Promotion of Cell Cycle Progression by Basic Helix-Loop-Helix E2A (original) (raw)
Abstract
Normal B-cell development requires the E2A gene and its encoded transcription factors E12 and E47. Current models predict that E2A promotes cell differentiation and inhibits G1 cell cycle progression. The latter raises the conundrum of how B cells proliferate while expressing high levels of E2A protein. To study the relationship between E2A and cell proliferation, we established a tissue culture-based model in which the activity of E2A can be modulated in an inducible manner using E47R, an E47-estrogen fusion construct, and E47ERT, a dominant negative E47-estrogen fusion construct. The two constructs were subcloned into retroviral vectors and expressed in the human pre-B-cell line 697, the human myeloid progenitor cell line K562, and the murine fibroblastic cell line NIH 3T3. In both B cells and non-B cells, suppression of E2A activity by E47ERT inhibited G1 progression and was associated with decreased expression of multiple cyclins including the G1-phase cyclin D2 and cyclin D3. Consistent with these findings, E2A null mice expressed decreased levels of cyclin D2 and cyclin D3 transcripts. In complementary experiments, ectopic expression of E47R promoted G1 progression and was associated with increased levels of multiple cyclins, including cyclin D2 and cyclin D3. The induction of some cyclin transcripts occurred even in the absence of protein synthesis. We conclude that, in some cells, E2A can promote cell cycle progression, contrary to the present view that E2A inhibits G1 progression.
The E2A gene and its alternatively spliced products E47 and E12 belong to the family of transcription factors characterized by a helix-loop-helix (HLH) dimerization motif (26). Structurally, E2A contains a C-terminal HLH motif with an adjacent basic (b) motif that is essential for DNA binding (40). E2A can homodimerize or heterodimerize with other transcription factors that also contain the HLH motif. The C terminus of E2A also contains a domain C, which contributes to dimerization in vivo (25). As a dimer, the bHLH motif recognizes the canonical sequence CANNTG, designated an E box. E47 but not E12 binds DNA well as a homodimer because E12 contains a different basic region as a result of alternative splicing (61). The bHLH motif also interacts directly with p300, a transcription cofactor that has histone acetyltransferase activity and can alter chromatin structure (18). The central portion of E2A contains the transcription activation domain AD2 with a loop-helix motif that is sufficient to transactivate plasmid reporters containing E boxes (2, 52). The N-terminal region encodes the transcription activation domain AD1, which contains an LDFS motif. This motif recruits the SAGA complex that is thought to remodel chromatin structure (35).
Expression of E2A is essential to normal B-cell development and regulates cell proliferation of some non-B cells. B cells, even at the earliest detectable developmental stage, do not develop in transgenic mice that lack E2A (4, 71). Mice that lack E2A have decreased numbers of normal precursor T cells and an increased incidence of T-cell lymphomas (3, 69). Ectopic expression of E2A in T-cell lymphomas/leukemias induces apoptosis, which is consistent with E2A-mediated suppression of abnormal T-cell proliferation (19, 45). In NIH 3T3 fibroblasts, ectopic expression of E2A appears to cause G1 arrest (47), supporting the present view that E2A suppresses cell proliferation.
Despite the importance of E2A in regulation of cell proliferation and in normal B-cell development, the identities of many of the important downstream target genes remain elusive. Candidate target genes have been identified, but these do not explain the complete absence of B cells in E2A null mice. The known target genes include immunoglobulin heavy chain (IgH), RAG-1, surrogate light chain lambda 5, terminal deoxynucleotidyltransferase (TdT), cyclin-dependent kinase (cdk) inhibitor p21, and transcription factor EBF (15, 28, 49, 55, 59). IgH, RAG-1, and lambda 5 are necessary to form the pre-B-cell receptor that is important for the proliferation and differentiation of pre-B cells (20). Mice that lack any of these genes have a defect in the pro-B-to-pre-B transition, resulting in a severe decrease to absence of pre-B cells with an increased number of pro-B cells (29, 30, 36). TdT null mice have normal numbers of B cells (24, 32). p21 null mice are viable with no reported defects in B-cell development (17). EBF null mice do not have any B cells (34), and thus, this may explain the E2A null phenotype. However, the known targets of EBF are also components of the pre-B-cell receptor (59). Furthermore, mice heterozygous for both E2A and EBF have decreased numbers of pro-B cells compared to those of mice heterozygous for either E2A or EBF, a finding that suggests that both transcription factors contribute independently to B-cell lymphopoiesis (42). Hence, additional target genes of E2A need to be identified in order to explain the complete absence of B cells in E2A null mice.
Of the known E2A target genes, only p21 directly regulates cell proliferation. Ectopic expression of E2A in the embryonic kidney cell line 293T increased the expression of p21 and may explain how E2A inhibits G1 progression of fibroblasts (49). However, the same group reported that ectopic expression of E2A in T-cell lymphoma/leukemia did not induce p21 (45), suggesting that E2A regulates p21 expression in a cell-type-specific manner. The relationship between E2A and p21 in B cells remains to be defined. Even the relationship between E2A and B-cell proliferation remains to be elucidated. If E2A does inhibit cell proliferation, then there is a paradox in that E2A is highly expressed in the most mitotically active B cells (54).
To better characterize the role of E2A in cell proliferation, we established an experimental system in which E2A activity could be increased or suppressed in an inducible manner. Surprisingly, suppression of E2A decreased cell proliferation while induction of E2A promoted cell proliferation of serum-deprived B cells and non-B cells. Consistent with these findings, suppression of E2A decreased the expression of multiple cyclins while induction of E2A induced a subset of these cyclins including cyclins D2 and D3. We conclude that E2A can promote cell cycle progression.
MATERIALS AND METHODS
Plasmids.
The plasmids MERT (63), MIGR1 (46), pMD-VSV-G and delta 8.2 (41), SV/E2-5 (15), and (523)4 luciferase reporter (12) were generously provided by Kathy Westin, CRC Centre for Cell and Molecular Biology, London, United Kingdom; Warren Pear, the University of Pennsylvania; Indera Verma, the Salk Institute; and Tom Kadesch, the University of Pennsylvania. The lentiviral vector hybrid human immunodeficiency virus (HIV)-murine stem cell virus (MSCV) has been previously reported (14). E47ERT was engineered by ligating the β-globin 5′ untranslated region, the bHLH domain of E47, and the ERT fragment of MERT. The β-globin 5′ untranslated region was isolated from the SV/E2-5 plasmid using _Bgl_II-_Nco_I digestion. The bHLH domain of E47 was amplified by PCR using the primers 5′CAT GAC ATG CCA TGG CGG CCG CCA GCG AGA TCA AG3′ and 5′ACA TCA CAT GCA TGC ATG TGC CCG GCG GGG TTG TG3′, and the product was digested with _Nco_I and _Sph_I restriction enzymes. The ERT fragment was isolated from MERT using _Sph_I-_Eco_RI digestion. The resulting construct was subcloned into the _Bgl_II-_Eco_RI site of the MIGR1 vector. E47R was engineered by ligating the E47 coding region of SV/E2-5 in frame with the estrogen receptor fragment of MERT, and the product was subcloned into the _Bgl_II site of the MIGR1 vector or into the _Bam_HI site of the hybrid HIV-MSCV vector. Thymidine kinase-Renilla luciferase plasmid was purchased from Promega (Madison, Wis.).
Tissue culture.
K562 cells were purchased from the American Type Culture Collection. 697, 293T, NIH 3T3, GP, and E2A wild-type and null murine embryonic fibroblasts (MEFs) were generously provided by Leslie Silberstein, Harvard University; Warren Pear, Tom Kadesch, and Garry Nolan, Stanford University Medical Center; and Cornelius Murre, University of California, San Diego. K562 and 697 cells were cultured in RPMI medium supplemented with 10% fetal bovine serum in 5% CO2. The remaining cells were cultured in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum. Tamoxifen and cycloheximide (Sigma, St. Louis, Mo.) were used at 1 μM and 0.1 mM concentrations, respectively.
Mouse tissue.
Liver tissues from day 18-19 E2A null and E2A heterozygous fetuses were generously provided by Yuan Zhuang, Duke University. Genomic DNA was isolated using the QIAamp DNA Mini kit (Qiagen Inc., Valencia, Calif.) following the manufacturer's protocol. The DNA was genotyped as previously described (69).
Luciferase assay.
293T cells were transiently transfected using calcium phosphate precipitate as previously described (15). The cells were harvested and analyzed using the dual luciferase assay kit from Promega following the manufacturer's protocol.
Generation of virus and transduction.
Retroviruses were generated using previously described methods (46). Briefly, MIGR1 and a vector that encodes the viral envelope protein VSV-G were transiently cotransfected by calcium precipitate into the packaging cell line GP. To generate lentiviruses, hybrid HIV-MSCV vector, expression plasmid for VSV-G, and helper plasmid delta 8.2 were transiently cotransfected by calcium precipitate into 293T cells as previously described (14). The culture media were collected 48 and 72 h posttransfection and stored in small aliquots at −80°C. Two million cells were transduced by spinoculation following published protocols (33). Cells, 1 ml of virus supernatant, and 8 μg of Polybrene (Sigma)/ml were centrifuged in a 24-well plate at 1,500 × g for 90 min at room temperature. Afterwards, the cells were washed once with 10 volumes of serum-free medium and then cultured.
FACS analysis.
Fluorescence-activated cell sorting (FACS) analysis was performed on a FACSTAR cell sorter, and data were analyzed using Cell-Quest software (Becton Dickinson, San Jose, Calif.) or Modfit LT software (Verity Software House, Topsham, Maine). Phycoerythrin-conjugated annexin V, fluorescein isothiocyanate-conjugated antibromodeoxyuridine (anti-BrdU; Pharmingen, San Diego, Calif.), and unconjugated anti-TdT (SuperTechs, Bethesda, Md.) antibodies were used following the manufacturers' protocols. Unconjugated antibody to c-myc epitope (9E10) was purchased from the Cell Center, University of Pennsylvania, and used following protocols for antibodies to TdT. Cell cycle analysis was performed using a propidium iodide (PI) staining kit (Sigma) following the manufacturer's protocol.
RNA analysis.
Total RNA was isolated using Ultraspec (BioTecx, Houston, Tex.) following the manufacturer's protocol. Reverse transcription-PCR (RT-PCR) for IgH, TdT, CD79b (B29), Rag-1, and β-actin was performed as previously described (15, 23). RNase protection assay (RPA) was performed as previously described (15). The DNA templates hCYC-1, hCC-1, and mCYC-1 were purchased from PharMingen and used to generate radioactive riboprobes.
Western blot analysis.
Western blotting was performed as previously described (57) using antibodies to E47 (N-649), β-actin (I-19), cyclin D2 (M-20), cyclin D3 (D-7), CDK4 (C-22), and CDK6 (C-21) (Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.). Briefly, cells were lysed in 10 mM HEPES–1 mM EDTA–60 mM KCl–0.5% NP-40–protease inhibitor cocktail (Sigma). Protein was quantified using the DC protein assay kit (Bio-Rad, Hercules, Calif.). Twenty micrograms of protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto nitrocellulose by electroblotting.
RESULTS
E47ERT suppresses E2A activity in an inducible manner.
An inducible inhibitor of E2A was engineered by modifying a preexisting inducible system that suppresses c-myb, a transcription factor important for early myeloid and peripheral T-cell proliferation (22, 37). This system utilizes MERT (myb-engrailed estrogen receptor-myc tag), an artificial chimeric transcription factor composed of the DNA binding domain of c-myb, the domain of the engrailed protein that represses transcription, and the domain of a mutated estrogen receptor that binds tamoxifen. When induced by tamoxifen, MERT binds to the DNA binding sites for c-myb and represses the transcription of c-myb-regulated genes (56, 63). To inhibit E2A, we engineered E47ERT, an artificial chimeric construct produced by replacing the c-myb DNA binding domain of MERT with the bHLH domain of E47, a major splice form of E2A (Fig. 1A). We reasoned that E47ERT would dimerize, bind E boxes, and suppress transcription only when induced with tamoxifen. Even as a heterodimer with native E47, E47ERT would suppress transcription because the engrailed repressor domain is dominant (63).
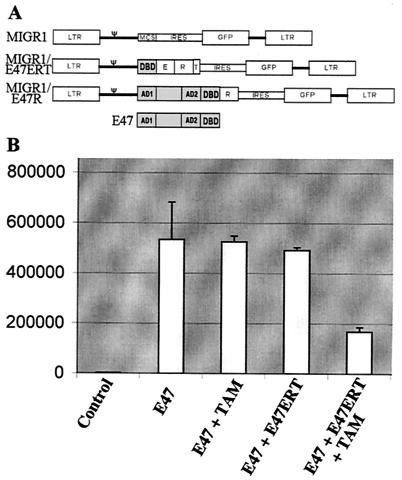
FIG. 1.
(A) Diagram of construct. LTR, long terminal repeats; ψ, viral packaging signal; MCS, multicloning site; IRES, internal ribosomal entry site; GFP, enhanced GFP; E, transcription repressor domain of engrailed protein; R, hormone-binding domain of estrogen receptor; T, myc tag; AD1, activation domain 1 of E47; AD2, activation domain 2 of E47; DBD, DNA binding domain of E47. Shaded regions represent E47 sequences. (B) Inducible inhibition of E2A activity by E47ERT. A luciferase reporter assay was performed on 293T cells that were transiently transfected and analyzed 3 days later. Control, E2A luciferase reporter plasmid that contains multiple E boxes in its enhancer; TAM, tamoxifen. Numbers on the y axis indicate luciferase activity.
To test the function of E47ERT, transient-transfection assays were performed using plasmids encoding E47 and E47ERT as well as the (523)4 reporter plasmid that contains multiple E boxes upstream of the luciferase gene (Fig. 1B). The (523)4 luciferase reporter is specific for active homodimers of E47 that are restricted to B cells or to non-B cells expressing ectopic E47 (53, 57, 66). In the presence of tamoxifen, E47ERT suppressed E47 activity by 50 to 75%; in contrast, E47ERT had no effect on E47 activity in the absence of tamoxifen. Induced E47ERT had no effect on the activity of other enhancers-promoters such as those of thymidine kinase, cytomegalovirus, and multimerized c-myc E boxes (data not shown), suggesting that the suppression by E47ERT is limited to E boxes normally bound by E47 and not due to nonspecific inhibition of transcription. These findings support the premise that E47ERT is an inducible inhibitor of E47 activity.
E47ERT suppresses known target genes.
To demonstrate that E47ERT can inhibit known endogenous target genes of E47, E47ERT was expressed in the pre-B-cell line 697. This cell line was chosen because it expresses many of the known target genes of E2A. E47ERT was subcloned into an MSCV-based retroviral vector that produces a bicistronic message that generates E47ERT and green fluorescent protein (GFP). The latter is cotranslated via an internal ribosomal entry site; hence, the level of E47ERT is linked to GFP, which can be easily quantified by FACS. Retrovirus stocks were generated and used to transduce 697 cells. The transduced cells, designated 697/E47ERT, were purified by FACS for GFP+ cells. Coexpression of GFP and E47ERT was verified by intracellular immunostaining of the cells using antibodies against the c-myc epitope of E47ERT (Fig. 2A).
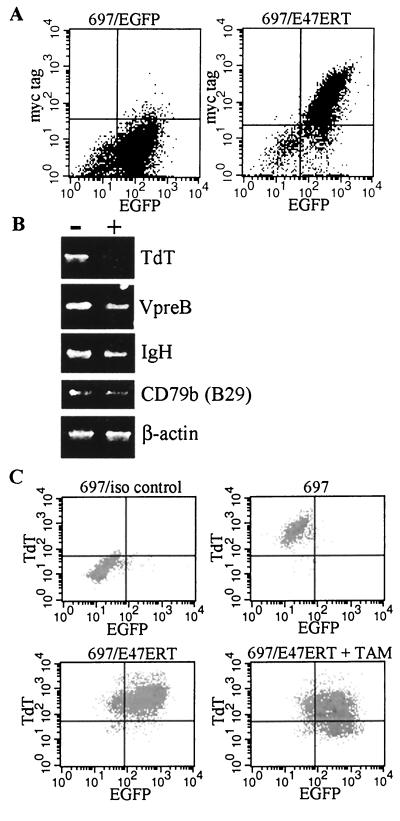
FIG. 2.
Inhibition of known target genes of E2A by E47ERT. (A) Two-color flow cytometry analysis for enhanced GFP (EGFP) and E47ERT expression. 697 stable lines that express enhanced GFP (697/EGFP) or both enhanced GFP and E47ERT (MIGR1/E47ERT) were stained for intracellular myc tag. (B) RT-PCR. 697/E47ERT cells were analyzed after treatment with 0.1% ethanol (−) or with tamoxifen (+) for 3 days. (C) Two-color flow cytometry for enhanced GFP and TdT expression. 697/E47ERT cells were analyzed after treatment with ethanol or tamoxifen (TAM) for 3 days.
697/E47ERT cells were treated with tamoxifen, total RNA was isolated, and specific messages were assessed by semiquantitative RT-PCR (Fig. 2B). The number of cycles was empirically determined to be in the linear range. RT-PCR results demonstrated that the message levels of known E2A target genes TdT, Vpre-B, and IgH were decreased. In contrast, β-actin and CD79b (B29) levels were not. We previously demonstrated that the expression of TdT was closely related to the activity of E2A (15). In addition, TdT protein has a relatively short half-life of approximately 4 to 8 h (7), making TdT an ideal candidate to measure the effects of E47ERT at the protein level. Induced E47ERT decreased anti-TdT antibody binding by 60% (Fig. 2C), a percentage similar to the decrease seen with the E2A luciferase reporter plasmid. These findings support the premise that E47ERT inhibits the activity of endogenous E47 in an inducible manner.
E47ERT decreases proliferation of B and non-B cells.
Current models of E2A function would predict that suppressing E2A would stimulate G1 progression and lead to an increase in cell proliferation; surprisingly, we found the opposite. As shown in Fig. 3A, 697/E47ERT cells treated with tamoxifen grew more slowly than 697/E47ERT cells without tamoxifen. Tamoxifen had no significant effect on the proliferation of the parental 697 cells.
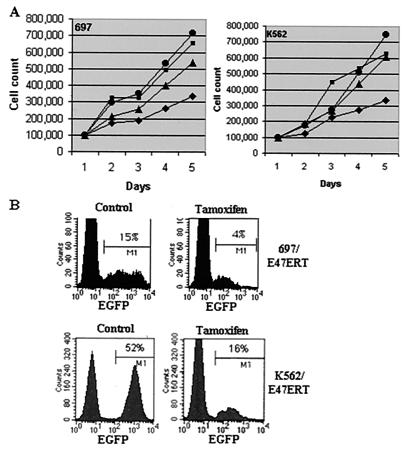
FIG. 3.
Inhibition of cell proliferation by E47ERT. (A) Growth curve. Parental 697 or K562 cells without tamoxifen (●) or with tamoxifen (▪) and stable lines expressing E47ERT without tamoxifen (▴) or with tamoxifen (⧫) were seeded at 100,000 cells/ml, and viable cells were counted daily for 5 days. The figure is representative of three independent experiments. (B) Single-color flow cytometry analysis for enhanced GFP+ cells. Parental enhanced GFP− cells were mixed with enhanced GFP+ cells expressing E47ERT, and the mixed cell population was treated with ethanol (control) or tamoxifen for 10 days.
To determine if the effects of E47ERT are specific to B cells, we introduced E47ERT into the immature myeloid cell line K562 and the murine fibroblast cell line NIH 3T3. E47ERT also suppressed the proliferation of K562 cells (Fig. 3A) and NIH 3T3 cells (data not shown). These findings suggest that E2A promotes cell proliferation in B and non-B cells.
To confirm this unexpected finding, we performed mixing experiments (Fig. 3B). Parental GFP− cells were mixed with E47ERT GFP+ cells and allowed to grow in a common flask in the presence or absence of tamoxifen. In both 697/E47ERT and K562/E47ERT cells, the percentages of GFP+ cells decreased in the presence of tamoxifen but not in its absence, confirming that suppressing E2A decreases cell proliferation. Mixing experiments with parent cells and cells expressing only GFP (697/GFP and K562/GFP) demonstrated that the percentage of GFP+ cells remained constant with or without tamoxifen (data not shown).
E47ERT inhibits cell cycle progression.
Cell proliferation is determined in part by the relative balance between cell cycle progression and apoptosis. To assess the relative contributions of these two processes in our system, 697/E47ERT and K562/E47ERT cells were analyzed by FACS. The 697/E47ERT cells were stained with PI and analyzed by FACS for cell cycle profile. Suppression of E2A activity increased the G0/G1 fraction and decreased the S fraction, indicating an inhibition of G1 cell cycle progression (Fig. 4B). The altered cell cycle profile was seen even with only 12 h of tamoxifen exposure (data not shown).
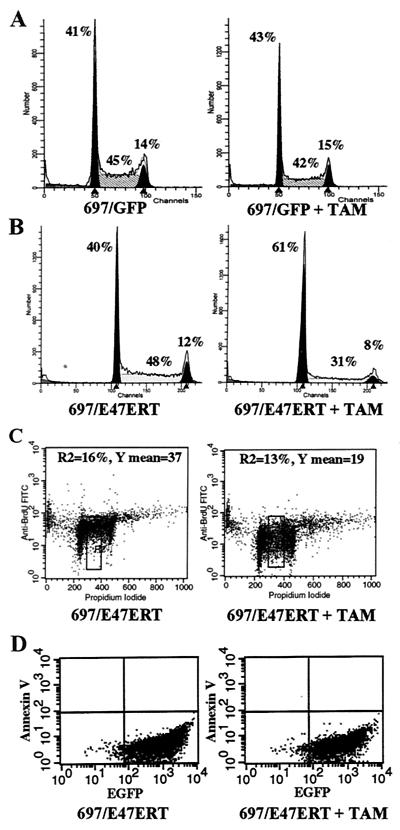
FIG. 4.
Effect of E47ERT on cell cycle progression and apoptosis. 697/GFP and 697/E47ERT cells were analyzed after treatment with ethanol or tamoxifen (TAM) for 1 day. (A) Cell cycle compartment analysis of 697/GFP cells by PI staining. (B) Cell cycle compartment analysis of 697/E47ERT cells by PI staining. (C) Two-color flow cytometry for BrdU incorporation and cell cycle compartmentation. FITC, fluorescein isothiocyanate. (D) Two-color flow cytometry analysis for GFP and annexin V binding.
No change in cell cycle profile was seen upon tamoxifen treatment of 697/GFP cells (Fig. 4A) and parental 697 cells (data not shown). Similar findings were seen with K562 cells (data not shown).
The decrease in cell cycle progression was also confirmed by BrdU incorporation studies (Fig. 4C). 697/E47ERT cells were exposed to BrdU for 1 h and then immunostained with anti-BrdU antibodies and costained with PI for cell cycle profile fractionation. As expected, the number of BrdU-positive cells decreased upon suppression of E2A, consistent with the decreased S-phase fraction. In addition, the average intensity of the BrdU staining of the cells in S phase decreased, suggesting that the rate of BrdU incorporation in S phase also decreased.
Apoptosis was measured by annexin V binding, an early apoptotic event. No change in annexin V binding was seen when E2A activity was suppressed (Fig. 4D). Similarly, no change in annexin V binding was seen in 697/GFP cells treated with tamoxifen (data not shown). In contrast, our positive control (serum starvation of the T-cell line Jurkat) demonstrated the expected increase in annexin V binding (data not shown).
E47ERT decreases the expression of cyclins.
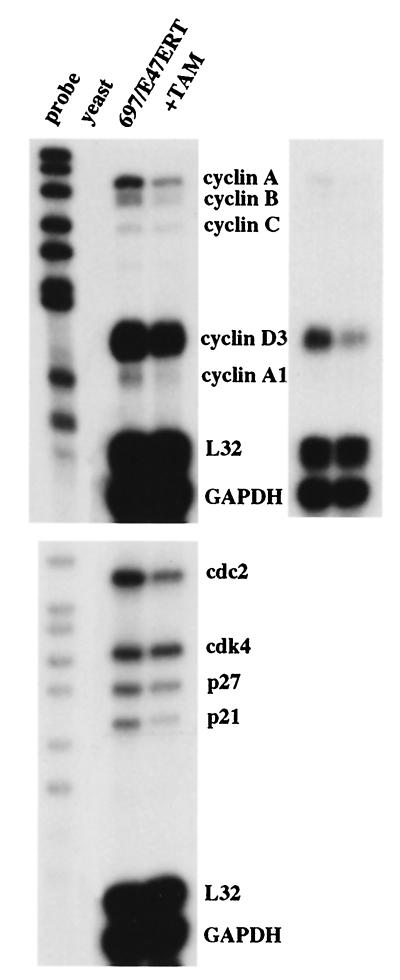
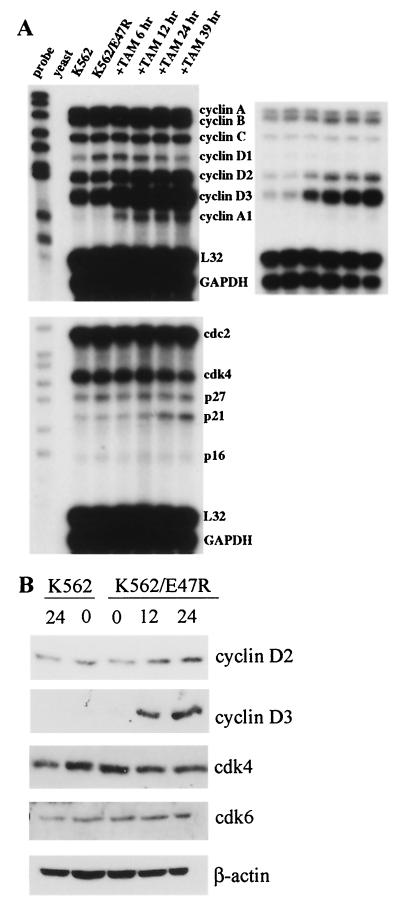
To confirm the effect of E2A on cell cycle progression, we examined the expression of multiple cell cycle regulatory genes. These include cyclins A, B, and D; the cdk's cdk4 and cdc2; and the cdk inhibitors p21, p27, and p16 (reviewed in reference 58). 697/E47ERT cells were exposed to tamoxifen for 12 to 16 h, and total RNA was analyzed by multiplex RPAs (Fig. 5). Suppression of E2A decreased the expression of cyclin A, cyclin A1, and cyclin B. These findings are consistent with inhibition of G1 progression, since the expression levels of these transcripts are low in G1 phase, increase throughout the S phase, and are at a maximum at mitosis. Suppression of E2A decreased the message levels of cyclin D3, cdc2, p27, and p21. The expression of these transcripts typically remains constant throughout the cell cycle, suggesting that E2A may regulate these transcripts independently of changes in cell cycle profile. No significant changes were seen in the mRNAs corresponding to cdk4 and the housekeeping genes L32 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Similar findings were seen in K562/E47ERT cells (data not shown). In contrast, no significant changes in transcript levels were seen in 697 cells that expressed only GFP (see Fig. 8A).
FIG. 5.
Decreased expression of multiple cell cycle regulatory genes by E47ERT. The figure shows results from an RPA. 697/E47ERT cells were analyzed after treatment with ethanol or tamoxifen (TAM) for 16 h. The upper right panel represents a shorter exposure of the upper left autoradiogram.
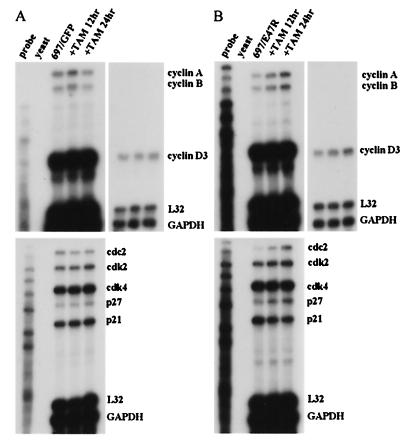
FIG. 8.
RPA. 697 cells that express GFP (697/GFP) (A) and 697 cells that express E47R (697/E47R) (B) were analyzed after treatment with tamoxifen (TAM) for 0, 12, and 24 h. The upper right autoradiograms represent shorter exposures of the upper left autoradiograms.
Decreased expression of cyclin D2 and cyclin D3 in E2A null mice.
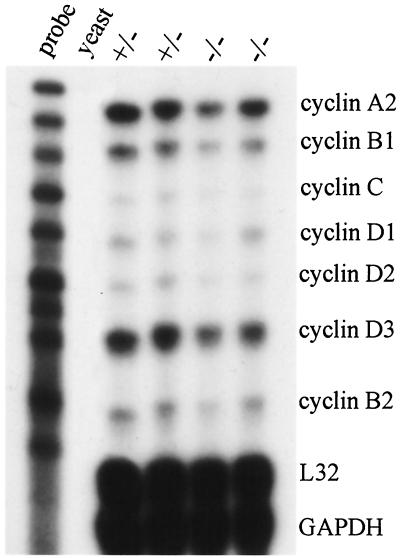
To investigate the relationship between E2A and cyclin D in primary cells, we analyzed the expression levels of cyclin D in E2A null mice. We analyzed the fetal livers of E18.5-stage embryos because the liver is the predominant organ for hematopoiesis at this stage, E2A null mice have apparently normal myeloid hematopoiesis (71), E2A is highly expressed in early myeloid progenitors (51, 68), and cyclin D2 and cyclin D3 are the major D cyclins expressed in hematopoietic progenitor cells (5, 62, 64). Since B cells represent only 1 to 2% of the fetal liver cells (71), the absence of B cells in the E2A null mice is unlikely to significantly alter levels of cyclin D2 and cyclin D3. The genotypes of nine fetuses were determined by PCR (data not shown). The fetal livers from E2A+/− (n = 5) and E2A−/− (n = 3) embryos were analyzed by RPA (Fig. 6). Only one E2A+/+ fetus was identified and was not further analyzed. The expression levels of cyclin D1, cyclin D2, and cyclin D3 for E2A+/− and E2A−/− embryos were quantified by phosphorimaging, normalized to the housekeeping transcript L32, and analyzed by an unpaired Student's t test. Expression of cyclins D2 and D3 decreased significantly by 27 and 18% with P < 0.018 and P < 0.009, respectively. In contrast, expression of cyclin D1 decreased by 10% with P < 0.21.
FIG. 6.
Decreased expression of multiple cell cycle regulatory genes in E2A null mice. Total RNA was isolated from livers of E2A heterozygous (+/−) and E2A null (−/−) fetuses and analyzed by RPA.
E47R, an E47-estrogen receptor fusion protein, increases cyclin levels.
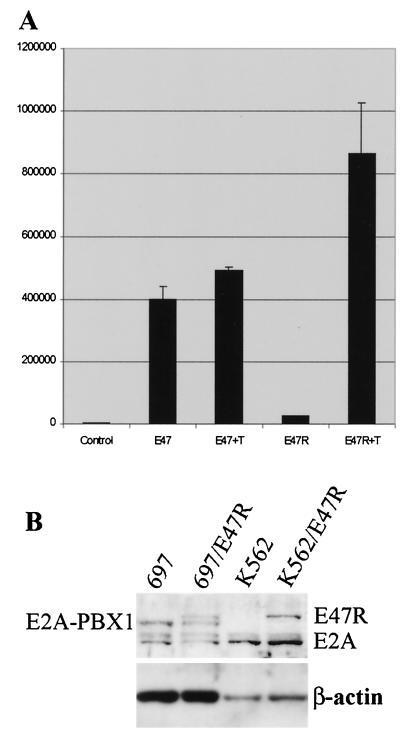
To confirm the unexpected findings resulting from suppression of E2A, we performed complementary studies in which E47 activity was induced. Constitutively active E47 appears to be toxic to cells, and stable lines overexpressing E47 have not been reported (28; unpublished data). To bypass the potential toxicity and generate a homogeneous cell population that can overexpress E47, an inducible E2A was engineered by ligating the full-length coding region of E47 in frame to the hormone-binding domain of the estrogen receptor. This construct, designated E47R, contains a point mutation in the hormone-binding region that is preferentially activated by the estrogen derivative tamoxifen. The function of E47R was tested by transient-transfection assay using the E2A reporter plasmid (Fig. 7A). The activity of E47R increased greater than 30-fold with the addition of tamoxifen. This activity was comparable to that of E47, suggesting that the E47 portion of E47R was fully active in the presence of tamoxifen. In the absence of tamoxifen, E2A activity was two- to threefold higher than background, indicating that the E47R construct is only slightly leaky and thus may have sufficiently reduced toxicity to permit generation of stable cell lines.
FIG. 7.
Properties of E47R, an inducible E47. (A) Luciferase reporter assay of E47R. 293T cells were transiently transfected and analyzed 3 days later. T, tamoxifen. Numbers on the y axis indicate luciferase activity. (B) Western blot analysis. Twenty micrograms of protein from stable lines of 697 and K562 cells that express E47R (697/E47R and K562/E47R, respectively) was analyzed using antibodies against E47.
E47R was subcloned into the MSCV-based retroviral vector (Fig. 1A). Hence, E47R and GFP are coexpressed off a bicistronic message. Retrovirus stocks were generated and used to transduce K562 cells that were then sorted for GFP+ cells using FACS and designated K562/E47R. Initial attempts to generate a stable line of 697 cells that express E47R were unsuccessful. We hypothesized that the slight leakiness of E47R was still toxic to 697 cells. To overcome this problem, E47R was subcloned into the hybrid HIV-MSCV lentiviral vector that expresses its transgene approximately 10-fold less than the MSCV-based vector (14). By using the hybrid HIV-MSCV vector, a stable line of 697/E47R cells was successfully isolated. Western blot analysis demonstrated that the stable lines expressed E47R at similar or decreased levels compared to the endogenous E2A protein levels in both 697 and K562 cells (Fig. 7B). 697 cells also express the oncogenic E2A-PBX1 fusion protein as a consequence of the chromosomal translocation t(1;19) involving the E2A and PBX1 genes (27).
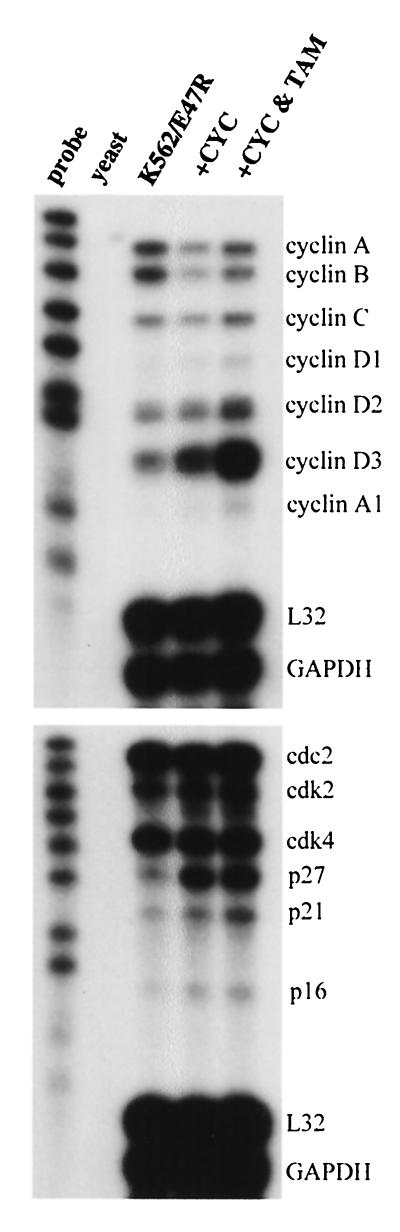
697/E47R cells were treated with tamoxifen, and RNA was isolated and analyzed by RPA (Fig. 8B). Induction of E47 increased the message levels of cyclin A, cyclin B, cyclin D3, cdc2, cdk2, and p27. No significant changes were seen in the levels of cdk4, p21, L32, and GAPDH transcripts. Similar findings were seen in K562/E47R cells treated with tamoxifen, although there were some differences (Fig. 9A). For example, message levels for cyclin A, cdc2, and p27 remained constant and the level for p21 increased. Furthermore, K562 cells expressed detectable levels of cyclin D1 and cyclin D2 transcripts that were not detected in 697 cells. E47 induced cyclin D2 but not cyclin D1 transcripts. Unlike results from the studies using E47ERT, E47R did not alter the cell cycle profile of K562/E47R or 697/E47R (data not shown), suggesting that the altered levels of transcripts are not secondary to altered cell cycle profile. No changes were seen in 697 cells expressing GFP (Fig. 8A), K562 cells expressing GFP, or the parental cells treated with tamoxifen (data not shown).
FIG. 9.
(A) RPA. K562 and K562/E47R cells were analyzed after treatment with tamoxifen (TAM) for various time intervals. The upper right represents a shorter exposure of the upper left autoradiogram. (B) The same cells were analyzed for changes in protein levels by Western blot analysis. The numbers above the autoradiogram represent time in hours.
The qualitative changes in the cell cycle profile and regulatory gene transcript levels upon suppression and induction of E2A activities are summarized in Table 1. Among these genes, cyclin D3, cyclin A1, and cyclin B expression levels correlated with E2A activities in both 697 and K562 cells. The expression levels of cyclin A, cyclin D2, cdc2, p27, and p21 also correlated with E2A activity but only in one of the two cell lines. For example, in 697 cells, cyclin A expression decreased with E47ERT and increased with E47R, but in K562 cells, cyclin A expression remained constant with E47R.
TABLE 1.
Summary of changes in cell cycle profile and expression levels of cell cycle regulatory transcriptsa
| Characteristic | Change in cell line: | |||
|---|---|---|---|---|
| 697/E47ERT | K562/E47ERT | 697/E47R | K562/E47R | |
| % S phase | ↓ | ↓ | ↔ | ↔ |
| Cyclin A | ↓ | ↓ | ↑ | ↔ |
| Cyclin B | ↓ | ↓ | ↑ | ↑ |
| Cyclin D1 | ND | ↔ | ND | ↔ |
| Cyclin D2 | ND | ↓ | ND | ↑ |
| Cyclin D3 | ↓ | ↓ | ↑ | ↑ |
| Cyclin A1 | ↓ | ↓ | ↑ | ↑ |
| cdc2 | ↓ | ↓ | ↑ | ↔ |
| cdk4 | ↔ | ↔ | ↔ | ↔ |
| p27 | ↓ | ↓ | ↑ | ↔ |
| p21 | ↓ | ↓ | ↔ | ↑ |
To determine if E2A also induces the protein levels of some cell cycle regulatory genes, K562/E47R cells were treated with tamoxifen and protein expression was analyzed by Western blot analysis (Fig. 9B). In agreement with the RPA, cyclins D2 and D3 increased while cdk4, cdk6, and β-actin remained constant. In agreement with the RPA, cyclin D3 was also induced in 697/E47R cells while cdk4, cdk6, and β-actin remained constant (data not shown).
E47 can induce some cyclins in the absence of protein synthesis.
Suppression of E47 and overexpression of E47 demonstrate that the expression of some cyclins may be regulated by E47. However, the mechanism by which E47 induces cyclin mRNA remains to be elucidated. E47 may directly bind the promoters of the various cyclin genes and activate their expression. Alternatively, E47 may induce other proteins, which in turn may regulate the expression of cyclin genes. To distinguish between these two possibilities, protein synthesis was inhibited using cycloheximide and levels of cyclin messages were measured upon posttranslational activation of E47R by tamoxifen (Fig. 10). Because of the potential toxicity of cycloheximide, protein synthesis inhibition was limited to 12 h, a time point in which induction of some cyclin messages could be easily detected in the K562/E47R cells. Cycloheximide treatment alone affected the levels of the various messages in different ways, presumably by increasing message stability or decreasing transcription due to depletion of labile factors. Cycloheximide alone decreased the transcript levels of cyclin A, cyclin B, and cyclin C but increased the transcript levels of cyclin D3 and p27. Compared to cycloheximide alone, activation of E47R in the presence of cycloheximide and increased expression of cyclin A, cyclin A1, cyclin B, cyclin C, cyclin D1, cyclin D2, cyclin D3, and p21 indicate that E47 can induce expression of these transcripts in the absence of protein synthesis. Although these findings would suggest that E47 might bind the promoters of these genes, we cannot exclude the possibility that E2A activates a cell regulatory pathway by an indirect mechanism.
FIG. 10.
Effect of E47R on expression of cell cycle regulatory genes in the absence of protein synthesis. K562/E47R cells were treated with tamoxifen (TAM) and cycloheximide (CYC) for 12 h and then analyzed by RPA.
E47 promotes entry into S phase.
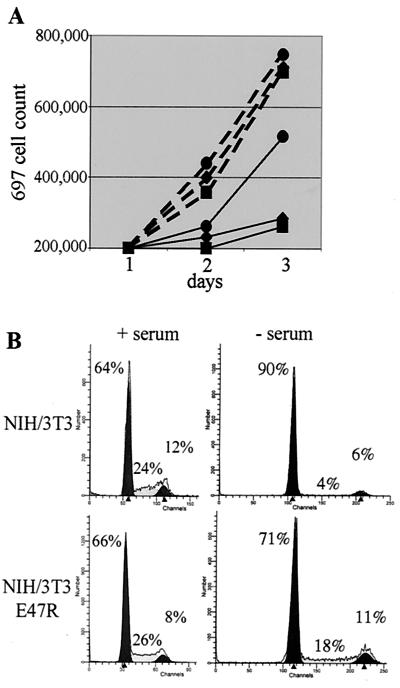
Overexpression of cyclin D decreases the dependence on serum for cell proliferation (10, 50). Since E47 induces cyclin D3, we hypothesized that 697/E47R cells would be less dependent on serum. To test this hypothesis, we analyzed the proliferation of parental 697, 697/GFP, and 697/E47R cells in RPMI medium with 10 and 0.6% serum. The growth curves of the three cells were similar at 10% serum (Fig. 11A). However, at 0.6% serum, 697/E47R cells grew faster than parental or 697/GFP cells. The increased growth rate of 697/E47R cells in low serum was confirmed by mixing GFP+ 697/E47R cells with parental 697 cells and demonstrating an increased percentage of GFP+ cells by flow cytometry (data not shown).
FIG. 11.
(A) E47 decreases serum dependence for cell proliferation. Parental 697 (⧫), 697/GFP (▪), and 697/E47R (●) cells were seeded at 200,000 cells/ml, and viable cells were counted daily for 3 days (dashed line, 10% serum; solid line, 0.6% serum). The figure is representative of three independent experiments. (B) E47 decreases serum dependence for G1 progression. Parental NIH 3T3 and stable cell lines expressing E47R were placed in 10% (+) or 0% (−) serum for 24 h and stained with PI, and the cell cycle compartment was analyzed by flow cytometry.
Ectopic expression of cyclin D promotes G1 progression. To determine if ectopic expression of E47 also promotes G1 progression, a stable line of NIH 3T3 cells that expressed E47R was engineered by retroviral transduction and sorting for GFP+ cells. NIH 3T3 cells were chosen because these cells can be easily synchronized in G0/G1 by serum starvation and transition to S phase can be promoted by the overexpression of cyclin D (50). Attempts to synchronize 697 or K562 cells in G0/G1 by serum starvation were unsuccessful. Although proliferation of 697 and K562 cells could be stopped by serum starvation, the cell cycle profile remained virtually identical (data not shown).
NIH 3T3 cells were serum starved for 24 h to arrest in G0/G1, and the cells were harvested and analyzed by FACS for cell cycle profile (Fig. 11B). In serum, NIH 3T3 and NIH 3T3/E47R cells had approximately a 20 to 30% S-phase fraction. With serum starvation, the NIH 3T3 cells were almost completely arrested in G0/G1 (G0/G1 fraction greater than 90% and S fraction less than 4%). In contrast, the NIH 3T3 cells expressing E47R had an S fraction of 12 to 18%, indicating persistent cell cycle progression and decreased requirement for growth factors.
DISCUSSION
E2A is hypothesized to promote B-cell differentiation and inhibit cell proliferation. However, present models of E2A function raise the conundrum of how B cells proliferate while expressing high levels of E2A. Possible explanations include (i) a B-cell-specific compensatory mechanism that can overcome E2A-mediated inhibition of proliferation, (ii) a B-cell-specific subversive mechanism that can convert E2A to a promoter of proliferation, and (iii) the possibility that our present models of E2A and cell proliferation are not generally true. To better characterize the relationship between E2A and cell proliferation, we developed an experimental system for modulating the activity of E2A in an inducible manner using two novel chimeric transcription factors, E47ERT and E47R. Suppression of E2A activity with E47ERT inhibited G1 progression and decreased the expression of multiple cyclins including cyclin D3. Consistent with these findings, ectopic expression of E47R promoted G1 progression and induced the expression of multiple cyclins including cyclin D2 and cyclin D3. Based on these findings, we propose a new function of E2A as a positive regulator of cell cycle progression in some B cells and non-B cells.
To confirm the regulation of cyclin D2 and cyclin D3 by E2A, we analyzed the E2A null mice. Given the high expression and the importance of E2A in B-cell development, B cells would have been the ideal cells to analyze. However, the absence of B cells in E2A null mice precluded such an analysis. Hence, we analyzed the hematopoietic cells in the fetal liver because myeloid progenitor cells express E2A (68), cyclin D2, and cyclin D3 (62). RPA demonstrated statistically significant decreases in the expression of cyclin D2 and cyclin D3 in the E2A null mice.
Although significant, the decreased expression of cyclin D2 and cyclin D3 was moderate to slight and appeared less than the decrease seen in 697 cells. This difference could be explained by multiple nonexclusive explanations. (i) In the E2A null fetus, there are compensatory mechanisms that can partially correct the defects induced by loss of E2A. (ii) E47ERT may be a more complete inhibitor of class A bHLH binding activity than knockout E2A because E47ERT can potentially heterodimerize and inhibit other bHLH factors such as HEB and E2-2. Since HEB can rescue B-cell development in E2A null mice (70), the target genes of HEB may be the same as those of E2A. If true, then HEB may also contribute to the regulation of cyclin D3. (iii) E2A contributes to regulation but is not the sole regulator of cyclin D2-cyclin D3. In some cells that do not express high levels of E2A, other transcription factors may be the major regulators of cyclin D3. This may be the case for the primary MEFs. Analysis of primary MEFs from wild-type and E2A null mice (generously provided by Cornelius Murre) shows low levels of E2A protein by Western blotting in the wild-type MEFs and no significant change in the levels of cyclin D2 and cyclin D3 by RPA in the E2A null MEFs (data not shown). These explanations could be tested once a conditional E2A knockout mouse is developed. Nevertheless, our studies indicate that, in some cell types, E2A can regulate the expression of cyclin D2 and cyclin D3.
Promotion of cell cycle proliferation, a new role for E2A.
There are numerous published results that are inconsistent with the present view that E2A suppresses cell proliferation. The E2A homologue in Drosophila daughterless is required for normal cell proliferation. Absence of daughterless causes defects in proliferation and abnormal loss of cyclin B expression in cells of the imaginal disk (11). Expression of E2A is highest in the proliferating B cells of the germinal centers of lymph nodes (54). Ectopic expression of E2A in the kidney embryonic cell line 293T causes an increase in the S-phase fraction (44). Ectopic expression of E47 did not decrease the S-phase fraction of the T-cell lymphoma cell line 1.F9 that was derived from an E2A null mouse (19). These findings and our findings are more consistent with our hypothesis that E2A can actually promote cell proliferation.
Our results appear to contradict the results of Peverali et al. (47). In this study, E2A was induced in NIH 3T3 cells as they recovered from serum starvation. The number of cells that incorporated BrdU was scored 24 h later. Induction of E2A prior to S-phase entry decreased the number of BrdU-positive cells. These results were interpreted as E2A-mediated G1 arrest. However, these results could be also seen if E2A promoted aberrant entry into S phase, leading to cell death and a decreased number of BrdU-positive cells 24 h later. This alternative hypothesis is more consistent with our findings and with the induction of apoptosis seen with ectopic expression of E2A in T-cell lymphoma-leukemia and 293T cells.
In 293T cells, ectopic expression of E2A induces the cdk inhibitor p21 (49), supporting the present view that E2A inhibits G1 progression. However, the relationship between E2A and p21 appears more complicated and may be cell type specific. For example, ectopic E2A does not appear to increase p21 in T-cell acute lymphoblastic leukemia (45). In our studies, ectopic E2A increased p21 in K562 cells but not in 697 cells. Even in cells in which E2A does induce p21, the effect on cell proliferation may not be easily predicted. Although p21 is often regarded as a cdk inhibitor, it can also promote S-phase entry by serving as a scaffold to assemble active cyclin D-cdk4 complexes (13). Hence, depending on the relative levels of cyclin D and cdk4, p21 could function as a promoter of cell proliferation or as an inhibitor. Given the large increase in cyclin D transcripts and the modest level of increase in p21 seen in K562 cells, we believe that these changes are more consistent with a transcription program to promote cell proliferation.
Cyclins as possible mediators of the effects of E2A on cell proliferation.
The mechanism by which E2A promotes cell proliferation probably involves induction of multiple cell cycle regulatory genes. Among these, E2A regulation of cyclin D2 and cyclin D3 may explain the effects of E2A on G1 progression and on serum dependence. The complex composed of cyclin D-cdk4 or -cdk6 phosphorylates the retinoblastoma tumor suppressor protein (Rb), which activates E2F to induce genes important for S phase (reviewed in reference 58) such as cyclin A, cdc2, cyclin E, thymidylate synthase, and DNA polymerase alpha (16). Suppression of cyclin D inhibits G1 progression (5), while ectopic expression of cyclin D promotes G1 progression and decreased dependence on serum (50). These effects are virtually identical to the effects seen with modulation of E2A activity and suggest that E2A promotes G1 progression by inducing cyclin D3. Induction of cyclin D3 by E2A is consistent with the observations that proliferating B cells express very high levels of E2A and cyclin D3 proteins (5, 54).
E2A also appears to regulate the expression of cyclin A, cyclin B, and cdc2, which are important regulatory proteins of S-phase entry and mitosis (31, 39, 43). The regulation of cell cycle progression by E2A may explain the difficulty in isolating stable lines that overexpress E2A. Overexpression of E2A may induce cyclins, leading to inappropriate cell cycle progression or phosphorylation of inappropriate substrates, which initiates the apoptotic pathway. In support of this hypothesis, apoptosis can be initiated in multiple cell types with ectopic expression of cyclin A (8, 38) or cyclin B (48).
While our experimental evidence strongly suggests that E2A induces the expression of cyclin A, cyclin B, and cdc2 in the 697 B-cell line, we wondered if E2A would function similarly under more biological conditions. For example, if E2A increased cyclin gene expression, then increased E2A activity may be one mechanism by which tumor cells have a proliferation advantage. The activity of E2A is regulated at the posttranscription level via oxidative state (6), phosphorylation (60), and interaction with Id (67). However, we hypothesized that, in some tumors, E2A may be increased at the message level and this would be accompanied by increased expression of its target genes. We queried a database that contains the transcript profiles of primary lymphomas (1) for 20 genes whose expression best correlated with expression of E2A, i.e., genes with high expression in lymphomas that express high levels of E2A and low expression in lymphomas that express low levels of E2A. In agreement with our results, cyclin A, cyclin B, and cdc2 genes are among the top 20 genes (Table 2). Many of the remaining genes either are implicated in cell cycle progression or have unknown function. These virtual microarray results are consistent with our experimental data and support our hypothesis that E2A regulates some cell cycle regulatory genes.
TABLE 2.
Expression levels of 20 genes that best correlate with expression of E2Aa
| Clone | Name |
|---|---|
| 114639 | E2A = E12/E47 HLH transcription factors |
| 124345 | CENP-F kinetochore protein = mitosin = cell-cycle-dependent 350K nuclear protein = AH antigen |
| 950690 | Cyclin A |
| 1286361 | Unknown UG Hs.104741 ESTs |
| 1185338 | Unknown UG Hs.104741 ESTs |
| 195630 | Topoisomerase II alpha (170 kDa) |
| 814701 | Mitotic feedback control protein Madp2 homologue |
| 137046 | nek2 protein kinase |
| 1368903 | Unknown |
| 712505 | CDC2 = cell division control protein 2 homologue = p34 protein kinase |
| 712157 | Aldehyde oxidase (AOX1) 5′-flanking region |
| 1341540 | BUB1 = putative mitotic checkpoint protein Ser/Thr kinase |
| 1422081 | nek2 protein kinase |
| 1306021 | ckshs1 = homologue of Cks1 = p34Cdc28/Cdc2- associated protein |
| 1355221 | Unknown UG Hs.124874 ESTs |
| 1352275 | pLK = homologue of Drosophila polo serine/ threonine kinase |
| 824060 | Cyclin B1 |
| 1356703 | Unknown |
| 684537 | p55CDC |
| 824709 | Cyclin A |
How E2A regulates the cell cycle regulatory genes remains to be elucidated. Possible mechanisms include (i) direct activation mediated by binding of E2A to the promoters of the cell cycle regulatory genes, (ii) indirect activation of the cell cycle regulatory genes that results from activation of cell proliferation pathways by E2A, and (iii) normal induction of cell cycle regulatory genes secondary to changes in the cell cycle profile. While the last possibility is consistent with the effects of E47ERT, it cannot explain the effects of E47R. Tamoxifen treatment of 697/E47R and K562/E47R cells growing in 10% serum induces many cell cycle regulatory genes without changing the cell cycle profile. Of the two remaining mechanisms, we favor a direct transcriptional activation by E2A because multiple cell cycle regulatory genes are induced even in the absence of protein synthesis (Fig. 10). Consistent with this hypothesis, the murine cyclin D3 contains three E boxes in the 5′-flanking region (65). The positions of two of the E boxes relative to the transcription start site appear conserved in the human cyclin D3 5′-flanking region (9). However, we cannot completely exclude an indirect effect mediated by changes in cyclin D3 message stability (21) or by stimulation of a signal transduction pathway leading to a proliferation transcription program. Future studies to characterize the mechanisms will increase our understanding of E2A function during normal and neoplastic cell proliferation.
ACKNOWLEDGMENTS
This work was supported by the McCabe Fellow Award and the ACS Pilot Project Award (J.K.C.). F.Z. was supported by NIH grant P01-DK52558 (to T. Kadesch).
We thank M. Carroll, A. DaCosta, W. El-Diery, J. Hess, T. Kadesch, W. Pear, and R. Wilson for critical reading of the manuscript.
REFERENCES
- 1.Alizadeh A A, Eisen M B, Davis R E, Ma C, Lossos I S, Rosenwald A, Boldrick J C, Sabet H, Tran T, Yu X, Powell J I, Yang L, Marti G E, Moore T, Hudson J, Jr, Lu L, Lewis D B, Tibshirani R, Sherlock G, Chan W C, Greiner T C, Weisenburger D D, Armitage J O, Warnke R, Staudt L M, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 2.Aronheim A, Shiran R, Rosen A, Walker M D. The E2A gene product contains two separable and functionally distinct transcription activation domains. Proc Natl Acad Sci USA. 1993;90:8063–8067. doi: 10.1073/pnas.90.17.8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bain G, Engel I, Robanus Maandag E C, te Riele H P, Voland J R, Sharp L L, Chun J, Huey B, Pinkel D, Murre C. E2A deficiency leads to abnormalities in αβ T-cell development and to rapid development of T-cell lymphomas. Mol Cell Biol. 1997;17:4782–4791. doi: 10.1128/mcb.17.8.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bain G, Maandag E C, Izon D J, Amsen D, Kruisbeek A M, Weintraub B C, Krop I, Schlissel M S, Feeney A J, van Roon M, et al. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell. 1994;79:885–892. doi: 10.1016/0092-8674(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 5.Bartkova J, Lukas J, Strauss M, Bartek J. Cyclin D3: requirement for G1/S transition and high abundance in quiescent tissues suggest a dual role in proliferation and differentiation. Oncogene. 1998;17:1027–1037. doi: 10.1038/sj.onc.1202016. [DOI] [PubMed] [Google Scholar]
- 6.Benezra R. An intermolecular disulfide bond stabilizes E2A homodimers and is required for DNA binding at physiological temperatures. Cell. 1994;79:1057–1067. doi: 10.1016/0092-8674(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 7.Bentolila L A, Fanton d'Andon M, Nguyen Q T, Martinez O, Rougeon F, Doyen N. The two isoforms of mouse terminal deoxynucleotidyl transferase differ in both the ability to add N regions and subcellular localization. EMBO J. 1995;14:4221–4229. doi: 10.1002/j.1460-2075.1995.tb00096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bortner D M, Rosenberg M P. Overexpression of cyclin A in the mammary glands of transgenic mice results in the induction of nuclear abnormalities and increased apoptosis. Cell Growth Differ. 1995;6:1579–1589. [PubMed] [Google Scholar]
- 9.Brooks A R, Shiffman D, Chan C S, Brooks E E, Milner P G. Functional analysis of the human cyclin D2 and cyclin D3 promoters. J Biol Chem. 1996;271:9090–9099. doi: 10.1074/jbc.271.15.9090. [DOI] [PubMed] [Google Scholar]
- 10.Brown J R, Nigh E, Lee R J, Ye H, Thompson M A, Saudou F, Pestell R G, Greenberg M E. Fos family members induce cell cycle entry by activating cyclin D1. Mol Cell Biol. 1998;18:5609–5619. doi: 10.1128/mcb.18.9.5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown N L, Paddock S W, Sattler C A, Cronmiller C, Thomas B J, Carroll S B. daughterless is required for Drosophila photoreceptor cell determination, eye morphogenesis, and cell cycle progression. Dev Biol. 1996;179:65–78. doi: 10.1006/dbio.1996.0241. [DOI] [PubMed] [Google Scholar]
- 12.Carter R S, Ordentlich P, Kadesch T. Selective utilization of basic helix-loop-helix-leucine zipper proteins at the immunoglobulin heavy-chain enhancer. Mol Cell Biol. 1997;17:18–23. doi: 10.1128/mcb.17.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng M, Olivier P, Diehl J A, Fero M, Roussel M F, Roberts J M, Sherr C J. The p21(Cip1) and p27(Kip1) CDK ‘inhibitors’ are essential activators of cyclin D-dependent kinases in murine fibroblasts. EMBO J. 1999;18:1571–1583. doi: 10.1093/emboj/18.6.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi J K, Hoang N, Vilardi A M, Conrad P, Emerson S G, Gewirtz A M. Hybrid HIV/MSCV LTR enhances transgene expression of lentiviral vectors in human CD34+ hematopoietic cells. Stem Cells. 2001;19:236–246. doi: 10.1634/stemcells.19-3-236. [DOI] [PubMed] [Google Scholar]
- 15.Choi J K, Shen C P, Radomska H S, Eckhardt L A, Kadesch T. E47 activates the Ig-heavy chain and TdT loci in non-B cells. EMBO J. 1996;15:5014–5021. [PMC free article] [PubMed] [Google Scholar]
- 16.DeGregori J, Kowalik T, Nevins J R. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol Cell Biol. 1995;15:4215–4224. doi: 10.1128/mcb.15.8.4215. . (Erratum, 15:5846–5847.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng C, Zhang P, Harper J W, Elledge S J, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 18.Eckner R, Yao T P, Oldread E, Livingston D M. Interaction and functional collaboration of p300/CBP and bHLH proteins in muscle and B-cell differentiation. Genes Dev. 1996;10:2478–2490. doi: 10.1101/gad.10.19.2478. [DOI] [PubMed] [Google Scholar]
- 19.Engel I, Murre C. Ectopic expression of E47 or E12 promotes the death of E2A-deficient lymphomas. Proc Natl Acad Sci USA. 1999;96:996–1001. doi: 10.1073/pnas.96.3.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fischer A, Malissen B. Natural and engineered disorders of lymphocyte development. Science. 1998;280:237–243. doi: 10.1126/science.280.5361.237. [DOI] [PubMed] [Google Scholar]
- 21.Garcia-Gras E A, Chi P, Thompson E A. Glucocorticoid-mediated destabilization of cyclin D3 mRNA involves RNA-protein interactions in the 3′-untranslated region of the mRNA. J Biol Chem. 2000;275:22001–22008. doi: 10.1074/jbc.m001048200. [DOI] [PubMed] [Google Scholar]
- 22.Gewirtz A M, Anfossi G, Venturelli D, Valpreda S, Sims R, Calabretta B. G1/S transition in normal human T-lymphocytes requires the nuclear protein encoded by c-myb. Science. 1989;245:180–183. doi: 10.1126/science.2665077. [DOI] [PubMed] [Google Scholar]
- 23.Ghia P, ten Boekel E, Sanz E, de la Hera A, Rolink A, Melchers F. Ordering of human bone marrow B lymphocyte precursors by single-cell polymerase chain reaction analyses of the rearrangement status of the immunoglobulin H and L chain gene loci. J Exp Med. 1996;184:2217–2229. doi: 10.1084/jem.184.6.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilfillan S, Dierich A, Lemeur M, Benoist C, Mathis D. Mice lacking TdT: mature animals with an immature lymphocyte repertoire. Science. 1993;261:1175–1178. doi: 10.1126/science.8356452. . (Erratum, 262:1957.) [DOI] [PubMed] [Google Scholar]
- 25.Goldfarb A N, Lewandowska K, Pennell C A. Identification of a highly conserved module in E proteins required for in vivo helix-loop-helix dimerization. J Biol Chem. 1998;273:2866–2873. doi: 10.1074/jbc.273.5.2866. [DOI] [PubMed] [Google Scholar]
- 26.Kadesch T. Helix-loop-helix proteins in the regulation of immunoglobulin gene transcription. Immunol Today. 1992;13:31–36. doi: 10.1016/0167-5699(92)90201-h. [DOI] [PubMed] [Google Scholar]
- 27.Kamps M P, Look A T, Baltimore D. The human t(1;19) translocation in pre-B ALL produces multiple nuclear E2A-Pbx1 fusion proteins with differing transforming potentials. Genes Dev. 1991;5:358–368. doi: 10.1101/gad.5.3.358. [DOI] [PubMed] [Google Scholar]
- 28.Kee B L, Murre C. Induction of early B cell factor (EBF) and multiple B lineage genes by the basic helix-loop-helix transcription factor E12. J Exp Med. 1998;188:699–713. doi: 10.1084/jem.188.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kitamura D, Kudo A, Schaal S, Muller W, Melchers F, Rajewsky K. A critical role of lambda 5 protein in B cell development. Cell. 1992;69:823–831. doi: 10.1016/0092-8674(92)90293-l. [DOI] [PubMed] [Google Scholar]
- 30.Kitamura D, Roes J, Kuhn R, Rajewsky K. A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin mu chain gene. Nature. 1991;350:423–426. doi: 10.1038/350423a0. [DOI] [PubMed] [Google Scholar]
- 31.Knudsen K E, Fribourg A F, Strobeck M W, Blanchard J M, Knudsen E S. Cyclin A is a functional target of retinoblastoma tumor suppressor protein-mediated cell cycle arrest. J Biol Chem. 1999;274:27632–27641. doi: 10.1074/jbc.274.39.27632. [DOI] [PubMed] [Google Scholar]
- 32.Komori T, Okada A, Stewart V, Alt F W. Lack of N regions in antigen receptor variable region genes of TdT-deficient lymphocytes. Science. 1993;261:1171–1175. doi: 10.1126/science.8356451. . (Erratum, 262:1957.) [DOI] [PubMed] [Google Scholar]
- 33.Kotani H, Newton III P B, Zhang S, Chiang Y L, Otto E, Weaver L, Blaese R M, Anderson W F, McGarrity G J. Improved methods of retroviral vector transduction and production for gene therapy. Hum Gene Ther. 1994;5:19–28. doi: 10.1089/hum.1994.5.1-19. [DOI] [PubMed] [Google Scholar]
- 34.Lin H, Grosschedl R. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature. 1995;376:263–267. doi: 10.1038/376263a0. [DOI] [PubMed] [Google Scholar]
- 35.Massari M E, Grant P A, Pray-Grant M G, Berger S L, Workman J L, Murre C. A conserved motif present in a class of helix-loop-helix proteins activates transcription by direct recruitment of the SAGA complex. Mol Cell. 1999;4:63–73. doi: 10.1016/s1097-2765(00)80188-4. [DOI] [PubMed] [Google Scholar]
- 36.Mombaerts P, Iacomini J, Johnson R S, Herrup K, Tonegawa S, Papaioannou V E. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 37.Mucenski M L, McLain K, Kier A B, Swerdlow S H, Schreiner C M, Miller T A, Pietryga D W, Scott W J, Jr, Potter S S. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell. 1991;65:677–689. doi: 10.1016/0092-8674(91)90099-k. [DOI] [PubMed] [Google Scholar]
- 38.Murphy M, Stinnakre M G, Senamaud-Beaufort C, Winston N J, Sweeney C, Kubelka M, Carrington M, Brechot C, Sobczak-Thepot J. Delayed early embryonic lethality following disruption of the murine cyclin A2 gene. Nat Genet. 1997;15:83–86. doi: 10.1038/ng0197-83. . (Erratum, 23:481, 1999.) [DOI] [PubMed] [Google Scholar]
- 39.Murray A W, Kirschner M W. Cyclin synthesis drives the early embryonic cell cycle. Nature. 1989;339:275–280. doi: 10.1038/339275a0. [DOI] [PubMed] [Google Scholar]
- 40.Murre C, McCaw P S, Vaessin H, Caudy M, Jan L Y, Jan Y N, Cabrera C V, Buskin J N, Hauschka S D, Lassar A B, et al. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- 41.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage F H, Verma I M, Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 42.O'Riordan M, Grosschedl R. Coordinate regulation of B cell differentiation by the transcription factors EBF and E2A. Immunity. 1999;11:21–31. doi: 10.1016/s1074-7613(00)80078-3. [DOI] [PubMed] [Google Scholar]
- 43.Pagano M, Pepperkok R, Verde F, Ansorge W, Draetta G. Cyclin A is required at two points in the human cell cycle. EMBO J. 1992;11:961–971. doi: 10.1002/j.1460-2075.1992.tb05135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pagliuca A, Gallo P, De Luca P, Lania L. Class A helix-loop-helix proteins are positive regulators of several cyclin-dependent kinase inhibitors' promoter activity and negatively affect cell growth. Cancer Res. 2000;60:1376–1382. [PubMed] [Google Scholar]
- 45.Park S T, Nolan G P, Sun X H. Growth inhibition and apoptosis due to restoration of E2A activity in T cell acute lymphoblastic leukemia cells. J Exp Med. 1999;189:501–508. doi: 10.1084/jem.189.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pear W S, Miller J P, Xu L, Pui J C, Soffer B, Quackenbush R C, Pendergast A M, Bronson R, Aster J C, Scott M L, Baltimore D. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood. 1998;92:3780–3792. [PubMed] [Google Scholar]
- 47.Peverali F A, Ramqvist T, Saffrich R, Pepperkok R, Barone M V, Philipson L. Regulation of G1 progression by E2A and Id helix-loop-helix proteins. EMBO J. 1994;13:4291–4301. doi: 10.1002/j.1460-2075.1994.tb06749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Porter L A, Singh G, Lee J M. Abundance of cyclin B1 regulates gamma-radiation-induced apoptosis. Blood. 2000;95:2645–2650. [PubMed] [Google Scholar]
- 49.Prabhu S, Ignatova A, Park S T, Sun X H. Regulation of the expression of cyclin-dependent kinase inhibitor p21 by E2A and Id proteins. Mol Cell Biol. 1997;17:5888–5896. doi: 10.1128/mcb.17.10.5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quelle D E, Ashmun R A, Shurtleff S A, Kato J Y, Bar-Sagi D, Roussel M F, Sherr C J. Overexpression of mouse D-type cyclins accelerates G1 phase in rodent fibroblasts. Genes Dev. 1993;7:1559–1571. doi: 10.1101/gad.7.8.1559. [DOI] [PubMed] [Google Scholar]
- 51.Quesenberry P J, Iscove N N, Cooper C, Brady G, Newburger P E, Stein G S, Stein J S, Reddy G P, Pearson-White S. Expression of basic helix-loop-helix transcription factors in explant hematopoietic progenitors. J Cell Biochem. 1996;61:478–488. doi: 10.1002/(SICI)1097-4644(19960601)61:3%3C478::AID-JCB15%3E3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 52.Quong M W, Massari M E, Zwart R, Murre C. A new transcriptional-activation motif restricted to a class of helix-loop-helix proteins is functionally conserved in both yeast and mammalian cells. Mol Cell Biol. 1993;13:792–800. doi: 10.1128/mcb.13.2.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ruezinsky D, Beckmann H, Kadesch T. Modulation of the IgH enhancer's cell type specificity through a genetic switch. Genes Dev. 1991;5:29–37. doi: 10.1101/gad.5.1.29. [DOI] [PubMed] [Google Scholar]
- 54.Rutherford M N, LeBrun D P. Restricted expression of E2A protein in primary human tissues correlates with proliferation and differentiation. Am J Pathol. 1998;153:165–173. doi: 10.1016/S0002-9440(10)65557-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schlissel M, Voronova A, Baltimore D. Helix-loop-helix transcription factor E47 activates germ-line immunoglobulin heavy-chain gene transcription and rearrangement in a pre-T-cell line. Genes Dev. 1991;5:1367–1376. doi: 10.1101/gad.5.8.1367. [DOI] [PubMed] [Google Scholar]
- 56.Schmidt M, Nazarov V, Stevens L, Watson R, Wolff L. Regulation of the resident chromosomal copy of c-myc by c-Myb is involved in myeloid leukemogenesis. Mol Cell Biol. 2000;20:1970–1981. doi: 10.1128/mcb.20.6.1970-1981.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shen C P, Kadesch T. B-cell-specific DNA binding by an E47 homodimer. Mol Cell Biol. 1995;15:4518–4524. doi: 10.1128/mcb.15.8.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sherr C J. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 59.Sigvardsson M, O'Riordan M, Grosschedl R. EBF and E47 collaborate to induce expression of the endogenous immunoglobulin surrogate light chain genes. Immunity. 1997;7:25–36. doi: 10.1016/s1074-7613(00)80507-5. [DOI] [PubMed] [Google Scholar]
- 60.Sloan S R, Shen C P, McCarrick-Walmsley R, Kadesch T. Phosphorylation of E47 as a potential determinant of B-cell-specific activity. Mol Cell Biol. 1996;16:6900–6908. doi: 10.1128/mcb.16.12.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun X H, Baltimore D. An inhibitory domain of E12 transcription factor prevents DNA binding in E12 homodimers but not in E12 heterodimers. Cell. 1991;64:459–470. doi: 10.1016/0092-8674(91)90653-g. . (Erratum, 66:423.) [DOI] [PubMed] [Google Scholar]
- 62.Suzuki R, Kuroda H, Komatsu H, Hosokawa Y, Kagami Y, Ogura M, Nakamura S, Kodera Y, Morishima Y, Ueda R, Seto M. Selective usage of D-type cyclins in lymphoid malignancies. Leukemia. 1999;13:1335–1342. doi: 10.1038/sj.leu.2401485. [DOI] [PubMed] [Google Scholar]
- 63.Taylor D, Badiani P, Weston K. A dominant interfering Myb mutant causes apoptosis in T cells. Genes Dev. 1996;10:2732–2744. doi: 10.1101/gad.10.21.2732. [DOI] [PubMed] [Google Scholar]
- 64.Teramoto N, Pokrovskaja K, Szekely L, Polack A, Yoshino T, Akagi T, Klein G. Expression of cyclin D2 and D3 in lymphoid lesions. Int J Cancer. 1999;81:543–550. doi: 10.1002/(sici)1097-0215(19990517)81:4<543::aid-ijc7>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 65.Wang Z, Sicinski P, Weinberg R A, Zhang Y, Ravid K. Characterization of the mouse cyclin D3 gene: exon/intron organization and promoter activity. Genomics. 1996;35:156–163. doi: 10.1006/geno.1996.0334. [DOI] [PubMed] [Google Scholar]
- 66.Weintraub H, Genetta T, Kadesch T. Tissue-specific gene activation by MyoD: determination of specificity by cis-acting repression elements. Genes Dev. 1994;8:2203–2211. doi: 10.1101/gad.8.18.2203. [DOI] [PubMed] [Google Scholar]
- 67.Wilson R B, Kiledjian M, Shen C P, Benezra R, Zwollo P, Dymecki S M, Desiderio S V, Kadesch T. Repression of immunoglobulin enhancers by the helix-loop-helix protein Id: implications for B-lymphoid-cell development. Mol Cell Biol. 1991;11:6185–6191. doi: 10.1128/mcb.11.12.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xin X Q, Nelson C, Collins L, Dorshkind K. Kinetics of E2A basic helix-loop-helix-protein expression during myelopoiesis and primary B cell differentiation. J Immunol. 1993;151:5398–5407. . (Erratum, 152:1635, 1994.) [PubMed] [Google Scholar]
- 69.Yan W, Young A Z, Soares V C, Kelley R, Benezra R, Zhuang Y. High incidence of T-cell tumors in E2A-null mice and E2A/Id1 double-knockout mice. Mol Cell Biol. 1997;17:7317–7327. doi: 10.1128/mcb.17.12.7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhuang Y, Barndt R J, Pan L, Kelley R, Dai M. Functional replacement of the mouse E2A gene with a human HEB cDNA. Mol Cell Biol. 1998;18:3340–3349. doi: 10.1128/mcb.18.6.3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhuang Y, Soriano P, Weintraub H. The helix-loop-helix gene E2A is required for B cell formation. Cell. 1994;79:875–884. doi: 10.1016/0092-8674(94)90076-0. [DOI] [PubMed] [Google Scholar]