated human T cells and macrophages, and both of these cell types are the primary targets of HIV, both having developed different receptors for HIV. This correlation is intriguing and suggests that CIITA, not unlike other known positive regulators of HIV LTR (i.e., Tat and NF-κB), regulates the transcription and replication of HIV. In contrast, mutations in the cysteine-rich portion of HIV Tat (C22S37G and C37G) decrease class II MHC transcription, leading to diminished class II MHC expression in the THP-1 (monocytic) and H-9 (T-lymphocyte) cell lines, an effect not seen with wild-type Tat (128). Interestingly these mutations can decrease class II MHC transcription without affecting CIITA expression (128), likely through disruptive interaction with transcription elongation factor b (49). This suggests a clever mechanism to evade immune detection (via loss of class II MHC) while potentially maintaining CIITA-dependent HIV LTR transcription. A seeming dilemma exists in the literature. Tosi et al. show that mutant HIV Tat, but not the wild type, interferes with class II MHC transcription and expression, whereas Kanazawa et al. present the opposite with a different mutation in the cysteine-rich region of HIV Tat (49, 128). This difference may be due simply to differential effects of HIV Tat and its mutants on expression of class II MHC in THP-1 and H-9 cells versus COS cells.
As discussed above, a lack or decrease in class II MHC can prove advantageous to invading pathogens. Conversely, induced class II MHC expression has an implicated role in inflammatory processes (reviewed in references 2, 27, 66, and 138) and the role of class II MHC in autoimmune disease has been extensively studied (reviewed in references 77, 90, and 124). The role of CIITA in these diseases is currently being studied. In nonobese diabetic mice it has been shown that the lack of CIITA prevented diabetes despite noticeable pancreatic infiltration (82), whereas class II MHC deficiency alone was insufficient to prevent cytotoxic T-cell-mediated disease pathology in a model of lymphocytic choriomeningitis virus-induced diabetes (55). In autoimmune thyroiditis, potential interactions between CIITA and single-strand binding protein 1, a regulator of thyroid-stimulating hormone gene expression, may contribute to increased class II MHC expression (5, 81), thus allowing thyroid cell antigen presentation to T cells (143).
Correction of the BLS defect is an obvious use of CIITA in gene therapy. Retroviral transfer of CIITA was recently employed to correct class II deficiency in cells from a BLS patient (11). Another significant application is the potential use of dominant-negative forms of CIITA in the production of class II MHC-deficient organs for transplantation (9, 22, 145). We have found that the lack of CIITA in heart donor grafts also greatly enhances graft survival in totally allogeneic hosts (W. J. Brickey, N. J. Felix, and J. P.-Y. Ting, unpublished data). This level of enhancement is beyond that produced by Aβ−/− grafts, perhaps owing to repression of all class II MHC expression including Ii and DM in CIITA knockout mice.
Tumors often lack class II MHC and thus have diminished immunogenicity. The expression of class II MHC and/or costimulatory molecules has shown promise in increasing tumor immunogenicity and tumor rejection (6, 94, 95). Loss of the retinoblastoma protein (Rb), a tumor suppressor, has been linked with loss of class II inducibility in retinoblastoma, non-small-cell lung carcinoma and bladder carcinoma (63, 65, 93). Rb is thought to in some way allow or enhance the accessibility of class II MHC promoters (63). Whether CIITA requires Rb to function is unclear and may depend on the relative expression of the individual proteins or other cell-type-specific factors (64, 129). In tumors with intact Rb, a failure to upregulate CIITA in response to IFN-γ is the most common explanation for absent class II MHC (64). CIITA is absent in small-cell lung cancer, and transfection of CIITA can restore class II expression (144). Restoring class II expression to tumor lines by introduction of either the IFN-γ gene (97) or the gene for CIITA (73) can improve the immunogenicity of the tumor. However, overexpression of CIITA leading to high levels of class II MHC correlated with increased tumorigenicity (73), suggesting that overexpression of CIITA may be detrimental in tumor therapy approaches. Other experiments suggest that class II-transfected tumors are more immunogenic than CIITA transfectants due to an ability of class II-transfected tumors to present endogenous antigen whereas CIITA expression also induces Ii, which favors presentation of exogenous antigens (3, 25).
As mentioned above, class I MHC and β2m can also be transcriptionally regulated by CIITA (40, 72, 104). Both class I and β2m promoters contain W, X, and Y motifs (40, 107), consistent with the requirement of these elements for transactivation by CIITA. In cell lines, CIITA has been observed to increase and in some cases induce class I expression following transfection of CIITA (78). However, class I MHC expression in CIITA-deficient mice is apparently normal. An answer to this conspicuous paradox may lie in the genetics of development. Trophoblasts lack expression of the classical class I and class II MHC (reviewed in references 56 and 134, expressing instead the nonclassical class I molecule HLA-G (53). CIITA fails to transactivate the HLA-G promoter in extravillous cytotrophoblast cells but does transactivate HLA-A and B promoters (39). Expression of CIITA can induce class I promoter activity in trophoblast-derived choriocarcinoma cells (58). Thus CIITA appears to be under tight control early in development. As CIITA-deficient mice develop normally, suggesting that the lack of CIITA does not interfere with development, one is left to propose that the effects of CIITA on class I transcription might have subtle roles during normal development. Alternatively, it could be easily argued that class I MHC expression in vivo is not regulated by CIITA. This interpretation is teleologically unsatisfying and begs the question of the necessity of obvious W, X, and Y elements within the class I MHC. Furthermore, recent chromatin immunoprecipitation experiments reveal that CIITA is bound to class I MHC and β2 promoters in vivo (76). It may be best to conclude that a physiologically relevant role for CIITA in regulating class I MHC expression will likely be subtle and possibly restricted to limited developmental or cell-specific events.
These early studies suggest that, in principle, modulation of CIITA, whether by suppression to generate better transplants or avoid autoimmune disease or by introduction to improve immune responses to pathogens or tumors, is a promising field of study with enormous potential.
SUMMARY
Great progress in understanding the relative importance of various portions of CIITA for transcriptional activation of class II MHC genes has been made since CIITA's discovery in 1993. Emerging from these studies is a fairly consistent picture where CIITA is expressed, binds GTP, translocates to the nucleus, and interacts with specific DNA-binding transcription factors and basal transcription components, thus opening and activating class II MHC and related promoters. Despite these strides, this model is essentially unchanged from that initially espoused. The observation that class II MHC promoters in some B cells are bound to X and Y box binding proteins and thus open even in the absence of CIITA, whereas these same promoters in non-B cells are closed until CIITA is present, is provocative. One potential explanation is that CIITA possesses two distinct functions, the ability to direct the opening of responsive promoters (presumably through some form of remodeling) and the ability to activate transcription through its activation domain and protein-protein interactions (132, 141). The presence of a locus control region responsive to a B-cell-specific factor is another possibility, yet CIITA must, in some fashion, be directing chromatin remodeling in cells which can be induced to express CIITA. While CBP is an obvious candidate for mediating remodeling, no conclusive experiments have shown that CBP is required for the remodeling of class II MHC promoters. The studies above support interactions between CIITA and transcription factors, but does CIITA merely bind these factors to place the activation domain appropriately? Why has it been difficult to demonstrate a role for CIITA in a transcription complex? Is GTP binding only essential for nuclear import? Is nuclear export of CIITA occurring and is it relevant? What aspect of class II MHC transcription requires that retinoblastoma protein Rb be present? Is CIITA a prototype for a family of transcriptional coactivators? Why is limited class II expression observed in the absence of CIITA? The evolutionary conservation of W-, X-, and Y-containing promoters in mammals, birds (104), amphibians (51), and fish (121) suggests that CIITA may be extremely old; what are its origins?
All remaining questions aside, CIITA is truly a remarkable protein. Controlled by up to four separate promoters, CIITA has been imparted a complex pattern of inducible and constitutive expression that can be regulated in developmental pathways. Through exercising specific control over the transcription of every major component of class II MHC antigen presentation pathway, CIITA gains the title of a master regulator. As CIITA appears to be class II MHC specific, it can be thought of as the core transcription factor of which all the remaining components are but cofactors. This is central to the concept of CIITA as a scaffolding protein or integrator and perhaps alters our view of transcriptional control away from promoters and individual factors towards a more unified enhanceosome perspective.
The view of CIITA as a master regulator has implications for practical applications that are staggering. Successful engineering of dominant-negative CIITAs may lead to the production of transplant tissues unable to express class II MHC and the associated self peptides which contribute so significantly to graft rejection. A thorough understanding of CIITA's molecular mechanisms may lead to therapeutics which allow temporary enhancement or suppression of class II MHC, thus favorably altering the immune response during critical events in pathogenesis, autoimmune disease, tumorigenesis, and neuroinflammation.
REFERENCES
- 1.Accolla R S, Jotterand-Bellomo M, Scarpellino L, Maffei A, Carra G, Guardiola J. aIr-1, a newly found locus on mouse chromosome 16 encoding a trans-acting activator factor for MHC class II gene expression. J Exp Med. 1986;164:369–374. doi: 10.1084/jem.164.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aisen P S. Inflammation and Alzheimer disease. Mol Chem Neuropathol. 1996;28:83–88. doi: 10.1007/BF02815208. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong T D, Clements V K, Martin B K, Ting J P Y, Ostrand-Rosenberg S. Major histocompatibility complex class II-transfected tumor cells present endogenous antigen and are potent inducers of tumor-specific immunity. Proc Natl Acad Sci USA. 1997;94:6886–6891. doi: 10.1073/pnas.94.13.6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnold H H, Braun T. Targeted inactivation of myogenic factor genes reveals their role during mouse myogenesis: a review. Int J Dev Biol. 1996;40:345–353. [PubMed] [Google Scholar]
- 5.Balducci-Silano P L, Suzuki K, Ohta M, Saito J, Ohmori M, Montani V, Napolitano G, Shong M, Taniguchi S I, Pietrarelli M, Lavaroni S, Mori A, Singer D S, Kohn L D. Regulation of major histocompatibility (MHC) class II human leukocyte antigen-DR alpha gene expression in thyrocytes by single strand binding protein-1, a transcription factor that also regulates thyrotropin receptor and MHC class I gene expression. Endocrinology. 1998;139:2300–2313. doi: 10.1210/endo.139.5.5991. [DOI] [PubMed] [Google Scholar]
- 6.Baskar S, Clements V K, Glimcher L H, Nabavi N, Ostrand-Rosenberg S. Rejection of MHC class II-transfected tumor cells requires induction of tumor-encoded B7-1 and/or B7-2 costimulatory molecules. J Immunol. 1996;156:3821–3827. [PubMed] [Google Scholar]
- 7.Benichou B, Strominger J L. Class II-antigen-negative patient and mutant B-cell lines represent at least three, and probably four, distinct genetic defects defined by complementation analysis. Proc Natl Acad Sci USA. 1991;88:4285–4288. doi: 10.1073/pnas.88.10.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beutler A S, Banck M S, Wedekind D, Hedrich H J. Tumor gene therapy made easy: allogeneic major histocompatibility complex in the C6 rat glioma model. Hum Gene Ther. 1999;10:95–101. doi: 10.1089/10430349950019228. [DOI] [PubMed] [Google Scholar]
- 9.Bontron S, Ucla C, Mach B, Steimle V. Efficient repression of endogenous major histocompatibility complex class II expression through dominant negative CIITA mutants isolated by a functional selection strategy. Mol Cell Biol. 1997;17:4249–4258. doi: 10.1128/mcb.17.8.4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boss J M. Regulation of transcription of MHC class II genes. Curr Opin Immunol. 1997;9:107–113. doi: 10.1016/s0952-7915(97)80166-5. [DOI] [PubMed] [Google Scholar]
- 11.Bradley M B, Fernandez J M, Ungers G, Diaz-Barrientos T, Steimle V, Mach B, O'Reilly R, Lee J S. Correction of defective expression in MHC class II deficiency (bare lymphocyte syndrome) cells by retroviral transduction of CIITA. J Immunol. 1997;159:1086–1095. [PubMed] [Google Scholar]
- 12.Brown A M, Linhoff M W, Stein B, Wright K L, Baldwin A S J, Basta P V, Ting J P. Function of NF-κB/Rel binding sites in the major histocompatibility complex class II invariant chain promoter is dependent on cell-specific binding of different NF-κB/Rel subunits. Mol Cell Biol. 1994;14:2926–2935. doi: 10.1128/mcb.14.5.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown A M, Wright K L, Ting J P. Human major histocompatibility complex class II-associated invariant chain gene promoter. Functional analysis and in vivo protein/DNA interactions of constitutive and IFN-gamma-induced expression. J Biol Chem. 1993;268:26328–26333. [PubMed] [Google Scholar]
- 14.Brown J A, Rogers E M, Boss J M. The MHC class II transactivator (CIITA) requires conserved leucine charged domains for interactions with the conserved W box promoter element. Nucleic Acids Res. 1998;26:4128–4136. doi: 10.1093/nar/26.18.4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buchanan S G, Gay N J. Structural and functional diversity in the leucine-rich repeat family of proteins. Prog Biophys Mol Biol. 1996;65:1–44. doi: 10.1016/s0079-6107(96)00003-x. [DOI] [PubMed] [Google Scholar]
- 16.Calvert I, Peng Z Q, Kung H F, Raziuddin Cloning and characterization of a novel sequence-specific DNA-binding protein recognizing the negative regulatory element (NRE) region of the HIV-1 long terminal repeat. Gene. 1991;101:171–176. doi: 10.1016/0378-1119(91)90408-4. [DOI] [PubMed] [Google Scholar]
- 17.Chang C-H, Guerder S, Hong S-C, van Ewijk W, Flavell R A. Mice lacking the MHC class II transactivator (CIITA) show tissue-specific impairment of MHC class II expression. Immunity. 1996;4:167–178. doi: 10.1016/s1074-7613(00)80681-0. [DOI] [PubMed] [Google Scholar]
- 18.Chang C H, Flavell R A. Class II transactivator regulates the expression of multiple genes involved in antigen presentation. J Exp Med. 1995;181:765–767. doi: 10.1084/jem.181.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang C H, Fontes J D, Peterlin M, Flavell R A. Class II transactivator (CIITA) is sufficient for the inducible expression of major histocompatibility complex class II genes. J Exp Med. 1994;180:1367–1374. doi: 10.1084/jem.180.4.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang C H, Hong S C, Hughes C C, Janeway C A, Jr, Flavell R A. CIITA activates the expression of MHC class II genes in mouse T cells. Int Immunol. 1995;7:1515–1518. doi: 10.1093/intimm/7.9.1515. [DOI] [PubMed] [Google Scholar]
- 21.Chapman H A. Endosomal proteolysis and MHC class II function. Curr Opin Immunol. 1998;10:93–102. doi: 10.1016/s0952-7915(98)80038-1. [DOI] [PubMed] [Google Scholar]
- 22.Chin K-C, Li G X, Ting J P-Y. Activation and transdominant suppression of MHC class II and HLA-DMB promoters by a series of C-terminal CIITA deletion mutants. J Immunol. 1997;159:2789–2794. [PubMed] [Google Scholar]
- 23.Chin K-C, Mao C, Skinner C, Riley J L, Wright K L, Moreno C S, Stark G R, Boss J M, Ting J P-Y. Molecular analysis of G1B and G3A IFN gamma mutants reveals that defects in CIITA or RFX result in defective class II MHC and Ii gene induction. Immunity. 1994;1:687–697. doi: 10.1016/1074-7613(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 24.Chin K C, Li G G, Ting J P. Importance of acidic, proline/serine/threonine-rich, and GTP-binding regions in the major histocompatibility complex class II transactivator: generation of transdominant-negative mutants. Proc Natl Acad Sci USA. 1997;94:2501–2506. doi: 10.1073/pnas.94.6.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clements V K, Baskar S, Armstrong T D, Ostrand-Rosenberg S. Invariant chain alters the malignant phenotype of MHC class II+ tumor cells. J Immunol. 1992;149:2391–2396. [PubMed] [Google Scholar]
- 26.Cogswell J P, Austin J, Ting J P. The W element is a positive regulator of HLA-DRA transcription in various DR+ cell types. J Immunol. 1991;146:1361–1367. [PubMed] [Google Scholar]
- 27.Collawn J F, Benveniste E N. Regulation of MHC class II expression in the central nervous system. Microbes Infect. 1999;1:893–902. doi: 10.1016/s1286-4579(99)00228-2. [DOI] [PubMed] [Google Scholar]
- 28.Collinge M, Pardi R, Bender J R. Class II transactivator-independent endothelial cell MHC class II gene activation induced by lymphocyte adhesion. J Immunol. 1998;161:1589–1593. [PubMed] [Google Scholar]
- 28a.Cressman D E, Chin K-C, Taxman D J, Ting J P-Y. A defect in the nuclear translocation of CIITA causes a form of type II bare lymphocyte syndrome. Immunity. 1999;10:163–171. doi: 10.1016/s1074-7613(00)80017-5. [DOI] [PubMed] [Google Scholar]
- 29.de Preval C, Lisowska-Grospierre B, Loche M, Griscelli C, Mach B. A trans-acting class II regulatory gene unlinked to the MHC controls expression of HLA class II genes. Nature. 1985;318:291. doi: 10.1038/318291a0. [DOI] [PubMed] [Google Scholar]
- 30.Dong Y, Rohn W M, Benveniste E N. IFN-gamma regulation of the type IV class II transactivator promoter in astrocytes. J Immunol. 1999;162:4731–4739. [PubMed] [Google Scholar]
- 31.Douhan J, Lieberson R, Knoll J H, Zhou H, Glimcher L H. An isotype-specific activator of major histocompatibility complex (MHC) class II genes that is independent of class II transactivator. J Exp Med. 1997;185:1885–1895. doi: 10.1084/jem.185.11.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Durand B, Sperisen P, Emery P, Barras E, Zufferey M, Mach B, Reith W. RFXAP, a novel subunit of the RFX DNA binding complex is mutated in MHC class II deficiency. EMBO J. 1997;16:1045–1055. doi: 10.1093/emboj/16.5.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Erickson F L, Dinesh-Kumar S P, Holzberg S, Ustach C V, Dutton M, Handley V, Corr C, Baker B J. Interactions between tobacco mosaic virus and the tobacco N gene. Philos Trans R Soc Lond B Biol Sci. 1999;354:653–658. doi: 10.1098/rstb.1999.0417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fontes J D, Jabrane-Ferrat N, Toth C R, Peterlin B M. Binding and cooperative interactions between two B cell-specific transcriptional coactivators. J Exp Med. 1996;183:2517–2521. doi: 10.1084/jem.183.6.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fontes J D, Jiang B, Peterlin B M. The class II trans-activator CIITA interacts with the TBP-associated factor TAFII32. Nucleic Acids Res. 1997;25:2522–2528. doi: 10.1093/nar/25.12.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fontes J D, Kanazawa S, Jean D, Peterlin B M. Interactions between class II transactivator and CREB binding protein increase transcription of major histocompatibility complex class II genes. Mol Cell Biol. 1999;19:941–947. doi: 10.1128/mcb.19.1.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fontes J D, Kanazawa S, Nekrep N, Peterlin B M. The class II transactivator CIITA is a transcriptional integrator. Microbes Infect. 1999;1:863–869. doi: 10.1016/s1286-4579(99)00232-4. [DOI] [PubMed] [Google Scholar]
- 38.Ghosh N, Piskurich J F, Wright G, Hassani K, Ting J P, Wright K L. A novel element and a TEF-2-like element activate the major histocompatibility complex class II transactivator in B-lymphocytes. J Biol Chem. 1999;274:32342–32350. doi: 10.1074/jbc.274.45.32342. [DOI] [PubMed] [Google Scholar]
- 39.Gobin S J, van den Elsen P J. The regulation of HLA class I expression: is HLA-G the odd one out? Semin Cancer Biol. 1999;9:55–59. doi: 10.1006/scbi.1998.0109. [DOI] [PubMed] [Google Scholar]
- 40.Gobin S J P, Peijnenburg A, Keijsers V, van den Elsen P J. Site alpha is crucial for two routes of IFN-gamma-induced MHC class I transactivation: the ISRE-mediated route and a novel pathway involving CIITA. Immunity. 1997;6:601–611. doi: 10.1016/s1074-7613(00)80348-9. [DOI] [PubMed] [Google Scholar]
- 41.Gourley T, Roys S, Lukacs N W, Kunkel S L, Flavell R A, Chang C H. A novel role for the major histocompatibility complex class II transactivator CIITA in the repression of IL-4 production. Immunity. 1999;10:377–386. doi: 10.1016/s1074-7613(00)80037-0. [DOI] [PubMed] [Google Scholar]
- 42.Han Y, Zhou Z H, Ransohoff R M. TNF-alpha suppresses IFN-gamma-induced MHC class II expression in HT1080 cells by destabilizing class II trans-activator mRNA. J Immunol. 1999;163:1435–1440. [PubMed] [Google Scholar]
- 43.Harton J A, Cressman D E, Chin K C, Der C J, Ting J P. GTP binding by class II transactivator: role in nuclear import. Science. 1999;285:1402–1405. doi: 10.1126/science.285.5432.1402. [DOI] [PubMed] [Google Scholar]
- 44.Hillig R C, Renault L, Vetter I R, Drell IV T, Wittinghofer A, Becker J. The crystal structure of rna1p: a new fold for a GTPase-activating protein. Mol Cell. 1999;3:781–791. doi: 10.1016/s1097-2765(01)80010-1. [DOI] [PubMed] [Google Scholar]
- 45.Hume C R, Lee J S. Congenital immunodeficiencies associated with absence of HLA class II antigens on lymphocytes results from distinct mutations in trans-acting factors. Hum Immunol. 1989;26:288–309. doi: 10.1016/0198-8859(89)90007-4. [DOI] [PubMed] [Google Scholar]
- 46.Inohara N, Koseki T, del Peso L, Hu Y, Yee C, Chen S, Carrio R, Merino J, Liu D, Ni J, Nunez G. Nod1, an Apaf-1-like activator of caspase-9 and nuclear factor-kappaB. J Biol Chem. 1999;274:14560–14567. doi: 10.1074/jbc.274.21.14560. [DOI] [PubMed] [Google Scholar]
- 47.Itoh-Lindstrom Y, Piskurich J F, Felix N J, Wang Y, Brickey W J, Platt J L, Koller B H, Ting J P. Reduced IL-4-, lipopolysaccharide-, and IFN-gamma-induced MHC class II expression in mice lacking class II transactivator due to targeted deletion of the GTP-binding domain. J Immunol. 1999;163:2425–2431. [PubMed] [Google Scholar]
- 48.Jabrane-Ferrat N, Fontes J D, Boss J M, Peterlin B M. Complex architecture of major histocompatibility complex class II promoters: reiterated motifs and conserved protein-protein interactions. Mol Cell Biol. 1996;16:4683–4690. doi: 10.1128/mcb.16.9.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kanazawa S, Okamoto T, Peterlin B M. Tat competes with CIITA for the binding to P-TEFb and blocks the expression of MHC class II genes in HIV infection. Immunity. 2000;12:61–70. doi: 10.1016/s1074-7613(00)80159-4. [DOI] [PubMed] [Google Scholar]
- 50.Kern I, Steimle V, Siegrist C A, Mach B. The two novel MHC class II transactivators RFX5 and CIITA both control expression of HLA-DM genes. Int Immunol. 1995;7:1295–1299. doi: 10.1093/intimm/7.8.1295. [DOI] [PubMed] [Google Scholar]
- 51.Kobari F, Sato K, Shum B P, Tochinai S, Katagiri M, Ishibashi T, Du Pasquier L, Flajnik M F, Kasahara M. Exon-intron organization of Xenopus MHC class II beta chain genes. Immunogen. 1995;42:376–385. doi: 10.1007/BF00179399. [DOI] [PubMed] [Google Scholar]
- 52.Kobe B, Deisenhofer J. Crystal structure of porcine ribonuclease inhibitor, a protein with leucine-rich repeats. Nature. 1993;366:751–756. doi: 10.1038/366751a0. [DOI] [PubMed] [Google Scholar]
- 53.Kovats S, Main E K, Librach C, Stubblebine M, Fisher S J, DeMars R. A class I antigen, HLA-G, expressed in human trophoblasts. Science. 1990;248:220–223. doi: 10.1126/science.2326636. [DOI] [PubMed] [Google Scholar]
- 54.Kretsovali A, Agalioti T, Spilianakis C, Tzortzakaki E, Merika M, Papamatheakis J. Involvement of CREB binding protein in expression of major histocompatibility complex class II genes via interaction with class II transactivator. Mol Cell Biol. 1998;18:6777–6783. doi: 10.1128/mcb.18.11.6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Laufer T M, von Herrath M G, Grusby M J, Oldstone M B, Glimcher L H. Autoimmune diabetes can be induced in transgenic major histocompatibility complex class II-deficient mice. J Exp Med. 1993;178:589–596. doi: 10.1084/jem.178.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Le Bouteiller P, Solier C, Proll J, Aguerre-Girr M, Fournel S, Lenfant F. Placental HLA-G protein expression in vivo: where and what for? Hum Reprod Update. 1999;5:223–233. doi: 10.1093/humupd/5.3.223. [DOI] [PubMed] [Google Scholar]
- 57.Lee Y J, Han Y, Lu H T, Nguyen V, Qin H, Howe P H, Hocevar B A, Boss J M, Ransohoff R M, Benveniste E N. TGF-beta suppresses IFN-gamma induction of class II MHC gene expression by inhibiting class II transactivator messenger RNA expression. J Immunol. 1997;158:2065–2075. [PubMed] [Google Scholar]
- 58.Lefebvre S, Moreau P, Dausset J, Carosella E D, Paul P. Downregulation of HLA class I gene transcription in choriocarcinoma cells is controlled by the proximal promoter element and can be reversed by CIITA. Placenta. 1999;20:293–301. doi: 10.1053/plac.1998.0380. [DOI] [PubMed] [Google Scholar]
- 59.Lennon A M, Ottone C, Rigaud G, Deaven L L, Longmire J, Fellous M, Bono R, Alcaide-Loridan C. Isolation of a B-cell-specific promoter for the human class II transactivator. Immunogen. 1997;45:266–273. doi: 10.1007/s002510050202. [DOI] [PubMed] [Google Scholar]
- 60.Le Roy E, Muhlethaler-Mottet A, Davrinche C, Mach B, Davignon J L. Escape of human cytomegalovirus from HLA-DR-restricted CD4+ T-cell response is mediated by repression of gamma interferon-induced class II transactivator expression. J Virol. 1999;73:6582–6589. doi: 10.1128/jvi.73.8.6582-6589.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lisowska-Grospierre B, Fondaneche M C, Rols M P, Griscelli C, Fischer A. Two complementation groups account for most cases of inherited MHC class II deficiency. Hum Mol Genet. 1994;3:953–958. doi: 10.1093/hmg/3.6.953. [DOI] [PubMed] [Google Scholar]
- 62.Lu H T, Riley J L, Babcock G T, Huston M, Stark G R, Boss J M, Ransohoff R M. Interferon (IFN) beta acts downstream of IFN-gamma-induced class II transactivator messenger RNA accumulation to block major histocompatibility complex class II gene expression and requires the 48-kD DNA-binding protein, ISGF3-gamma. J Exp Med. 1995;182:1517–1525. doi: 10.1084/jem.182.5.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lu Y, Boss J M, Hu S X, Xu H J, Blanck G. Apoptosis-independent retinoblastoma protein rescue of HLA class II messenger RNA IFN-gamma inducibility in non-small cell lung carcinoma cells. Lack of surface class II expression associated with a specific defect in HLA-DRA induction. J Immunol. 1996;156:2495–2502. [PubMed] [Google Scholar]
- 64.Lu Y, Tschickardt M E, Schmidt B J, Blanck G. IFN-gamma inducibility of class II transactivator is specifically lacking in human tumour lines: relevance to retinoblastoma protein rescue of IFN-gamma inducibility of the HLA class II genes. Immunol Cell Biol. 1997;75:325–332. doi: 10.1038/icb.1997.50. [DOI] [PubMed] [Google Scholar]
- 65.Lu Y, Ussery G D, Muncaster M M, Gallie B L, Blanck G. Evidence for retinoblastoma protein (RB) dependent and independent IFN-gamma responses: RB coordinately rescues IFN-gamma induction of MHC class II gene transcription in noninducible breast carcinoma cells. Oncogene. 1994;9:1015–1019. [PubMed] [Google Scholar]
- 66.MacDermott R P. Alterations of the mucosal immune system in inflammatory bowel disease. J Gastroenterol. 1996;31:907–916. doi: 10.1007/BF02358624. [DOI] [PubMed] [Google Scholar]
- 67.Mach B, Steimle V, Martinez-Soria E, Reith W. Regulation of MHC class II genes: lessons from a disease. Annu Rev Immunol. 1996;14:301–331. doi: 10.1146/annurev.immunol.14.1.301. [DOI] [PubMed] [Google Scholar]
- 68.Mahanta S K, Scholl T, Yang F C, Strominger J L. Transactivation by CIITA, the type II bare lymphocyte syndrome-associated factor, requires participation of multiple regions of the TATA box binding protein. Proc Natl Acad Sci USA. 1997;94:6324–6329. doi: 10.1073/pnas.94.12.6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maity S N, Sinha S, Ruteshouser E C, de Crombrugghe B. Three different polypeptides are necessary for DNA binding of mammalian heteromeric CCAAT binding factor. J Biol Chem. 1992;267:16574–16580. [PubMed] [Google Scholar]
- 70.Mantovani R. The molecular biology of the CCAAT-binding factor NF-Y. Gene. 1999;239:15–27. doi: 10.1016/s0378-1119(99)00368-6. [DOI] [PubMed] [Google Scholar]
- 71.Mantovani R, Pessara U, Tronche F, Li X Y, Knapp A M, Pasquali J L, Benoist C, Mathis D. Monoclonal antibodies to NF-Y define its function in MHC class II and albumin gene transcription. EMBO J. 1992;11:3315–3322. doi: 10.1002/j.1460-2075.1992.tb05410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Martin B K, Chin K-C, Skinner C A, Olsen J C, Dey A, Ozato K, Ting J P-Y. Induction of MHC class I expression by the MHC class II transactivator (CIITA) Immunity. 1997;6:591–600. doi: 10.1016/s1074-7613(00)80347-7. [DOI] [PubMed] [Google Scholar]
- 73.Martin B K, Frelinger J G, Ting J P. Combination gene therapy with CD86 and the MHC class II transactivator in the control of lung tumor growth. J Immunol. 1999;162:6663–6670. [PubMed] [Google Scholar]
- 74.Masternak K, Barras E, Zufferey M, Conrad B, Corthals G, Aebersold R, Sanchez J C, Hochstrasser D F, Mach B, Reith W. A gene encoding a novel RFX-associated transactivator is mutated in the majority of MHC class II deficiency patients. Nat Genet. 1998;20:273–277. doi: 10.1038/3081. [DOI] [PubMed] [Google Scholar]
- 75.Masternak, K., A. Muhlethaler-Mottet, J. Villard, M. Peretti, and W. Reith. Molecular genetics of the bare lymphocyte syndrome. Rev. Immunogenet., in press. [PubMed]
- 76.Masternak K, Muhlethaler-Mottet A, Villard J, Zufferey M, Steimle V, Reith W. CIITA is a transcriptional coactivator that is recruited to MHC class II promoters by multiple synergistic interactions with an enhanceosome complex. Genes Dev. 2000;14:1156–1166. [PMC free article] [PubMed] [Google Scholar]
- 77.McDevitt H O. The role of MHC class II molecules in susceptibility and resistance to autoimmunity. Curr Opin Immunol. 1998;10:677–681. doi: 10.1016/s0952-7915(98)80088-5. [DOI] [PubMed] [Google Scholar]
- 78.Mermod N, O'Neill E A, Kelly T J, Tjian R. The proline-rich transcriptional activator of CTF/NF-I is distinct from the replication and DNA binding domain. Cell. 1989;58:741–753. doi: 10.1016/0092-8674(89)90108-6. [DOI] [PubMed] [Google Scholar]
- 79.Miller D M, Rahill B M, Boss J M, Lairmore M D, Durbin J E, Waldman J W, Sedmak D D. Human cytomegalovirus inhibits major histocompatibility complex class II expression by disruption of the Jak/Stat pathway. J Exp Med. 1998;187:675–683. doi: 10.1084/jem.187.5.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mitchell P J, Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989;245:371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- 81.Montani V, Taniguchi S I, Shong M, Suzuki K, Ohmori M, Giuliani C, Napolitano G, Saji M, Fiorentino B, Reimold A M, Ting J P, Kohn L D, Singer D S. Major histocompatibility class II HLA-DR alpha gene expression in thyrocytes: counter regulation by class II transactivator and the thyroid Y box protein. Endocrinology. 1998;139:280–289. doi: 10.1210/endo.139.1.5673. [DOI] [PubMed] [Google Scholar]
- 82.Mora C, Wong F S, Chang C H, Flavell R A. Pancreatic infiltration but not diabetes occurs in the relative absence of MHC class II-restricted CD4 T cells: studies using NOD/CIITA-deficient mice. J Immunol. 1999;162:4576–4588. [PubMed] [Google Scholar]
- 83.Moreno C S, Beresford G W, Louis-Plence P, Morris A C, Boss J M. CREB regulates MHC class II expression in a CIITA-dependent manner. Immunity. 1999;10:143–151. doi: 10.1016/s1074-7613(00)80015-1. [DOI] [PubMed] [Google Scholar]
- 84.Moreno C S, Emery P, West J E, Durand B, Reith W, Mach B, Boss J M. Purified X2BP cooperatively binds class II MHC X box region in the presence of purified RFX, the X box factor deficient in the bare lymphocyte syndrome. J Immunol. 1995;155:4313–4321. [PubMed] [Google Scholar]
- 85.Morris A C, Riley J L, Fleming W H, Boss J M. MHC class II gene silencing in trophoblast cells is caused by inhibition of CIITA expression. Am J Reprod Immunol. 1998;40:385–394. doi: 10.1111/j.1600-0897.1998.tb00423.x. [DOI] [PubMed] [Google Scholar]
- 86.Muhlethaler-Mottet A, Otten L A, Steimle V, Mach B. Expression of MHC class II molecules in different cellular and functional compartments is controlled by differential usage of multiple promoters of the transactivator CIITA. EMBO J. 1997;16:2851–2860. doi: 10.1093/emboj/16.10.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nagarajan U M, Louis-Plence P, DeSandro A, Nilsen R, Bushey A, Boss J M. RFX-B is the gene responsible for the most common cause of the bare lymphocyte syndrome, an MHC class II immunodeficiency. Immunity. 1999;10:153–162. doi: 10.1016/s1074-7613(00)80016-3. . (Erratum, 10:399.) [DOI] [PubMed] [Google Scholar]
- 88.Nandan D, Reiner N E. TGF-beta attenuates class II transactivator and reveals an accessory pathway of IFN-gamma action. J Immunol. 1997;158:1095–1101. [PubMed] [Google Scholar]
- 89.Navarrete Santos A, Kehlen A, Schutte W, Langner J, Riemann D. Regulation by transforming growth factor-beta1 of class II mRNA and protein expression in fibroblast-like synoviocytes from patients with rheumatoid arthritis. Int Immunol. 1998;10:601–607. doi: 10.1093/intimm/10.5.601. [DOI] [PubMed] [Google Scholar]
- 90.Nepom G T, Erlich H. MHC class II molecules and autoimmunity. Annu Rev Immunol. 1991;9:493–525. doi: 10.1146/annurev.iy.09.040191.002425. [DOI] [PubMed] [Google Scholar]
- 91.Nikcevich K M, Piskurich J F, Hellendall R P, Wang Y, Ting J P. Differential selectivity of CIITA promoter activation by IFN-gamma and IRF-1 in astrocytes and macrophages: CIITA promoter activation is not affected by TNF-alpha. J Neuroimmunol. 1999;99:195–204. doi: 10.1016/s0165-5728(99)00117-4. [DOI] [PubMed] [Google Scholar]
- 92.O'Keefe G M, Nguyen V T, Benveniste E N. Class II transactivator and class II MHC gene expression in microglia: modulation by the cytokines TGF-beta, IL-4, IL-13 and IL-10. Eur J Immunol. 1999;29:1275–1285. doi: 10.1002/(SICI)1521-4141(199904)29:04<1275::AID-IMMU1275>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 93.Osborne A, Tschickardt M, Blanck G. Retinoblastoma protein expression facilitates chromatin remodeling at the HLA-DRA promoter. Nucleic Acids Res. 1997;25:5095–5102. doi: 10.1093/nar/25.24.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ostrand-Rosenberg S, Clements V K, Thakur A, Cole G A. Transfection of major histocompatibility complex class I and class II genes causes tumour rejection. J Immunogenet (Oxford) 1989;16:343–349. doi: 10.1111/j.1744-313x.1989.tb00481.x. [DOI] [PubMed] [Google Scholar]
- 95.Ostrand-Rosenberg S, Thakur A, Clements V. Rejection of mouse sarcoma cells after transfection of MHC class II genes. J Immunol. 1990;144:4068–4071. [PubMed] [Google Scholar]
- 96.Otten L A, Steimle V, Bontron S, Mach B. Quantitative control of MHC class II expression by the transactivator CIITA. Eur J Immunol. 1998;28:473–478. doi: 10.1002/(SICI)1521-4141(199802)28:02<473::AID-IMMU473>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 97.Panelli M C, Wang E, Shen S, Schluter S F, Bernstein R M, Hersh E M, Stopeck A, Gangavalli R, Barber J, Jolly D, Akporiaye E T. Interferon (IFN)-gamma gene transfer of an EMT6 tumor that is poorly responsive to IFN-gamma stimulation: increase in tumor immunogenicity is accompanied by induction of a mouse class II transactivator and class II MHC. Cancer Immunol Immunother. 1996;42:99–107. doi: 10.1007/s002620050258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Papageorgiou A C, Shapiro R, Acharya K R. Molecular recognition of human angiogenin by placental ribonuclease inhibitor—an X-ray crystallographic study at 2.0 A resolution. EMBO J. 1997;16:5162–5177. doi: 10.1093/emboj/16.17.5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pieters J. MHC class II restricted antigen presentation. Curr Opin Immunol. 1997;9:89–96. doi: 10.1016/s0952-7915(97)80164-1. [DOI] [PubMed] [Google Scholar]
- 100.Piskurich J F, Linhoff M W, Wang Y, Ting J P. Two distinct gamma interferon-inducible promoters of the major histocompatibility complex class II transactivator gene are differentially regulated by STAT1, interferon regulatory factor 1, and transforming growth factor β. Mol Cell Biol. 1999;19:431–440. doi: 10.1128/mcb.19.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Piskurich J F, Wang Y, Linhoff M W, White L C, Ting J P. Identification of distinct regions of 5′ flanking DNA that mediate constitutive, IFN-gamma, STAT1, and TGF-beta-regulated expression of class II transactivator gene. J Immunol. 1998;160:233–240. [PubMed] [Google Scholar]
- 102.Quan V, Towey M, Sacks S, Kelly A P. Absence of MHC class II gene expression in a patient with a single amino acid substitution in class II transactivator protein CIITA. Immunogen. 1999;49:957–963. doi: 10.1007/s002510050579. [DOI] [PubMed] [Google Scholar]
- 103.Ramassar V, Goes N, Hobart M, Halloran P F. Evidence for the in vivo role of class II transactivator in basal and IFN-gamma induced class II expression in mouse tissue. Transplantation. 1996;62:1901–1907. doi: 10.1097/00007890-199612270-00038. [DOI] [PubMed] [Google Scholar]
- 104.Riegert P, Andersen R, Bumstead N, Dohring C, Dominguez-Steglich M, Engberg J, Salomonsen J, Schmid M, Schwager J, Skjodt K, Kaufman J. The chicken beta 2-microglobulin gene is located on a non-major histocompatibility complex microchromosome: a small, G+C-rich gene with X and Y boxes in the promoter. Proc Natl Acad Sci USA. 1996;93:1243–1248. doi: 10.1073/pnas.93.3.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rigaud G, Barbaro A D, Nicolis M, Cestari T, Ramarli D, Riviera A-P, Accolla R S. Induction of CIITA and modification of in vivo HLA-DRA promoter occupancy in normal thymic epithelial cells treated with IFN-gamma. J Immunol. 1996;156:4254–4258. [PubMed] [Google Scholar]
- 106.Riley J L, Westerheide S D, Price J A, Brown J A, Boss J M. Activation of class II MHC genes requires both the X box region and class II transactivator (CIITA) Immunity. 1995;2:533–543. doi: 10.1016/1074-7613(95)90033-0. [DOI] [PubMed] [Google Scholar]
- 107.Rohn W, Tang L P, Dong Y, Benveniste E N. IL-1 beta inhibits IFN-gamma-induced class II MHC expression by suppressing transcription of class II transactivator gene. J Immunol. 1999;162:886–896. [PubMed] [Google Scholar]
- 108.Saifuddin M, Roebuck K A, Chang C H, Ting J P Y, Spear G T. Cutting edge: activation of HIV-1 transcription by the MHC class II transactivator. J Immunol. 2000;164:3941–3945. doi: 10.4049/jimmunol.164.8.3941. [DOI] [PubMed] [Google Scholar]
- 109.Sartoris S, Tosi G, De Lerma Barbaro A, Cestari T, Accolla R S. Active suppression of class II transactivator-encoding AIR-1 locus is responsible for the lack of major histocompatibility complex class II gene expression observed during differentiation from B cells to plasma cells. Eur J Immunol. 1996;26:2456–2460. doi: 10.1002/eji.1830261028. [DOI] [PubMed] [Google Scholar]
- 110.Sartoris S, Valle M T, Barbaro A L, Tosi G, Cestari T, D'Agostino A, Megiovanni A M, Manca F, Accolla R S. HLA class II expression in uninducible hepatocarcinoma cells after transfection of AIR-1 gene product CIITA: acquisition of antigen processing and presentation capacity. J Immunol. 1998;161:814–820. [PubMed] [Google Scholar]
- 111.Scholl T, Mahanta S K, Strominger J L. Specific complex formation between the type II bare lymphocyte syndrome-associated transactivators CIITA and RFX5. Proc Natl Acad Sci USA. 1997;94:6330–6334. doi: 10.1073/pnas.94.12.6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Seidl C, Saraiya C, Osterweil Z, Fu Y P, Lee J S. Genetic complexity of regulatory mutants defective for HLA class II gene expression. J Immunol. 1992;148:1576–1584. [PubMed] [Google Scholar]
- 113.Shani M, Faerman A, Emerson C P, Pearson-White S, Dekel I, Magal Y. The consequences of a constitutive expression of MyoD1 in ES cells and mouse embryos. Symp Soc Exp Biol. 1992;46:19–36. [PubMed] [Google Scholar]
- 114.Silacci P, Mottet A, Steimle V, Reith W, Mach B. Developmental extinction of major histocompatibility complex class II gene expression in plasmocytes is mediated by silencing of the transactivator gene CIITA. J Exp Med. 1994;180:1329–1336. doi: 10.1084/jem.180.4.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sims T N, Goes N B, Ramassar V, Urmson J, Halloran P F. In vivo class II transactivator expression in mice is induced by a non-interferon-gamma mechanism in response to local injury. Transplantation. 1997;64:1657–1664. doi: 10.1097/00007890-199712270-00005. [DOI] [PubMed] [Google Scholar]
- 116.Sims T N, Halloran P F. MHC class II regulation in vivo in the mouse kidney. Microbes Infect. 1999;1:903–912. doi: 10.1016/s1286-4579(99)00227-0. [DOI] [PubMed] [Google Scholar]
- 117.Steimle V, Durand B, Barras E, Zufferey M, Hadam M R, Mach B, Reith W. A novel DNA-binding regulatory factor is mutated in primary MHC class II deficiency (bare lymphocyte syndrome) Genes Dev. 1995;9:1021–1032. doi: 10.1101/gad.9.9.1021. [DOI] [PubMed] [Google Scholar]
- 118.Steimle V, Otten L A, Zufferey M, Mach B. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency (or bare lymphocyte syndrome) Cell. 1993;75:135–146. [PubMed] [Google Scholar]
- 119.Steimle V, Siegrist C A, Mottet A, Lisowska-Grospierre B, Mach B. Regulation of MHC class II expression by interferon-gamma mediated by the transactivator gene CIITA. Science. 1994;265:106–109. doi: 10.1126/science.8016643. [DOI] [PubMed] [Google Scholar]
- 120.Sugawara M, Scholl T, Ponath P D, Strominger J L. A factor that regulates class II major histocompatibility complex gene DPA is a member of a subfamily of zinc finger proteins that includes a Drosophila developmental control protein. Mol Cell Biol. 1994;14:8438–8450. doi: 10.1128/mcb.14.12.8438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sultmann H, Mayer W E, Figueroa F, O'Huigin C, Klein J. Organization of MHC class II B genes in the zebrafish (Brachydanio rerio) Genomics. 1994;23:1–14. doi: 10.1006/geno.1994.1452. [DOI] [PubMed] [Google Scholar]
- 122.Tai A K, Zhou G, Chau K, Ono S J. Cis-element dependence and occupancy of the human invariant chain promoter in CIITA-dependent and -independent transcription. Mol Immunol. 1999;36:447–460. doi: 10.1016/s0161-5890(99)00061-9. [DOI] [PubMed] [Google Scholar]
- 123.Tanaka M, Herr W. Differential transcriptional activation by Oct-1 and Oct-2: interdependent activation domains induce Oct-2 phosphorylation. Cell. 1990;60:375. doi: 10.1016/0092-8674(90)90589-7. [DOI] [PubMed] [Google Scholar]
- 124.Taneja V, David C S. HLA class II transgenic mice as models of human diseases. Immunol Rev. 1999;169:67–79. doi: 10.1111/j.1600-065x.1999.tb01307.x. [DOI] [PubMed] [Google Scholar]
- 125.Taxman, D. J., D. E. Cressman, and J. P. Y. Ting. Identification of CIITA-regulated genes by representational difference analysis: discoordinate regulation of the DN/DO heterdodimer. J. Immunol., in press. [DOI] [PubMed]
- 126.Ting J P-Y, Wright K L, Chin K-C, Brickey W J, Li G. The DMB promoter: delineation, in vivo footprint, trans-activation and transdominant-suppression. J Immunol. 1997;159:5457–5462. [PubMed] [Google Scholar]
- 127.Ting J P-Y, Zhu X S. Class II MHC genes: a model gene regulatory system with great biologic consequences. Microbes Infect. 1999;1:855–861. [PubMed] [Google Scholar]
- 128.Tosi G, De Lerma Barbaro A, D'Agostino A, Valle M T, Megiovanni A M, Manca F, Caputo A, Barbanti-Brodano G, Accolla R S. HIV-1 Tat mutants in the cysteine-rich region downregulate HLA class II expression in T lymphocytic and macrophage cell lines. Eur J Immunol. 2000;30:19–28. doi: 10.1002/1521-4141(200001)30:1<19::AID-IMMU19>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 129.Tschickardt M E, Lu Y, Jacim M, Ussery G D, Steimle V, Mach B, Blanck G. RB and a novel E2F-1 binding protein in MHC class II deficient B-cell lines and normal IFN-gamma induction of the class IL transactivator CIITA in class II non-inducible RB-defective tumor lines. Int J Cancer. 1995;62:461–465. doi: 10.1002/ijc.2910620417. [DOI] [PubMed] [Google Scholar]
- 130.Vilen B J, Cogswell J P, Ting J P. Stereospecific alignment of the X and Y elements is required for major histocompatibility complex class II DRA promoter function. Mol Cell Biol. 1991;11:2406–2415. doi: 10.1128/mcb.11.5.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Vilen B J, Penta J F, Ting J P-Y. Structural constraints within a trimeric transcriptional regulatory region: constitutive and interferon-gamma inducible expression of the HLA-DRA gene. J Biol Chem. 1992;267:23728–23734. [PubMed] [Google Scholar]
- 132.Villard J, Muhlethaler-Mottet A, Bontron S, Mach B, Reith W. CIITA-induced occupation of MHC class II promoters is independent of the cooperative stabilization of the promoter-bound multi-protein complexes. Int Immunol. 1999;11:461–469. doi: 10.1093/intimm/11.3.461. [DOI] [PubMed] [Google Scholar]
- 133.Villard J, Peretti M, Masternak K, Barras E, Caretti G, Mantovani R, Reith W. A functionally essential domain of RFX5 mediates activation of major histocompatibility complex class II promoters by promoting cooperative binding between RFX and NF-Y. Mol Cell Biol. 2000;20:3364–3376. doi: 10.1128/mcb.20.10.3364-3376.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Weetman A P. The immunology of pregnancy. Thyroid. 1999;9:643–646. doi: 10.1089/thy.1999.9.643. [DOI] [PubMed] [Google Scholar]
- 135.Weintraub H. The MyoD family and myogenesis: redundancy, networks, and thresholds. Cell. 1993;75:1241–1244. doi: 10.1016/0092-8674(93)90610-3. [DOI] [PubMed] [Google Scholar]
- 136.Weintraub H, Davis R, Tapscott S, Thayer M, Krause M, Benezra R, Blackwell T K, Turner D, Rupp R, Hollenberg S. The myoD gene family: nodal point during specification of the muscle cell lineage. Science. 1991;251:761–766. doi: 10.1126/science.1846704. [DOI] [PubMed] [Google Scholar]
- 137.Westerheide S D, Louis-Plence P, Ping D, He X F, Boss J M. HLA-DMA and HLA-DMB gene expression functions through the conserved S-X-Y region. J Immunol. 1997;158:4812–4821. [PubMed] [Google Scholar]
- 138.Weyand C M, Goronzy J J. HLA polymorphisms and T cells in rheumatoid arthritis. Int Rev Immunol. 1999;18:37–59. doi: 10.3109/08830189909043018. [DOI] [PubMed] [Google Scholar]
- 139.Williams G S, Malin M, Vremec D, Chang C H, Boyd R, Benoist C, Mathis D. Mice lacking the transcription factor CIITA—a second look. Int Immunol. 1998;10:1957–1967. doi: 10.1093/intimm/10.12.1957. [DOI] [PubMed] [Google Scholar]
- 140.Wojciechowski W, DeSanctis J, Skamene E, Radzioch D. Attenuation of MHC class II expression in macrophages infected with Mycobacterium bovis bacillus Calmette-Guerin involves class II transactivator and depends on the Nramp1 gene. J Immunol. 1999;163:2688–2696. [PubMed] [Google Scholar]
- 141.Wright K L, Chin K C, Linhoff M, Skinner C, Brown J A, Boss J M, Stark G R, Ting J P. CIITA stimulation of transcription factor binding to major histocompatibility complex class II and associated promoters in vivo. Proc Natl Acad Sci USA. 1998;95:6267–6272. doi: 10.1073/pnas.95.11.6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wright K L, White L C, Kelly A, Beck S, Trowsdale J, Ting J P-Y. Coordinate regulation of the human TAP1 and LMP2 genes from a shared bi-directional promoter. J Exp Med. 1995;181:1459–1471. doi: 10.1084/jem.181.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wu Z, Biro P A, Mirakian R, Hammond L, Curcio F, Ambesi-Impiombato F S, Bottazzo G F. HLA-DMB expression by thyrocytes: indication of the antigen-processing and possible presenting capability of thyroid cells. Clin Exp Immunol. 1999;116:62–69. doi: 10.1046/j.1365-2249.1999.00831.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yazawa T, Kamma H, Fujiwara M, Matsui M, Horiguchi H, Satoh H, Fujimoto M, Yokoyama K, Ogata T. Lack of class II transactivator causes severe deficiency of HLA-DR expression in small cell lung cancer. J Pathol. 1999;187:191–199. doi: 10.1002/(SICI)1096-9896(199901)187:2<191::AID-PATH206>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 145.Yun S, Gustafsson K, Fabre J W. Suppression of human anti-porcine T-cell immune responses by major histocompatibility complex class II transactivator constructs lacking the amino terminal domain. Transplantation. 1998;66:103–111. doi: 10.1097/00007890-199807150-00016. [DOI] [PubMed] [Google Scholar]
- 146.Zeleznik-Le N J, Azizkhan J C, Ting J P. Affinity-purified CCAAT-box-binding protein (YEBP) functionally regulates expression of a human class II major histocompatibility complex gene and the herpes simplex virus thymidine kinase gene. Proc Natl Acad Sci USA. 1991;88:1873–1877. doi: 10.1073/pnas.88.5.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zhong G, Fan T, Liu L. Chlamydia inhibits interferon gamma-inducible major histocompatibility complex class II expression by degradation of upstream stimulatory factor 1. J Exp Med. 1999;189:1931–1938. doi: 10.1084/jem.189.12.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhou H, Glimcher L H. Human MHC class II gene transcription directed by the carboxyl terminus of CIITA, one of the defective genes in type II MHC combined immune deficiency. Immunity. 1995;2:545–553. doi: 10.1016/1074-7613(95)90034-9. [DOI] [PubMed] [Google Scholar]
- 149.Zhou H, Su H S, Zhang X, Douhan III J, Glimcher L H. CIITA-dependent and -independent class II MHC expression revealed by a dominant negative mutant. J Immunol. 1997;158:4741–4749. [PubMed] [Google Scholar]
- 150.Zhu, X.-S., M. W. Linhoff, G. Li, K.-C. Chin, S. N. Maity, and J. P.-Y. Ting. 2000. Transcriptional scaffold: CIITA interacts with NF-Y, RFX, and CREB to cause stereospecific regulation of class II major histocompatibility promoter. **20:**6051–6061. [DOI] [PMC free article] [PubMed]