Forkhead Transcription Factors Are Critical Effectors of Cell Death and Cell Cycle Arrest Downstream of PTEN (original) (raw)
Abstract
PTEN acts as a tumor suppressor, at least in part, by antagonizing phosphoinositide 3-kinase (PI3K)/Akt signaling. Here we show that Forkhead transcription factors FKHRL1 and FKHR, substrates of the Akt kinase, are aberrantly localized to the cytoplasm and cannot activate transcription in PTEN-deficient cells. Restoration of PTEN function restores FKHR to the nucleus and restores transcriptional activation. Expression of a constitutively active form of FKHR that cannot be phosphorylated by Akt produces the same effect as reconstitution of PTEN on PTEN-deficient tumor cells. Specifically, activated FKHR induces apoptosis in cells that undergo PTEN-mediated cell death and induces G1 arrest in cells that undergo PTEN-mediated cell cycle arrest. Furthermore, both PTEN and constitutively active FKHR induce p27KIP1 protein but not p21. These data suggest that Forkhead transcription factors are critical effectors of PTEN-mediated tumor suppression.
The PTEN/MMAC/TEP-1 tumor suppressor gene (hereafter referred to as PTEN) is a common target of somatic mutation in a number of malignancies including prostate and endometrial cancers, glioblastoma, and melanoma (6, 26, 34, 36, 38, 54, 63, 67, 69, 73). In addition, germ line mutations in the PTEN gene are associated with the development of Cowden disease, an inherited hamartoma syndrome associated with an elevated risk of breast and thyroid cancers (37, 45). The PTEN protein product (PTEN) functions as both a protein and lipid phosphatase (39, 44). The former activity is associated with inhibition of cell spreading and dephosphorylation of focal adhesion kinase (65). PTEN lipid phosphatase activity is specific for the 3 position of phosphatidylinositol-3,4,5-trisphosphate and phosphatidylinositol-3,4-bisphosphate, both of which are by-products of the lipid kinase activity of the phosphoinositide 3-kinase (PI3K) (39). This latter PTEN activity is associated with the ability of PTEN to antagonize signaling through the PI3K pathway and hence to block inappropriate activation of the serine threonine kinase Akt (reviewed in references 7 and 71).
Reintroduction of PTEN into certain PTEN-null tumor cells, such as U87-MG and 786-O, leads to the induction of a G1 arrest (21, 33, 53). This arrest requires the lipid phosphatase activity of PTEN and can be overridden by a constitutively active form of the Akt, a downstream effector of PI3K (21, 53). In keeping with these data, PTEN heterozygosity results in excessive proliferation in murine prostate and thyroid tissues; _PTEN_−/− embryos have widespread excess bromodeoxyuridine incorporation, and _PTEN_−/− embryonic stem ES cells show abnormal cell cycle kinetics and reduced p27KIP1 (p27) levels (19, 62, 64). These data demonstrate a necessary role for PTEN in cell cycle regulation.
Introduction of PTEN into certain other PTEN-null tumor cells such as LNCaP, MDA-MB-468, and U251 results in the induction of apoptosis or anoikis (14, 15, 35, 43). This induction is also tied to inhibition of PI3K and Akt (35, 43). Further, the study of murine PTEN loss-of-function alleles has revealed defects in apoptosis. _PTEN_−/− murine fibroblasts are impaired in their response to apoptotic stimuli such as UV irradiation and osmotic stress (62). PTEN+/− mice have abnormal lymphoid aggregates, and lymphocytes from these mice have reduced annexin V staining, a marker of apoptosis (52). Finally, PTEN+/− mice also develop a lymphoproliferative syndrome that results from, and phenocopies, defects in Fas signaling (18). Collectively, these data support a necessary role for PTEN in mediating apoptosis in fibroblasts and lymphocytes. PTEN, like p53, is therefore a regulator of both cell cycle progression and apoptosis.
Potential effectors of PI3K signaling, downstream of PTEN, include a number of identified Akt substrates such as BAD, caspase 9, IKKα, and the Forkhead transcription factors FKHR, FKHRL1, and AFX (4, 5, 8, 13, 31, 48, 66). Each of these substrates is implicated in cell survival. Other downstream targets of Akt include nitric oxide synthetase, GSK3, and 4E-BP1/Phas-I (12, 24, 41). While each of these proteins is a known Akt substrate, with respect to the function of PTEN as a tumor suppressor it is not known which substrates are necessary and/or sufficient for enacting cell cycle control or for inducting apoptosis. One possibility is that different Akt substrates are responsible for enacting cell cycle control and regulating apoptosis. Alternatively, it is possible that one Akt target might be critical for both functions. Therefore, we sought to determine whether one or more of these substrates was deregulated in PTEN-null tumors and, in addition, to determine whether any one target was either necessary or sufficient for PTEN to regulate the cell cycle or to induce apoptosis.
Here we show for the first time that members of the forkhead transcription factor family are deregulated and inactive in PTEN null cells. Furthermore, a form of the Forkhead factor FKHR (FKHR;AAA) that cannot be phosphorylated by Akt is sufficient to induce apoptosis in PTEN-null cells. In addition, this constitutively active form of FKHR induced a cell cycle arrest rather than apoptosis in PTEN-null cells that likewise undergo a G1 arrest following restoration of PTEN function. As shown before for PTEN, the phosphosite mutant form of FKHR was also capable of inducing p27 and not p21. These data suggest that an active form of FKHR can complement PTEN deficiency in both the cell cycle and apoptotic pathways and suggest that FKHR may function as regulator of both proliferation and cell survival in the PI3K signaling pathway. These results further suggest that FKHR or its related family members AFX and FKHRL1 are critical proteins downstream of PTEN and that restoration of forkhead function might suppress tumorigenesis in PTEN-deficient tumor cells.
MATERIALS AND METHODS
Plasmids.
pCD19, pSG5L, pSG5L-HA-PTEN, pSG5L-HA-PTEN;G129R, pSG5L-HA-PTEN;G129E, pSG5L-HA-PTEN;1–353, pBABE-puroL, pBABE-puroL-HA-PTEN; pBABE-puroL-HA-PTEN;G129R and pGL3-promoter (Promega) were described previously (53, 57, 68). pcDNA3-Flag-FKHR, pcDNA3-Flag-FKHR;H215, pcDNA3-Flag-FKHR;AAA, and pGL2promoter-3×IRS were gifts of E. Tang, F. Barr, and K. Guan (66). The inserts from pcDNA3-Flag-FKHR or the mutant derivatives, restricted with _Bam_HI and _Xba_I, were ligated to the vector from similarly restricted pcDNA3-GFP to give pcDNA3-GFP-FKHR, pcDNA3-GFP-FKHR;H215R and pcDNA3-GFP-FKHR;AAA. Oligonucleotides 5′-GCGCGGATCCATGGCCGAGGCGCCTCAGGTG-3′ and 5′-CGCGCTCGAGGAATTCTCAGCCTGACACCCAGCTATG-3′ were used to PCR amplify the FKHR cDNAs. The PCR products, restricted with _Bam_HI and _Xho_I, were ligated to similarly restricted pBABE-puroL to give pBABE-puroL-FKHR, pBABE-puroL-FKHR;H215R, and pBABE-puroL-FKHR;AAA. Oligonucleotides 5′-GCGCGCTAGCGTGACAGAGTGAGACTCTGTCTCTATTTAAATAAATAAGTAAATAAATAAAC-3′ and 5′-GGGG AGATCTGCTTTGTATTTCACAATGTTTTCATTTTCATTGTTTGCCCAG TTTATTTATTT-3′, containing the forkhead site of the FasL promoter, were phosphorylated, annealed, and ligated to pGL3-promoter restricted with _Bgl_II and _Nhe_I to give pGL3-promoter-FasL. This plasmid was subsequently restricted with _Bgl_II and _Hin_dIII, blunted, and ligated to remove the simian virus 40 promoter and give pGL3-FasL. pAdTrack-CMV and pAdEasy-1 were the gifts of B. Vogelstein and K. Polyak (28). The insert from pcDNA3-Flag-FKHR;AAA liberated by restriction with _Xba_I and partial digestion with _Hin_dIII was ligated to similarly restricted pAdTrack-CMV vector to give pAd-FKHR;AAA.
Cell lines, cell culture, transfection, and MTS assay.
LNCaP cells were maintained at 37°C in a humidified 5% CO2 atmosphere in RPMI 1640 containing 10% fetal calf serum (FCS) (HyClone), penicillin and streptomycin (PS), 2.5 g of glucose per liter, 10 mM HEPES, 1 mM sodium pyruvate, and 2 mM l-glutamine. DU-145 cells were maintained at 37°C in a humidified 10% CO2 atmosphere in Dulbecco's modified Eagle's medium containing 10% FCS and PS. ACHN, 786-O, and U2-OS cells were maintained as previously described (53). Phoenix-ampho (φX-A) cells were maintained at 37°C in a humidified 10% CO2 atmosphere in Dulbecco's modified Eagle's medium containing 10% FCS and PS. 786-O cells were transfected using Fugene reagent (Boehringer Mannheim) as previously described (70). U2-OS, ACHN, and φX-A cells were transfected by the _N,N_-bis-(2-hydroxyethyl)-2-aminoethanesulfuric acid-buffered saline (BBS)–calcium phosphate method as previously described (10, 57).
LNCaP and 786-O cell viability was assayed using the Cell Titer 96 aqueous nonradioactive cell proliferation assay (Promega) as specified by the manufacturer. Briefly, cells were detached with trypsin and collected in 10 ml of complete medium. A 100-μl volume of cells was aliquoted in triplicate into 96-well plates. Then 20 μl of a 1:20 dilution of phenazine methosulfate in 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2_H_-tetrazolium, inner salt (MTS), reagent was added to each well. The plates were incubated at 37°C in 10% CO2 for 15 min. Formazan product was detected by measuring the absorbance at 490 nm.
Antibodies, immunoblotting, protein extraction, cell fractionation.
Anti-PTEN(C54) (53), anti-HA (BabCo), anti-FKHRL1, anti-phospho-FKHRL1 (Upstate Biotechnology), anti-GSK3 (New England Biolabs [NEB]), anti-phospho-GSK3 (NEB), anti-phospho-Akt (NEB), anti-Akt (NEB), anti-p27 (Transduction Laboratories), anti-p70S6K (Santa Cruz Biotechnology), and 245 anti-RB (Pharmingen) antibodies were used at a dilution of 1:1,000. M5 anti-flag antibody (Sigma) was used at 10 μg/ml. Anti-p21 antibody (Transduction Laboratories) was used at 1:500. Anti-tubulin antibody (ICN) was used at 1:2,000. Anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Biodesign International) was used at 1:5,000.
Cell lysates were prepared and immunoblot analyses were performed as previously described (70).
786-O and ACHN cells were fractionated by swelling for 10 min in RBS buffer (10 mM HEPES [pH 7.2], 10 mM NaCl, 1.5 mM MgCl2) containing 5 μg of leupeptin per ml, 2 μg of aprotinin per ml, 50 μg of phenylmethylsulfonyl fluoride per ml, 5 mM NaF, and 0.5 mM sodium orthovanadate. The cells were disrupted by 60 (786-O) or 10 (ACHN) manual strokes of a Dounce homogenizer. Nuclei were pelleted by centrifugation at 2,700 rpm for 5 min, washed three times in RBS buffer, and lysed in RIPA buffer (10 mM NaPO4, 150 mM NaCl, 1% NP-40, 0.1% deoxycholate, 0.1% sodium dodecyl sulfate). Cytoplasmic proteins were precipitated with 10% trichloroacetic acid, washed with 80% acetone, washed with double-distilled H2O, and solubilized in 1× protein sample buffer.
FACS and cell cycle analysis.
Cell cycle analysis was performed as previously described (57, 70). Briefly, 786-O cells grown on p100 plates were transfected with 4 μg of pCD19 plasmid and the amounts of either pSG5 or pcDNA3 expression plasmid indicated in the figure legends. At 48 h after transfection, the cells were harvested, stained with fluorescein isothiocyanate-conjugated anti-CD19 antibody and propidium iodide, and analyzed by two-color fluorescence-activated cell sorting (FACS) (Beckton-Dickinson).
Retrovirus production and infection.
Amphotrophic retroviral supernatants were produced as previously described (51). Briefly, φX-A cells, split 1:4 the previous day, were transfected with 25 μg of the indicated pBABEpuroL plasmid DNA. After 16 h the medium was changed and the cells were incubated in a 10% CO2 incubator for 48 h. The medium was harvested and stored at −70°C until needed. 786-O and LNCaP cells were incubated with 5 ml of thawed viral supernatant containing 5 μg of Polybrene (hexadimethine bromide) (Sigma H9268) per ml and incubated at 37°C for 4 h. A 5-ml volume of complete medium was added, and the cells were maintained for 40 h under standard growth conditions, after which the medium was changed to complete medium supplemented with 2 μg of puromycin per ml. Drug-resistant cells were selected and harvested after 72 h. Typically, 85% of the LNCaP or 786-O cells infected with the pBABE-puroL retrovirus were drug resistant.
Adenovirus production and infection.
Recombinant FKHR;AAA adenovirus was generated as previously described (28). Briefly, pAdTrack-CMV and pAd-FKHR;AAA were linearized and individually cotransformed into electrocompetent BJ5183 cells (Quantum Biotechnologies) along with pAdEasy-1. Next, recombinant adenovirus DNA, isolated from kanamycin-resistant colonies, was amplified in Top10 cells (Invitrogen), purified by CsCl2 density gradient centrifugation, linearized with Pac1, and transfected into 293 cells with Lipofectamine (Life Technologies). After 7 to 10 days, packaged virus was collected and used to infect 20 p150 plates of 293 cells. The amplified virus was isolated by freeze-thaw extraction, purified by CsCl2 density gradient centrifugation, and subjected to titer determination by lysis of 293 cells. LNCaP and 786-O cells were infected with Ad-vector at a multiplicity of infection of 50 and with Ad-FKHR;AAA at a multiplicity of infection of 100.
Reporter assays.
Transfections for reporter assays were carried out in 6- or 24-well plates. At 36 h after transfection, cells were lysed in 1× reporter lysis buffer as specified by the manufacturer (Promega). Cleared lysates were used in luciferase and β-galactosidase assays as described previously (57). Relative light units were normalized to β-galactosidase activity. The fold activation was obtained by dividing corrected luciferase values by the corrected luciferase value obtained in the presence of the vector and reporter plasmids alone.
Real-time quantitative PCR.
RNA was prepared using the RNeasy RNA isolation kit (Qiagen) as specified by the manufacturer, including DNase treatment. Total RNA (1 μg) was reverse transcribed at 42°C for 45 min in a 20-μl reaction mixture containing 250 μM each deoxynucleoside triphosphate, 20 U of RNase inhibitor, 50 U of murine leukemia virus reverse transcriptase (RT), 2.5 μM random hexamers, and 1× RT buffer (1.5 mM MgCl2) and then denatured at 99°C for 5 min. An RT-minus reaction was also performed for each sample. Specific primers and fluorogenic probe for human p27 (Fw, 5′-GCAATGCGCAGGAATAAGGA-3′; Rev, 5′-TCCACAGAACCGGCATTTG-3′; probe, 5′-CGACCTGCAACCGACGATTCTTCTACTCA-3′) were designed using Primer Express 1.0 software. Amplification of the GAPDH gene was used to standardize the amount of RNA in each reaction mixture (Taqman GAPDH control reagents). PCR was performed using an ABI Prism 7700 sequence detector. The Taqman PCR core reagent kit was used as specified by the manufacturer with the following modifications: dUTP was replaced by dTTP and incubation with AmpErase was omitted. PCR mixtures each contained 1 μl of cDNA (equivalent o 50 ng of template RNA), 2.5 U of AmpliTaq Gold, and 100 nM (each) oligonucleotide primers and fluorogenic probe in a volume of 50 μl. Amplifications consisted of 60 cycles of 94°C for 45 s, 58°C for 45 s, and 72°C for 1 min. All reagents for real-time PCR were purchased from Perkin-Elmer Applied Biosystems.
In each experiment, additional reactions with seven serial twofold dilutions of 786-O cDNA as template were performed with each set of primers and probes on the same 96-well plate to generate standard curves, which related the threshold cycle (CT) to the log input amount of template. All samples were amplified in triplicate. The relative amount of p27 transcripts in each sample was determined by using the standard-curve method and by normalizing for GAPDH mRNA expression levels, as previously described (20; Applied Biosystems, ABI Prism 7700 Sequence Detection System User Bulletin, vol. 2, p. 1–35, 1997).
Protein half-life determination by cycloheximide treatment.
At 20 h after adenovirus infection, 786-O cells were treated with 25 μg of cycloheximide per ml. At the indicated times, the cells were washed, scrape harvested into 500 μl of phosphate-buffered saline, pelleted by centrifugation at 400 × g for 5 min, and stored at −70°C. Cell extracts were prepared as described above and immunoblotted with the indicated antibodies. Multiple exposures were obtained and then digitized using a Scanmaker III flatbed scanner. The resulting immunoblot signals were quantified using ImageQuant software (Molecular Dynamics). Only radiographs where the peak quantification showed nonsaturating signals were used. The half-life was calculated from exponential curve fits to the data plotted in log-linear fashion, as previously described (70).
RESULTS
FKHRL1 protein levels and phosphorylation are deregulated in the absence of PTEN.
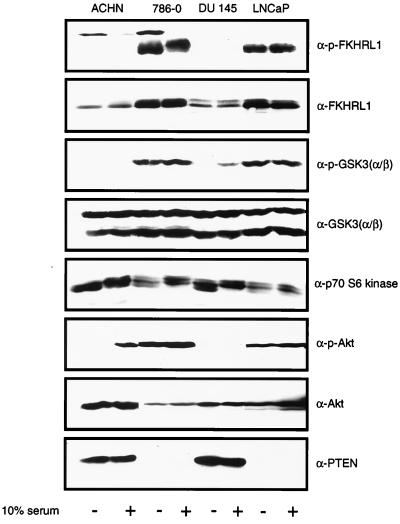
A survey of the activation state of downstream targets of Akt was undertaken using antibodies against specific phosphopeptides. Two pairs of cell lines were used, ACHN and 786-O renal carcinoma cells and DU145 and LNCaP prostate carcinoma cells. ACHN and DU145 both retain wild-type PTEN alleles and express an intact PTEN protein, while 786-O and LNCaP cells fail to express any full-length PTEN protein (53) (Fig. 1). Whole-cell extracts were prepared from serum-starved cells or from starved cells that were stimulated with serum. As previously shown, in these _PTEN_−/− cells the phosphorylated and activated form of Akt is overabundant (53) (Fig. 1). Extracts were immunoblotted with antibodies that detect phosphorylation of GSK3 and FKHRL1. In the absence of serum, deregulation of GSK3-α phosphorylation was noted in the two PTEN-null cell lines (Fig. 1). Immunoblotting also demonstrated a marked increase in phosphorylated FKHRL1 in the PTEN-null cell lines; however, the total amount of FKHRL1 was also elevated in these cells. While both endogenous AFX and FKHR were detected in all of these cells (data not shown), phosphospecific antibodies were, in our hands, incapable of recognizing endogenous phosphorylated AFX or FKHR. In contrast to the results obtained with FKHRL1 and GSK3, phosphorylated Bad was not detected in these cells and p70S6K was not consistently hyperphosphorylated in a manner that reflected the loss of PTEN (Fig. 1 and data not shown).
FIG. 1.
Immunoblot detection of Akt substrates in PTEN-null cells. Whole-cell extracts were prepared from the indicated serum-starved or serum-stimulated cells and separated by gel electrophoresis. Separated proteins were transferred to nitrocellulose. Membranes were incubated with the indicated immune reagents, and bound antibody was detected by enhanced chemiluminescence. For GSK3, serum stimulation was carried out for 10 min, while for the remaining blots, the cells were stimulated for 90 min. In certain instances the lane order was change for clarity.
FKHR localization is constitutively cytoplasmic in PTEN−/− cells.
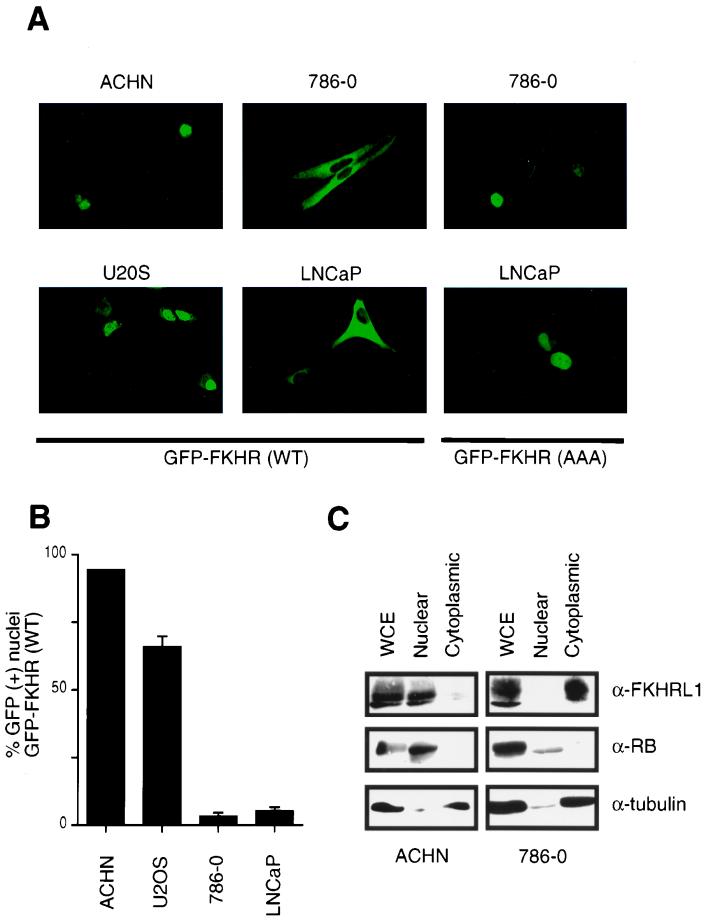
Previous data showed that Akt-dependent inhibition of FKHR or FKHRL1 is mediated, at least in part, by phosphorylation-dependent localization of these transcription factors to the cytoplasm (4, 5). These considerations and the data in Fig. 1 led us to ask whether Forkhead factors might be aberrantly localized in PTEN-null cells. To this end, 786-O and ACHN cells were fractionated into cytoplasmic and nuclear fractions. Anti-FKHRL1 immunoblotting demonstrated that FKHRL1 was indeed cytoplasmic in PTEN-null 786-O cells while it was primarily localized to the nucleus in ACHN cells (Fig. 2C). As controls for fractionation, immunoblotting demonstrated that β-tubulin was found in the cytoplasm and the retinoblastoma protein (pRB) was found in the nucleus (Fig. 2C).
FIG. 2.
FKHR is mislocalized in PTEN-null cells. (A) Fluorescence microscopy of GFP-FKHR in PTEN-plus and PTEN-null cells. The indicated cells were transiently transfected, as described in Materials and Methods, with a plasmid encoding GFP-FKHR or GFP-FKHR;AAA. At 24 h after transfection, GFP-FKHR was detected by fluorescence microscopy in living cells. (B) Quantification of the results obtained with GFP-FKHR as shown in panel A. The percentage of cells with nuclear localization of GFP-FKHR was determined by manual counting of GFP-positive nuclei. Data shown are the mean and standard error of duplicate experiments and are representative of two independent experiments. (C) Localization of FKHRL1 in 786-O and ACHN cells. 786-O and ACHN cells were fractionated into cytoplasmic and nuclear compartments by hypotonic lysis and Dounce homogenization. Equivalent cell fractions were loaded on the gel and immunoblotted with anti-FKHRL1, anti-β-tubulin, and anti-pRB antibodies as indicated.
To examine the localization of Forkhead factors in living cells, plasmids encoding green fluorescent protein (GFP)-FKHR fusion proteins were introduced into cells containing or lacking PTEN. Here, we chose to use FKHR as a representative of the class of forkhead transcription factors that include FKHR, FKHRL1, and AFX. After 24 h, the localization of GFP-FKHR in living cells was determined by direct visualization using fluorescence microscopy. In cells that have PTEN, GFP-FKHR was found primarily in the nucleus (ACHN cells) or in both the nucleus and cytoplasm (U2-OS cells). In contrast, GFP-FKHR was localized exclusively in the cytoplasm in cells lacking PTEN (786-O and LNCaP cells) (Fig. 2A). These data were quantified by manual counting of cells (Fig. 2B). In contrast, a FKHR mutant (FKHR;AAA) lacking the three Akt phosphoacceptor sites (T24A, S256A, and S319A) (66) was found primarily in the nucleus in PTEN-null cells (Fig. 2A). These data suggest that in PTEN-null cells, FKHR is mislocalized to the cytoplasm due to persistent activation of the PI3K pathway and hence persistent FKHR phosphorylation.
PTEN expression relocalizes GFP-FKHR to the nucleus in PTEN null cells.
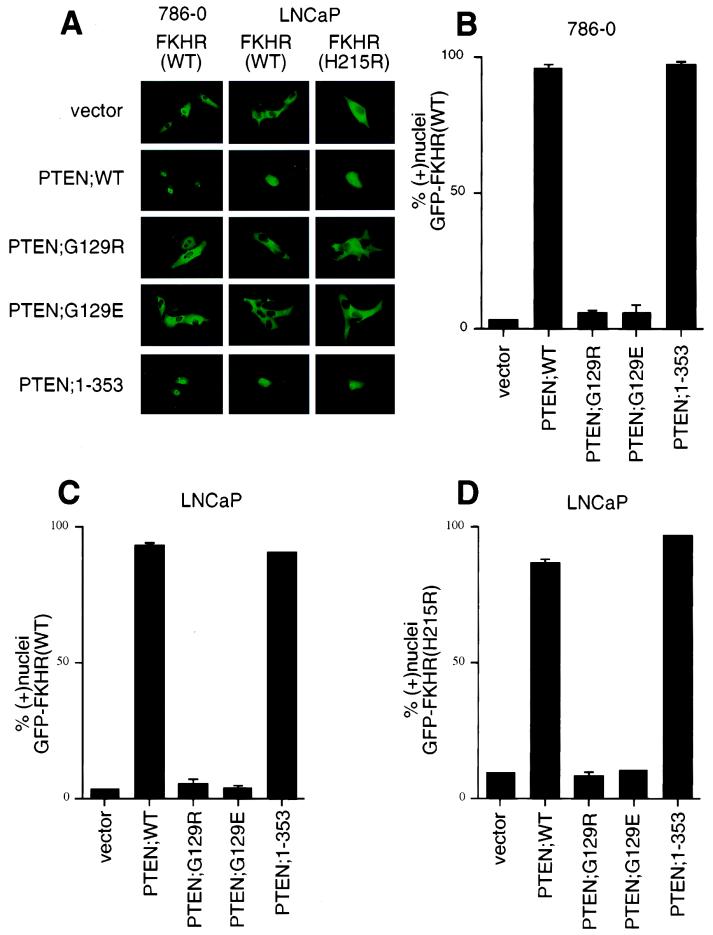
To determine whether reexpression of PTEN protein could effect a change in the localization of GFP-FKHR, plasmids encoding either wild-type or mutant PTEN derivatives were transfected into both 786-O and LNCaP cells along with the plasmid encoding GFP-FKHR. In greater than 90% of LNCaP or 786-O cells cotransfected with PTEN;WT, GFP-FKHR was localized to the nuclei (Fig. 3A to C). In contrast, GFP-FKHR remained cytoplasmic when coproduced with either PTEN mutant (PTEN;G129R or PTEN;G129E) (Fig. 3A to C). PTEN;G129E retains protein but not lipid phosphatase activity, whereas PTEN;G129R lacks both these activities (21, 43, 53). Thus, PTEN protein phosphatase activity is not sufficient for the induction of nuclear localization of GFP-FKHR. PTEN;1–353 is a truncated form of PTEN that retains lipid and protein phosphatase activity and can inhibit cell cycle progression and Akt kinase activity comparably to wild-type PTEN (Fig. 3A to C) (32, 70; S. Ramaswamy and W. R. Sellers, unpublished data). In keeping with these data, expression of PTEN;1–353 led to the nuclear accumulation of GFP-FKHR;WT. Together, these data suggest that FKHR is aberrantly localized in PTEN-null cells and that reconstitution of PTEN lipid phosphatase activity is sufficient for localizing FKHR to the nucleus.
FIG. 3.
Reconstitution of PTEN relocalizes GFP-FKHR to the nucleus. (A) Cellular localization of GFP-FKHR in 786-O and LNCaP cells. 786-O cells were transiently transfected with pCDNA3-GFP-FKHR along with pSG5L or pSG5L-PTEN or plasmids encoding the indicated mutant derivatives. Similarly, LNCaP cells were transiently cotransfected with pCDNA3-GFP-FKHR or GFP-FKHR;H215R along with pSG5L plasmid encoding the indicated PTEN cDNAs. At 24 h after transfection, GFP-FKHR was detected by fluorescence microscopy in living cells. (B) Quantitation of the results from panel A (left). The percentage of cells with nuclear GFP-FKHR was determined as in Fig. 2B. The mean and standard error for experimental duplicates are shown, and the results obtained are representative of the results obtained in three independent experiments. (C) Quantification of the results from panel A (middle). Data are shown as in panel B. (D) Quantification of the results from panel A (right). Data are shown as in panel B.
FKHR transcriptional activity is defective in PTEN-null cells.
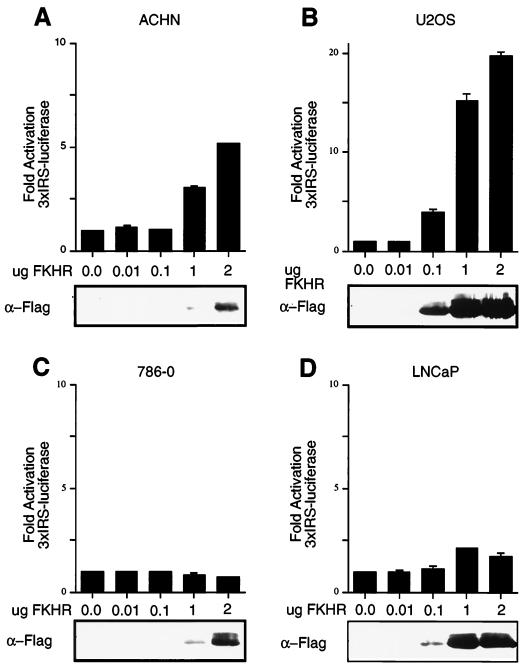
FKHR can activate transcription from a minimal promoter element contained within the IGFBP-1 promoter (27, 66). Likewise, FKHRL1 can activate transcription from a sequence derived from the FasL promoter (5). The localization data obtained using GFP-FKHR fusion proteins suggested that FKHR might not activate transcription in a PTEN-null cell. To test this, PTEN+/+ ACHN and U2-OS cell were transfected with a luciferase reporter plasmid containing a 3×IRS element or a FasL promoter element, along with a plasmid encoding Flag-tagged FKHR. In these cells, FKHR transfection resulted in a dose-dependent increase in transcription (Fig. 4A and B and data not shown). In these cells, FKHR;H215R, harboring a point mutation in the DNA-binding domain, had no effect (data not shown). In contrast to these results, transfection of wild-type FKHR in the PTEN-null 786-O and LNCaP cells did not activate transcription from either the 3×IRS or FasL promoter elements (Fig. 4C and D and data not shown). These data demonstrate that the ability of FKHR to activate transcription is defective in PTEN-null cells. Note that neither reporter used in these experiments was capable of assaying endogenous forkhead activity. Specifically, in the absence of exogenous FKHR, when these reporters were compared to the same reporters lacking an intact Forkhead DNA binding, there was no significant difference in overall transcriptional activation (data not shown). This presumably indicates that other elements in these synthetic promoters contribute to the relatively high level of basal activity.
FIG. 4.
FKHR-dependent transcriptional activation is defective in PTEN-null cells. (A and B) FKHR transactivates the 3×IRS promoter in ACHN and U2-OS cells. ACHN (A) and U2-OS (B) cells were transiently cotransfected with 3×IRS-luciferase reporter plasmids along with either pCDNA3 vector or with the indicated amounts of pCDNA3-Flag-FKHR. In each transfection, a constant amount of pCMX-βGal plasmid was included. At 36 h after transfection, luciferase activity was determined as described in Materials and Methods. Fold activation was calculated by normalizing the measured light units by the measured β-galactosidase activity and then normalizing to the activity of the 3×IRS-promoter luciferase construct when transfected with vector alone. Data shown are the mean and standard error of independent duplicate experiments and are representative of three independent experiments. (C and D) FKHR fails to activate transcription in 786-O and LNCaP cells. 786-O (C) and LNCaP (D) cells were transiently cotransfected with a 3×IRS-luciferase reporter plasmid and either vector or pCDNA3-Flag-FKHR as in panels A and B. Data are shown as for panels A and B.
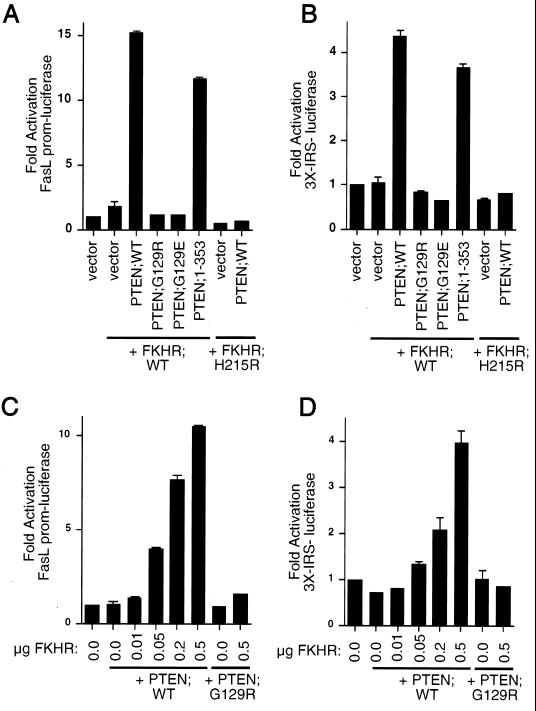
While wild-type FKHR failed to activate transcription in these cells, the phosphorylation site mutant FKHR;AAA was capable of activating transcription in the PTEN-null cells (data not shown). We next asked whether PTEN, as an antagonist of PI3K/Akt signaling, could rescue FKHR transcriptional activation. To this end, 786-O cells were cotransfected with the FasL promoter-luciferase reporter plasmid along with either empty vector or wild-type FKHR. In keeping with the data in Fig. 4, wild-type FKHR did not activate transcription from this promoter (Fig. 5A and B). Likewise, cotransfection of wild-type FKHR with plasmids encoding the PTEN mutant PTEN;G129R or PTEN;G129E failed to activate the FasL promoter (Fig. 5A). In contrast, cotransfection of plasmids encoding either PTEN;WT or PTEN;1–353 rescued transcriptional activity (Fig. 5A). As a control, production of PTEN;WT along with the DNA-binding-defective mutant FKHR;H215R had no effect on transcription (Fig. 5A) even though PTEN relocalizes the GFP-FKHR;H215R mutant efficiently to the nucleus (Fig. 3A and D). To ask whether these observations held with other FKHR responsive reporters, we performed similar experiments using the 3×IRS-luciferase reporter plasmid (Fig. 5B and D). Here, FKHR again was incapable of activating transcription when overexpressed. Cotransfection of FKHR along with either PTEN;WT or PTEN;1–353 restored FKHR-dependent activation, while cotransfection with PTEN;G129R and PTEN;G129E did not. Finally, PTEN restored the dose-dependent transcriptional activity of FKHR;WT when measured on both the FasL promoter and the 3×IRS promoter, while PTEN;G129R had no effect at the highest doses of FKHR tested (Fig. 5C and D). Taken together, the above data suggest that PTEN allows for appropriate localization of FKHR and for appropriate transcriptional function.
FIG. 5.
Reconstitution of PTEN restores FKHR transcriptional activation. (A and B) Wild-type PTEN and PTEN;1–353 restore transcriptional activation of FKHR in 786-O cells. 786-O cells were transiently cotransfected with either the FasL promoter-luciferase reporter plasmid (A) or the 3×IRS promoter-luciferase reporter plasmid (B) along with plasmids encoding the indicate proteins. Data are shown in Fig. 4A. (C and D) Wild-type PTEN, but not PTEN;G129R, rescues dose-dependent FKHR transactivation. 786-O cells were cotransfected with either the FasL promoter-luciferase reporter plasmid (C) or the 3×IRS promoter luciferase plasmid (D) along with plasmids encoding the indicated proteins. Data are shown as for Fig. 4A.
Activated FKHR induces cell death in LNCaP cells.
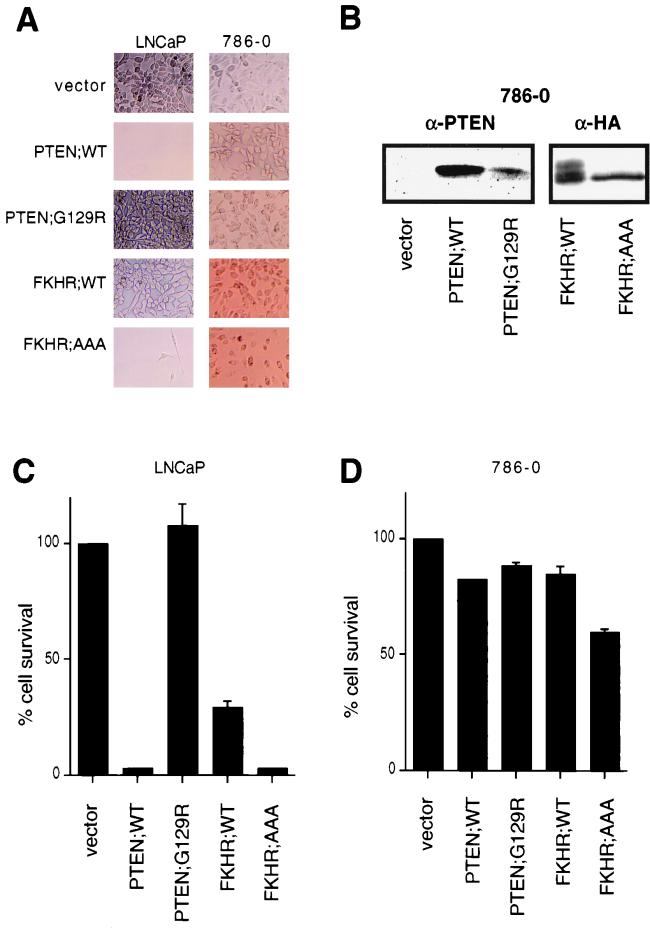
Certain PTEN-null cells, such as PTEN−/− mouse embryo fibroblasts are resistant to apoptotic stimuli (62), and PTEN reconstitution to or treatment with PI3K inhibitors of certain PTEN-null tumor cells (e.g., UMG-251 or LNCaP cells) results in the induction of cell death that is, at least in part, mediated through apoptosis (9, 14, 15, 35). Likewise, FKHRL1 and FKHR can both induce apoptosis (5, 66). Thus, we next asked whether FKHR or the FKHR;AAA mutant could induce cell death in PTEN-null LNCaP or 786-O cells. To test this, PTEN-null 786-O or LNCaP cells were incubated with culture supernatants containing amphotrophic retroviruses encoding PTEN;WT, PTEN;G129R, FKHR;WT, and FKHR;AAA and were then selected with puromycin. In keeping with previously reported results (14, 43), PTEN induced cell death in LNCaP cells and completely suppressed the emergence of puromycin-resistant cells (Fig. 6A and C). On the other hand, infection with retroviruses producing PTEN;G129R had no effect on cell viability. Cells infected with FKHR;WT were more prone to cell death than were vector-infected controls; however, puromycin-resistant populations expressing FKHR;WT were obtained (data not shown). In comparison, infection with retroviruses producing FKHR;AAA, like PTEN, led to marked suppression of cell viability and completely suppressed the emergence of puromycin-resistant clones. Thus, the activated form of FKHR complemented PTEN deficiency in these cells.
FIG. 6.
FKHR induces cell death in PTEN-null LNCaP cells but not PTEN-null 786-O cells. (A) 786-O and LNCaP cells were infected with retroviruses directing the expression of the indicated proteins. Infected cells were then grown in the presence of puromycin, and viable cells were photographed by light microscopy after 4 days of selection. Magnification, ×95. (B) Protein expression in puromycin-resistant populations of 786-O cells. Whole-cell extracts were prepared from puromycin-resistant 786-O cells infected as in panel A and immunoblotted with either anti-PTEN or anti-HA antibodies. (C and D) MTS assays were performed, as described in Materials and Methods, to quantify the results shown in panel A. To normalize the results between the two cell lines, cell viability is expressed as a percentage of the vector controls. The data shown are the mean and standard error of independent duplicates and are representative of two independent infections.
In contrast to the results obtained with LNCaP cells, retroviral transduction of PTEN into 786-O cells did not induce cell death (Fig. 6A, B, and D). 786-O cultures were likewise infected with viruses leading to the production of FKHR or FKHR;AAA. Here, surprisingly, FKHR and FKHR;AAA had little overall effect on cell viability (Fig. 6A and D). Furthermore, puromycin-resistant polyclonal lines expressing these proteins were derived (Fig. 6B). Thus, activated FKHR can induce apoptosis in a cell line in which PTEN induces apoptosis but does not induce apoptosis in a cell line immune to PTEN-induced apoptosis.
FKHR induces a cell cycle block in PTEN-null cells.
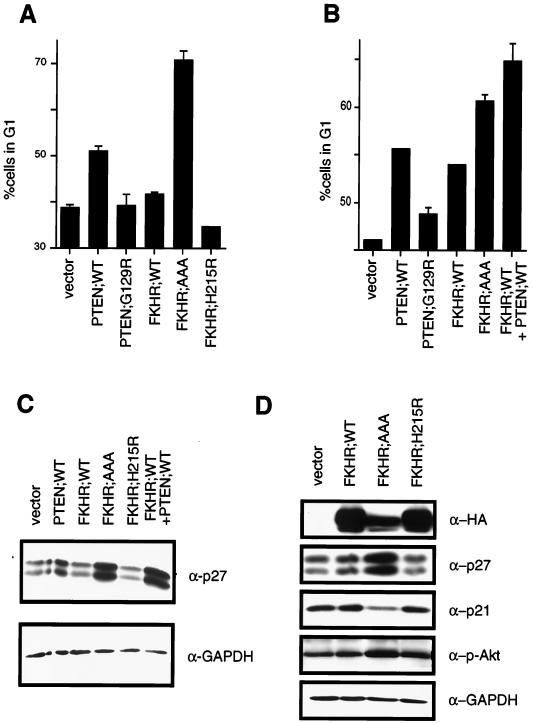
In U87-MG and 768-O cells, reintroduction of PTEN by adenovirus infection or transient transfection induces a cell cycle arrest in G1 rather than inducing apoptosis (21, 35, 53). One possibility, among many, for the lack of apoptosis in these cells is that additional genetic alterations in these cells render PTEN incapable of inducing apoptosis. If FKHR is a critical downstream activator of apoptosis in these cells, perhaps this putative defect is a defect in FKHR function. If so, this might account for the lack of effect of the FKHR;AAA mutant in the cell death assay performed with 786-O cells (Fig. 6A). Alternatively, FKHR or other Forkhead factors might function in both the apoptotic and cell cycle function of the PI3K/PTEN/Akt pathway. To test this hypothesis, a transient cell cycle assay was used. 786-O cells were transiently transfected with a plasmid encoding the cell surface marker CD19 along with plasmids encoding PTEN;WT or PTEN;G129R. PTEN;WT, but not PTEN;G129R, induced a modest cell cycle block. While FKHR;WT had a minimal effect on the G1 population, FKHR;AAA induced a robust G1 arrest (Fig. 7A) but FKHR;H215R did not. Thus, activated FKHR can complement the loss of PTEN in 786-O cells. We next asked whether PTEN could “rescue” the apparent defect in FKHR;WT-mediated cell cycle arrest. In keeping with the ability of PTEN to relocalize FKHR and to restore transcriptional activation, cotransfection of PTEN;WT along with FKHR;WT led to an increase in the G1 population comparable to that induced by the FKHR;AAA mutant (Fig. 7B). These data suggest that restoration of functional FKHR to these cells, by cotransfection of PTEN or by rendering FKHR immune to Akt phosphorylation, is sufficient to arrest PTEN-null cells in G1.
FIG. 7.
FKHR induces a G1 arrest in PTEN-null 786-O cells. (A) FKHR;AAA induces a G1 arrest in PTEN-null 786-O cells. 786-O cells were transiently transfected with pCD19 and plasmids directing the expression of the indicated proteins. At 48 h after transfection, the cell cycle distribution of the transfected cells was determined by combined fluorescein isothiocyanate-conjugated anti-CD19 and propidium iodide staining. The data shown are the mean and standard error of replicates and are representative of two independent experiments. (B) PTEN cooperates with FKHR to induce a cell cycle arrest. 786-O cells were transfected with pCD19 and plasmids directing the expression of the indicated proteins. The data shown are as in panel A. (C) PTEN and FKHR;AAA induce p27 in 786-O cells. 786-O cells were transiently transfected with a plasmid encoding the cell surface marker CD19, along with either vector plasmid, or plasmids encoding the indicated proteins. At 36 h after transfection, cells were harvested by trypsinization and collected on anti-CD19-coated magnetic beads. A 75-μg portion of whole-cell extracts, derived from the isolated cells, was separated by gel electrophoresis and immunoblotted with the indicated antibody reagents. (D) FKHR;AAA induces p27 but does not induce p21. 786-O cells were infected with retroviruses encoding the indicated proteins and selected with puromycin. Following selection, protein extracts were prepared and immunoblotted with the indicated antibody reagents.
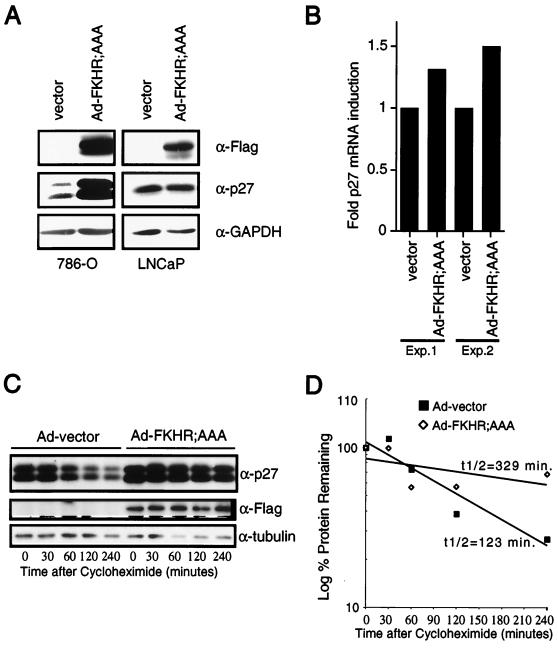
Loss of PTEN in cells leads to a reduction in p27 protein levels (33, 64). Thus, to begin to characterize the G1 arrest induced by FKHR;AAA, we first examined p27 protein levels. To do this, 786-O cells were transiently transfected with a plasmid encoding the cell surface marker CD19 along with the vector plasmid or with plasmids encoding PTEN;WT, FKHR;WT, or FKHR;AAA. The CD19+ and hence transfected cells were collected on anti-CD19-coated magnetic beads. Protein extracts were prepared and immunoblotted with an antiserum specific for p27. Here, wild-type PTEN and constitutively active FKHR;AAA both induced p27 protein (Fig. 7C). In this cell line, immunoblots for p27 consistently showed a doublet that is recognized by multiple independent anti-p27 antisera (data not shown). Anti-GAPDH immunoblotting served to confirm equivalent protein loading. In addition, while PTEN induced a modest increase in p27 levels, wild-type FKHR cotransfected along with wild-type PTEN induced p27 levels comparably to those induced by FKHR;AAA (Fig. 7C). These data were confirmed and extended using retroviral delivery of FKHR;AAA to 786-O cells and adenovirus delivery of FKHR;AAA (Fig. 8). While p27 was again induced by FKHR;AAA, p21 was not (Fig. 7D). These data suggest that p27 is a specific downstream target of FKHR. Finally, infection of 786-O but not LNCaP cells with adenovirus directing the expression of FKHR;AAA (Ad-FKHR;AAA) induced p27 protein.
FIG. 8.
FKHR;AAA induces p27 mRNA and prolongs the half-life of p27 protein. (A) FKHR;AAA induces p27 in 786-O cells but not in LNCaP cells. 786-O and LNCaP cells were infected with either Ad-vector or Ad-FKHR;AAA as indicated. At 36 h after infection, protein extracts were prepared and immunoblotted with the indicated antibody reagents. (B) FKHR;AAA induces p27 mRNA in 786-O cells. 786-O cells were infected with Ad-vector or Ad-FKHR;AAA as indicated. At 24 h after infection, the cells were harvested, mRNA was collected and protein extracts were prepared from duplicate plates. p27 mRNA was measured by real-time quantitative PCR using an ABI Prism 7700 sequence detector as described in Materials and Methods. Each sample was assayed in triplicate for both p27 mRNA and GAPDH mRNA. Fold induction of mRNA indicates the p27/GAPDH ratio of each sample normalized to the ratio obtained with Ad-vector. The data obtained from two independent infections performed on different days are shown. (C) FKHR;AAA prolongs the half-life of the p27 protein. 786-O cells were infected with Ad-vector or Ad-FKHR;AAA as for panel B. At 24 h after infection, cycloheximide was added to a final concentration of 25 μg/ml. The cycloheximide was present for the entire course of the experiment. Protein extracts were prepared at the indicated time points and immunoblotted with the indicated antibody reagents. (D) Calculation of the half-life of the p27 protein. Multiple radiographic exposures of the products of the experiment in panel C were obtained (data not shown). Quantitation of the p27 signal intensity was obtained from exposures in which the signal was nonsaturating for the entire time course. Signal intensities were normalized to the signal intensity obtained at time zero. The percent signal remaining was plotted on a log-linear plot, and an exponential curve fit was applied. The calculated half-life is shown. These results are representative of two independent experiments.
While our data and the genetic evidence in Caenorhabditis elegans suggest that Forkhead factors are critical downstream targets of PTEN, it is possible that our results reflect not a downstream effect of FKHR but, rather, a negative regulation of Akt by FKHR, perhaps through feedback inhibition. To ask whether such a mechanism might account for the actions of FKHR in these cells, we examined the state of phosphorylation and hence activation of Akt. The immunoblots described above were stripped and reprobed with an antiserum specific to the serine 473 phosphorylation on Akt. Here, in keeping with previously published data, production of wild-type PTEN led to an ablation of phospho-Akt (data not shown), whereas FKHR;AAA production was associated with a modest increase in the level of phosphorylated Akt while the total level of Akt remained unchanged (Fig. 7C and data not shown). These data suggest that feedback inhibition of Akt is not the mechanism by which FKHR promotes either apoptosis or a G1 arrest.
FKHR;AAA induces p27 mRNA and prolongs the p27 protein half-life.
To begin to address the mechanism through which FKHR regulates p27, p27 mRNA levels were determined in 786-O cells following adenovirus infection with Ad-FKHR;AAA. At 24 h, cells infected with Ad-FKHR;AAA demonstrated a modest (1.3- to 1.5-fold) induction of the p27 mRNA compared to that in cells infected with Ad-vector (Fig. 8B). Consistent with these results, we have found that Ad-PTEN induces a 1.8-fold induction in p27 mRNA (S. Ramaswamy, S. Signoretti, M. Loda and W. R. Sellers, unpublished data). Next, 786-O cells were again infected with Ad-vector or Ad-FKHR;AAA. At 24 h after infection, the cells were treated continuously with cycloheximide (25 μg/ml). At specific time points, the p27 protein level was determined by immunoblotting in both the Ad-vector- and Ad-FKHR;AAA-infected cells. Here, we found that the protein half-life was increased from 123 to 329 min in the Ad-FKHR;AAA infection. Thus, FKHR;AAA induces a modest change in the p27 mRNA level and a significant increase in the p27 protein half-life.
DISCUSSION
Our data show that localization and transcriptional activity of FKHR is aberrant in PTEN-null cells. Reconstitution of wild-type PTEN, but not lipid phosphatase-inactive mutants, restores both localization and transcriptional activation of FKHR in these cells. While wild-type FKHR is relatively inactive in PTEN null cells, a phosphosite mutant of FKHR (FKHR;AAA) that is no longer phosphorylated by Akt can still localize to the nucleus and activate transcription in such cells. This mutant induces death in a cell line susceptible to PTEN-mediated cell death. Surprisingly, it does not induce apoptosis but, rather, induces a G1 arrest in cells that likewise arrest with wild-type PTEN. Together, the data derived from the cell death assays and the cell cycle arrest assays support the notion that an intact and active FKHR protein is capable of carrying out PTEN function in its absence. That is, activated FKHR complements the loss of PTEN in two different functional assays. These data support the idea that FKHR is sufficient for PTEN function in cells. Finally, previous data have shown that PTEN-null cells have low levels of p27 and that reintroduction of PTEN up regulates p27 levels (33, 64). We find that FKHR;AAA dramatically induces p27 levels in PTEN-null cells. These data suggest that the finding of aberrant p27 levels in the absence of PTEN might arise as a consequence of the lack of FKHR function in such cells. In keeping with these data, Medema et al. recently reported similar data which demonstrated a role for Forkhead factors as regulators of cell cycle progression and, using defined genetic cells, showed that such regulation does indeed depend on the induction of p27 (40).
The PI3K/Akt pathway is a well-known oncogenic signaling pathway (75). Cell survival and cell proliferation have been linked to this pathway in multiple systems. For example, interleukin-3-dependent cell lines require Akt for survival, as do cells in which anoikis is blocked by Ras activation (17, 29, 61). On the other hand, expression of activated PI3K in the absence of serum can induce DNA synthesis (30). Furthermore, PTEN is capable of inducing apoptosis or a cell cycle arrest, and loss of PTEN in primary cells leads to either excessive proliferation or defects in apoptosis. In mammalian cell-based assays, a diverse group of substrates have been linked to Akt activation. In C. elegans, on the other hand, the insulin/PI3K/Akt signaling pathway that regulates aging, while conserved with mammalian cells, has thus far yielded only the Forkhead homologue daf-16 as a downstream target (46). It is possible that the deregulated activity of multiple Akt substrates contributes to the neoplastic properties inherent to a PTEN-null tumor cell and that certain substrates might individually contribute to the regulation of apoptosis and cell proliferation. Our data, however, support the notion that the pathway linking PI3K and PTEN to transformation of mammalian cells is essentially identical to the pathway regulating aging in C. elegans. This pathway is comprised of a receptor tyrosine kinase such as IGF-IR (daf-2), PI3K (ageI), Akt-1 (akt1) and Akt-2 (akt2), PDK-1 (pdk1), PTEN (daf-18), and the daf-16 homologues (FKHR, FKHRL1, and AFX) (23, 25, 42, 46, 47, 49, 50, 55).
It is interesting that elements of this pathway that are linked genetically in C. elegans are the same elements of the pathway that have been associated with genetic alterations in human tumors. The PI3KCA gene is amplified in ovarian cancer and is also found as a retroviral oncogene (1, 59). Akt-1 and Akt-2 are amplified in a limited number of tumors, and Akt-1 is the cellular homologue of v-Akt (2, 3, 11, 56). Finally, PTEN is widely mutated in cancer, and FKHR has been the target of translocation in rhabdomyosarcoma. Interestingly, in this tumor, two different translocations give rise to the fusion proteins PAX3-FKHR or PAX7-FKHR (16, 22, 58). Our data support the notion that FKHR could act as a tumor suppressor; thus, one untested possibility is that these translocations might produce chimeric proteins that could act in a dominant negative manner to inactivate FKHR function.
The notion that a transcription factor might induce a G1 arrest or induce cell death is not new. Indeed, this is precisely the case for p53. The parallels between these pathways are striking. p53 receives signals that reflect the state of the genome (DNA damage) at least in part from a PI3K family member, ATM. This signal may be transmitted through phosphorylation of p53. p53 can then enact a G1 arrest through transcriptional regulation of p21. p53 induces apoptotic cell death through both transcription-dependent and -independent mechanisms. FKHR, on the other hand, receives signals primarily from the environment external to the cell. These signals are transmitted through a type I PI3K and result in the phosphorylation of FKHR and its subsequent inactivation. In its active state FKHR, can promote a G1 arrest through the induction of p27 and can induce apoptosis perhaps through regulation of Fas signaling or through regulation of FasL itself.
How does FKHR regulate p27? p27 is primarily regulated posttranscriptionally, both through ubiquitin-mediated proteolysis and through translation controls. There is limited information to suggest that transcriptional regulation of p27 is important. Furthermore, PTEN did not alter p27 mRNA levels (33). On the other hand, Medema et al. (40) have demonstrated activation of the p27 promoter by AFX, and we have shown that both wild-type PTEN and wild-type FKHR, but not mutant controls, were capable of inducing activation of the p27 promoter (data not shown). In addition, Medema et al. reported a modest induction in p27 mRNA levels (40). We have also seen a modest (1.3- to 1.5-fold induction in mRNA upon adenovirus expression of FKHR;AAA (Fig. 8B) and upon adenovirus expression of PTEN. In addition, however, the half-life of p27 protein is significantly prolonged. Here, it is possible that a modest increase in p27 levels induced through transcription might lead to inhibition of cyclin-dependent kinase activity followed by a decrease in p27 phosphorylation and then a change in the half-life of p27 protein. Since this process involves a catalytic mechanism, a small change in p27 mRNA levels could lead to a large difference in protein half-life. For example, an increase in the transcription of p27 could alter the balance between the two proposed complexes of p27 and cyclin E-cdk2, one inhibitory and one in which p27 is degraded (60, 72). Alternatively, it is possible that FKHR;AAA directly alters or regulates components of the p27 degradation apparatus. Specifically, it will be of interest to know whether Forkhead factors can alter the levels of any of the components of the Skp-Cul-F box (SCF) complex.
The mechanism that underlies FKHR induction of apoptosis is likewise not yet clear. FKHRL1 can regulate the FasL promoter, suggesting that these transcription factors might directly regulate the levels of this death effector (5). In keeping with this notion, PTEN+/− mice develop an autoimmune lymphoid hyperplasia syndrome that phenocopies mutations in the murine Fas gene (18, 74). On the other hand, cells from the PTEN+/− animals did not demonstrate defects in FasL or Fas but, rather, were defective in the apoptotic response to Fas (18). In either case, it would appear that PTEN-mediated and, by extension, FKHR-mediated apoptosis probably involves the Fas pathway.
Finally, our data support the notion that, as is the case in C. elegans, signaling pathways might be more linear, at least with respect to transformation, than is commonly suspected. This would lead one to further suspect that PTEN-null cells might be particularly sensitive to inhibitors directed against members of this pathway; if true, such dependence would bode well for the future success of therapeutics aimed at intervening in PI3K signaling.
ACKNOWLEDGMENTS
This work was supported by grants from the Department of Defense (DAMD17-98-1-8596), NIH (RO1CA85912), Gillette Women's Cancer Program, and CaPCURE foundation to W.R.S.; from the NIH (RO1CA81755) to M.L.; and from the Department of Defense to F.V.
We thank E. Tang, F. Barr, K. Guan, K. Polyak, and B. Vogelstein for the generous gift of plasmid reagents; Kornelia Polyak for assistance in adenovirus production; and Myles Brown, Bill Kaelin, Mark Ewen, David Livingston, Matt Meyerson, Kornelia Polyak, and Barrett Rollins for their critical review of the manuscript. N.N. thanks Takehisa Iwai for scientific advice.
REFERENCES
- 1.Aoki M, Batista O, Bellacosa A, Tsichlis P, Vogt P K. The akt kinase: molecular determinants of oncogenicity. Proc Natl Acad Sci USA. 1998;95:14950–14955. doi: 10.1073/pnas.95.25.14950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bellacosa A, de Feo D, Godwin A K, Bell D W, Cheng J Q, Altomare D A, Wan M, Dubeau L, Scambia G, Masciullo V, et al. Molecular alterations of the AKT2 oncogene in ovarian and breast carcinomas. Int J Cancer. 1995;64:280–285. doi: 10.1002/ijc.2910640412. [DOI] [PubMed] [Google Scholar]
- 3.Bellacosa A, Testa J R, Staal S P, Tsichlis P N. A retroviral oncogene, akt, encoding a serine-threonine kinase containing an SH2-like region. Science. 1991;254:274–277. doi: 10.1126/science.254.5029.274. [DOI] [PubMed] [Google Scholar]
- 4.Biggs W H, III, Meisenhelder J, Hunter T, Cavenee W K, Arden K C. Protein kinase B/Akt-mediated phosphorylation promotes nuclear exclusion of the winged helix transcription factor FKHR1. Proc Natl Acad Sci USA. 1999;96:7421–7426. doi: 10.1073/pnas.96.13.7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunet A, Bonni A, Zigmond M J, Lin M Z, Juo P, Hu L S, Anderson M J, Arden K C, Blenis J, Greenberg M E. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 6.Cairns P, Okami K, Halachmi S, Halachmi N, Esteller M, Herman J G, Jen J, Isaacs W B, Bova G S, Sidransky D. Frequent inactivation of PTEN/MMAC1 in primary prostate cancer. Cancer Res. 1997;57:4997–5000. [PubMed] [Google Scholar]
- 7.Cantley L C, Neel B G. New insights into tumor suppression: PTEN suppresses tumor formation by restraining the phosphoinositide 3-kinase/AKT pathway. Proc Natl Acad Sci USA. 1999;96:4240–4245. doi: 10.1073/pnas.96.8.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cardone M H, Roy N, Stennicke H R, Salvesen G S, Franke T F, Stanbridge E, Frisch S, Reed J C. Regulation of cell death protease caspase-9 by phosphorylation. Science. 1998;282:1318–1321. doi: 10.1126/science.282.5392.1318. [DOI] [PubMed] [Google Scholar]
- 9.Carson J P, Kulik G, Weber M J. Antiapoptotic signaling in LNCaP prostate cancer cells: a survival signaling pathway independent of phosphatidylinositol 3′-kinase and Akt/protein kinase B. Cancer Res. 1999;59:1449–1453. [PubMed] [Google Scholar]
- 10.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng J Q, Ruggeri B, Klein W M, Sonoda G, Altomare D A, Watson D K, Testa J R. Amplification of AKT2 in human pancreatic cells and inhibition of AKT2 expression and tumorigenicity by antisense RNA. Proc Natl Acad Sci USA. 1996;93:3636–3641. doi: 10.1073/pnas.93.8.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cross D A, Alessi D R, Cohen P, Andjelkovich M, Hemmings B A. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 13.Datta S R, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg M E. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell. 1997;91:231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 14.Davies M A, Koul D, Dhesi H, Berman R, McDonnell T J, McConkey D, Yung W K, Steck P A. Regulation of Akt/PKB activity, cellular growth, and apoptosis in prostate carcinoma cells by MMAC/PTEN. Cancer Res. 1999;59:2551–2556. [PubMed] [Google Scholar]
- 15.Davies M A, Lu Y, Sano T, Fang X, Tang P, LaPushin R, Koul D, Bookstein R, Stokoe D, Yung W K, Mills G B, Steck P A. Adenoviral transgene expression of MMAC/PTEN in human glioma cells inhibits Akt activation and induces anoikis. Cancer Res. 1998;58:5285–5290. [PubMed] [Google Scholar]
- 16.Davis R J, D'Cruz C M, Lovell M A, Biegel J A, Barr F G. Fusion of PAX7 to FKHR by the variant t(1;13)(p36;q14) translocation in alveolar rhabdomyosarcoma. Cancer Res. 1994;54:2869–2872. [PubMed] [Google Scholar]
- 17.del Peso L, Gonzalez-Garcia M, Page C, Herrera R, Nunez G. Interleukin-3-induced phosphorylation of BAD through the protein kinase Akt. Science. 1997;278:687–689. doi: 10.1126/science.278.5338.687. [DOI] [PubMed] [Google Scholar]
- 18.Di Cristofano A, Kotsi P, Peng Y F, Cordon-Cardo C, Elkon K B, Pandolfi P P. Impaired fas response and autoimmunity in Pten(+/−) mice. Science. 1999;285:2122–2125. doi: 10.1126/science.285.5436.2122. [DOI] [PubMed] [Google Scholar]
- 19.Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi P P. Pten is essential for embryonic development and tumour suppression. Nat Genet. 1998;19:348–355. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- 20.Fink L, Seeger W, Ermert L, Hanze J, Stahl U, Grimminger F, Kummer W, Bohle R M. Real-time quantitative RT-PCR after laser-assisted cell picking. Nat Med. 1998;4:1329–1333. doi: 10.1038/3327. [DOI] [PubMed] [Google Scholar]
- 21.Furnari F B, Huang H J, Cavenee W K. The phosphoinositol phosphatase activity of PTEN mediates a serum-sensitive G1 growth arrest in glioma cells. Cancer Res. 1998;58:5002–5008. [PubMed] [Google Scholar]
- 22.Galili N, Davis R J, Fredericks W J, Mukhopadhyay S, Rauscher F J D, Emanuel B S, Rovera G, Barr F G. Fusion of a fork head domain gene to PAX3 in the solid tumour alveolar rhabdomyosarcoma. Nat Genet. 1993;5:230–235. doi: 10.1038/ng1193-230. [DOI] [PubMed] [Google Scholar]
- 23.Gil E B, Malone Link E, Liu L X, Johnson C D, Lees J A. Regulation of the insulin-like developmental pathway of Caenorhabditis elegans by a homolog of the PTEN tumor suppressor gene. Proc Natl Acad Sci USA. 1999;96:2925–2930. doi: 10.1073/pnas.96.6.2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gingras A C, Kennedy S G, O'Leary M A, Sonenberg N, Hay N. 4E-BP1, a repressor of mRNA translation, is phosphorylated and inactivated by the Akt(PKB) signaling pathway. Genes Dev. 1998;12:502–513. doi: 10.1101/gad.12.4.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gottlieb S, Ruvkun G. daf-2, daf-16 and daf-23: genetically interacting genes controlling Dauer formation in Caenorhabditis elegans. Genetics. 1994;137:107–120. doi: 10.1093/genetics/137.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guldberg P, thor Straten P, Birck A, Ahrenkiel V, Kirkin A F, Zeuthen J. Disruption of the MMAC1/PTEN gene by deletion or mutation is a frequent event in malignant melanoma. Cancer Res. 1997;57:3660–3663. [PubMed] [Google Scholar]
- 27.Guo S, Rena G, Cichy S, He X, Cohen P, Unterman T. Phosphorylation of serine 256 by protein kinase B disrupts transactivation by FKHR and mediates effects of insulin on insulin-like growth factor-binding protein-1 promoter activity through a conserved insulin response sequence. J Biol Chem. 1999;274:17184–17192. doi: 10.1074/jbc.274.24.17184. [DOI] [PubMed] [Google Scholar]
- 28.He T C, Zhou S, da Costa L T, Yu J, Kinzler K W, Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci USA. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khwaja A, Rodriguez-Viciana P, Wennstrom S, Warne P H, Downward J. Matrix adhesion and Ras transformation both activate a phosphoinositide 3-OH kinase and protein kinase B/Akt cellular survival pathway. EMBO J. 1997;16:2783–2793. doi: 10.1093/emboj/16.10.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klippel A, Escobedo M A, Wachowicz M S, Apell G, Brown T W, Giedlin M A, Kavanaugh W M, Williams L T. Activation of phosphatidylinositol 3-kinase is sufficient for cell cycle entry and promotes cellular changes characteristic of oncogenic transformation. Mol Cell Biol. 1998;18:5699–5711. doi: 10.1128/mcb.18.10.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kops G J, de Ruiter N D, De Vries-Smits A M, Powell D R, Bos J L, Burgering B M. Direct control of the Forkhead transcription factor AFX by protein kinase B. Nature. 1999;398:630–634. doi: 10.1038/19328. [DOI] [PubMed] [Google Scholar]
- 32.Lee J O, Yang H, Georgescu M M, Di Cristofano A, Maehama T, Shi Y, Dixon J E, Pandolfi P, Pavletich N P. Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell. 1999;99:323–334. doi: 10.1016/s0092-8674(00)81663-3. [DOI] [PubMed] [Google Scholar]
- 33.Li D, Sun H. PTEN/MMAC1/TEP1 suppresses the tumorigenicity and induces G1 cell cycle arrest in human glioblastoma cells. Proc Natl Acad Sci USA. 1998;95:15406–15411. doi: 10.1073/pnas.95.26.15406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li D M, Sun H. TEP1, encoded by a candidate tumor suppressor locus, is a novel protein tyrosine phosphatase regulated by transforming growth factor beta. Cancer Res. 1997;57:2124–2129. [PubMed] [Google Scholar]
- 35.Li J, Simpson L, Takahashi M, Miliaresis C, Myers M P, Tonks N, Parsons R. The PTEN/MMAC1 tumor suppressor induces cell death that is rescued by the AKT/protein kinase B oncogene. Cancer Res. 1998;58:5667–5672. [PubMed] [Google Scholar]
- 36.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang S I, Puc J, Miliaresis C, Rodgers L, McCombie R, Bigner S H, Giovanella B C, Ittmann M, Tycko B, Hibshoosh H, Wigler M H, Parsons R. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 37.Liaw D, Marsh D J, Li J, Dahia P L, Wang S I, Zheng Z, Bose S, Call K M, Tsou H C, Peacocke M, Eng C, Parsons R. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat Genet. 1997;16:64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- 38.Liu W, James C D, Frederick L, Alderete B E, Jenkins R B. PTEN/MMAC1 mutations and EGFR amplification in glioblastomas. Cancer Res. 1997;57:5254–5257. [PubMed] [Google Scholar]
- 39.Maehama T, Dixon J E. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-triphosphate. J Biol Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 40.Medema R H, Kops G J P, Bos J L, Burgering B M T. AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27Kip1. Nature. 2000;404:782–787. doi: 10.1038/35008115. [DOI] [PubMed] [Google Scholar]
- 41.Michell B J, Griffiths J E, Mitchelhill K I, Rodriguez-Crespo I, Tiganis T, Bozinovski S, de Montellano P R, Kemp B E, Pearson R B. The Akt kinase signals directly to endothelial nitric oxide synthase. Curr Biol. 1999;12:845–848. doi: 10.1016/s0960-9822(99)80371-6. [DOI] [PubMed] [Google Scholar]
- 42.Mihaylova V T, Borland C Z, Manjarrez L, Stern M J, Sun H. The PTEN tumor suppressor homolog in Caenorhabditis elegans regulates longevity and dauer formation in an insulin receptor-like signaling pathway. Proc Natl Acad Sci USA. 1999;96:7427–7432. doi: 10.1073/pnas.96.13.7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Myers M P, Pass I, Batty I H, Van der Kaay J, Stolarov J P, Hemmings B A, Wigler M H, Downes C P, Tonks N K. The lipid phosphatase activity of PTEN is critical for its tumor suppressor function. Proc Natl Acad Sci USA. 1998;95:13513–13518. doi: 10.1073/pnas.95.23.13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Myers M P, Stolarov J P, Eng C, Li J, Wang S I, Wigler M H, Parsons R, Tonks N K. P-TEN, the tumor suppressor from human chromosome 10q23, is a dual-specificity phosphatase. Proc Natl Acad Sci USA. 1997;94:9052–9057. doi: 10.1073/pnas.94.17.9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nelen M R, van Staveren W C, Peeters E A, Hassel M B, Gorlin R J, Hamm H, Lindboe C F, Fryns J P, Sijmons R H, Woods D G, Mariman E C, Padberg G W, Kremer H. Germline mutations in the PTEN/MMAC1 gene in patients with Cowden disease. Hum Mol Genet. 1997;6:1383–1387. doi: 10.1093/hmg/6.8.1383. [DOI] [PubMed] [Google Scholar]
- 46.Ogg S, Paradis S, Gottlieb S, Patterson G I, Lee L, Tissenbaum H A, Ruvkun G. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 47.Ogg S, Ruvkun G. The C. elegans PTEN homolog, DAF-18, acts in the insulin receptor-like metabolic signaling pathway. Mol Cell. 1998;2:887–893. doi: 10.1016/s1097-2765(00)80303-2. [DOI] [PubMed] [Google Scholar]
- 48.Ozes O N, Mayo L D, Gustin J A, Pfeffer S R, Pfeffer L M, Donner D B. NF-KappaB activation by tumor necrosis factor requires the Akt serine-threonine kinase. Nature. 1999;401:82–85. doi: 10.1038/43466. [DOI] [PubMed] [Google Scholar]
- 49.Paradis S, Ailion M, Toker A, Thomas J H, Ruvkun G. A PDK1 homolog is necessary and sufficient to transduce AGE-1 PI3 kinase signals that regulate diapause in Caenorhabditis elegans. Genes Dev. 1999;13:1438–1452. doi: 10.1101/gad.13.11.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paradis S, Ruvkun G. Caenorhabditis elegans Akt/PKB transduces insulin receptor-like signals from AGE-1 PI3 kinase to the DAF-16 transcription factor. Genes Dev. 1998;12:2488–2498. doi: 10.1101/gad.12.16.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pear W S, Nolan G P, Scott M L, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Podsypanina K, Ellenson L H, Nemes A, Gu J, Tamura M, Yamada K M, Cordon-Cardo C, Catoretti G, Fisher P E, Parsons R. Mutation of Pten/Mmac1 in mice causes neoplasia in multiple organ systems. Proc Natl Acad Sci USA. 1999;96:1563–1568. doi: 10.1073/pnas.96.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramaswamy S, Nakamura N, Vazquez F, Batt D B, Perera S, Roberts T M, Sellers W R. Regulation of G1 progression by the PTEN tumor suppressor protein is linked to inhibition of the phosphatidylinositol 3-kinase/Akt pathway. Proc Natl Acad Sci USA. 1999;96:2110–2115. doi: 10.1073/pnas.96.5.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Risinger J I, Hayes A K, Berchuck A, Barrett J C. PTEN/MMAC1 mutations in endometrial cancers. Cancer Res. 1997;57:4736–4738. [PubMed] [Google Scholar]
- 55.Rouault J P, Kuwabara P E, Sinilnikova O M, Duret L, Thierry-Mieg D, Billaud M. Regulation of dauer larva development in Caenorhabditis elegans by daf-18, a homologue of the tumour suppressor PTEN. Curr Biol. 1999;9:329–332. doi: 10.1016/s0960-9822(99)80143-2. [DOI] [PubMed] [Google Scholar]
- 56.Ruggeri B A, Huang L, Wood M, Cheng J Q, Testa J R. Amplification and overexpression of the AKT2 oncogene in a subset of human pancreatic ductal adenocarcinomas. Mol Carcinog. 1998;21:81–86. [PubMed] [Google Scholar]
- 57.Sellers W R, Novitch B G, Miyake S, Heith A, Otterson G A, Kaye F J, Lassar A B, Kaelin W G., Jr Stable binding to E2F is not required for the retinoblastoma protein to activate transcription, promote differentiation, and suppress tumor cell growth. Genes Dev. 1998;12:95–106. doi: 10.1101/gad.12.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shapiro D N, Sublett J E, Li B, Downing J R, Naeve C W. Fusion of PAX3 to a member of the forkhead family of transcription factors in human alveolar rhabdomyosarcoma. Cancer Res. 1993;53:5108–5112. [PubMed] [Google Scholar]
- 59.Shayesteh L, Lu Y, Kuo W L, Baldocchi R, Godfrey T, Collins C, Pinkel D, Powell B, Mills G B, Gray J W. PIK3CA is implicated as an oncogene in ovarian cancer. Nat Genet. 1999;21:99–102. doi: 10.1038/5042. [DOI] [PubMed] [Google Scholar]
- 60.Sheaff R J, Groudine M, Gordon M, Roberts J M, Clurman B E. Cyclin E-CDK2 is a regulator of p27Kip1. Genes Dev. 1997;11:1464–1478. [Google Scholar]
- 61.Songyang Z, Baltimore D, Cantley L C, Kaplan D R, Franke T F. Interleukin 3-dependent survival by the Akt protein kinase. Proc Natl Acad Sci USA. 1997;94:11345–11350. doi: 10.1073/pnas.94.21.11345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stambolic V, Suzuki A, de la Pompa J L, Brothers G M, Mirtsos C, Sasaki T, Ruland J, Penninger J M, Siderovski D P, Mak T W. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 1998;95:29–39. doi: 10.1016/s0092-8674(00)81780-8. [DOI] [PubMed] [Google Scholar]
- 63.Steck P A, Pershouse M A, Jasser S A, Yung W K, Lin H, Ligon A H, Langford L A, Baumgard M L, Hattier T, Davis T, Frye C, Hu R, Swedlund B, Teng D H, Tavtigian S V. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15:356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 64.Sun H, Lesche R, Li D M, Liliental J, Zhang H, Gao J, Gavrilova N, Mueller B, Liu X, Wu H. PTEN modulates cell cycle progression and cell survival by regulating phosphatidylinositol 3,4,5-triphosphate and Akt/protein kinase B signaling pathway. Proc Natl Acad Sci USA. 1999;96:6199–6204. doi: 10.1073/pnas.96.11.6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tamura M, Gu J, Matsumoto K, Aota S, Parsons R, Yamada K M. Inhibition of cell migration, spreading, and focal adhesion by tumor suppressor PTEN. Science. 1998;280:1614–1617. doi: 10.1126/science.280.5369.1614. [DOI] [PubMed] [Google Scholar]
- 66.Tang E D, Nunez G, Barr F G, Guan K L. Negative regulation of the forkhead transcription factor FKHR by Akt. J Biol Chem. 1999;274:16741–16746. doi: 10.1074/jbc.274.24.16741. [DOI] [PubMed] [Google Scholar]
- 67.Tashiro H, Blazes M S, Wu R, Cho K R, Bose S, Wang S I, Li J, Parsons R, Ellenson L H. Mutations in PTEN are frequent in endometrial carcinoma but rare in other common gynecological malignancies. Cancer Res. 1997;57:3935–3940. [PubMed] [Google Scholar]
- 68.Tedder T F, Isaacs C M. Isolation of cDNAs encoding the CD19 antigen of human and mouse B lymphocytes. J Immunol. 1989;143:712–717. [PubMed] [Google Scholar]
- 69.Teng D H, Hu R, Lin H, Davis T, Iliev D, Frye C, Swedlund B, Hansen K L, Vinson V L, Gumpper K L, Ellis L, El-Naggar A, Frazier M, Jasser S, Langford L A, Lee J, Mills G B, Pershouse M A, Pollack R E, Tornos C, Troncoso P, Yung W K, Fujii G, Berson A, Steck P A. MMAC1/PTEN mutations in primary tumor specimens and tumor cell lines. Cancer Res. 1997;57:5221–5225. [PubMed] [Google Scholar]
- 70.Vazquez F, Ramaswamy S, Nakamura N, Sellers W R. Phosphorylation of the PTEN tail regulates protein stability and function. Mol Cell Biol. 2000;20:5010–5018. doi: 10.1128/mcb.20.14.5010-5018.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vazquez F, Sellers W R. The PTEN tumor suppressor protein: an antagonist of phosphoinositide 3-kinase signaling. Biochim Biophys Acta. 2000;1470:M21–M35. doi: 10.1016/s0304-419x(99)00032-3. [DOI] [PubMed] [Google Scholar]
- 72.Vlach J, Hennecke S, Amati B. Phosphorylation-dependent degradation of the cyclin-dependent kinase inhibitor p27. EMBO J. 1997;16:5334–5344. doi: 10.1093/emboj/16.17.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang S I, Puc J, Li J, Bruce J N, Cairns P, Sidransky D, Parsons R. Somatic mutations of PTEN in glioblastoma multiforme. Cancer Res. 1997;57:4183–4186. [PubMed] [Google Scholar]
- 74.Watanabe-Fukunaga R, Brannan C I, Copeland N G, Jenkins N A, Nagata S. Lymphoproliferation disorder in mice explained by defects in Fas antigen that mediates apoptosis. Nature. 1992;356:314–317. doi: 10.1038/356314a0. [DOI] [PubMed] [Google Scholar]
- 75.Whitman M, Kaplan D R, Schaffhausen B, Cantley L, Roberts T M. Association of phosphatidylinositol kinase activity with polyoma middle-T competent for transformation. Nature. 1985;315:239–242. doi: 10.1038/315239a0. [DOI] [PubMed] [Google Scholar]