Strong Functional Interactions of TFIIH with XPC and XPG in Human DNA Nucleotide Excision Repair, without a Preassembled Repairosome (original) (raw)
Abstract
In mammalian cells, the core factors involved in the damage recognition and incision steps of DNA nucleotide excision repair are XPA, TFIIH complex, XPC-HR23B, replication protein A (RPA), XPG, and ERCC1-XPF. Many interactions between these components have been detected, using different physical methods, in human cells and for the homologous factors in Saccharomyces cerevisiae. Several human nucleotide excision repair (NER) complexes, including a high-molecular-mass repairosome complex, have been proposed. However, there have been no measurements of activity of any mammalian NER protein complex isolated under native conditions. In order to assess relative strengths of interactions between NER factors, we captured TFIIH from cell extracts with an anti-cdk7 antibody, retaining TFIIH in active form attached to magnetic beads. Coimmunoprecipitation of other NER proteins was then monitored functionally in a reconstituted repair system with purified proteins. We found that all detectable TFIIH in gently prepared human cell extracts was present in the intact nine-subunit form. There was no evidence for a repair complex that contained all of the NER components. At low ionic strength TFIIH could associate with functional amounts of each NER factor except RPA. At physiological ionic strength, TFIIH associated with significant amounts of XPC-HR23B and XPG but not other repair factors. The strongest interaction was between TFIIH and XPC-HR23B, indicating a coupled role of these proteins in early steps of repair. A panel of antibodies was used to estimate that there are on the order of 105 molecules of each core NER factor per HeLa cell.
Nucleotide excision repair (NER) removes damage to mammalian DNA caused by UV light and some chemical mutagens (10, 31). The complete NER reaction involves damage recognition and formation of an open DNA structure around the lesion, followed by dual incision, excision of an oligonucleotide of 24 to 32 residues, and gap-filling DNA synthesis to restore an undamaged DNA molecule. In human cells the minimal set of components involved in performing this reaction comprises replication protein A (RPA), XPA, XPC-HR23B, XPG, ERCC1-XPF, TFIIH, replication factor C (RFC), PCNA, DNA polymerase δ or ɛ, and DNA ligase I (2). These factors can be divided into two groups, the six core factors necessary for damage recognition and dual incision (RPA, XPA, XPC-HR23B, XPG, ERCC1-XPF, and TFIIH) and those for repair DNA synthesis in the gap-filling step.
In a current NER model, the XPC-HR23B complex is likely to be the initial damage recognition factor if lesions are situated in a nontranscribed strand (5, 59). Subsequently, the DNA around the site of the lesion is opened asymmetrically in an ATP-dependent manner, employing the TFIIH complex with its two helicases, XPB and XPD (13, 14). This open DNA complex creates the substrate for cleavage by the two structure-specific endonucleases XPG (3′ of the lesion) and ERCC1-XPF (5′ of the lesion), which cut near the junction between the single-and the double-stranded DNA (13, 36, 43, 57). Once the incisions have been placed, a damage-containing oligonucleotide of approximately 24 to 32 nucleotides is released and the DNA structure is restored by replicative DNA synthesis.
An array of different protein-protein interactions between the factors involved in the first steps of NER has been detected by methods that range from immunoprecipitation and affinity chromatography to Saccharomyces cerevisiae two-hybrid systems (3). Most investigations have been concerned only with individual interactions between the proteins involved in NER and have used detection methods that assess physical interactions but not function. Complexes between NER proteins have been reported with compositions that vary according to the isolation and the detection methods used. For example, in S. cerevisiae, all of the core NER factors have been detected after partial purification using His-tagged TFIIH or Rad14 subunits, in an assembly designated a repairosome (49, 61). An alternative view of the NER mechanism in S. cerevisiae is that particular subcomplexes of a few factors are sequentially assembled on DNA (16).
To date, a systematic comparison of relative strengths of interactions has not been made. In this study we used immunoprecipitation from a human cell extract active in NER in order to assess which core NER proteins interact with one another most readily. Functionally relevant interactions were analyzed by testing directly for NER activity.
MATERIALS AND METHODS
Immunoprecipitation.
Whole-cell extracts from lymphoblastoid or fibroblast cells were made from approximately 109 cells according to the method of Manley and coworkers (33) with modifications as indicated in reference 66. Extracts had a concentration of 20 to 40 μg of protein per μl in extract dialysis buffer (25 mM HEPES-KOH [pH 7.9], 0.1 M KCl, 17% glycerol [vol/vol], 1 mM EDTA, 1 mM dithiothreitol, and 12 mM MgCl2). M-450 paramagnetic Dynabeads (goat anti-mouse immunoglobulin G [IgG]; DYNAL) were washed with extract dialysis buffer and incubated with an anti-cdk7 monoclonal antibody (MO1-1; Novocastra Laboratories) or an anti-XPG monoclonal antibody (8H7) at a ratio of about 0.5 μg of antibody per 107 beads overnight at 4°C. Cell extracts were diluted as necessary to 20 mg of protein per ml with extract dialysis buffer and incubated with the MO1.1 beads for 2 h at room temperature. For each 10 μg of extract protein, 1 μl (4 × 105) of beads was used. Beads were collected on a magnetic particle concentrator (DYNAL MPC), and the supernatant was removed for analysis. Beads were then washed three times in 10 volumes of buffer W (25 mM HEPES–KOH [pH 7.6], 10% glycerol, and 0.01% Triton X-100) containing the KCl concentration indicated in the figures. Beads were resuspended in buffer W containing 50 mM KCl and used for sodium dodecyl sulfate-polyacrylamide gel electrophoresis–immunoblot analysis or in vitro NER assays.
Immunoblotting.
Proteins were separated on sodium dodecyl sulfate–10% polyacrylamide gels and transferred to Immobilon P polyvinylidene difluoride (Millipore) membranes. Primary rabbit polyclonal or mouse monoclonal antibodies were as follows: for XPA, a 1/1,000 dilution of polyclonal antibody AHP452 (Serotec), raised against recombinant human XPA protein (30); for XPC, a 1/2,000 dilution of polyclonal antibody RW028 raised against residues 96 to 299 of human XPC protein (5); for HR23B, a 1/10,000 dilution of polyclonal antibody against Rad23 (49); for XPG, a 1/250 dilution of monoclonal antibody 8H7 (13); for XPB, a 1/1,000 dilution of monoclonal antibody 1B3; for cyclin H, a 1/2,000 dilution of monoclonal antibody 2D4; for p62, a 1/10,000 dilution of monoclonal antibody 3C9 (the last three were provided by J.-M. Egly); for the RPA p34 subunit, a 1/250 dilution of monoclonal antibody 34A (22); for XPF, a 1/3,000 dilution of polyclonal antibody RA1 raised against residues 571 to 905 of human XPF protein (24); and for ERCC1, a 1/1,500 dilution of polyclonal antibody RW017 (24). The membranes were incubated with the primary antibody for 1 to 2 h, followed by incubation for 1 h with either a 1/25,000 dilution of peroxidase-labeled anti-mouse IgG or a 1/50,000 dilution of peroxidase-labeled anti-rabbit IgG (both from Sigma). Bands were visualized by chemiluminescence (Amersham Pharmacia Biotech). The intensity of the chemiluminescent signal was quantified using NIH Image software after the X-ray film was scanned. Density units were plotted against protein concentration, and the amount of each protein in the immunoprecipitated fraction was estimated.
Dual-incision NER assay.
Reconstituted repair reactions (mixtures, 8.5 μl) were carried out in a buffer containing 45 mM HEPES–KOH (pH 7.8), 70 mM KCl, 7 mM MgCl2, 1 mM dithiothreitol, 0.3 mM EDTA, 12.5% (vol/vol) glycerol, 2.5 μg of bovine serum albumin, 0.025% (vol/vol) NP-40, and 2 mM ATP. Unless indicated otherwise, each reconstituted reaction mixture contained 50 ng of RPA, 22.5 ng of XPA, 10 ng of the XPC-HR23B complex, 50 ng of XPG, 20 ng of the ERCC1-XPF complex, and 1.5 μl of Hep TFIIH (heparin-Sepharose fraction IV from HeLa cells [34]). Following preincubation for 10 min at 30°C, 50 ng of Pt-GTG DNA (56) was added and reaction mixtures were incubated for 90 min at 30°C. Reactions were stopped by rapid freezing. Six nanograms of an oligonucleotide complementary to the excised DNA fragment was added to the reaction mixture. This oligonucleotide contains four extra G residues at the 5′ end and was annealed to the excised products by heating at 95°C and gradually cooling the mixtures to 20°C. Excision products were radiolabeled with 0.1 U of Sequenase version 2.0 polymerase (U.S. Biochemicals) and 1 μCi of [α-32P]dCTP (3,000 Ci/mmol), separated on a denaturing 14% polyacrylamide gel, and visualized by autoradiography and with a phosphorimager as described previously (56). Phosphorimager data were used to quantify the results.
RESULTS
Immunoprecipitation of NER proteins from HeLa cell extracts with an antibody against TFIIH.
Our object was to search for functional interactions between NER proteins. We thought it important to use a minimally disruptive but specific method under conditions where all factors were present at relative concentrations similar to those in the cell. The starting point was a human cell extract active in NER and with no overproduced factors. TFIIH was the initial target factor for several reasons. TFIIH is a core component of NER (14), and there was some physical evidence that it interacts with other core factors such as XPA (41, 46) and XPG (21, 39). TFIIH is composed of nine subunits, designated XPB, XPD, p62, p52, p44, p34, cdk7, cyclin H, and Mat1. The last three subunits comprise the cdk-activating kinase (CAK) subcomplex that phosphorylates the C-terminal domain of RNA polymerase II during transcription (60). The MO1.1 antibody raised against the cdk7 subunit of TFIIH was a particularly attractive tool because this antibody can be used to isolate transcriptionally active TFIIH from HeLa cells, even while the TFIIH is still attached to the antibody-resin beads (44). Further, the cdk7 subunit of TFIIH is not required for the core NER reaction, and so an antibody attached to cdk7 was unlikely to interfere with repair (2).
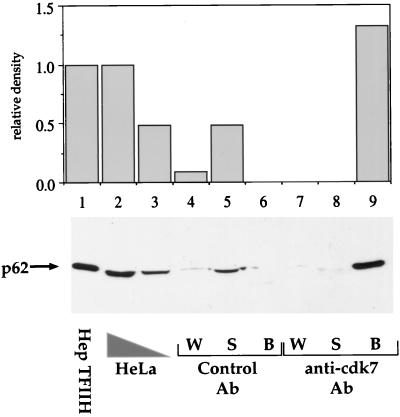
TFIIH from HeLa cell extracts was immunoprecipitated by cdk7 antibodies bound to paramagnetic beads, and the input (HeLa cell extract), the supernatant (HeLa cell extract after immunoprecipitation), and the beads (the TFIIH-bound fraction) were analyzed by immunoblotting using an antibody against the p62 subunit of TFIIH. Mouse whole IgG was used as a precipitation control. TFIIH complex can be recovered in a specific manner from HeLa cells in this way (Fig. 1, compare lanes 6 to 9). Essentially all of the TFIIH present in HeLa cells could be immunoprecipitated by this antibody, showing that the great majority of TFIIH includes CAK subunits (Fig. 1, lanes 8 and 9). With control antibody, TFIIH was detected in the supernatant but not in the immunoprecipitated fraction (Fig. 1, lanes 5 and 6).
FIG. 1.
Quantitative immunoprecipitation of TFIIH from HeLa cell extracts with a cdk7 antibody. TFIIH was immunoprecipitated from a HeLa whole-cell extract with a cdk7 antibody and detected using monoclonal antibody 3C9 against the p62 subunit. Lane 1 contains 5 μl of Hep TFIIH (fraction IV from HeLa cells), and lanes 2 and 3 contain 100 and 50 μg of HeLa whole-cell extract protein, respectively. HeLa cell extract protein (400 μg in 20 μl) was added to 40 μl of antibody (Ab) beads. Lanes 4 to 9 contain 20 μl of the supernatant (S) and first-wash (W) fractions and 10 μl of the bead (B) fraction. The control antibody was mouse whole IgG.
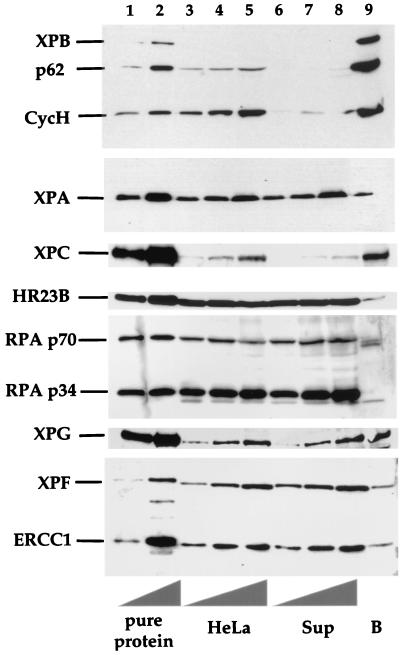
We asked whether the immunoprecipitated TFIIH complex was associated with other NER factors after washing was carried out under relatively mild conditions (50 mM KCl). TFIIH was immunoprecipitated from HeLa cell extract (Fig. 2, top panel, compare lanes 3 to 5 with lanes 6 to 8). After being washed, the TFIIH attached to the beads remained in the nine-subunit form and contained, for example, the core subunits XPB and p62 as well as the CAK subunit cyclin H (lane 9).
FIG. 2.
Coimmunoprecipitation of TFIIH and other NER factors. TFIIH was immunoprecipitated from a HeLa whole-cell extract, and TFIIH bound fractions were analyzed by immunoblotting. Detection was performed by using the indicated antibodies against proteins involved in the first steps of NER. Lanes 1 and 2 contain pure proteins (from top to bottom) as follows: 2.5 and 5 μl of Hep TFIIH, 22.5 and 45 ng of XPA, 75 and 150 ng of XPC-HR23B, 125 and 250 ng of RPA, 125 and 250 ng of XPG, and 50 and 100 ng of ERCC1-XPF. Lanes 3 to 5 contain 17.5, 35, and 70 μg of HeLa cell extract protein, respectively; lanes 6 to 8 contain 17.5, 35, and 70 μg of the supernatant protein from the same extract after immunoprecipitation (Sup); and lane 9 contains 20 μl of antibody beads (B) with the immunoprecipitate.
The presence of other NER factors in the immunoprecipitate was assayed by immunoblotting. In the fraction bound to the beads, some XPA, XPC, HR23B, XPG, and ERCC1-XPF were found (Fig. 2, lane 9). A significant amount of RPA was not detected; only weak cross-reacting bands were seen, migrating at positions different from those of the p70 and p34 subunits. The immunoblots were quantified to estimate the fractions of NER proteins that were immunoprecipitated (with the amount of TFIIH recovered from HeLa cell extracts being considered 100%). The fractions of NER proteins (in relation to the total amount in the HeLa cell extract) immunoprecipitated with the anti-cdk7 antibody are shown in Table 1. About 36% of the total XPC and 15% of the total XPG present in HeLa cell extracts were found in the TFIIH fraction. The amounts of XPA and ERCC1-XPF present in this fraction were below 5% of the total amount present in HeLa cells. XPC exists in a tightly bound complex with HR23B, but the fraction of HR23B precipitated was only about 1%. This amount is consistent with the approximately 10-fold excess of HR23B over XPC in cells, where most of the HR23B is not bound to XPC (62). The immunoblotting measurements, quantified with standard curves, also yielded estimates of the number of molecules of each core NER factor per HeLa cell. It is interesting that in nearly all cases there are on the order of 105 molecules per cell, with XPC possibly being the limiting factor (Table 1).
TABLE 1.
Number of NER proteins per HeLa cell and fraction of each that can bind to TFIIH in HeLa cell extracts
| NER factor | Relative amt immunoprecipitated (%) | Amt immunoprecipitated (fmol) per μl of immunoprecipitatea | Amt (fmol) per reconstituted reaction mixtureb | Approx no. of molecules per HeLa cell (this study)c | Approx no. of molecules per HeLa cell (reference) | |
|---|---|---|---|---|---|---|
| 1 μl | 3 μl | |||||
| TFIIH | 100 | 270 | 810 | 500 | 1.1 × 105 (23) | |
| RPA | 450 | 2 × 105 | 0.3 × 105 to 2 × 105 (65) | |||
| XPA | 4 | 15 | 45 | 700 | 2 × 105 | |
| ERCC1 | 4 | 15 | 45 | 60 | 1 × 105 | |
| XPF | 4 | 15 | 45 | 60 | 1 × 105 | |
| HR23B | 1 | 20 | 60 | 80 | 2.5 × 105 | 2 × 105 to 4 × 105 (62) |
| XPC | 36 | 20 | 60 | 80 | 2.5 × 104 | 4 × 104 to 8 × 104 (62) |
| XPG | 15 | 35 | 100 | 340 | 8 × 104 |
Interactions between TFIIH and other NER components at low ionic strength.
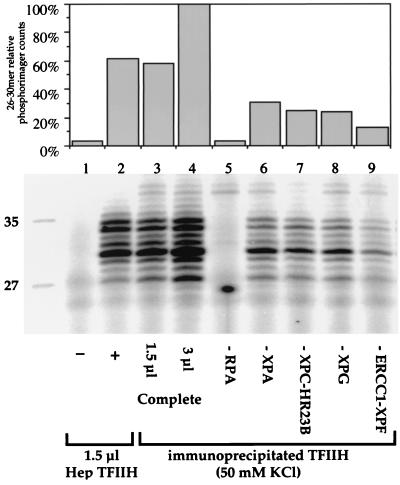
The NER activity of the immunoprecipitates was tested. We found that the TFIIH bound to the cdk7 antibody beads was active in an assay that detects the damaged oligonucleotides released by dual incision during NER (Fig. 3, lanes 3 and 4). Repair was carried out in reaction mixtures including the purified core factors XPA, XPC-HR23B, RPA, XPG, and ERCC1-XPF. Reaction mixtures containing immunoprecipitated TFIIH had activity comparable to that of reactions with purified TFIIH (Fig. 3, compare lane 2 with lane 3). To test whether functionally significant amounts of other NER factors had coprecipitated with the TFIIH complex, each purified factor was individually omitted from a reconstituted reaction mixture containing the immunoprecipitate. Significant dual incision, about 20 to 25% of the full reaction, was found in the absence of either XPA, XPC-HR23B, or XPG (Fig. 3, lanes 6 to 8). Limited repair, about 10% of the full reaction, was found when ERCC1-XPF was omitted (Fig. 3, lane 9). This result indicates that some XPA, XPC-HR23B, XPG, and ERCC1-XPF can bind to TFIIH and are functional in NER after immunoprecipitates are washed with 50 mM KCl. No dual-incision products were detected, however, when RPA was omitted from reaction mixtures (Fig. 3, lane 5), showing that no functionally significant amounts of this protein are present in the immunoprecipitate.
FIG. 3.
Functional interactions of TFIIH with other NER factors in HeLa cell extracts. cdk7 antibody beads containing TFIIH and associated proteins immunoprecipitated from HeLa cells were added to dual-incision assays reconstituted with purified factors. Protein-protein interactions were tested by omission of individual repair factors as indicated. Lanes 1 to 4 contain XPA, RPA, XPC-HR23B, XPG, and ERCC1-XPF, with no TFIIH (lane 1), 1.5 μl of Hep TFIIH (lane 2), or TFIIH beads (lanes 3 and 4). Excised fragments were detected by a direct end-labeling procedure which extends the 24 to 32-nucleotide products by 4 nucleotides. The quantification shown at the top was normalized to lane 4, containing 3 μl of TFIIH beads. TFIIH interactions in lanes 3 to 9 were measured on beads washed with buffer containing 50 mM KCl. Lanes 5 to 9 contain 3 μl of beads.
In each case in Fig. 3, lanes 6 to 9, where a single factor was omitted, the NER activity was lower than in the reaction mixture containing immunoprecipitate supplemented with all of the purified proteins. This lower activity may largely be due to the fact that in each case, omitting the factor significantly reduced its concentration in the reaction mixture so that it became rate limiting (Table 1).
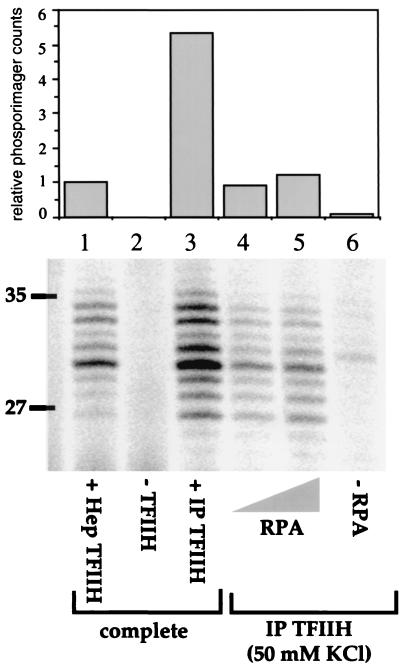
Because the only core factor that was completely absent under these conditions was RPA, we tested the NER activities of the immunoprecipitates after addition of purified RPA only (Fig. 4). The TFIIH immunoprecipitate was indeed active for dual incision when supplemented with RPA. A level of NER activity was attained (Fig. 4, lanes 4 and 5) that was similar to the level found in a reaction mixture reconstituted with purified components (Fig. 4, lane 1). A severalfold-higher level of repair could be attained by adding the full set of purified NER proteins to the immunoprecipitate (Fig. 4, lane 3).
FIG. 4.
RPA restores NER activity to HeLa immunoprecipitates (IP). The results of dual-incision assays are presented as in Fig. 3. Lanes 1 to 3 contain XPA, RPA, XPC-HR23B, XPG, and ERCC1-XPF, with 1.5 μl of Hep TFIIH (lane 1) or no TFIIH (lane 2). Magnetic beads (10 μl) containing TFIIH and associated factors were washed with buffer containing 50 mM KCl and used in reconstituted dual-incision assays either with all the other repair factors added (lane 3), with only RPA added (lanes 4 to 5), or with no additions (lane 6). Lanes 4 and 5 contain 50 ng (0.45 pmol) and 125 ng (1.1 pmol) of RPA, respectively.
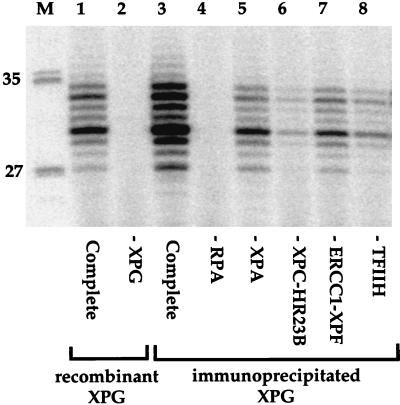
As an independent test for interactions between these proteins, similar experiments were performed using an antibody against XPG protein. The 8H7 antibody can recover XPG in a form active as a nuclease, without inhibiting DNA repair (8). In an immunoprecipitate from HeLa cell extracts with the 8H7 antibody, some XPA, XPC-HR23B, ERCC1-XPF, and TFIIH activities were also detected (Fig. 5, lanes 5 to 8). As found with the anti-cdk7 immunoprecipitates, there was no activity in the absence of RPA (Fig. 5, lane 4). Significantly, although the relative amounts of the recovered NER factors were different with the 8H7 antibody (for example, less XPC-HR23B and more ERCC1-XPF), the same overall functional group of factors were present in immunoprecipitates after the beads were washed with buffer containing 50 mM KCl.
FIG. 5.
Functional interactions between XPG and other NER factors. XPG was immunoprecipitated from HeLa cell extracts, and the 8H7 antibody beads containing XPG and associated proteins were added to reconstituted dual-incision assays. Functional interactions were tested by omission of individual repair factors as indicated. Lanes marked “Complete” contain all the repair factors. Lane 1 contains 45 ng of purified recombinant XPG protein; lanes 3 to 8 contain 3 μl of XPG magnetic beads; and lane M contains molecular size markers, with lengths (in bases) noted at left.
Strong interaction between TFIIH and XPC-HR23B.
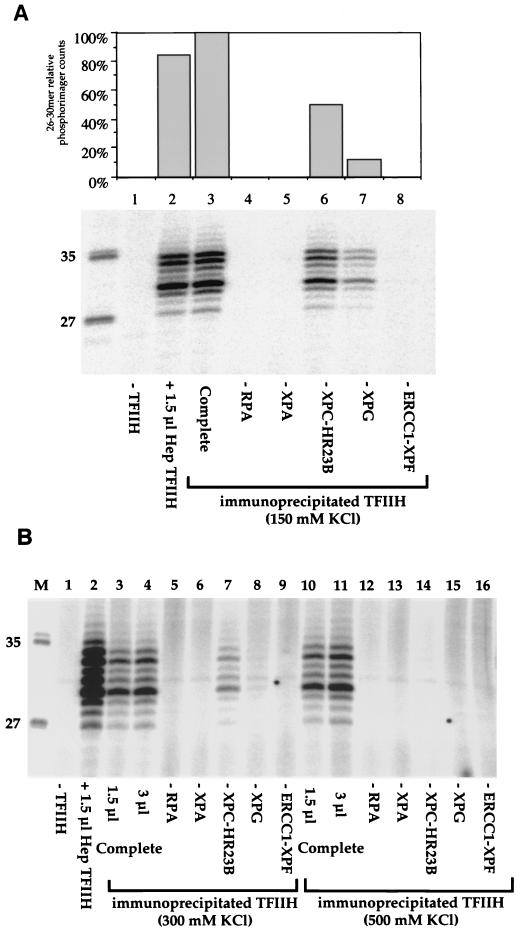
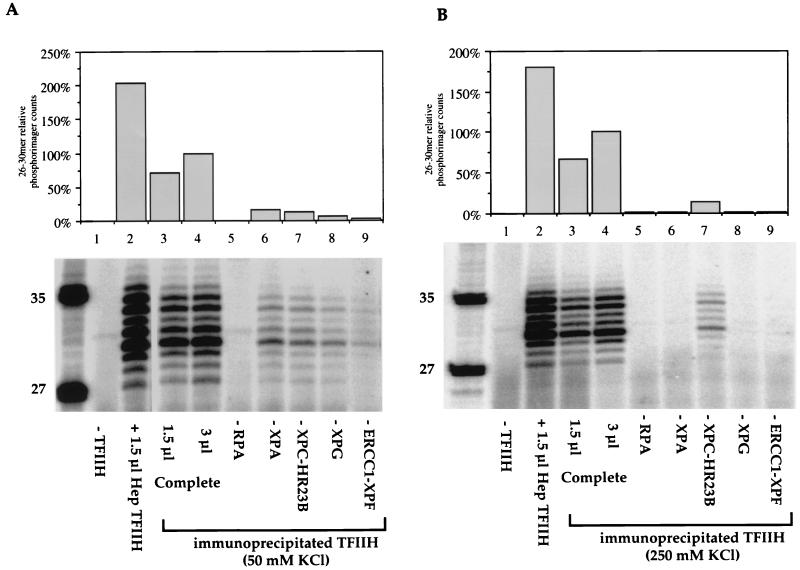
The results presented so far show that five of the core protein factors involved in the first stages of NER could be coimmunoprecipitated after washing was carried out with relatively mild ionic-strength buffer (50 mM KCl). The immunoprecipitation approach offers the potential to discriminate between stronger and weaker functional interactions by using higher-stringency conditions. Figure 6A shows an experiment using a wash buffer containing 150 mM KCl, near physiological ionic strength. The immunoprecipitated TFIIH remained as active as the complex purified from HeLa cells (Fig. 6A, lanes 2 and 3). However, when each of the other NER factors was sequentially omitted, NER activity was detected only in the absence of the XPC-HR23B complex and in the absence of XPG (Fig. 6A, lanes 6 and 7). This finding indicates that interactions between TFIIH and XPC-HR23B or XPG are stronger than those between TFIIH and XPA or ERCC1-XPF. When the salt concentration in the wash buffer was increased to 300 mM KCl, the only remaining functionally significant interaction was with TFIIH-XPC (Fig. 6B, lanes 5 to 9). The interaction between TFIIH and XPC was finally disrupted by washing immunoprecipitate beads with 500 mM KCl (Fig. 6B, lanes 12 to 16).
FIG. 6.
Sensitivity of TFIIH interactions to ionic strength. cdk7 antibody beads containing TFIIH and associated proteins immunoprecipitated from HeLa cells were added to dual-incision assays reconstituted with purified factors. Protein-protein interactions were tested by omission of individual repair factors as indicated. (A) TFIIH interactions on beads washed with buffer containing 150 mM KCl. Lanes 1 to 3 contain XPA, RPA, XPC-HR23B, XPG, and ERCC1-XPF, with no TFIIH (lane 1), 1.5 μl of Hep TFIIH (lane 2), or TFIIH beads (lane 3). The quantification shown at the top was normalized to lane 3, containing 3 μl of TFIIH beads. Lanes 4 to 8 contain 3 μl of beads. (B) TFIIH interactions on beads washed with buffer containing 300 mM KCl (lanes 3 to 9) or 500 mM KCl (lanes 10 to 16). Lanes 1 to 4, 10, and 11 contain XPA, RPA, XPC-HR23B, XPG, and ERCC1-XPF, with no TFIIH (lane 1), 1.5 μl of Hep TFIIH (lane 2), 1.5 μl of TFIIH beads (lanes 3 and 10), or 3 μl of TFIIH beads (lanes 4 and 11). Lanes 5 to 16 contain 3 μl of beads. Lane M contains molecular size markers, with lengths (in bases) noted at left.
Interactions between NER proteins in XPA-deficient cell extracts.
In the experiments described above, HeLa cells were used to examine interactions between NER proteins. In order to examine interactions in situations where one component was missing, a convenient and readily available resource is NER-defective lymphoblastoid cell lines from XP patients. In control experiments, TFIIH was immunoprecipitated from an extract of an NER-proficient lymphoblastoid cell line, 705ori (45). As in the previous experiments, it was possible to immunoprecipitate TFIIH from this cell extract in active form (Fig. 7, lanes 2 to 4). Interactions were analyzed by separately omitting each of the repair factors. The results were similar to those obtained with HeLa cell extracts under the same conditions (50 mM KCl). TFIIH from lymphoblastoid cells coimmunoprecipitated with some XPA, XPC-HR23B, XPG, and ERCC1-XPF activities. When immunoprecipitated TFIIH was washed with 250 mM KCl buffer, only XPC-HR23B activity was detected (Fig. 7B).
FIG. 7.
Functional interactions of TFIIH with other NER factors in extracts from normal lymphoblastoid cells. cdk7 magnetic beads containing TFIIH and associated proteins immunoprecipitated from 705ori cells were added to dual-incision assay mixtures reconstituted with purified factors. Protein-protein interactions were tested by omission of individual repair factors as indicated. Lanes marked “Complete” contain all the factors. (A) TFIIH interactions on beads washed with buffer containing 50 mM KCl. (B) TFIIH interactions on beads washed with buffer containing 250 mM KCl. In each case, lane 2 contains 1.5 μl of Hep TFIIH; lanes 3 and 4 contain 1.5 and 3 μl of cdk7 magnetic beads, respectively; and lanes 5 to 9 contain 3 μl of beads.
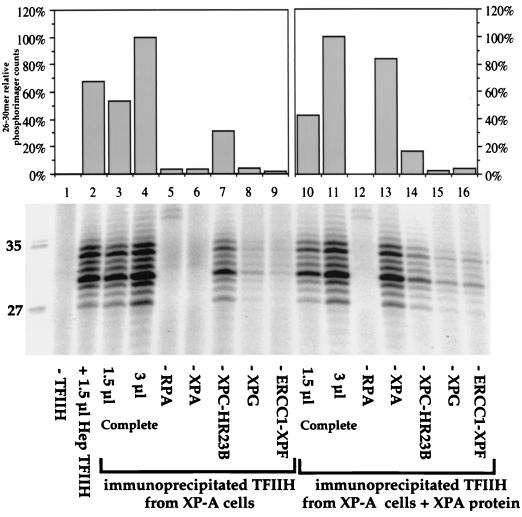
XPA binds damaged DNA and plays a central role in NER, interacting with many core repair factors (reviewed in reference 3). To study the influence of XPA in interactions between NER proteins, we used the lymphoblastoid cell line GM2345, derived from patient XP2OS. This cell line is completely defective in NER, showing lower levels of reduced-size XPA mRNA (52) and no detectable XPA activity (reference 37 and D. Batty, unpublished results). TFIIH immunoprecipitated from GM2345 lymphoblastoid cell extract had an activity in NER similar to the activity of TFIIH purified from HeLa cells and normal lymphoblastoid cells (Fig. 8, lanes 2 to 4). As expected, the immunoprecipitate was inactive when XPA was omitted (Fig. 8, lane 6). However, a strong interaction between TFIIH and XPC complex was still evident (Fig. 8, lane 7). Small amounts of XPG and ERCC1-XPF were also coimmunoprecipitated (Fig. 8, lanes 8 and 9).
FIG. 8.
Functional interactions of TFIIH with other NER factors in extracts from XP-A cells. cdk7 magnetic beads containing TFIIH and associated proteins immunoprecipitated from GM2345 XP-A cells were washed with buffer containing 50 mM KCl and added to dual-incision assays reconstituted with purified factors. Protein-protein interactions were tested by omission of individual repair factors as indicated. Lanes marked “Complete” contain all the repair factors. Lanes 3 to 9 contain reaction mixtures with TFIIH immunoprecipitated from the XP-A cell extracts; lanes 10 to 16 contain reaction mixtures with TFIIH immunoprecipitated from the XP-A extracts after addition of 1 ng of purified XPA per μg of cell extract protein and incubation for 30 min at 30°C. Lane 2 contains 1.5 μl of Hep TFIIH, lanes 3 and 10 contain 1.5 μl of TFIIH antibody beads, and lanes 4 to 9 and 11 to 16 contain 3 μl of beads. Quantification was performed on a phosphorimager relatively to the lane that contains 3 μl of TFIIH beads (lane 4 for the first set of values and lane 11 for the second set).
To further investigate the influence of XPA on interactions between NER components, pure recombinant XPA was incubated with XP-A cell extract to give an XPA concentration of 600 nM, the same as used previously for complementation for NER activity (24). Immunoprecipitation was carried out with cdk7 antibody, and the beads were washed with buffer containing 50 mM KCl. In this case, XPA immunoprecipitated with TFIIH, indicating that an interaction formed with recombinant XPA (Fig. 8, lane 13). Relatively more XPA activity was found on the TFIIH beads after incubation with exogenous XPA in this way (compare Fig. 7A, lane 6, with Fig. 8, lane 13). This would be the expected consequence of a shift in the equilibrium towards TFIIH-XPA association when excess XPA is present (normal cell extracts contain about 300 nM XPA). Interactions with the XPC complex and XPG were detected as previously shown (Fig. 8, lanes 14 and 15). The amounts of recovered ERCC1-XPF complex were slightly increased. Comparison of the complexes isolated before and after XPA complementation (lanes 9 and 16) suggests that the presence of XPA may provide a link between TFIIH and ERCC1-XPF.
DISCUSSION
Interactions between components of human NER.
One objective of our experiments was to look for evidence for a preassembled active NER repairosome in human cells. The approach taken here had several features. First, we used proteins at their native relative concentrations in cell extracts active for repair, with no factors being overproduced, tagged, or concentrated on a column. Second, a gentle procedure to isolate a core factor was employed, using an antibody against a part of TFIIH that is not necessary for repair. Third, a functional assay for interactions was used for the first time, and relative strengths of associations were assessed by adjusting the ionic strength. No evidence was found for a preassembled human repairosome complex. We did find that at low ionic strength, five of the core factors could associate with one another (TFIIH, XPA, XPG, XPC-HR23B, and ERCC1-XPF). With increasingly stringent washing in higher-salt buffers, all of these interactions could be disrupted, with the interaction between TFIIH and XPC-HR23B being the most stable and that between TFIIH and XPG being the next most stable.
The existence of interactions of various strengths between all these components is anticipated from the results of a study that has looked for pairwise interactions between them using affinity tagging and two-hybrid or antibody methods (3). Many of the pairwise interactions have been characterized to the point of mapping interaction domains to defined regions of the proteins. Thus, there are specific contacts between components, many of which may reflect those contacts made between the core factors as damage is recognized and an incision complex assembles on a damaged site.
One somewhat unexpected finding of our experiments was that significant amounts of RPA are not associated with the other factors, even at low ionic strength. It is known, however, that RPA can interact with XPA (18, 25, 28, 35, 51, 58). This interaction is most significant in a ternary complex with damaged DNA (18) where the principal DNA contacts are made by RPA (54). Our results simply indicate that in solution at normal concentrations, the association of RPA with other repair factors is the weakest interaction. This was foreseeable, perhaps, from the earlier observation that when human whole-cell extracts are passed over a phosphocellulose column in buffer containing 0.1 M salt, all of the NER recombination and incision factors except for RPA are bound to the matrix. RPA quantitatively flows through the column (55).
Some previous investigations have failed to distinguish clearly between the tight, salt-resistant interactions that hold together the subunits of core factors and the looser interactions between different core factors. The strong interactions between XPC and HR23B, between the three subunits of RPA, or between ERCC1 and XPF appear to be dominated by hydrophobic forces, and in most cases the associations are formed as the subunits fold together soon after they are synthesized. As shown here, interactions between the core factors are dominated by ionic forces which nevertheless involve specific regions of the interacting pairs.
A consequence of not making a distinction between the strengths of different interactions has been the occasional report that many human DNA repair proteins can be found together in very high-molecular-weight complexes, accompanied by numerous DNA replication, transcription, and recombination factors from other pathways (32, 63). Neither the functional activity nor the relative stability of such associations has been investigated.
He and Ingles (19) loaded HeLa cell extracts onto an affinity column made with XPA and found that unquantified amounts of many NER proteins could be found bound to the matrix after the column was washed with 0.1 M salt. The XPA column may have worked partially by ion exchange, as this is essentially the same result as that obtained with a phosphocellulose column (55). The difference is that in this case some RPA was bound. This would be the consequence of a very high local XPA concentration, which would shift the equilibrium to favor XPA-RPA complexes.
At the other extreme, XPB was tagged with a hemagglutinin epitope and TFIIH was isolated from whole-cell extracts with a hemagglutinin antibody attached to antibody beads. Following extensive washing with 0.1 M KCl buffer, no XPC, XPG, or any other NER factors were detected by immunoblotting (64). This is consistent with the fact that at low concentration, XPC and XPG will eventually dissociate from TFIIH. It also seems possible that the use of a tagged XPB may not always be ideal for capturing a TFIIH-XPC interaction under native conditions.
We believe that most evidence points towards a mechanism for human NER in which individual core factors come together to be assembled at a site of DNA damage, rather than being part of a larger preassembled compex. In living cells, for example, the ERCC1-XPF factor diffuses freely and rapidly until a site of damage is encountered, where it remains bound for several minutes before dissociating upon completion of repair. The molecular mass of freely diffusing ERCC1-XPF matches that of its two subunits, indicating that ERCC1-XPF is not normally resident in a high-molecular-weight complex (20).
TFIIH in cell extracts is in the nine-subunit form and active in NER.
It is noteworthy that almost all of the TFIIH in a HeLa whole-cell extract was immunoprecipitated with cdk7 antibody, showing that nearly 100% of the TFIIH present in the cell includes the CAK components. We found that this nine-subunit TFIIH complex is active in NER even when anchored to beads. TFIIH immunoprecipitated in a similar way has also been shown to be active in transcription (44). However, several previous studies have detected other versions of TFIIH, including a six-subunit form, a nonfunctional five-subunit form lacking XPD, and a separate complex of XPD with CAK (1, 11, 48, 50, 67). Indeed, a six-subunit form of TFIIH lacking the CAK subunits cdk7, cyclin H, and MAT1 can function in NER in a reconstituted system when isolated from HeLa cells (38) or as a recombinant protein complex (2). By analogy with some studies of yeast, it has been suggested that this six-subunit form of TFIIH is the one that normally functions in NER. This is because a form of yeast TFIIH lacking the kinase components is found preferentially associated with other NER factors in a high-molecular-mass fraction from yeast cell extracts (15, 61).
However, the current situation with regard to yeast TFIIH is as follows. Transcriptionally active S. cerevisiae TFIIH is also composed of nine subunits, each of which has an ortholog in human TFIIH (shown in parentheses), as follows: Tfb1 (p62), Tfb2 (p55), Tfb3 (Mat1), Tfb4 (p34), Rad3 (XPD), Ssl1 (p44), Ssl2 (XPB), Kin28 (cdk7), and Ccl1 (cyclin H). The Kin28 and Ccl1 subunits contain CAK kinase activity and dissociate more readily from the other seven subunits (15, 61). Upon further purification of yeast TFIIH, the Ssl2 and Tfb4 subunits are lost, leaving a five-subunit core of Tfb1, Tfb2, Tfb3, Rad3, and Ssl1 (7). This core is quite different from the composition of smaller forms of human TFIIH. The six-subunit form of human TFIIH lacks MAT1, while its homolog Tfb3 is, in contrast, a tightly bound component of five-subunit yeast TFIIH (7). Further, a five-subunit subcomplex of the most firmly associated human TFIIH subunits can be formed (53) comprising XPB, p62, p52, p44, and p34. Only three of these correspond to components of the five-subunit yeast TFIIH (Tfb1, Tfb2, and Ssl1).
These major differences seem unlikely to reflect basic alterations in the mechanism of TFIIH action in yeast versus mammalian cells. Instead it seems likely that in general the different TFIIH subcomplexes in mammalian cells and yeast arise during purification, as subunits gradually dissociate. It is possible that none of the smaller forms is physiologically relevant. We find that the nine-subunit form is the predominant form in cell extracts, that a nine-subunit form is functional in NER (2), and that this form of TFIIH can readily associate with other NER factors.
Functional interactions of TFIIH with XPC and XPG.
Interactions detected between core components in cell extracts do not necessarily represent preformed complexes in the cell. Instead they may reflect the interactions between protein components which normally take place on DNA. Nevertheless, the firmer interactions, between TFIIH and XPC and between TFIIH and XPG, have special functional significance. About 36% of all the XPC and 15% of all the XPG present in a HeLa whole-cell extract is complexed with TFIIH at 50 mM KCl. For the other NER factors, only fractions smaller than 10% were detected in the TFIIH bound fraction. When the ionic strength was increased to the more physiological level of 150 mM, the only functionally detectable interactions were between TFIIH and XPC-HR23B and TFIIH and XPG. The TFIIH-XPC complex interaction is the strongest. This is consistent with a model of the preincision opening reaction where the XPC complex, as the primary damage recognition factor, binds to a distorted site and then recruits TFIIH (14). In that study, both of these factors were required to see the earliest DNA opening or “bubble” formation around an adduct. XPC-HR23B as a primary recognition factor may bring the TFIIH onto the DNA, where the other factors may bind and perform opening of the DNA around the lesion. Recently, support for this model was presented by Yokoi et al. (68), who found that although TFIIH by itself has little DNA-binding activity, XPC could recruit TFIIH to a DNA-bound form. Further, Li et al. (30) showed that in a cell extract, the binding of XPA and TFIIH to damaged DNA is dependent on the presence of XPC. As shown in Fig. 8, the functional TFIIH-XPC interaction is independent of the presence of XPA and so XPA is most likely brought into the complex after XPC and TFIIH.
The fact that TFIIH interacts more strongly with XPC and XPG than with other NER factors probably accounts for the observation that XPC and XPG are sometimes found in partially purified TFIIH preparations (12, 39), depending on the particular chromatography steps. The mechanistic significance of the TFIIH-XPG interaction is not entirely clear. Like TFIIH, XPA, and RPA, XPG is a component of the NER preincision complex and must be in place before ERCC1-XPF can act (8, 40). On the other hand, mutation data indicate that XPG has at least one additional function (42) so the TFIIH-XPG interaction may be relevant to an entirely different process. Both TFIIH and XPG also act in a separate pathway of transcription-coupled repair of oxidative damage (9, 26). It is noteworthy that in S. cerevisiae, the Rad4 and Rad2 proteins (yeast homologs of XPC and XPG, respectively) can associate with yeast TFIIH under some conditions (4, 17).
Sequence of events in NER in human cells.
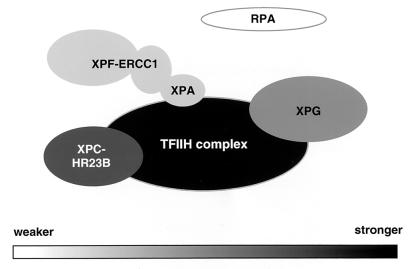
Figure 9 shows a schematic drawing of the interactions between TFIIH and other core factors, where interactions are shaded according to strength. The strongest interaction is between XPC-HR23B and TFIIH, followed by the interaction of XPG with TFIIH complex. Weaker interactions are between XPA and ERCC1-XPF. XPA can interact with ERCC1 (6, 19, 27, 29, 47, 51) and with TFIIH complex (41, 46). In XPA-defective cell extracts, we detected hardly any ERCC1-XPF in the absence of XPA protein (Fig. 8). So far no direct interaction between ERCC1-XPF and TFIIH has been found in mammalian cells, and this is reflected in the figure. It seems likely that encounters between the NER factors and damaged DNA encourage the formation of new and sequential interactions and that weaker interactions cooperate to form strong temporary associations on DNA. During the present study, all of the interactions were studied in extracts from nonirradiated cells. It is possible of course that other transient NER complexes form after a cell is damaged by UV irradiation or a chemical agent, and this is an area for future study.
FIG. 9.
Protein-protein interactions between human NER factors. Presented is a summary of the most stable interactions between core NER factors in human cell extracts; darker tones represent stronger interactions with TFIIH, and lighter tones represent weaker interactions with TFIIH, as detected by functional assays in the present study.
REFERENCES
- 1.Adamczewski J P, Rossignol M, Tassan J P, Nigg E A, Moncollin V, Egly J M. MAT1, cdk7 and cyclin H form a kinase complex which is UV light-sensitive upon association with TFIIH. EMBO J. 1996;15:1877–1884. [PMC free article] [PubMed] [Google Scholar]
- 2.Araújo S J, Tirode F, Coin F, Pospiech H, Syväoja J E, Stucki M, Hübscher U, Egly J-M, Wood R D. Nucleotide excision repair of DNA with recombinant human proteins: definition of the minimal set of factors, active forms of TFIIH and modulation by CAK. Genes Dev. 2000;14:349–359. [PMC free article] [PubMed] [Google Scholar]
- 3.Araújo S J, Wood R D. Protein complexes in nucleotide excision repair. Mutat Res. 1999;435:23–33. doi: 10.1016/s0921-8777(99)00042-7. [DOI] [PubMed] [Google Scholar]
- 4.Bardwell A J, Bardwell L, Iyer N, Svejstrup J Q, Feaver W J, Kornberg R D, Friedberg E C. Yeast nucleotide excision repair proteins Rad2 and Rad4 interact with RNA polymerase II basal transcription factor b (TFIIH) Mol Cell Biol. 1994;14:3569–3576. doi: 10.1128/mcb.14.6.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Batty D P, Otrin V R, Levine A S, Wood R D. Stable binding of human XPC-hHR23B complex to irradiated DNA confers strong discrimination for damaged sites. J Mol Biol. 2000;300:275–290. doi: 10.1006/jmbi.2000.3857. [DOI] [PubMed] [Google Scholar]
- 6.Bessho T, Sancar A, Thompson L H, Thelen M P. Reconstitution of human excision nuclease with recombinant XPF-ERCC1 complex. J Biol Chem. 1997;272:3833–3837. doi: 10.1074/jbc.272.6.3833. [DOI] [PubMed] [Google Scholar]
- 7.Chang W H, Kornberg R D. Electron crystal structure of the transcription factor and DNA repair complex, core TFIIH. Cell. 2000;102:609–613. doi: 10.1016/s0092-8674(00)00083-0. [DOI] [PubMed] [Google Scholar]
- 8.Constantinou A, Gunz D, Evans E, Lalle P, Bates P A, Wood R D, Clarkson S G. Conserved residues of human XPG protein important for nuclease activity and function in nucleotide excision repair. J Biol Chem. 1999;274:5637–5648. doi: 10.1074/jbc.274.9.5637. [DOI] [PubMed] [Google Scholar]
- 9.Cooper P K, Nouspikel T, Clarkson S G, Leadon S A. Defective transcription-coupled repair of oxidative base damage in Cockayne Syndrome patients from XP group G. Science. 1997;275:990–993. doi: 10.1126/science.275.5302.990. [DOI] [PubMed] [Google Scholar]
- 10.de Laat W L, Jaspers N G J, Hoeijmakers J H J. Molecular mechanism of nucleotide excision repair. Genes Dev. 1999;13:768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- 11.Drapkin R, Le Roy G, Cho H, Akoulitchev S, Reinberg D. Human cyclin-dependent kinase-activating kinase exists in three distinct complexes. Proc Natl Acad Sci USA. 1996;93:6488–6493. doi: 10.1073/pnas.93.13.6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drapkin R, Reardon J T, Ansari A, Huang J C, Zawel L, Ahn K J, Sancar A, Reinberg D. Dual role of TFIIH in DNA excision-repair and in transcription by RNA-polymerase-II. Nature. 1994;368:769–772. doi: 10.1038/368769a0. [DOI] [PubMed] [Google Scholar]
- 13.Evans E, Fellows J, Coffer A, Wood R D. Open complex formation around a lesion during nucleotide excision repair provides a structure for cleavage by human XPG protein. EMBO J. 1997;16:625–638. doi: 10.1093/emboj/16.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans E, Moggs J G, Hwang J R, Egly J-M, Wood R D. Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. EMBO J. 1997;16:6559–6573. doi: 10.1093/emboj/16.21.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feaver W J, Huang W Y, Gileadi O, Myers L, Gustafsson C M, Kornberg R D, Friedberg E C. Subunit interactions in yeast transcription/repair factor TFIIH—requirement for Tfb3 subunit in nucleotide excision repair. J Biol Chem. 2000;275:5941–5946. doi: 10.1074/jbc.275.8.5941. [DOI] [PubMed] [Google Scholar]
- 16.Guzder S N, Sung P, Prakash L, Prakash S. Nucleotide excision-repair in yeast is mediated by sequential assembly of repair factors and not by a pre-assembled repairosome. J Biol Chem. 1996;271:8903–8910. doi: 10.1074/jbc.271.15.8903. [DOI] [PubMed] [Google Scholar]
- 17.Habraken Y, Sung P, Prakash S, Prakash L. Transcription factor TFIIH and DNA endonuclease Rad2 constitute yeast nucleotide excision-repair factor-3—implications for nucleotide excision-repair and Cockayne-syndrome. Proc Natl Acad Sci USA. 1996;93:10718–10722. doi: 10.1073/pnas.93.20.10718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He Z, Henricksen L A, Wold M S, Ingles C J. RPA involvement in the damage-recognition and incision steps of nucleotide excision repair. Nature. 1995;374:566–569. doi: 10.1038/374566a0. [DOI] [PubMed] [Google Scholar]
- 19.He Z G, Ingles C J. Isolation of human complexes proficient in nucleotide excision repair. Nucleic Acids Res. 1997;25:1136–1141. doi: 10.1093/nar/25.6.1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Houtsmuller A B, Rademakers S, Nigg A L, Hoogstraten D, Hoeijmakers J H J, Vermeulen W. Action of DNA repair endonuclease ERCC1/XPF in living cells. Science. 1999;284:958–961. doi: 10.1126/science.284.5416.958. [DOI] [PubMed] [Google Scholar]
- 21.Iyer N, Reagan M S, Wu K J, Canagarajah B, Friedberg E C. Interactions involving the human RNA-polymerase-II transcription/nucleotide excision-repair complex TFIIH, the nucleotide excision repair protein XPG, and Cockayne syndrome group-B (CSB) protein. Biochemistry. 1996;35:2157–2167. doi: 10.1021/bi9524124. [DOI] [PubMed] [Google Scholar]
- 22.Kenny M K, Schlegel U, Furneaux H, Hurwitz J. The role of human single-stranded DNA binding protein and its individual subunits in simian virus 40 DNA replication. J Biol Chem. 1990;265:7693–7700. [PubMed] [Google Scholar]
- 23.Kimura H, Tao Y, Roeder R G, Cook P R. Quantitation of RNA polymerase II and its transcription factors in a HeLa cell: little soluble holoenzyme but significant amounts of polymerases attached to the nuclear substructure. Mol Cell Biol. 1999;19:5383–5392. doi: 10.1128/mcb.19.8.5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Köberle B, Masters J R W, Hartley J A, Wood R D. Defective repair of cisplatin-induced DNA damage caused by reduced XPA protein in testicular germ cell tumours. Curr Biol. 1999;9:273–276. doi: 10.1016/s0960-9822(99)80118-3. [DOI] [PubMed] [Google Scholar]
- 25.Lee S H, Kim D K, Drissi R. Human xeroderma-pigmentosum group-A protein interacts with human replication protein-A and inhibits DNA-replication. J Biol Chem. 1995;270:21800–21805. doi: 10.1074/jbc.270.37.21800. [DOI] [PubMed] [Google Scholar]
- 26.LePage F, Kwoh E E, Avrutskaya A, Gentil A, Leadon S A, Sarasin A, Cooper P K. Transcription-coupled repair of 8-oxoguanine: requirement for XPG, TFIIH, and CSB and implications for Cockayne syndrome. Cell. 2000;101:159–171. doi: 10.1016/s0092-8674(00)80827-2. [DOI] [PubMed] [Google Scholar]
- 27.Li L, Elledge S J, Peterson C A, Bales E S, Legerski R J. Specific association between the human DNA repair proteins XPA and ERCC1. Proc Natl Acad Sci USA. 1994;91:5012–5016. doi: 10.1073/pnas.91.11.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li L, Lu X Y, Peterson C A, Legerski R J. An interaction between the DNA repair factor XPA and replication protein A appears essential for nucleotide excision repair. Mol Cell Biol. 1995;15:5396–5402. doi: 10.1128/mcb.15.10.5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li L, Peterson C A, Lu X Y, Legerski R J. Mutations in XPA that prevent association with ERCC1 are defective in nucleotide excision repair. Mol Cell Biol. 1995;15:1993–1998. doi: 10.1128/mcb.15.4.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li R Y, Calsou P, Jones C J, Salles B. Interactions of the transcription/DNA repair factor TFIIH and XP repair proteins with DNA lesions in a cell-free repair assay. J Mol Biol. 1998;281:211–218. doi: 10.1006/jmbi.1998.1949. [DOI] [PubMed] [Google Scholar]
- 31.Lindahl T, Wood R D. Quality control by DNA repair. Science. 1999;286:1897–1905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- 32.Maldonado E, Shiekhattar R, Sheldon M, Cho H, Drapkin R, Rickert P, Lees E, Anderson C W, Linn S, Reinberg D. A human RNA-polymerase-II complex-associated with srb and DNA-repair proteins. Nature. 1996;381:86–89. doi: 10.1038/381086a0. [DOI] [PubMed] [Google Scholar]
- 33.Manley J L, Fire A, Samuels M, Sharp P A. In vitro transcription: whole cell extract. Methods Enzymol. 1983;101:568–582. doi: 10.1016/0076-6879(83)01038-1. [DOI] [PubMed] [Google Scholar]
- 34.Marinoni J C, Rossignol M E, Egly J M. Purification of the transcription/repair factor TFIIH and evaluation of its associated activities in vitro. Methods. 1997;12:235–253. doi: 10.1006/meth.1997.0476. [DOI] [PubMed] [Google Scholar]
- 35.Matsuda T, Saijo M, Kuraoka I, Kobayashi T, Nakatsu Y, Nagai A, Enjoji T, Masutani C, Sugasawa K, Hanaoka F, Yasui A, Tanaka K. DNA-repair protein XPA binds replication protein-A (RPA) J Biol Chem. 1995;270:4152–4157. doi: 10.1074/jbc.270.8.4152. [DOI] [PubMed] [Google Scholar]
- 36.Matsunaga T, Mu D, Park C H, Reardon J T, Sancar A. Human DNA-repair excision nuclease—analysis of the roles of the subunits involved in dual incisions by using anti-XPG and anti-ERCC1 antibodies. J Biol Chem. 1995;270:20862–20869. doi: 10.1074/jbc.270.35.20862. [DOI] [PubMed] [Google Scholar]
- 37.Miura N, Miyamoto I, Asahina H, Satokata I, Tanaka K, Okada Y. Identification and characterization of XPAC protein, the gene product of the human XPAC (xeroderma pigmentosum group A complementing) gene. J Biol Chem. 1991;266:19786–19789. [PubMed] [Google Scholar]
- 38.Mu D, Hsu D S, Sancar A. Reaction-mechanism of human DNA-repair excision nuclease. J Biol Chem. 1996;271:8285–8294. doi: 10.1074/jbc.271.14.8285. [DOI] [PubMed] [Google Scholar]
- 39.Mu D, Park C H, Matsunaga T, Hsu D S, Reardon J T, Sancar A. Reconstitution of human DNA-repair excision nuclease in a highly defined system. J Biol Chem. 1995;270:2415–2418. doi: 10.1074/jbc.270.6.2415. [DOI] [PubMed] [Google Scholar]
- 40.Mu D, Wakasugi M, Hsu D S, Sancar A. Characterization of reaction intermediates of human excision-repair nuclease. J Biol Chem. 1997;272:28971–28979. doi: 10.1074/jbc.272.46.28971. [DOI] [PubMed] [Google Scholar]
- 41.Nocentini S, Coin F, Saijo M, Tanaka K, Egly J M. DNA-damage recognition by XPA protein promotes efficient recruitment of transcription factor IIH. J Biol Chem. 1997;272:22991–22994. doi: 10.1074/jbc.272.37.22991. [DOI] [PubMed] [Google Scholar]
- 42.Nouspikel T, Lalle P, Leadon S A, Cooper P K, Clarkson S G. A common mutational pattern in xeroderma pigmentosum group G/Cockayne syndrome patients: implications for a second XPG function. Proc Natl Acad Sci USA. 1997;94:3116–3121. doi: 10.1073/pnas.94.7.3116. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.O'Donovan A, Davies A A, Moggs J G, West S C, Wood R D. XPG endonuclease makes the 3′ incision in human DNA nucleotide excision repair. Nature. 1994;371:432–435. doi: 10.1038/371432a0. [DOI] [PubMed] [Google Scholar]
- 44.Ossipow V, Tassan J P, Nigg E A, Schibler U. A mammalian RNA polymerase II holoenzyme containing all components required for promoter-specific transcription initiation. Cell. 1995;83:137–146. doi: 10.1016/0092-8674(95)90242-2. [DOI] [PubMed] [Google Scholar]
- 45.Otrin V, Kuraoka I, Nardo T, McLenigan M, Eker A, Stefanini M, Levine A, Wood R. Relationship of the xeroderma pigmentosum group E DNA repair defect to the chromatin and DNA binding proteins UV-DDB and replication protein A. Mol Cell Biol. 1998;18:3182–3190. doi: 10.1128/mcb.18.6.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park C H, Mu D, Reardon J T, Sancar A. The general transcription-repair factor TFIIH is recruited to the excision-repair complex by the XPA protein independent of the TFIIE transcription factor. J Biol Chem. 1995;270:4896–4902. doi: 10.1074/jbc.270.9.4896. [DOI] [PubMed] [Google Scholar]
- 47.Park C H, Sancar A. Formation of a ternary complex by human XPA, ERCC1, and ERCC4(XPF) excision-repair proteins. Proc Natl Acad Sci USA. 1994;91:5017–5021. doi: 10.1073/pnas.91.11.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reardon J T, Ge H, Gibbs E, Sancar A, Hurwitz J, Pan Z Q. Isolation and characterization of 2 human transcription factor IIH (TFIIH)-related complexes—ERCC2/CAK and TFIIH. Proc Natl Acad Sci USA. 1996;93:6482–6487. doi: 10.1073/pnas.93.13.6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rodriguez K, Talamantez J, Huang W, Reed S H, Wang Z, Chen L, Feaver W J, Friedberg E C, Tomkinson A E. Affinity purification and partial characterization of a yeast multiprotein complex for nucleotide excision repair using histidine-tagged Rad14 protein. J Biol Chem. 1998;273:34180–34189. doi: 10.1074/jbc.273.51.34180. [DOI] [PubMed] [Google Scholar]
- 50.Rossignol M, Kolb-Cheynel I, Egly J M. Substrate specificity of the cdk-activating kinase (CAK) is altered upon association with TFIIH. EMBO J. 1997;16:1628–1637. doi: 10.1093/emboj/16.7.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saijo M, Kuraoka I, Masutani C, Hanaoka F, Tanaka K. Sequential binding of DNA-repair proteins RPA and ERCC1 to XPA in vitro. Nucleic Acids Res. 1996;24:4719–4724. doi: 10.1093/nar/24.23.4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Satokata I, Tanaka K, Miura N, Miyamoto I, Satoh Y, Kondo S, Okada Y. Characterization of a splicing mutation in group-A xeroderma pigmentosum. Proc Natl Acad Sci USA. 1990;87:9908–9912. doi: 10.1073/pnas.87.24.9908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schultz P, Fribourg S, Poterszman A, Mallouh V, Moras D, Egly J M. Molecular structure of human TFIIH. Cell. 2000;102:599–607. doi: 10.1016/s0092-8674(00)00082-9. [DOI] [PubMed] [Google Scholar]
- 54.Schweizer U, Hey T, Lipps G, Krauss G. Photocrosslinking locates a binding site for the large subunit of human replication protein A to the damaged strand of cisplatin-modified DNA. Nucleic Acids Res. 1999;27:3183–3189. doi: 10.1093/nar/27.15.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shivji M K K, Kenny M K, Wood R D. Proliferating cell nuclear antigen is required for DNA excision repair. Cell. 1992;69:367–374. doi: 10.1016/0092-8674(92)90416-a. [DOI] [PubMed] [Google Scholar]
- 56.Shivji M K K, Moggs J G, Kuraoka I, Wood R D. Dual incision assays for nucleotide excision repair using DNA with a lesion at a specific site. In: Henderson D S, editor. DNA repair protocols: eukaryotic systems. Vol. 113. Totowa, N.J: Humana Press; 1999. pp. 373–392. [DOI] [PubMed] [Google Scholar]
- 57.Sijbers A M, de Laat W L, Ariza R R, Biggerstaff M, Wei Y-F, Moggs J G, Carter K C, Shell B K, Evans E, de Jong M C, Rademakers S, de Rooij J, Jaspers N G J, Hoeijmakers J H J, Wood R D. Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell. 1996;86:811–822. doi: 10.1016/s0092-8674(00)80155-5. [DOI] [PubMed] [Google Scholar]
- 58.Stigger E, Drissi R, Lee S. Functional-analysis of human replication protein-A in nucleotide excision-repair. J Biol Chem. 1998;273:9337–9343. doi: 10.1074/jbc.273.15.9337. [DOI] [PubMed] [Google Scholar]
- 59.Sugasawa K, Ng J M Y, Masutani C, S. I, van der Spek P J, Eker A P M, Hanaoka F, Bootsma D, Hoeijmakers J H J. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol Cell. 1998;2:223–232. doi: 10.1016/s1097-2765(00)80132-x. [DOI] [PubMed] [Google Scholar]
- 60.Svejstrup J, Vichi P, Egly J-M. The multiple roles of transcription/repair factor TFIIH. Trends Biochem Sci. 1996;21:346–350. [PubMed] [Google Scholar]
- 61.Svejstrup J Q, Wang Z, Feaver W J, Wu X, Bushnell D A, Donahue T F, Friedberg E C, Kornberg R D. Different forms of TFIIH for transcription and DNA repair: holo-TFIIH and a nucleotide excision repairosome. Cell. 1995;80:21–28. doi: 10.1016/0092-8674(95)90447-6. [DOI] [PubMed] [Google Scholar]
- 62.van der Spek P J, Eker A, Rademakers S, Visser C, Sugasawa K, Masutani C, Hanaoka F, Bootsma D, Hoeijmakers J H J. XPC and human homologs of Rad23—intracellular localization and relationship to other nucleotide excision repair complexes. Nucleic Acids Res. 1996;24:2551–2559. doi: 10.1093/nar/24.13.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Y, Cortez D, Yazdi P, Neff N, Elledge S J, Qin J. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000;14:927–939. [PMC free article] [PubMed] [Google Scholar]
- 64.Winkler G S, Vermeulen W, Coin F, Egly J M, Hoeijmakers J H J, Weeda G. Affinity purification of human DNA-repair transcription factor TFIIH using epitope-tagged xeroderma-pigmentosum B-protein. J Biol Chem. 1998;273:1092–1098. doi: 10.1074/jbc.273.2.1092. [DOI] [PubMed] [Google Scholar]
- 65.Wold M S. Replication protein A: a heterotrimeric single-stranded DNA binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 66.Wood R D, Biggerstaff M, Shivji M K K. Detection and measurement of nucleotide excision repair synthesis by mammalian cell extracts in vitro. Methods. 1995;7:163–175. [Google Scholar]
- 67.Yankulov K Y, Bentley D L. Regulation of CDK7 substrate specificity by MAT1 and TFIIH. EMBO J. 1997;16:1638–1646. doi: 10.1093/emboj/16.7.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yokoi M, Masutani C, Maekawa T, Sugasawa K, Ohkuma Y, Hanaoka F. The xeroderma pigmentosum group C protein complex XPC-HR23B plays an important role in the recruitment of transcription factor IIH to damaged DNA. J Biol Chem. 2000;275:9870–9875. doi: 10.1074/jbc.275.13.9870. [DOI] [PubMed] [Google Scholar]