Multiple Splicing Defects in an Intronic False Exon (original) (raw)
Abstract
Splice site consensus sequences alone are insufficient to dictate the recognition of real constitutive splice sites within the typically large transcripts of higher eukaryotes, and large numbers of pseudoexons flanked by pseudosplice sites with good matches to the consensus sequences can be easily designated. In an attempt to identify elements that prevent pseudoexon splicing, we have systematically altered known splicing signals, as well as immediately adjacent flanking sequences, of an arbitrarily chosen pseudoexon from intron 1 of the human hprt gene. The substitution of a 5′ splice site that perfectly matches the 5′ consensus combined with mutation to match the CAG/G sequence of the 3′ consensus failed to get this model pseudoexon included as the central exon in a dhfr minigene context. Provision of a real 3′ splice site and a consensus 5′ splice site and removal of an upstream inhibitory sequence were necessary and sufficient to confer splicing on the pseudoexon. This activated context also supported the splicing of a second pseudoexon sequence containing no apparent enhancer. Thus, both the 5′ splice site sequence and the polypyrimidine tract of the pseudoexon are defective despite their good agreement with the consensus. On the other hand, the pseudoexon body did not exert a negative influence on splicing. The introduction into the pseudoexon of a sequence selected for binding to ASF/SF2 or its replacement with β-globin exon 2 only partially reversed the effect of the upstream negative element and the defective polypyrimidine tract. These results support the idea that exon-bridging enhancers are not a prerequisite for constitutive exon definition and suggest that intrinsically defective splice sites and negative elements play important roles in distinguishing the real splicing signal from the vast number of false splicing signals.
A major question in mammalian pre-mRNA splicing is how exons (or introns) are recognized. Mammalian transcripts are typically tens of thousands of bases long, with large introns alternating with internal exons that are usually less than 300 bases long. How are these small exons recognized within this sea of intronic sequences? Sequences defining a consensus are found at virtually all exon-intron joints (59). At the upstream side of an exon, the 15-nucleotide (nt) 3′ splice site consensus is Y10NCAG/G, and at the downstream side, the 5′ splice site is MAG/GURAGU (M equals A or C; boldface type indicates invariant nucleotides). At a variable distance upstream of the exon (often ∼30 nt) lies the branch point, with the very loose consensus YNYURAY. The AG dinucleotide immediately preceding the exon and the GU dinucleotide immediately following the exon are almost always present. However, considerable variation is found at the other positions: the highest frequency of occurrence of a particular base at a given position ranges from about 35 to 80%. As a result, less than 5% of actual 3′ or 5′ splice sites represent a perfect match to the consensuses described above (our analysis of a database of 2,800 human exons compiled by Reese et al. [http://www.fruitfly.org/sequence/human-datasets.html]).
In a typical mammalian transcript, there are many sequences that match these consensuses as well as or better than the sequences at real splice sites yet they are not used for splicing (47, 71). We will refer to these false sites as pseudosites. Real exons are recognized and spliced cotranscriptionally (5, 6, 44, 48, 91) with a half-time of about 5 min in vivo (44); the pseudosites are efficiently ignored during this process. There must be additional signals that distinguish real splice sites from pseudosites or vice versa. These additional recognition elements could act either positively or negatively, and examples of either type have been demonstrated over the last few years. Positive elements, or splicing enhancers, were first recognized as purine-rich sequences that promote splicing when present within some exons (90); more recently, the list of such enhancers has expanded to include AC-rich sequences (24) and intronic locations (13, 38, 58, 76). Despite the demonstration of specific interactions between enhancers and mammalian _trans_-acting mediators (10, 20, 41, 56, 84), at best only very degenerate motifs have emerged to define the enhancer sequence (51). Enhancers are better understood from the alternative splicing examples for the sex determination genes in drosophila, where several of these sequence elements, along with their _trans_-acting mediators, having been genetically characterized (for a review, see reference 52). Most enhancers have been identified in the context of alternative splicing; it is not yet clear whether they play an important role in constitutive splicing. The characteristics of the SR family of proteins support this idea, since some of these proteins bind to specific enhancer elements (51, 57, 82) and can also act as essential splicing factors (46, 89). Multiple sequences in constitutively spliced human β-globin exon 2 have been shown to act as enhancers when tested in an alternative splicing framework, and it has been proposed that they function in constitutive exon definition (70). However, their physiological significance within the globin gene is not clear.
Our knowledge of the role and mechanism of action of splicing silencers is more limited. It is known that these sequences can act from within an exon (28, 41) or an intron (16, 33, 35, 42), but no consensus sequence for such elements is apparent. In mammals, a silencer-binding protein has been identified in only a few cases (12, 16, 25, 42, 49). A well-studied example in drosophila is the transposase transcript, where the protein PSI has been shown to interact with 5′ pseudosplice sites near the real 5′ splice site to block the alternative splicing of an internal exon (74). Once again, it is not clear that these cases of silencing represent general inhibitory mechanisms operating in constitutive splicing decisions as well. However, the identification of PTB (hnRNP I) (17, 49, 60) and hnRNP A1 (8, 12, 25) as splicing inhibitors raises the possibility of such a role, given the abundance of these proteins as part of hnRNP complexes (30).
The effects of premature termination codons on nuclear RNA processing has prompted proposals that the translatability of exons plays a role in their recognition during nuclear RNA processing (15, 88; H. C. Dietz, Letter, Am. J. Hum. Genet. **60:**729–730, 1997). In this scenario, the absence of in-frame stop codons in a potential exon could provide the defining criterion for distinguishing real splice sites from pseudosplice sites. However, there is no direct evidence for a cellular mechanism that could capitalize on this information, i.e., recognize the translatability of an exon before it is spliced. Moreover, there are many examples of premature termination codons that do not affect pre-mRNA splicing (e.g., reference 88). A real exon sequence is constrained by its obligation to code for a protein, and so exon sequences do differ statistically significantly from intron sequences, e.g., in the frequency of particular hexanucleotides (94). Recognition of this constraint and a requirement for an open reading frame (ORF) together with the splice consensus sequences allows the prediction of exons within large genes with fairly high reliability (77, 78, 93). However, it is difficult to see how these statistical differences could be exploited to afford molecular recognition. The more likely mechanism is that exon recognition proceeds through the binding of protein or RNA factors to specific sequences or structures.
Site-directed mutagenesis of cloned genes and the analysis of mutations in endogenous genes have revealed _cis_-acting sequences that are necessary for constitutive splicing. In addition to the usual necessity for the conserved GU and AG dinucleotides, these studies have shown absolute or partial dependence on consensus nucleotides in other positions. However, the severity of the phenotype resulting from changes at a given position varies from one splice site to the next (14, 47, 64). In addition, mutations in the exon or the intron outside of the consensus sequence can also have strong effects (14, 64, 67). Some of these sequences have been characterized as enhancers or silencers, as mentioned above, and some of these have been shown to bind specific proteins or snRNPs. In other cases, secondary structure has been shown to play a role in either promoting or inhibiting splicing (3, 32, 34, 40). The general picture that has emerged from these studies is the recognition of the consensus sequences by different snRNPs, with this binding being stabilized either by exon- or intron-bridging interactions with SR and other proteins or by secondary or higher-order structures. However, the specificity and exact nature of these interactions are not clear enough to allow the formulation of rules for exon recognition.
Although the requirements for splicing of exons and introns have been extensively investigated, little attention has been paid to the other side of the coin: why are the many sites that match the consensus splice sites, the pseudosites, not used? Do they have perfectly functional splice sites but lack enhancer sequences? Are the false sites defective despite their agreement with the consensus? Are there silencing elements that keep an otherwise functional site from being recognized? We have used a mutational approach to address these questions. Examining the sequence of the large first intron of the human hprt gene, we chose a 3′ pseudosplice site followed by a downstream 5′ pseudosplice site to define a pseudoexon. Rather than using mutation to knock out the splicing function, we asked what it takes to “knock in” splicing, that is, to convert the pseudoexon into a functional exon. We found that despite their apparent similarity to what we understand as splice sites, the intronic pseudosplice sites bordering this pseudoexon were defective. In addition, we found the 3′ pseudosplice site to be associated with an intronic splicing silencer. A requirement for exonic enhancers was not evident in our studies.
MATERIALS AND METHODS
Cell lines and cell culture.
DG44 is a CHO cell line with a double deletion at the dhfr locus (87), and U1S is a CHO cell line with a double deletion of aprt (9). These cells were grown in monolayer culture in Ham's F-12 medium (GIBCO/Life Technologies) supplemented with 10% fetal calf serum (Atlanta Biologicals). All cells were cultured at 37°C in a humidified atmosphere of 95% air and 5% CO2. Generally, the medium was changed every 3 days.
Criteria for selecting hprt pseudoexons.
Consensus matrices for the 5′ splice site (SAG/GURAGU, S equals C or A) and the 3′ splice site (Y10NCAG/G) were derived from a database of 2,800 exons from a set of unique human genes (http://www.fruitfly.org/sequence/human-datasets.html). These matrices were similar to those reported by Senapathy et al. for primate genes (71) and are available upon request. Splice site consensus scores were calculated essentially as described by Shapiro and Senapathy (72). A score of 100 represents the best match to the consensus, 0 represents the worst possible match, and the GU and AG dinucleotides at the intron borders were an absolute requirement. For a pseudoexon, we asked for a minimum 3′ splice site consensus score of 69, that being the lowest 3′ score for a real exon in this gene, and a minimum 5′ splice site score of 75, which is the lowest 5′ score among the real hprt exons. We then asked that a high-scoring 3′ splice site be followed by a downstream high-scoring 5′ splice site to give a minimum combined score of 150 for a pseudoexon, which is the lowest combined score for a real internal hprt exon. We demanded that the minimum exon size be 50 nt, since decreasing larger exons below this size reduces splicing efficiency (29) and natural small exons below this size may be initially recognized as larger entities (81). The maximum exon size was set rather arbitrarily to 200: 85% of human exons are <200 nt, the average exon size being about 135 nt (39, 94). We also required the presence of a possible branch point within 60 nt upstream of an exon with a bulged A at position 6 and at least 3 of the remaining 6 nt capable of base pairing to U2 snRNA. Here again, we were guided by the nature of the real splice sites in the hprt gene: increasing the stringency of this requirement by one additional base pair would have caused the loss of six of the eight real 3′ splice sites. Finally, we purged pseudoexons (60%) from sets that shared a 3′ or 5′ pseudosplice site, choosing the candidate that produced the highest combined score. One 141-nt pseudoexon (pseudoexon 1), starting a position 6106 (31) near the center of hprt intron 1, was chosen for study. For some experiments, we utilized a second pseudoexon from hprt intron 1, i.e., pseudoexon 2, which is 123 nt long and starts at position 15862.
Plasmid constructions.
A more detailed description of the methods used for plasmid constructions is available from us upon request. For diagrams of the plasmids, see Fig. 4, 5, and 6.
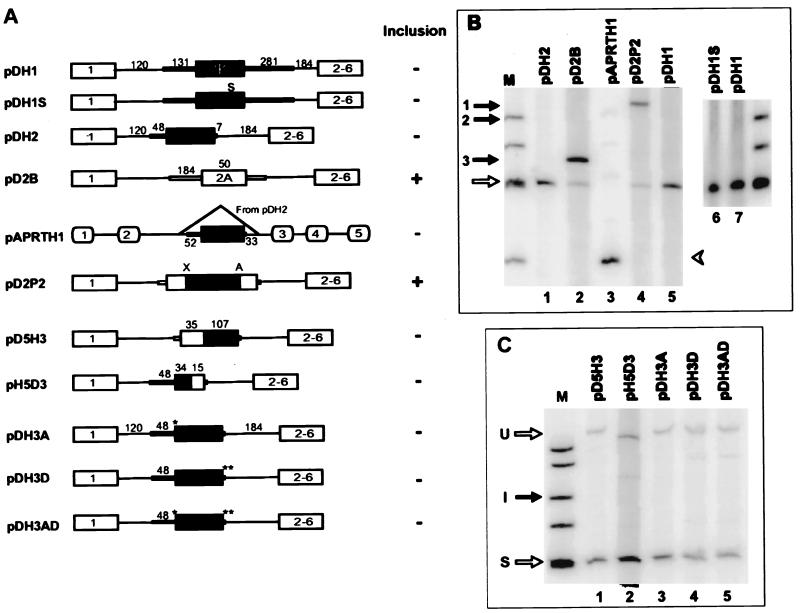
FIG. 4.
Splicing characteristics of constructs carrying various configurations of pseudoexon 1. (A) Schematic representations of pseudoexon 1 constructs. Individual constructs are described in the text. Ψ, hprt pseudoexon 1; S, mutation of a single nonsense codon to sense in the body of the pseudoexon (all constructs other than pDH1 contain this change); ∗, mutation of the original CAG/C sequence at the 3′ pseudosplice site to the consensus CAG/G; ∗∗, mutation of the original CAGG/GUGUGA sequence at the 5′ pseudosplice site to the consensus CAG/GUGAGU; X and A, _Xho_I and _Apa_I restriction sites, respectively. (B and C) Phosphorimages of RT-PCR products from total RNA extracted from permanently transfected cells. The closed arrow indicates exon inclusion (I) and the open arrow indicates exon skipping (S) in dhfr minigene transcripts; closed arrow 1, 2, or 3 indicates inclusion of the inserted exon in the construct pD2P2, pDH2, or pD2B, respectively. The closed arrowhead indicates inclusion and the open arrowhead indicates skipping of pseudoexon 1 in aprt transcripts. The markers (lanes M) were φX174 _Hin_fI fragments. U, unspliced transcripts.
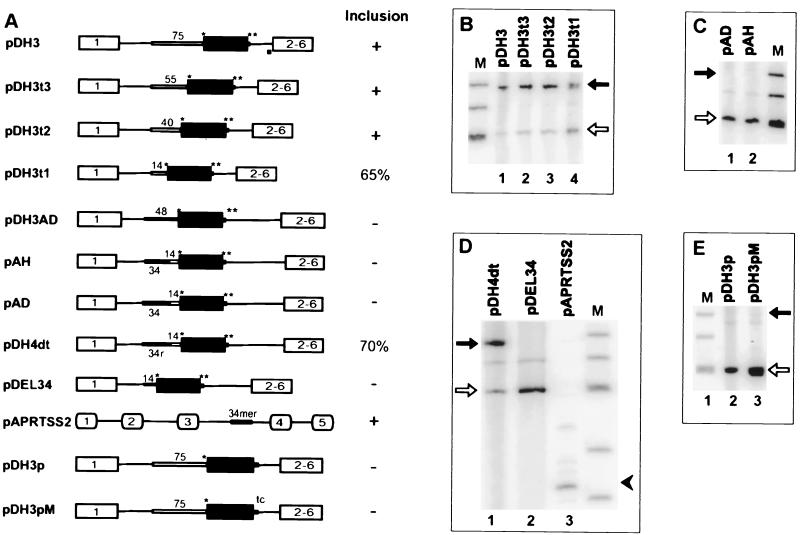
FIG. 5.
Effect of altering the upstream flank and 3′ and 5′ splice sites on the splicing of pseudoexon 1. (A) Schematic representations of pseudoexon constructs. Individual constructs are described in the text. The asterisks represent consensus CAG/G and CAG/GUGAGU sequences as described in the legend to Fig. 4. An open bar indicates a sequence derived from the 3′ end of hprt intron 1 (an authentic 3′ splice site), except for pAD, where it was derived from dhfr intron 1. The 34r in construct pDH4dt denotes the reverse sequence of the 34-mer intronic splicing silencer. The tc in the diagram of pDH3pM denotes a GU-to-TC mutation that removes a potential competing GU dinucleotide at position +7. In the column labeled Inclusion, +means >90% inclusion of the central exon, − means skipping of the central exon, and a number indicates percent inclusion of the central exon (percent included/included + skipped). (B, C, D, and E) Phosphorimages of RT-PCR products from total RNA extracted from permanently transfected cells (B, C, and D) or transiently transfected cells (E). The closed arrows indicate exon inclusion and the open arrows indicate exon skipping in the dhfr minigene transcripts. The closed arrowhead indicates inclusion of aprt exon 4 in the pAPRTSS2 context. Lanes M, φX174 _Hin_fI fragments.
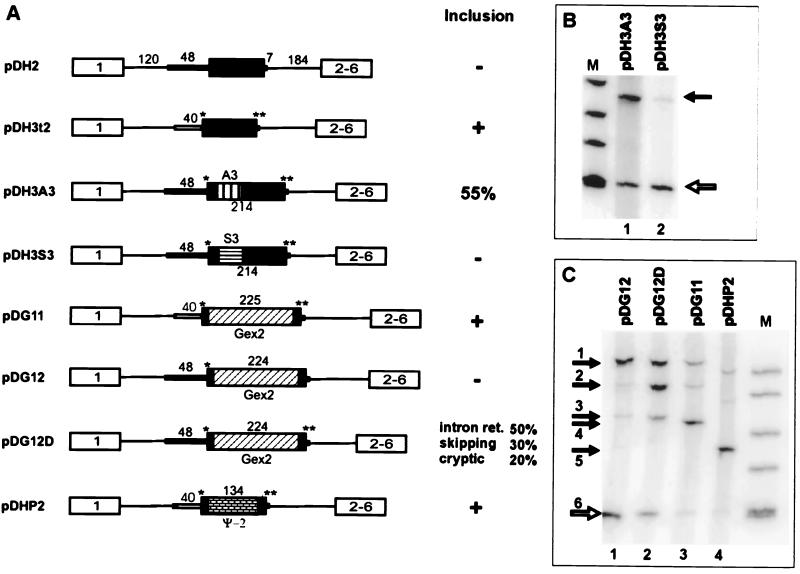
FIG. 6.
Effect of exon body sequence alteration on splicing. (A) Schematic representations of pseudoexon constructs and derivatives. Individual constructs are described in the text. A single asterisk indicates mutation to the CAG/G of a consensus 3′ splice site, and a double asterisk indicates mutation to a perfect CAG/GUGAGU 5′ consensus splice site. A3 denotes a 72-nt insert containing three tandem repeats of an ASF/SF2-binding SELEX winning sequence, and S3 denotes an analogous insert containing three tandem repeats of an SC35-binding SELEX winning sequence (84). Gex2 indicates human β-globin exon 2, and Ψ-2 indicates hprt pseudoexon 2. (B and C) Phosphorimages of RT-PCR products of RNA extracted from permanently transfected cells. The closed arrows indicate exon inclusion and the open arrows indicate exon skipping in the dhfr minigene context. Lanes M, φX174 _Hin_fI fragments. Closed arrows in panel C indicate the following: 1, size of unspliced RNA (or DNA) in constructs containing the globin exon; 2, size of RNA that has retained intron 1 in pDG12D (confirmed by sequencing); 3, size of RNA produced by splicing of the central exon at an upstream cryptic 3′ splice site in pDG12 and pDG12D (confirmed by sequencing); 4, size corresponding to inclusion of β-globin exon 2 in pDG11 (confirmed by sequencing); 5, size corresponding to inclusion of pseudoexon 2 in pDHP2; 6, size corresponding to skipping of the central exon in all constructs of the dhfr minigene. Lanes M, φX174 _Hin_fI fragments.
Pseudoexon 1 was cloned together with its flanks from human placental DNA by PCR amplification of the region from 131 nt upstream to 281 nt downstream of the pseudoexon using primers with _Pst_I restriction site tails. pDH1 was constructed by inserting this sequence into a unique _Pst_I site in the sole intron of the Chinese hamster dhfr minigene pDCH1P11 (63). pDH1S was constructed from pDH1 using mutagenic primers that converted the sole potential TGA stop codon at position 101 within the pseudoexon to CGA. To create pDH2, the pseudoexon 1 flanking sequences were trimmed by PCR amplification of pDH1S with _Pst_I-tailed primers designed to include just 48 nt of the upstream flank and 7 nt of the downstream flank and insertion of the PCR product into the _Pst_I site of pDCH1P11. Plasmid pDH3A was constructed by PCR-based site-directed mutagenesis to substitute a G for the C at position +1 of pseudoexon 1, resulting in the CAG/G sequence of a consensus 3′ splice site sequence. pDH3D was similarly constructed to change the sequence downstream of pseudoexon 1 from CAG/GUGUGA to the consensus 5′ splice site sequence CAG/GUGAGU. Plasmid pDH3AD, containing both the optimized 3′ splice site and 5′ splice site sequences, was made by cloning a 415-nucleotide _Bst_XI fragment from pDH3D that spanned the pseudoexon 5′ splice site sequence into pDH3A. The pseudoexon 1/dhfr exon 2 chimeric plasmid pH5D3 was constructed by a PCR-ligation-PCR method (1). pH5D3A was constructed by PCR amplifying a sequence extending through the first 34 nt of pseudoexon 1 and ligation to a fragment bearing sequences from dhfr exon 2 and intron 2 from pD2B (18) at a position 15 nt upstream of the 3′ end of the exon. A dhfr exon2/pseudoexon 1 chimeric plasmid was similarly constructed by ligating a PCR product extending through the first 35 nt of dhfr exon 2 to pseudoexon 1 at a position 107 nt upstream of the 3′ end, yielding pD5H3. Plasmid pAPRTH1 was constructed by PCR amplification of a pDH2 fragment containing pseudoexon 1 from 52 nt upstream of the 3′ pseudosplice site to 33 nt downstream of the 5′ pseudosplice site and inserting this 246-nt PCR product into the _Eco_RI site in intron 2 of pWTaprt (44). Plasmid pD2P2 was constructed by inserting the 141-nt pseudoexon 1 sequence between the _Xho_I and _Apa_I sites of exon 2A in pD2C3, a close derivative of pD2B constructed by Will Fairbrother (32a). Plasmid pDH3 was constructed by replacing the putative polypyrimidine tract (PPT) and branch point upstream of pseudoexon 1 in pDH3D with 76 nt (−75 to +1) comprising the assumed branch point region, PPT, and 3′ splice site at the 3′ end of the real intron 1 of the human hprt gene; thus, pDH3 has a real 3′ splice site and an optimized 5′ splice site. In the course of this construction, an _Afl_II site was introduced downstream of the _Pst_I site at the upstream end of the 76-mer. Plasmids pDH3t3, pDH3t2, and pDH3t1 were constructed by deleting 20 nt (−75 to −56), 35 nt (−75 to −41), and 61 nt (−75 to −15), respectively, from the 76-nt hprt intron 1 fragment in pDH3. Plasmid pAH was derived by replacing 10 nt of the PPT (−14 to −5) upstream of pseudoexon 1 in pDH3AD with its hprt intron 1 counterpart. Plasmid pAD was constructed similarly, by replacing the same 10 nt upstream of pseudoexon 1 with the dhfr intron 1 counterpart (a PPT from another real 3′ splice site). pDH4dt was constructed by inserting a synthetic double-stranded oligonucleotide that included the reverse complement of the original 34-mer into the _Afl_II site 14 nt upstream of the pseudoexon in pDH3t1. pDEL34 was created by deleting 34 nt upstream of the PPT in pDH3AD using PCR methodology. Thus, pDEL34 retains only 14 nt of the upstream sequence originally flanking pseudoexon 1. pAPRTSS2 was constructed by inserting a 34-mer from position −48 to position −15 of the pseudoexon 1 upstream flank into a position 14 nt upstream of exon 4 in the aprt gene; thus, the 34-mer occupies the same position relative to the downstream aprt or hprt pseudoexon. Plasmid pDH3A3 was constructed by inserting three tandem copies of the A3 enhancer sequence (responsive to ASF/SF2 [84]) into a _Nar_I site 24 nt downstream of the 5′ end of pseudoexon I in pDH3AD; PCR amplification with _Nar_I-tailed primers and a template provided by R. Tacke and J. Manley were used. Similarly, plasmid pDH3S3 was constructed by inserting three tandem repeats of sequence S3, a SELEX winner binding to SC35 (84) into the _Nar_I site of pseudoexon 1.
To make plasmid DG11, pseudoexon 1 in pDH3t2 was first replaced with a 9-nt-long exon containing a _Bcl_I restriction site to produce pDH3EL. Human β-globin gene exon 2 (from +1 to the _Bam_HI site at +211) was PCR amplified with _Bcl_I-tailed primers and inserted into the _Bcl_I site of pDH3EL. To make plasmid DG12, pseudoexon 1 in pDH3A was first replaced with an 8-nt-long exon containing a _Bcl_I restriction site to produce pDEL. To create pDG12, human β-globin gene exon 2 (from +1 to the _Bam_HI site at +211) was PCR amplified with _Bcl_I-tailed primers and inserted into the _Bcl_I site of pDEL. pDG12D was created from pDG12 by mutating the 5′ splice site to the consensus sequence by PCR mutagenesis. pDHP2 was similarly constructed by insertion of a pseudoexon 2 sequence from the human hprt gene (123 nt starting at position 15862) into pDH3EL.
PCR amplification of DNA templates.
PCR from DNA templates was performed with 1 to 2 ng of plasmid DNA or 1 to 2 μg of genomic DNA and Taq DNA polymerase (Perkin Elmer) in accordance with the supplier's recommendations. A typical cycle consisted of initial denaturation at 94°C for 5 min, followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 61°C for 30 s, and extension at 72°C for 60 s. After completion of the final cycle, a final extension was done at 72°C for an additional 7 min.
Transfection.
For transient transfections, monolayers of cells (3 × 106 in a 100-mm-diameter dish) were transfected with 10 μg of plasmid DNA using Lipofectamine (GIBCO/Life Technologies). Ten micrograms of plasmid DNA was added to 0.6 ml of Opti-MEM medium (GIBCO/Life Technologies) and mixed with 0.6 ml of Opti-MEM medium containing 30 μl of Lipofectamine. This mixture was incubated at room temperature for 30 min. After rinsing of the cells successively with phosphate-buffered saline and serum-free Opti-MEM medium. 4.5 ml of Opti-MEM medium was added to the above-described mixture and used to cover the cells. After incubation at 37°C for 5 h, 5 ml of alpha-MEM medium supplemented with 10% fetal bovine serum was added and incubation was continued. Total RNA was extracted 48 h after the transfection. For permanent transfections, a similar procedure was used but 1 μg of a plasmid harboring a neo gene (a derivative of pEGFP-N1; Clontech) was included for cotransfection. At 72 h after transfection, one-fifth of the cells were passaged into selective medium containing 400 μg of active G418 (Geneticin; GIBCO/Life Technologies) per ml. After 8 to 10 days, transfectant colonies were pooled and expanded and total RNA was extracted. Each transfection experiment was first carried out by transient transfection and then confirmed by permanent transfections.
RNA analysis.
Total RNA was extracted from exponentially growing cells as follows (65). Cells were lysed in a solution of 2% sodium dodecyl sulfate; 200 mM Tris-HCl (pH 7.5), and 1 mM EDTA. DNA and proteins were precipitated with 1.5 M potassium acetate and centrifuged for 10 min at 4°C in a microcentrifuge. The supernatant was extracted twice with chloroform-isoamyl alcohol, and RNA was precipitated with 0.65 volume of isopropanol. RNA extracted from a nearly confluent 100-mm dish of cells was treated with 30 U of RNase-free DNase I (Boehringer Mannheim), 3 mM MgCl2, and 100 U of RNasin (Promega). We used reverse transcriptase (RT), followed by PCR (RT-PCR), to quantify the splicing products. For the RT reaction, a 20-μl reaction mixture contained 1 μg of RNA (dissolved in water), 0.4 μg of random hexamer, 10 mM dithiothreitol, 40 U of RNasin, 0.5 mM all four deoxynucleoside triphosphates, 4 μl of 5 × RT buffer, and 200 U of SuperScript RT (all from Promega except the RT, which was from GIBCO/Life Technologies). The reaction was carried out at 37°C for 1 h, followed by 5 min of heating at 95°C to inactivate the enzyme. Three microliters of the RT reaction mixture was used for a 50-μl PCR mixture. PCR products were labeled with [α-32P]dATP to allow quantitation by phosphorimaging (18). The PCR conditions were as follows: denaturing at 95°C for 30 s, annealing at 61°C for 30 s, and extension at 72°C for 60 s. After 25 cycles, a 7-min extension at 72°C was carried out. A 6-μl sample of the PCR mixture was electrophoresed in a 5% nondenaturing polyacrylamide gel.
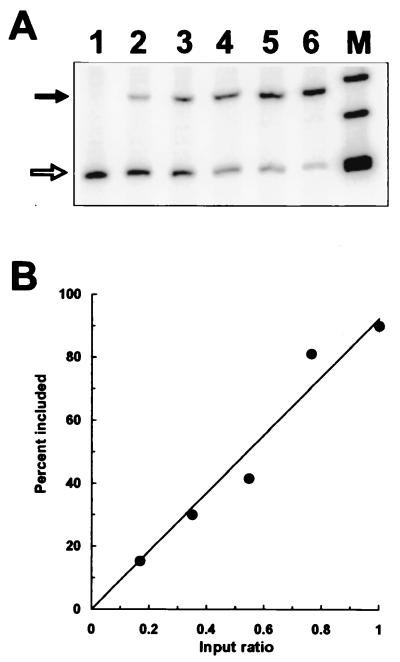
To establish the quantitative nature of the RT-PCR method in the determination of ratios of different RNA molecules, we prepared mixtures of mRNA that had included (DH3t3) or skipped (DH2) pseudoexon 1 in the dhfr minigene context. These mRNA samples were combined in proportions of 0:10, 2:8, 4:6, 6:4, 8:2, and 10:0. The mixtures were subjected to RT-PCR using the conditions described above. Phosphorimaging (Molecular Dynamics) resulted in good quantitative agreement between the input ratios and the output of the PCR, as shown in Fig. 1.
FIG. 1.
Quantitation of the RT-PCR assay for exon skipping. An RNA preparation from a cell population exhibiting complete pseudoexon skipping (DH2) was mixed in various ratios with an RNA preparation from a cell population exhibiting mostly (90%) pseudoexon inclusion (DH3t3). The sample amounts were chosen so that the unmixed samples (lanes 1 and 6) produced signals similar in intensity. After RT-PCR, the radioactive PCR products were separated by polyacrylamide gel electrophoresis. (A) Phosphorimage of the PCR products. The closed arrow indicates exon inclusion, and the open arrow indicates exon skipping. Lanes 1 to 6 represent mixtures containing the RNA exhibiting 90% exon inclusion in the following proportions of the total: 0, 0.17, 0.35, 0.55, 0.76, and 1, respectively. Lane M, φX174 _Hin_fI fragments. (B) Graphical representation of the data in panel A.
The principal primers used have already been described by Kessler et al. (44): for the dhfr context, primers 19 and 28; for the aprt context, primers 1 and 8. Additional primers used to analyze pAPRTSS2 products for aprt were AEx3FDL (forward) in exon 3(CCACAGTGTCAGCCTCCTAT) and A3exon5B (reverse) in exon 5(GGAGAGAGAAGAATGGTACT).
RESULTS
Pseudosplice sites are abundant in the human hprt gene.
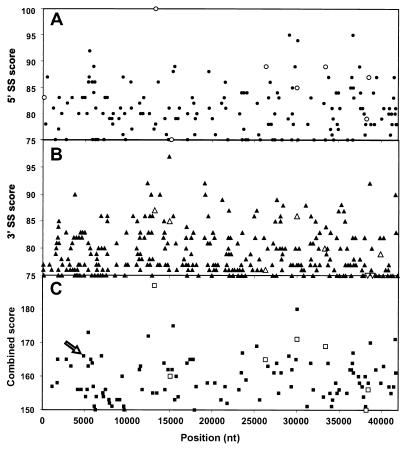
Although conserved elements are important for the selection of splice sites, they do not seem to be sufficient to account for the accuracy of exon-intron discrimination. Splice sites typically contain several mismatches with the consensus; sequences with similar degrees of mismatching are abundant. We term these unused yet seemingly “good” sequences (close conformity to the consensus) pseudosplice sites. We did a computer search for such sites in the 42-kb human hprt gene, which contains nine exons and eight introns. We used a consensus matrix for 5′ and 3′ human splice sites calculated from a set of 2,400 of each type found in a database of nonredundant human genes (http://www.fruitfly.org/sequence/human-datasets.html) and a scoring formula adapted from that of Shapiro and Senapathy (72). Many pseudosplice sites were found distributed throughout the gene. As shown in Fig. 2A, there are eight real 5′ splice site in the human hprt gene but there are over 100 5′ pseudosplice sites that have scores higher than the lowest-scoring real internal 5′ splice site. The case is even worse for 3′ splice sites, where 683 pseudosites were found with higher scores than the lowest-scoring real site (Fig. 2B). Expressing sequence scores as information content (80) or using a search algorithm based on a neural network comparison (http://www.fruitfly.org/seq_tools/splice.html) did not change the basic observation that such pseudosites outnumber real sites by an order of magnitude (data not shown). The recognition problem of choosing the right splice sites, and only the right ones, seems formidable. Part of the solution might be recognition of an entire exon by the splicing machinery, thereby ignoring the isolated pseudosplice sites. To determine whether this approach is sufficient to resolve the recognition problem, we searched the hprt gene for a combination of sequence elements that resemble entire exons, including a potential branch point, using the criteria specified in the legend to Fig. 2C and in Materials and Methods. As shown in Fig. 2C, we found 103 good-looking exons that are ignored by the cell (which we call pseudoexons), all of which have higher combined 3′ splice site and 5′ splice site scores than the lowest scoring of the seven real internal hprt exons. There must be additional signals that distinguish real exons from pseudoexons or vice versa. This additional information could be acting positively to promote the recognition of real exons or acting negatively to repress the recognition of pseudoexons.
FIG. 2.
Pseudosplice sites and pseudoexons in the human hprt gene. (A) Locations and consensus scores of 9-nt sequences resembling 5′ splice sites in the human hprt gene. The open symbols indicate the eight real 5′ splice sites (SS). Only those sites having scores equal to or higher than the lowest real 5′ splice site scores (75 for intron 3) are shown. (B) Locations and consensus scores of 15-nt sequences resembling 3′ splice sites. The open symbols indicate the eight real 5′ splice sites. Only those 285 sites having scores equal to or higher than 75 are shown. The lowest scores of real 3′ splice site are 71 and 69 for introns 6 and 7, respectively; these two points are plotted as 75 and indicated by downward-pointing open triangles. There are 675 sequences that would meet the lower cutoff of 69. (C) Locations and combined 3′ and 5′ scores of 103 pseudoexons in the hprt gene. The criteria for a pseudoexon were a 3′ pseudosplice site scoring at least 69, followed within at least 50 nt but no more than 200 nt by a 5′ pseudosplice site scoring at least 75, plus the presence of a sequence resembling a branch point within 60 nt upstream of the 3′ pseudosplice site (see Materials and Methods). If more than one pseudoexon shared a 3′ or 5′ pseudosplice site, only the highest-scoring candidate was chosen. The arrow points to the score of pseudoexon 1, which was selected for further study.
We have approached the problem of exon recognition by determining what changes are necessary to turn a pseudoexon into a real one. In this way, we hoped to define some of the sequence elements involved. We chose a model pseudoexon (pseudoexon 1) located in large (13-kb) intron 1 of the human hprt gene (Fig. 2C). Pseudoexon 1 has a 5′ splice site score of 83 (higher than those of two of the seven internal real hprt exons), a 3′ splice site score of 83 (higher than those of four of the seven internal exons), and a combined score of 166 (higher than those of four of the seven internal exons). The comparable values for the average of 1,980 internal exons in the human gene database are 84, 82, and 165. Pseudoexon 1 is 141 nt long and has possible branch points located 15, 39, and 45 nt upstream of its 3′ pseudosplice site (Fig. 3, line 1). By these criteria, then, pseudoexon 1 has the appearance of an average exon.
FIG. 3.
Sequence changes at the 3′ pseudosplice site of hprt pseudoexon 1. The slash indicates the predicted potential 3′ splice site. Upstream dhfr host minigene sequences are in italics. Point mutations are in lowercase. The intronic splicing silencer 34-mer and its truncated and mutated versions are underlined. An inverted 34-mer sequence is overlined. A 10-nt PPT taken from the authentic 3′ splice site of hprt intron 1 is shaded. The original 5′ splice site sequence downstream of the pseudoexon was CAG/GUGUGA, which is present here only in pDH2. All of the other constructs in this list contained the 5′ splice site consensus sequence CAG/GUGaGu.
No autonomous negative element lies within the pseudoexon 1 body or distal flanking sequences.
One way of explaining the nonutilization of pseudosplice sites is that they reside in a negative context, being masked either by secondary structures (7, 21, 26, 45, 50) or by steric or mechanistic hindrance from proteins binding to nearby sites (42, 73). To test whether or not the skipping of pseudoexon 1 is dependent on the larger context of the hprt gene, we placed the pseudoexon together with only 131 nt of the upstream and 281 nt of the downstream flanking sequences into the sole 300-nt intron of a DHFR minigene (pDH1, Fig. 4A). This construct was then transfected into a CHO dhfr deletion mutant (DG44). In this and all of the similar analyses described below, total RNA was isolated from pooled permanent transfectants and analyzed for splicing by RT-PCR. In all cases, transient transfections were also carried out, with essentially the same results. In pDH1 transcripts, dhfr exons 1 and 2 were spliced together without inclusion of pseudoexon 1 (Fig. 4B, lane 5). Thus, this sequence had retained its inability to be spliced in this more limited and foreign minigene context. In contrast, when a real exon (exon 2A, a second copy of dhfr exon 2) was inserted into the same position in this minigene intron (pD2B, Fig. 4A), it was efficiently included (Fig. 4B, lane 2, and reference 18).
If pseudoexon 1 had been spliced in, a stop codon would have interrupted the ORF. Such premature translation terminations have been shown to decrease mRNA levels (56, 88), either by destabilization (43) or by interference with splicing (27, 61). To eliminate the possibility of such effects in this system, we mutated the single in-frame stop codon in pseudoexon 1 to a sense codon (see Materials and Methods). This modification (pDH1S, Fig. 4A) did not change the nonsplicing phenotype (data not shown). This no-nonsense construct was used as the starting point for all subsequent modifications.
To search for negative elements within the hprt sequences surrounding pseudoexon 1, we further deleted the pseudoexon flanks, leaving only 48 nt upstream of the 3′ pseudosplice site and 7 nt downstream of the 5′ pseudosplice site (pDH2, Fig. 4A). This truncation did not change the skipping phenotype of pseudoexon 1 (Fig. 4B, lane 1), suggesting that no negative elements had been removed. Similarly truncated real dhfr exon 2A was efficiently included when placed in this same context (pD2ID; data not shown). Thus, the host dhfr intron flanks did not exert a negative influence at this proximity.
We also tested the ability of pseudoexon 1 to be spliced in another gene context, moving the trimmed pseudoexon into intron 2 of the hamster aprt gene, yielding pAPRTH1 (Fig. 4A). This construct was transfected into CHO cell line U1S with aprt deleted (9). This truncated version of pseudoexon 1 failed to be spliced in this foreign context as well (Fig. 4B, lane 3).
To test for the presence of negative elements within pseudoexon 1 body, we inserted the pseudoexon 1 sequence, exclusive of flanking intron sequences, into dhfr exon 2A in a derivative of splicing-permissive construct pD2B to create pD2P2 (Fig. 4A). The resulting 191-nt exon was efficiently spliced (Fig. 4B, lane 4), suggesting that no autonomously acting negative element was harbored pseudoexon 1.
We next separated the upstream and downstream halves of pseudoexon 1 region in an attempt to isolate the splicing defect to the 3′ or 5′ pseudosplice site. Two chimeric plasmids were constructed. pD5H3 contained a downstream segment pseudoexon 1 and an upstream segment from dhfr exon 2A, including the real 3′ splice site that precedes exon 2A (Fig. 4A). pH5D3 contained an upstream segment pseudoexon 1 and a downstream segment from dhfr exon 2A, including the real 5′ splice site that follows exon 2A (Fig. 4A). Neither of these two constructs allowed splicing of the pseudoexon (Fig. 4C, lanes 1 and 2). We concluded that there are defective or negative elements in both moieties of the pseudoexon.
Consensus 5′ and 3′ splice sites fail to transform pseudoexon 1 into a real exon.
Although the splice sites of pseudoexon 1 exhibit reasonable agreement with the consensus, it is possible that they are nevertheless defective, with better agreement required in this particular context. We therefore made constructs that optimized the 3′ splice site (without altering the PPT), the 5′ splice site, or both. Each of these constructs was derived from pDH2. In pDH3A, the CAG/C sequence at the 3′ pseudosplice site was changed to the consensus CAG/G (Fig. 4A); this change did not bring about splicing of pseudoexon (Fig. 4C, lane 3). In pDH3D, the original CAG/GUGUGA sequence at the 5′ pseudosplice site was changed to the consensus CAG/GUGAGU (Fig. 4A); although a small amount of intron 2 splicing was now apparent, the major product was still the exon-skipped species (Fig. 4C, lane 4). Both of these changes were then incorporated into pDH3AD (Fig. 3, line 2; Fig. 4A); the combination likewise resulted in predominant exon skipping (Fig. 4C, lane 5). These results suggested that the less than perfect pseudosplice sites are not, or at least not fully, responsible for keeping the pseudoexon silent.
Provision of a 3′ region from a real intron can promote splicing of the pseudoexon.
To test the idea that the inability of pseudoexon 1 to be spliced comes from its 48-nt upstream flank, we replaced this suspected region with a 75-nt sequence from the 3′ end of a real intron, hprt intron 1. To maximize the chance of a positive result, we used pDH3AD, with the optimized 3′ and 5′ splice site sequences described above for this substitution. The splicing phenotype of the resultant construct, pDH3 (Fig. 5A), showed that these changes were indeed sufficient to convert pseudoexon 1 into a real exon (Fig. 5B, lane 1). A series of 5′ truncations was then carried out to determine whether shorter sequences from hprt intron 1 could also suffice. pDH3t3, pDH3t2, and pDH3t1 (Fig. 3, line 4) retain 55, 40, and 14 nt from the 3′ end of hprt intron 1, respectively (Fig. 5A). In all of these truncated versions, the pseudoexon was included much more than it was skipped (Fig. 5B, lanes 2, 3, and 4). Even in pDH3t1, with only 14 nt of the hprt intron 1 sequence, pseudoexon inclusion was the predominant (65%) phenotype (Fig. 5B, lane 4). Sequencing of the pDH3 RT-PCR products corresponding to the inclusion of pseudoexon 1 confirmed that splicing had taken place at the expected sites. In pDH3t1, the hprt PPT is joined to the upstream dhfr minigene intron sequence. Apparently a branch point sequence is being recruited from that region (e.g., possibly via a TAGGGAC sequence 52 nt upstream of the 3′ splice site).
A sequence upstream of the pseudoexon acts as an intronic splicing silencer.
The splicing-positive construct pDH3t1 differs from its splicing-negative counterpart pDH3AD in that it has a different PPT and lacks the remaining 34 nt of the upstream sequence from the 48-nt pseudoexon 1 flank. Thus, the failure of pDH3AD transcripts to be spliced could be due to (i) a defective PPT, (ii) a defective branch point, (iii) a negative element present in the original pseudoexon 1 upstream flank, or (iv) any combination of these three. To test for a negative element in the upstream flank, we inserted the 34-nt sequence from −48 to −15 into pDH3t1 just upstream of the hprt intron 1 PPT, forming pAH (Fig. 3, line 3; Fig. 5A). The pseudoexon in pAH transcripts was no longer spliced (Fig. 5C, lane 1), implying that this upstream sequence does play a negative role. To test the sequence specificity of the 34-mer, it was reversed in its position upstream of the PPT, forming pDH4dt (Fig. 3, line 5; Fig. 5A). Splicing of the pseudoexon in pDH4dt was greatly improved (to 69%) (Fig. 5D, lane 1). Thus, it is the sequence of the 34-mer, rather than its increase of the spacing between upstream (e.g., a branch point) and downstream elements (54), that is responsible for most of the splicing inhibition. We also tested a PPT taken from another real intron, dhfr intron 1, in the presence of the 34-mer. Transcripts from this construct, pAD (Fig. 5A), also failed to splice the pseudoexon (Fig. 5C, lane 2), suggesting that the inhibition by the 34-mer is not specific for the hprt intron 1 PPT. The generality of the inhibition was put to a more rigorous test by placing the 34-mer in a completely different splicing context, 14 nt upstream of exon 4 in the hamster aprt gene, to form pAPRTSS2 (Fig. 5A). Exon 4 was not skipped; rather, a longer form of exon 4 was produced (Fig. 5D, lane 3). Sequencing of the RT-PCR product showed that a cryptic 3′ splice site 17 nt upstream of the normal 3′ splice site had been used. Thus, splicing at the normal aprt 3′ splice site was, in fact, inhibited by the 34-mer, notwithstanding the fact that a new 3′ splice site was recruited within the 34-mer itself (position 31 of the 34-mer). We concluded that this 34-nt region contains an intronic splicing silencer and this silencer prevented pseudoexon 1 from being included in the final mRNA.
The PPT upstream of the pseudoexon is defective.
The silencer may be sufficient for inhibiting the 3′ pseudosplice site; alternatively, the PPT of the 3′ pseudosplice site may be intrinsically defective. To distinguish between these possibilities, we deleted the 34-mer from pDH3AD (Fig. 4A). The resulting plasmid, pDEL34 (Fig. 5A), retains the original PPT upstream of the pseudoexon, the change to CAG/G at the 3′ pseudosplice site, and the optimized downstream 5′ splice site. Pseudoexon 1 was still skipped in pDEL34 transcripts (Fig. 5D, lane 2). pDEL34 and pDH3t1 differ only in the 14-nt PPT, yet the pseudoexon is skipped in the former and included in the latter. We conclude that the 14-nt PPT flanking pseudoexon 1 is defective or plays some active negative role in splicing. Thus, at least two independent defective or negative elements are present in the immediate upstream flanking sequence of pseudoexon 1, more than accounting for its inability to be spliced.
The 5′ pseudosplice site is defective.
Having established a defective 3′ splice site and the presence of an intronic silencer sequence in the 3′ splice site region, we returned to the 5′ pseudosplice site. For the study of the 3′ splice site region, the 5′ splice site sequence had been optimized to the consensus CAG/GUGAGU. We investigated whether this optimization is necessary when a functional 3′ splice site is present. The original 5′ pseudosplice site (CAG/GUGUGA) was combined with the functional 3′ splice site from hprt intron 1 to form pDH3p (Fig. 5A). Pseudoexon 1 was ignored in cells transiently transfected with pDH3p (Fig. 5E, lane 2). Thus, despite its reasonable agreement (consensus agreement score of 84) with the consensus, the 5′ pseudosplice site is defective even when paired across the exon with a functional 3′ splice site. There is a second GU dinucleotide just downstream of and adjacent to the proposed 5′ pseudosite GU; we thought the sequence resembling a 5′ splice site based on this GU (GGU/GUGAGU, consensus agreement score of 72) might interfere with the proposed site (73). To test this possibility, we mutated the last two nucleotides within this possible interfering site. The GU-to-UC change reduced the consensus agreement score to 58 (GGU/GUGAUC) without changing the originally proposed 5′ pseudosplice site sequence. The pseudoexon was still skipped in transcripts of the resulting plasmid, pDH3pM (Fig. 5A and E). Thus, in addition to the deficiency of the 3′ pseudosplice site, the 5′pseudosplice site is defective.
Contribution of the exon body to splicing recognition.
In several cases of alternative splicing, so-called weak splice sites can be activated by the presence of a splicing enhancer. The best characterized of these are exonic splicing enhancers (23, 41, 82, 84–86, 90), but enhancing sequences can also be found in introns (13, 38, 58, 76). The sequences surrounding pseudoexon 1 were not chosen to appear especially weak, yet they are not functional; it is possible that they simply lack necessary positive information within the exon body. We therefore investigated whether pseudoexon 1 can be activated by the presence of a sequence that might act as an exonic splicing enhancer. First, we introduced a known strong enhancer into pseudoexon 1 in pDH3AD. We chose a sequence selected by Tacke and Manley (84) for its ability to bind to the splicing factor ASF/SF2. Three tandem repeats of the SELEX winning sequence A3 were inserted 20 nt from the 5′ end of pseudoexon 1 (Fig. 6A).
RT-PCR results showed that the A3 sequences resulted in inclusion of pseudoexon 1 in 55% of the spliced transcripts. (Fig. 6B, lane 1). In contrast, the introduction of the SC35-selected sequence S3 did not have a detectable effect on splicing of pseudoexon 1 (Fig. 6B, lane 2). Thus, it appeared that this particular enhancer (A3) was able to activate splicing at the 3′ pseudosplice site. However, sequencing of the pertinent PCR product showed that splicing of intron 1 actually took place at a cryptic site 17 nt upstream of the proposed 3′ pseudosplice site (data not shown). This site is the same site activated when the upstream 34-mer was inserted into the aprt gene. Ironically, this site lies within a sequence that acts negatively in other contexts. The originally chosen 3′ splice site, defined here as including the PPT, remained refractory to splicing.
This result suggests that enhancer sequences could contribute to the definition of a constitutive exon. In fact, Schaal and Maniatis have suggested that multiple distinct splicing enhancers are present within exon 2 of the constitutively spliced human β-globin gene, where they function to specify the 3′ splice site (69). To test the role of these β-globin exon 2 sequences in the pseudoexon system, we first we replaced pseudoexon 1 in pDH3t2 with β-globin exon 2 to form pDG11. This plasmid contains a functional hprt intron 1-derived 3′ splice site, as well as an optimized 5′ splice site (Fig. 6A). Like pseudoexon 1 (Fig. 5B, lane 3), the β-globin exon was efficiently included in this case (Fig. 6C, lane 3), indicating that it is capable of being spliced in this permissive context. We then swapped the human β-globin exon 2 sequence for the pseudoexon 1 sequence of the nonpermissive plasmid pDH3A. The resulting construct, pDG12, has the slightly improved (3′ splice site CAG/G) original 3′ pseudosplice site, the β-globin exon 2 body, and the original 5′ pseudosplice site (Fig. 6A). In this context, the β-globin exon 2 sequence was no better than the pseudoexon 1 sequence it replaced, as there was no splicing to either the originally proposed 3′ splice site or to the cryptic site 17 nt upstream (Fig. 6C, lane 1). Thus, the enhancer elements that are present in this β-globin exon are unable to convert this pseudoexon into an exon. The enhancement exhibited by the A3 sequences described above was realized in a more permissive context, in that an optimized 5′ splice site was present. Our final test of the β-globin sequences was to replace the original 5′ splice site of pDG12 with this optimized sequence, forming pDG12D (Fig. 6A). Multiple species resulted from these transcripts, including three spliced species, as well as some unspliced transcripts. The exact nature of these RNA molecules was determined by sequencing of the RT-PCR products. The most abundant product had spliced intron 2 but retained intron 1. Almost as frequent were exon skipping and inclusion of the exon spliced at the cryptic 3′ splice site. Thus, the improvement of the 5′ splice site influenced the enhancing action of the β-globin sequences but in a complex way.
Are exonic splicing enhancers required for exon definition? The result described above obtained with pDH3 and its derivatives argues against this idea. In these plasmids, the 3′ splice site was taken from the 3′ end of a working intron, the 5′ splice site matched the consensus, and the pseudoexon body lying between these two endpoints was efficiently spliced. It should be remembered that the pseudoexon body is actually an intron sequence and so would not be expected to contain any enhancer elements. To confront the possibility that an enhancer was fortuitously present, we substituted another hprt intronic sequence for pseudoexon 1. This 123-nt sequence, termed pseudoexon 2, was also selected from hprt intron 1 with the constraint that it not include any sequences resembling splice sites. The resulting construct, pDHP2 (Fig. 6A), gave rise to RNA that predominantly included this pseudoexon 2. This result supports the idea that it is defective splice sites, rather than the lack of an exonic enhancer, that account for the nonsplicing of pseudoexon sequences.
DISCUSSION
Pseudoexons.
It has long been recognized that consensus sequences alone must be insufficient to identify either introns or exons, since higher eukaryotic transcripts generally contain many more sequences with good agreement with the consensus than there are bona fide splice sites (71). In this study, we surveyed the human hprt gene as a model and found a good example of this incongruity: hundreds of sequences resembling 5′ and 3′ splice sites were found in this 42-kb transcript. Moreover, the stipulation that exons be of limited length and that a short branch point sequence be present upstream of the 3′ pseudosplice sites did not solve the problem. Using criteria based on the characteristics of the seven real internal exons in this gene, 103 exon-like sequences were found. We called these sequences pseudoexons (79) without any implication of whether or not they were used as real exons at some point during evolution.
Reading frames are without effect on splicing.
Computer programs can predict the locations of exons within gene sequences with reasonable accuracy (77, 78, 93). However, efficient exon-finding programs use the protein coding information of exons as a guide. Could the cell also be using this type of information to differentiate real exons? Interruptions in the ORF of an internal exon usually result in lower levels of the nucleus-associated mRNA (see reference 56 for a review), and in some cases internal exons bearing nonsense mutations are skipped (27) or fail to splice (53). These results have suggested the idea of nuclear scanning of pre-mRNA for translatable exons (15, 27, 88). The intronic (pseudoexons 1 and 2) and exonic (β-globin) sequences we introduced here produced stop codons near the start of the central exon, yet we saw no evidence of either decreased mRNA levels or increased exon skipping associated with these disruptions of translatability (pDG11, pDHP2, pDH3A3, and pDG12D). These results add to previous cases indicating normal splicing despite the presence of nonsense mutations in the tpi (19) and aprt (43) genes and argue against a general mechanism for exon identification based on ORFs.
The 5′ pseudosplice site.
Pseudoexon 1 is not spliced even when placed in a favorable context within a dhfr minigene, i.e., a context in which a real exon with similarly trimmed minimal flanks is efficiently spliced. Moreover, we found no evidence that the pseudoexon sequence proper provides a negative influence. The 5′ pseudosplice site at the downstream end of pseudoexon 1 was not used even when the upstream 3′ pseudosite was replaced with a functional 3′ splice site. These data suggest that the 5′ pseudosplice site is defective. The sequence of the 5′ pseudosplice site is CAG/GUGUGA; it has a consensus score of 83, which is higher than those of 42% of the 2,400 authentic 5′ splice sites in the database we have used. However, this particular sequence was not found among the 5′ splice sites in the database. The first position of the 5′ consensus, at −3 relative to the splice site, is weakly conserved and is sometimes not considered part of the consensus. The score and rank of the pseudosplice site are similar (82 and higher than 37% of real sites) if this 8-mer version of the consensus (AG/GUGUGA) is considered. Unlike the 9-mer, this specific 8-mer sequence appears three times as a real 5′ splice site in this database. This last result reinforces the idea that consensus sequences by themselves are inadequate to specify correct splicing (41, 62, 71) and implies that context still played a role in our experiments. This context could include some as yet undefined enhancer sequence close to the 5′ splice site in question. Alternatively, the context effect could result from a local secondary or higher-order structure. Whatever the deficiency of the context, it can be overridden by providing a perfect consensus 5′ splice site sequence to go with the functional 3′ splice site. Perhaps this particular sequence requires the action of an enhancer-bound splicing factor either to recruit U1 snRNP or to position the snRNP so that it avoids being steered into a dead-end complex. For instance, Nelson and Green found that a perfect consensus 5′ splice site was less sensitive to negative context effects than was a less-than-perfect β-globin 5′ splice site (62). A more detailed mutagenic analysis of the 5′ pseudosplice site and its real counterparts should resolve some of these questions.
The 3′ pseudosplice site.
Similarly, a 3′ splice site with a high score of agreement with the consensus is insufficient for splicing. The 3′ pseudosplice site has a sequence (UUCUCCUGCCUCAG/C) consensus score of 83, higher than the score of 53% of the 2,400 actual 3′ splice sites in the database. We can consider three possibilities to explain why this site is not used: (i) it is masked by some local secondary or higher-order structure; (ii) it is a weak site that needs promotion by a strong 5′ splice site, a strong branch point, or exonic enhancers; or (iii) this sequence is somehow intrinsically defective (for example, an essential splicing factor cannot efficiently bind to it), and so it fails to function regardless of the context. We did not include the branch point as one of the elements that we varied in these experiments, since the consensus sequence for the branch point is quite degenerate (36) and mutations in branch points often result in the recruitment of a nearby cryptic site (66, 68). Our inability to activate splicing at this 3′ pseudosplice site supports the third possibility. Our attempts to activate splicing included, in combination, the substitution of a G at position +1 to form a CAG/G consensus at the potential splice site, the provision of known enhancer sequences within the body of the downstream exon, and the placement of an optimized 5′ splice site sequence downstream of the pseudoexon. Splicing was indeed activated by a combination of these three changes, but it occurred instead at a nearby upstream cryptic site (CUCCUGGGUUCUAG/C) with a lower consensus matching score of 69. On the other hand, the database of real 3′ splice sites contains two sequences similar (allowing only C-to-U or U-to-C changes in the PPT region) to this optimized (with CAG/G) 3′ pseudosplice site. The exact sequence of the 3′ pseudosplice site is not present, but this result is not unexpected since 99% of real 3′ splice site sequences (taken as 15-mers) are represented only once. This result implies that the exact C and U placement within the PPT is important. There is evidence in support of this idea; for example, Singh et al. (75) found that, in vitro, sequences selected for binding to U2AF65 were enriched in U but interrupted by two or three C's, the consensus being UUUUU(U/C)CC(C/U)UUUUUUUCC. In a survey of 3′ sequences that function in splicing, Coolidge et al. found U tracts to be the most effective, but their placement relative to the splice site was critical (22). In an analysis of in vivo mutations, we previously demonstrated a 50% splicing decrease brought about by a single U-to-C change in a PPT (18). Here again, a detailed mutational analysis of the 3′ pseudosplice site sequence should be revealing.
An intronic splicing silencer.
Splicing to the original candidate 3′ splice site was effected when a bona fide PPT (from hprt intron 1) was placed just upstream of a CAG/G sequence and when an optimized 5′ splice site followed the pseudoexon. However, even in this permissive situation, a 34-nt intronic silencer sequence upstream of the pseudoexon prevented it from being included in the mRNA. This sequence also inhibited splicing at the 3′ splice site of an aprt intron into which it had been inserted. Ironically, splicing to a cryptic 3′ splice site within the 34-mer was promoted in the aprt case. This cryptic site was also activated in the dhfr minigene when ASF/SF2 or β-globin exonic splicing enhancers were included in the pseudoexon. It is possible that the inhibition is exerted through this cryptic site, which could act as a competitor for U2 snRNP, tying it up in a dead-end complex that precludes any nearby potential 3′ splice site, similar to what has been proposed for the immunoglobulin M2 exonic silencer (41). A second model for splicing inhibition based on secondary or higher-order structures that sequester the 3′ splice sites seems less likely in view of the fact that the 34-mer inhibits splicing of at least three different downstream 3′ sites: the hprt intron 1 site joined to pseudoexon 1 and hprt intron 1 joined to β-globin exon 2 and aprt exon 4.
A search for the occurrence of the 34-mer silencer sequence in the human genome revealed that it is homologous to a region in an Alu repeat. The Alu homology extends through the body of pseudoexon 1: the 141-nt pseudoexon is homologous to the sequence from position 179 to position 23 (reverse of the standard orientation) of the Alu Sc family consensus, although it lacks an internal stretch of 16 nt (4). It is not surprising that pseudoexon 1 is a repeated sequence, since they represent about a third to a half of the human genome. Using RepeatMasker (http://ftp.genome.washington.edu/cgi-bin/RepeatMasker) to search just for Alu repeats in the 41,109-bp hprt gene we have analyzed, we found 32 instances, comprising 23% of the gene sequence. Moreover, Alu sequences in the reverse orientation contain several sequences resembling 3′ and 5′ splice sites. Although several cases of splicing at an Alu sequence have been reported (55), the vast majority of Alu sites contain pseudosites; i.e., they are not used. This coincidence raised the possibility that Alu repeats were the source of the majority of pseudoexons detected in our computer analysis. We therefore repeated our search for pseudoexons in an hprt sequence that had been divested of Alu repeats. Only 20% (21 of 103) of the pseudoexons were eliminated in this reanalysis, the same proportion as the number of nucleotides removed (23%). Thus, Alu sequences are no more likely to contribute to a pseudoexon than nonrepeated sequences. The question of why pseudosplice sites in Alu sequences are not used is, at this point, no different than the question of why pseudosplice sites in general are not used.
Exon inclusion without a recognized exonic splicing enhancer.
It is possible that real exons are recognized because they contain exonic splicing enhancers and pseudoexons are not recognized because they lack them. Exonic splicing enhancers have been demonstrated, for the most part, in alternatively spliced exons. However, mutations in exons outside of the consensus sequence can disrupt the splicing of constitutive exons (18, 64, 67) and some constitutively spliced exons contain sequences that can provide splicing enhancement to alternatively spliced exons (69, 92). In particular, human β-globin gene exon 2 has been shown to harbor at least three such sequences, which bind to different SR proteins (57, 69). Substitution of this β-globin exon 2 sequence for the pseudoexon 1 body did promote splicing. However, a complex pattern resulted, leading to intron retentions and exon skipping, as well as partial exon inclusion. Moreover, the 3′ splicing that did take place did so at a site upstream from the candidate we had identified on the basis of agreement with the 3′ consensus. Insertion of ASF/SF2 binding sequences into the pseudogene body also stimulated splicing and again to the lower-scoring upstream 3′ splice site sequence. Exon inclusion still required the presence of an improved 5′ splice site. The exclusive use of the distal lower-scoring 3′ splice site in both cases suggests either a topological constraint or, as discussed above, an intrinsic defectiveness of the proximal sequence despite its better agreement with the consensus. Interestingly, an SC35 binding sequence did not promote splicing here. Such context specificity for exonic splicing enhancers has been seen before, whereby some exons respond to an inserted SC35 binding sequence, others respond to an ASF/SF2 sequence, and yet others respond to both (57, 69).
The results of the enhancer experiments described above are consistent with the idea that pseudoexons are not used because they lack enhancer sequences, but they still do not explain why some potential splice sites can be recruited in this way and others can not. When functional splice sites were joined to pseudoexon 1 and the 34-mer inhibitory sequence was deleted, pseudoexon 1 was efficiently spliced despite its apparent lack of an enhancer. Moreover, substitution of a different intronic sequence for the exon body, termed pseudoexon 2, also resulted in exon inclusion. The inclusion of both pseudoexons in this context supports the idea that exon-bridging enhancers are not a prerequisite for constitutive exon definition or recognition. It may be, as is often stated for cases of alternative splicing, that enhancers are needed only when the splice sites are weak. However, our understanding of what distinguishes a weak from a strong splice site is incomplete. Quantitative agreement with the consensus is not a reliable guide, as evidenced here by the recruitment of a poorer-scoring upstream 3′ splice site over our original choice when enhancers were included in the exon. Taken together, our data are also consistent with another model, one in which the function of an enhancer is to counteract the effect of a nearby splicing inhibitor, as has been reported for several specific systems (2, 11, 41, 95). In line with this idea, we have found that sequences that can act as exonic splicing inhibitors are common in the human genome, occurring at a frequency of one per several hundred nucleotides (Fairbrother, submitted).
Our data also speak to a more ingenious model for the recognition of intronic splice site-like sites. It has been suggested that large introns may be removed by a process in which smaller sections are first extracted via intermediate splicing events (37). Hatton et al. demonstrated the stepwise removal of a large intron in the Drosophila ultrabithorax transcript by resplicing at the junction between certain joined exons. An extension of this strategy would be to drop the requirement for resplicing: proximally located intermediate exons would be spliced only to be removed by subsequent splicing of external exons. The final splice would remove a now much-abbreviated intron, facilitated by the new-found proximity of the final 5′ and 3′ splice sites. In this piecemeal splicing scenario, the pseudosites are not false sites at all but function rather as the functional boundaries of intermediate introns. Our data do not support such a model, since the 5′ and 3′ pseudosplice sites represented by the ends of pseudoexon 1 do not function when placed in the apparently favorable context of the small dhfr minigene.
ACKNOWLEDGMENTS
This work was supported by NIH grant GM22629.
We thank Will Fairbrother and Jim Manley for useful discussions.
REFERENCES
- 1.Ali S A, Steinkasserer A. PCR-ligation-PCR mutagenesis: a protocol for creating gene fusions and mutations. BioTechniques. 1995;18:746–750. [PubMed] [Google Scholar]
- 2.Amendt B A, Hesslein D, Chang L -J, Stoltzfus C M. Presence of negative and positive cis-acting RNA splicing elements within and flanking the first tat coding exon of human immunodeficiency virus type 1. Mol Cell Biol. 1994;14:3960–3970. doi: 10.1128/mcb.14.6.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balvay L, Libri D, Fiszman M Y. Pre-mRNA secondary structure and the regulation of splicing. Bioessays. 1993;15:165–169. doi: 10.1002/bies.950150304. [DOI] [PubMed] [Google Scholar]
- 4.Batzer M A, Deininger P L, Hellmann-Blumberg U, Jurka J, Labuda D, Rubin C M, Schmid C W, Zietkiewicz E, Zuckerkandl E. Standardized nomenclature for Alu repeats. J Mol Evol. 1996;42:3–6. doi: 10.1007/BF00163204. [DOI] [PubMed] [Google Scholar]
- 5.Bauren G, Wieslander L. Splicing of Balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell. 1994;76:183–192. doi: 10.1016/0092-8674(94)90182-1. [DOI] [PubMed] [Google Scholar]
- 6.Beyer A L, Osheim Y N. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988;2:754–765. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- 7.Blanchette M, Chabot B. A highly stable duplex structure sequesters the 5′ splice site region of hnRNP A1 alternative exon 7B. RNA. 1997;3:405–419. [PMC free article] [PubMed] [Google Scholar]
- 8.Blanchette M, Chabot B. Modulation of exon skipping by high-affinity hnRNP A1-binding sites and by intron elements that repress splice site utilization. EMBO J. 1999;18:1939–1952. doi: 10.1093/emboj/18.7.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bradley W E, Letovanec D. High-frequency nonrandom mutational event at the adenine phosphoribosyltransferase (aprt) locus of sib-selected CHO variants heterozygous for aprt. Somatic Cell Genet. 1982;8:51–66. doi: 10.1007/BF01538650. [DOI] [PubMed] [Google Scholar]
- 10.Burnette J M, Hatton A R, Lopez A J. Trans-acting factors required for inclusion of regulated exons in the Ultrabithorax mRNAs of Drosophila melanogaster. Genetics. 1999;151:1517–1529. doi: 10.1093/genetics/151.4.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caputi M, Casari G, Guenzi S, Tagliabue R, Sidoli A, Melo C A, Baralle F E. A novel bipartite splicing enhancer modulates the differential processing of the human fibronectin EDA exon. Nucleic Acids Res. 1994;22:1018–1022. doi: 10.1093/nar/22.6.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caputi M, Mayeda A, Krainer A R, Zahler A M. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 1999;18:4060–4067. doi: 10.1093/emboj/18.14.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carlo T, Sterner D A, Berget S M. An intron splicing enhancer containing a G-rich repeat facilitates inclusion of a vertebrate micro-exon. RNA. 1996;2:342–353. [PMC free article] [PubMed] [Google Scholar]
- 14.Carothers A M, Urlaub G, Grunberger D, Chasin L A. Splicing mutants and their second-site suppressors at the dihydrofolate reductase locus in Chinese hamster ovary cells. Mol Cell Biol. 1993;13:5085–5098. doi: 10.1128/mcb.13.8.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carter M S, Li S, Wilkinson M F. A splicing-dependent regulatory mechanism that detects translation signals. EMBO J. 1996;15:5965–5975. [PMC free article] [PubMed] [Google Scholar]
- 16.Chabot B, Blanchette M, Lapierre I, La Branche H. An intron element modulating 5′ splice site selection in the hnRNP A1 pre-mRNA interacts with hnRNP A1. Mol Cell Biol. 1997;17:1776–1786. doi: 10.1128/mcb.17.4.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan R C, Black D L. The polypyrimidine tract binding protein binds upstream of neural cell-specific c-src exon N1 to repress the splicing of the intron downstream. Mol Cell Biol. 1997;17:4667–4676. doi: 10.1128/mcb.17.8.4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen I T, Chasin L A. Direct selection for mutations affecting specific splice sites in a hamster dihydrofolate reductase minigene. Mol Cell Biol. 1993;13:289–300. doi: 10.1128/mcb.13.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng J, Maquat L E. Nonsense codons can reduce the abundance of nuclear mRNA without affecting the abundance of pre-mRNA or the half-life of cytoplasmic mRNA. Mol Cell Biol. 1993;13:1892–1902. doi: 10.1128/mcb.13.3.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chou M-Y, Rooke N, Turck C W, Black D L. hnRNP H is a component of a splicing enhancer complex that activates a c-src alternative exon in neuronal cells. Mol Cell Biol. 1999;19:69–77. doi: 10.1128/mcb.19.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clouet d'Orval B, d'Aubenton Carafa Y, Sirand-Pugnet P, Gallego M, Brody E, Marie J. RNA secondary structure repression of a muscle-specific exon in HeLa cell nuclear extracts. Science. 1991;252:1823–1828. doi: 10.1126/science.2063195. [DOI] [PubMed] [Google Scholar]
- 22.Coolidge C J, Seely R J, Patton J G. Functional analysis of the polypyrimidine tract in pre-mRNA splicing. Nucleic Acids Res. 1997;25:888–896. doi: 10.1093/nar/25.4.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cooper T A. Muscle-specific splicing of a heterologous exon mediated by a single muscle-specific splicing enhancer from the cardiac troponin T gene. Mol Cell Biol. 1998;18:4519–4525. doi: 10.1128/mcb.18.8.4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coulter L R, Landree M A, Cooper T A. Identification of a new class of exonic splicing enhancers by in vivo selection. Mol Cell Biol. 1997;17:2143–2150. doi: 10.1128/mcb.17.4.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Del Gatto-Konczak F, Olive M, Gesnel M -C, Breathnach R. hnRNP A1 recruited to an exon in vivo can function as an exon splicing silencer. Mol Cell Biol. 1999;19:251–260. doi: 10.1128/mcb.19.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deshler J O, Rossi J J. Unexpected point mutations activate cryptic 3′ splice sites by perturbing a natural secondary structure within a yeast intron. Genes Dev. 1991;5:1252–1263. doi: 10.1101/gad.5.7.1252. [DOI] [PubMed] [Google Scholar]
- 27.Dietz H C, Kendzior R J., Jr Maintenance of an open reading frame as an additional level of scrutiny during splice site selection. Nat Genet. 1994;8:183–188. doi: 10.1038/ng1094-183. [DOI] [PubMed] [Google Scholar]
- 28.Dominski Z, Kole R. Identification of exon sequences involved in splice site selection. J Biol Chem. 1994;269:23590–23596. [PubMed] [Google Scholar]
- 29.Dominski Z, Kole R. Selection of splice sites in pre-mRNAs with short internal exons. Mol Cell Biol. 1991;11:6075–6083. doi: 10.1128/mcb.11.12.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dreyfuss G M, Matunis J, Pinol-Roma S, Burd C G. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 31.Edwards A, Voss H, Rice P, Civitello A, Stegemann J, Schwater C, Zimmermann J, Erfle H, Caskey C T, Ansorge W. Automated DNA sequencing of the human HPRT locus. Genomics. 1990;6:593–608. doi: 10.1016/0888-7543(90)90493-e. [DOI] [PubMed] [Google Scholar]
- 32.Eperon L P, Graham I R, Griffiths A D, Eperon I C. Effects of RNA secondary structure on alternative splicing of pre-mRNA: is folding limited to a region behind the transcribing RNA polymerase? Cell. 1988;54:393–401. doi: 10.1016/0092-8674(88)90202-4. [DOI] [PubMed] [Google Scholar]
- 32a.Fairbrother, W. G., and L. A. Chasin. Human genomic sequences that inhibit splicing. Mol. Cell. Biol., in press. [DOI] [PMC free article] [PubMed]
- 33.Gilbert E, Del Gatto F, Champion-Arnaud P, Gesnel M C, Breathnach R. Control of BEK and K-SAM splice sites in alternative splicing of the fibroblast growth factor receptor 2 pre-mRNA. Mol Cell Biol. 1993;13:5461–4568. doi: 10.1128/mcb.13.9.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goguel V, Rosbash M. Splice site choice and splicing efficiency are positively influenced by pre-mRNA intramolecular base pairing in yeast. Cell. 1993;72:893–901. doi: 10.1016/0092-8674(93)90578-e. [DOI] [PubMed] [Google Scholar]
- 35.Gooding C, Roberts G C, Moreau G, Nadal-Ginard B, Smith C W. Smooth muscle-specific switching of alpha-tropomyosin mutually exclusive exon selection by specific inhibition of the strong default exon. EMBO J. 1994;13:3861–3872. doi: 10.1002/j.1460-2075.1994.tb06697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Green M R. Pre-mRNA splicing. Annu Rev Genet. 1986;20:671–708. doi: 10.1146/annurev.ge.20.120186.003323. [DOI] [PubMed] [Google Scholar]
- 37.Hatton A R, Subramaniam V, Lopez A J. Generation of alternative Ultrabithorax isoforms and stepwise removal of a large intron by resplicing at exon-exon junctions. Mol Cell. 1998;2:787–796. doi: 10.1016/s1097-2765(00)80293-2. [DOI] [PubMed] [Google Scholar]
- 38.Haut D D, Pintel D J. Inclusion of the NS2-specific exon in minute virus of mice mRNA is facilitated by an intronic splicing enhancer that affects definition of the downstream small intron. Virology. 1999;258:84–94. doi: 10.1006/viro.1999.9696. [DOI] [PubMed] [Google Scholar]
- 39.Hawkins J D. A survey on intron and exon lengths. Nucleic Acids Res. 1988;16:9893–9908. doi: 10.1093/nar/16.21.9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Howe K J, Ares M., Jr Intron self-complementarity enforces exon inclusion in a yeast pre-mRNA. Proc Natl Acad Sci USA. 1997;94:12467–12472. doi: 10.1073/pnas.94.23.12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kan J L, Green M R. Pre-mRNA splicing of IgM exons M1 and M2 is directed by a juxtaposed splicing enhancer and inhibitor. Genes Dev. 1999;13:462–471. doi: 10.1101/gad.13.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanopka A, Muhlemann O, Akusjarvi G. Inhibition by SR proteins of splicing of a regulated adenovirus pre-mRNA. Nature. 1996;381:535–538. doi: 10.1038/381535a0. [DOI] [PubMed] [Google Scholar]
- 43.Kessler O, Chasin L A. Effects of nonsense mutations on nuclear and cytoplasmic adenine phosphoribosyltransferase RNA. Mol Cell Biol. 1996;16:4426–4435. doi: 10.1128/mcb.16.8.4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kessler O, Jiang Y, Chasin L A. Order of intron removal during splicing of endogenous adenine phosphoribosyltransferase and dihydrofolate reductase pre-mRNA. Mol Cell Biol. 1993;13:6211–6222. doi: 10.1128/mcb.13.10.6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kister L, Domenjoud L, Gallinaro H, Monique J. A cis-acting selector of a 5′ splice site. Cooperation between the sequence of the site and an upstream exonic element. J Biol Chem. 1993;268:21955–21961. [PubMed] [Google Scholar]
- 46.Krainer A R, Conway G C, Kozak D. Purification and characterization of pre-mRNA splicing factor SF2 from HeLa cells. Genes Dev. 1990;4:1158–1171. doi: 10.1101/gad.4.7.1158. [DOI] [PubMed] [Google Scholar]
- 47.Krawczak M, Reiss J, Cooper D N. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum Genet. 1992;90:41–54. doi: 10.1007/BF00210743. [DOI] [PubMed] [Google Scholar]
- 48.LeMaire M F, Thummel C S. Splicing precedes polyadenylation during Drosophila E74A transcription. Mol Cell Biol. 1990;10:6059–6063. doi: 10.1128/mcb.10.11.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin C H, Patton J G. Regulation of alternative 3′ splice site selection by constitutive splicing factors. RNA. 1995;1:234–245. [PMC free article] [PubMed] [Google Scholar]
- 50.Liu H X, Goodall G J, Kole R, Filipowicz W. Effects of secondary structure on pre-mRNA splicing: hairpins sequestering the 5′ but not the 3′ splice site inhibit intron processing in Nicotiana plumbaginifolia. EMBO J. 1995;14:377–388. doi: 10.1002/j.1460-2075.1995.tb07012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu H X, Zhang M, Krainer A R. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev. 1998;12:1998–2012. doi: 10.1101/gad.12.13.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lopez A J. Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu Rev Genet. 1998;32:279–305. doi: 10.1146/annurev.genet.32.1.279. [DOI] [PubMed] [Google Scholar]
- 53.Lozano F, Maertzdorf B, Pannell R, Milstein C. Low cytoplasmic mRNA levels of immunoglobulin kappa light chain genes containing nonsense codons correlate with inefficient splicing. EMBO J. 1994;13:4617–4622. doi: 10.1002/j.1460-2075.1994.tb06783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luukkonen B G, Seraphin B. The role of branchpoint-3′ splice site spacing and interaction between intron terminal nucleotides in 3′ splice site selection in Saccharomyces cerevisiae. EMBO J. 1997;16:779–792. doi: 10.1093/emboj/16.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Makalowski W, Mitchell G A, Labuda D. Alu sequences in the coding regions of mRNA: a source of protein variability. Trends Genet. 1994;10:188–193. doi: 10.1016/0168-9525(94)90254-2. [DOI] [PubMed] [Google Scholar]
- 56.Maquat L E. When cells stop making sense: effects of nonsense codons on RNA metabolism in vertebrate cells. RNA. 1995;1:453–465. [PMC free article] [PubMed] [Google Scholar]
- 57.Mayeda A, Screaton G R, Chandler S D, Fu X D, Krainer A R. Substrate specificities of SR proteins in constitutive splicing are determined by their RNA recognition motifs and composite pre-mRNA exonic elements. Mol Cell Biol. 1999;19:1853–1863. doi: 10.1128/mcb.19.3.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Modafferi E F, Black D L. A complex intronic splicing enhancer from the c-src pre-mRNA activates inclusion of a heterologous exon. Mol Cell Biol. 1997;17:6537–6545. doi: 10.1128/mcb.17.11.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mount S M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982;10:459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mulligan G J, Guo W, Wormsley S, Helfman D M. Polypyrimidine tract binding protein interacts with sequences involved in alternative splicing of beta-tropomyosin pre-mRNA. J Biol Chem. 1992;267:25480–25487. [PubMed] [Google Scholar]
- 61.Naeger L K, Schoborg R V, Zhao Q, Tullis G E, Pintel D J. Nonsense mutations inhibit splicing of MVM RNA in cis when they interrupt the reading frame of either exon of the final spliced product. Genes Dev. 1992;6:1107–1119. doi: 10.1101/gad.6.6.1107. [DOI] [PubMed] [Google Scholar]
- 62.Nelson K K, Green M R. Splice site selection and ribonucleoprotein complex assembly during in vitro pre-mRNA splicing. Genes Dev. 1988;2:319–329. doi: 10.1101/gad.2.3.319. [DOI] [PubMed] [Google Scholar]
- 63.Noe V, Ciudad C, Chasin L. Effect of differential polyadenylation and cell growth phase on dihydrofolate reductase mRNA stability. J Biol Chem. 1999;274:27807–27814. doi: 10.1074/jbc.274.39.27807. [DOI] [PubMed] [Google Scholar]
- 64.O'Neill J P, Rogan P K, Cariello N, Nicklas J A. Mutations that alter RNA splicing of the human HPRT gene: a review of the spectrum. Mutat Res. 1998;411:179–214. doi: 10.1016/s1383-5742(98)00013-1. [DOI] [PubMed] [Google Scholar]
- 65.Peppel K, Baglioni C. A simple and fast method to extract RNA from tissue culture cells. BioTechniques. 1990;9:711–713. [PubMed] [Google Scholar]
- 66.Reed R, Maniatis T. Intron sequences involved in lariat formation during pre-mRNA splicing. Cell. 1985;41:95–105. doi: 10.1016/0092-8674(85)90064-9. [DOI] [PubMed] [Google Scholar]
- 67.Reed R, Maniatis T. A role for exon sequences and splice-site proximity in splice-site selection. Cell. 1986;46:681–690. doi: 10.1016/0092-8674(86)90343-0. [DOI] [PubMed] [Google Scholar]
- 68.Ruskin B, Greene J M, Green M R. Cryptic branch point activation allows accurate in vitro splicing of human beta-globin intron mutants. Cell. 1985;41:833–844. doi: 10.1016/s0092-8674(85)80064-7. [DOI] [PubMed] [Google Scholar]
- 69.Schaal T D, Maniatis T. Multiple distinct splicing enhancers in the protein-coding sequences of a constitutively spliced pre-mRNA. Mol Cell Biol. 1999;19:261–273. doi: 10.1128/mcb.19.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schaal T D, Maniatis T. Selection and characterization of pre-mRNA splicing enhancers: identification of novel SR protein-specific enhancer sequences. Mol Cell Biol. 1999;19:1705–1719. doi: 10.1128/mcb.19.3.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Senapathy P, Shapiro M B, Harris N L. Splice junctions, branch point sites, and exons: sequence statistics, identification, and applications to genome project. Methods Enzymol. 1990;183:252–278. doi: 10.1016/0076-6879(90)83018-5. [DOI] [PubMed] [Google Scholar]
- 72.Shapiro M B, Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987;15:7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Siebel C W, Fresco L D, Rio D C. The mechanism of somatic inhibition of Drosophila P-element pre-mRNA splicing: multiprotein complexes at an exon pseudo-5′ splice site control U1 snRNP binding. Genes Dev. 1992;6:1386–1401. doi: 10.1101/gad.6.8.1386. [DOI] [PubMed] [Google Scholar]
- 74.Siebel C W, Kanaar R, Rio D C. Regulation of tissue-specific P-element pre-mRNA splicing requires the RNA-binding protein PSI. Genes Dev. 1994;8:1713–1725. doi: 10.1101/gad.8.14.1713. [DOI] [PubMed] [Google Scholar]
- 75.Singh R, Valcarcel J, Green M R. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- 76.Sirand-Pugnet P, Durosay P, Brody E, Marie J. An intronic (A/U)GGG repeat enhances the splicing of an alternative intron of the chicken beta-tropomyosin pre-mRNA. Nucleic Acids Res. 1995;23:3501–3507. doi: 10.1093/nar/23.17.3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Snyder E E, Stormo G D. Identification of protein coding regions in genomic DNA. J Mol Biol. 1995;248:1–18. doi: 10.1006/jmbi.1995.0198. [DOI] [PubMed] [Google Scholar]
- 78.Solovyev V, Salamov A. The Gene-Finder computer tools for analysis of human and model organisms genome sequences. Intelligent Syst Mol Biol. 1997;5:294–302. [PubMed] [Google Scholar]
- 79.Solovyev V V, Salamov A A, Lawrence C B. The prediction of human exons by oligonucleotide composition and discriminant analysis of spliceable open reading frames. ISMB. 1994;2:354–362. [PubMed] [Google Scholar]
- 80.Stephens R M, Schneider T D. Features of spliceosome evolution and function inferred from an analysis of the information at human splice sites. J Mol Biol. 1992;228:1124–1136. doi: 10.1016/0022-2836(92)90320-j. [DOI] [PubMed] [Google Scholar]
- 81.Sterner D A, Berget S M. In vivo recognition of a vertebrate mini-exon as an exon-intron-exon unit. Mol Cell Biol. 1993;13:2677–2687. doi: 10.1128/mcb.13.5.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sun Q, Hampson R K, Rottman F M. In vitro analysis of bovine growth hormone pre-mRNA alternative splicing. Involvement of exon sequences and trans-acting factor(s) J Biol Chem. 1993;268:15659–15666. [PubMed] [Google Scholar]
- 83.Sun Q, Mayeda A, Hampson R K, Krainer A R, Rottman F M. General splicing factor SF2/ASF promotes alternative splicing by binding to an exonic splicing enhancer. Genes Dev. 1993;7:2598–2608. doi: 10.1101/gad.7.12b.2598. [DOI] [PubMed] [Google Scholar]
- 84.Tacke R, Manley J L. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J. 1995;14:3540–3551. doi: 10.1002/j.1460-2075.1995.tb07360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tian H, Kole R. Selection of novel exon recognition elements from a pool of random sequences. Mol Cell Biol. 1995;15:6291–6298. doi: 10.1128/mcb.15.11.6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tian M, Maniatis T. A splicing enhancer complex controls alternative splicing of doublesex pre-mRNA. Cell. 1993;74:105–114. doi: 10.1016/0092-8674(93)90298-5. [DOI] [PubMed] [Google Scholar]
- 87.Urlaub G, Kas E, Carothers A M, Chasin L A. Deletion of the diploid dihydrofolate reductase locus from cultured mammalian cells. Cell. 1983;33:405–412. doi: 10.1016/0092-8674(83)90422-1. [DOI] [PubMed] [Google Scholar]
- 88.Urlaub G, Mitchell P J, Ciudad C J, Chasin L A. Nonsense mutations in the dihydrofolate reductase gene affect RNA processing. Mol Cell Biol. 1989;9:2868–2880. doi: 10.1128/mcb.9.7.2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang J, Takagaki Y, Manley J L. Targeted disruption of an essential vertebrate gene: ASF/SF2 is required for cell viability. Genes Dev. 1996;10:2588–2599. doi: 10.1101/gad.10.20.2588. [DOI] [PubMed] [Google Scholar]
- 90.Watakabe A, Tanaka K, Shimura Y. The role of exon sequences in splice site selection. Genes Dev. 1993;7:407–418. doi: 10.1101/gad.7.3.407. [DOI] [PubMed] [Google Scholar]
- 91.Wuarin J, Schibler U. Physical isolation of nascent RNA chains transcribed by RNA polymerase II: evidence for cotranscriptional splicing. Mol Cell Biol. 1994;14:7219–7225. doi: 10.1128/mcb.14.11.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yeakley J M, Morfin J P, Rosenfeld M G, Fu X D. A complex of nuclear proteins mediates SR protein binding to a purine-rich splicing enhancer. Proc Natl Acad Sci USA. 1996;93:7582–7587. doi: 10.1073/pnas.93.15.7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang M Q. Identification of protein coding regions in the human genome by quadratic discriminant analysis. Proc Natl Acad Sci USA. 1997;94:565–568. doi: 10.1073/pnas.94.2.565. . (Erratum, 94:5495.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang M Q. Statistical features of human exons and their flanking regions. Hum Mol Genet. 1998;7:919–932. doi: 10.1093/hmg/7.5.919. [DOI] [PubMed] [Google Scholar]
- 95.Zheng Z M, He P, Baker C C. Selection of the bovine papillomavirus type 1 nucleotide 3225 3′ splice site is regulated through an exonic splicing enhancer and its juxtaposed exonic splicing suppressor. J Virol. 1996;70:4691–4699. doi: 10.1128/jvi.70.7.4691-4699.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]