Multicenter Comparison Trial of DNA Extraction Methods and PCR Assays for Detection of Chlamydia pneumoniae in Endarterectomy Specimens (original) (raw)
Abstract
The reported rate of detection of Chlamydia pneumoniae DNA within atherosclerotic lesions by PCR varies between 0 and 100%. In this study, identical sets of coded experimental atheroma samples (n = 15) and spiked controls (n = 5) were analyzed by 16 test methods in nine centers by means of PCR. The positive controls were correctly identified to levels of 1, 0.1, and 0.01 inclusion bodies of C. pneumoniae/ml of tissue homogenate by 16 (100%), 11 (69%), and 3 (19%) of the test methods, respectively. Three out of 16 negative controls (19%) were rated positive. Positivity rates for atheroma samples varied between 0 and 60% for the different test methods, with the maximum concordant result for positivity being only 25% for one carotid artery sample. There was no consistent pattern of positive results among the various laboratories, and there was no correlation between the detection rates and the sensitivity of the assay used.
Accumulating evidence has been supporting a role of Chlamydia pneumoniae, an important respiratory pathogen in humans (9), in the pathogenesis of atherosclerosis. Seroepidemiological studies suggesting an association between C. pneumoniae and coronary artery disease (18) were followed by the direct demonstration of C. pneumoniae in the affected tissue by means of PCR (1, 8, 10, 11, 15, 16, 19), which was found to be more sensitive than cell culturing for this purpose.
PCR has the potential to detect a single organism, but its application to clinical specimens must be considered with caution for a number of reasons (2, 4). PCR results can vary widely; for the detection of C. pneumoniae in atheromatous lesions, some study groups reported positivity rates of up to 100%, while others reported that PCR completely failed to detect the organism (7, 22).
To date, there are no standardized PCR or other nucleic acid amplification methods for the detection of C. pneumoniae. Current PCRs are all in-house assays using different primers, reaction conditions, and methods of detection. Furthermore, DNA extraction procedures prior to PCR vary between different laboratories. Apart from differences in methodology, comparison of data proves difficult because of the different clinical materials analyzed within individual studies.
Hence, it remains to be clarified whether these discrepancies in positivity rates for C. pneumoniae in atheromas are the result of methodological differences or whether they actually reflect differences in the amounts of C. pneumoniae in the materials studied. The aim of this study was to compare established DNA extraction methods and PCR protocols used by different centers by using specimens as identical as possible.
MATERIALS AND METHODS
Study design.
Nine laboratories with experience in the detection of C. pneumoniae in atheromas by PCR received panels consisting of aliquots of 20 samples identical in composition but numbered by a participant-specific code. Each laboratory was free to choose the preferred technique of DNA extraction and PCR.
Experimental panels.
Informed consent was obtained from patients, and all procedures were done in accordance with institutional human experimentation guidelines. Specimens (n = 15) obtained from patients suffering from severe atherosclerosis and therefore undergoing vascular surgery (carotid artery, n = 6; femoral artery, n = 1; aneurysm of the abdominal aorta, n = 5; aortic valve, n = 2; and coronary artery, n = 1) were directly transferred from the operating theater to the laboratory and frozen immediately at −80°C. All clinical samples were thawed in one session, decalcified and homogenized to 30 mg/ml in Dulbecco's phosphate-buffered saline (PBS; Gibco Life Technologies, Paisley, Scotland) using sterile tissue grinders, vortexed, immediately divided into 2-ml aliquots, and refrozen at −80°C. Manipulations of negative controls (fresh porcine aortic tissue, homogenized to 30 mg/ml in Dulbecco's PBS) and surgical specimens were done in a vertical laminar flow cabinet. Both the laboratory and the pipettes had never been used for _C. pneumoniae_-related work before.
After the processing of negative controls and clinical specimens, positive controls were prepared in a laminar flow cabinet located in a physically separated room. For preparation of the positive controls (n = 4), the number of HEp-2 cells (CCL-23; American Type Culture Collection) containing C. pneumoniae inclusions (strain MUL-1; provided by M. Maass) was counted 72 h after infection by means of immunofluorescence microscopy. In addition, the number of corresponding chlamydial particles was estimated by staining serial dilutions of purified chlamydial particles by means of direct immunofluorescence. Mock-infected tissue controls were prepared as follows. Infected cells were harvested in PBS by scraping, and the cells were disrupted by vortexing with glass beads for 20 s to ensure homogeneity. To provide identical conditions for all participants, a dilution series containing 103, 102, 101, and 1 inclusions/ml (corresponding to 5 × 105 to 5 × 102 particles) was prepared for spiking stocks of mock-infected positive controls. To each 30 ml of porcine tissue homogenate (30 mg/ml), 300 μl of the above-described C. pneumoniae dilution series was added to final concentrations calculated to contain 10, 1, 0.1, and 0.01 inclusions/ml or, alternatively, 5 × 103 to 5 chlamydial particles/ml of porcine aortic homogenate. Controls were vortexed prior to division into 2-ml aliquots. Regardless of the participant-specific code, the negative control was always given the code number following that of the strongest positive control.
Experimental panels were stored at −80°C until they were shipped on dry ice by air freight to collaborating laboratories within 48 h. Participants were requested to keep samples at −80°C until analysis was performed.
DNA extraction methods and PCR assays.
Each laboratory followed at least one of their established procedures for C. pneumoniae detection in atheromas by PCR. Table 1 displays applied laboratory methodologies and summarizes the key variables of the different testing methods.
TABLE 1.
Summary of testing methods for the detection of C. pneumoniae in endarterectomy samples
| Parameter | Details for the following laboratory: | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | Da | E | F | Ga | H | Ia | ||||||||
| 1a | 1b | 2a | 2b | 1 | 2 | 3 | 1 | 2 | 3 | |||||||
| Amt of sample used for DNA extraction (mg) | 6 | 6 | 18 | 1.5 | 1.5 | 6 | 6 | 6 | 60 | 30 | 30 | 30 | 30 | 6 | 30 | 30 |
| DNA extraction | High Pure PCR template preparation kitb | High Pure PCR template preparation kitb | Proteinase K digestion, PCIc, ethanol precipitation | Proteinase K digestion, PCIc, ethanol precipitation | Proteinase K digestion, PCIc, ethanol precipitation | QIAmp-Blood kitd | QIAmp-Blood kitd | QIAmp-DNA mini kit— tissue protocold | Genomic DNA preparation kit for tissued | EASY-DNA kite + silica particles | EASY-DNA kite + silica particles | EASY-DNA kite + silica particles | Genomic DNA preparation kit for tissued | Proteinase K digestion, PCIc, ethanol precipitation | Proteinase K digestion, PCIc, ethanol precipitation | Proteinase K digestion, PCIc, ethanol precipitation |
| PCR target gene | omp-1 | omp-1 | Cloned _Pst_I fragment | omp-1 | omp-1 | omp-1 | omp-1 | omp-1 | 16S rRNA | 16S rRNA | omp-1 | omp-1 | 16S rRNA | 16S rRNA | 16S rRNA | omp-1 |
| PCR reference(s) | 21 | Not published | 3, 11 | 21 | 20 | 21 | 20 | 21 | Not published | 14 | 14 | Not published | 15 | 6 nested with 5 | 12 | 21 (slightly modified) |
| Relative amt of original sample used in first PCR (mg) | 0.3 | 0.3 | 9 | 0.3 | 0.3 | 0.3 | 0.3 | 0.9 | 3 | 1.5 | 1.5 | 1.5 | 7.5 | 0.75 | 0.95 | 0.95 |
| PCR setup | Nested | Nested | Nested | Nested | Single | Nested | Single | Nested | Real time | Single | Single | Nested | Nested | Nested | Single | Nested |
| Amt of amplicon transferred in nested PCR (μl) | 5 | 0.2f | 5 | 1g | 0 | 1g | 0 | 5 | 0 | 0 | 0 | 1 | 0.5 | 5 | 0 | 10 |
| Touchdown technique | Yes | No | No | Yes | No | Yes | No | Yes | No | Yes | Yes | Yes | No | No | Yes | Yes |
| Product detection method | Agarose gel electrophoresis | Agarose gel electrophoresis | Southern blotting | Agarose gel electrophoresis | EIAh | Agarose gel electrophoresis | EIAh | Agarose gel electrophoresis | Light detectioni | Southern blotting | Southern blotting | Southern blotting | Southern blotting | PAGEj | PAGEj | PAGEj |
Statistical analysis.
A mathematical model was applied based on the straightforward assumption that the probability of a positive reaction (Pr) depends only on the amount of DNA present in the specimen and the sensitivity of the method to detect it. The only model which allows for a comparison of methods which is stochastically independent of the specific selection of tissue samples is the two-parametric logistic model:
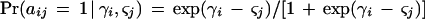
In this equation, aij is the realization of a random variable taking on the value 0 if the result of method j with tissue sample i is negative and the value 1 if it is positive; parameter γ_i_ reflects the amount of DNA present in tissue i, and ς_j_ reflects the sensitivity of method j. To simplify computations, the parameter ς_j_ is set proportional (factor α) to the detection limit or, more specifically, to its logarithm sj:
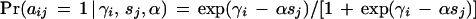
The advantage of this model lies in the fact that it makes a conditional maximum-likelihood estimation of the parameters possible. Estimation of method parameters is independent of the tissue parameters and vice versa.
We have applied this model to estimate the expected number of positive results for each method, assuming average proportions of 30 and 40% of true positive specimens. Furthermore, the probability distribution of the number of methods with 0, 1, 2, … positive results under the same assumptions was computed and compared to the observed frequencies by chi-square tests.
RESULTS
Blinded, otherwise identical panels consisting of endarterectomy specimens and controls were analyzed in nine laboratories by 16 different PCR testing methods.
In three laboratories, the materials were analyzed more than once, with the same or different primers and with single-round or nested PCR. For analysis and discussion, each testing method was considered a separate experiment; therefore, each attempt was treated as a separate analysis.
The sample volume used for DNA extraction was in the range of 50 μl (1.5 mg) to 2 ml (60 mg) of tissue homogenate, and five different commercially available kits as well as standard phenol-chloroform DNA preparation methods were used. Three different species-specific gene regions of C. pneumoniae served as target sequences for PCR: the 16S rRNA gene (5 protocols), the omp-1 gene (10 protocols), and a cloned _Pst_I fragment (1 protocol). PCR setups were as follows: single-round PCR, 5 protocols; nested PCR, 10 protocols; and real-time PCR, 1 protocol. Some laboratories performed a touchdown technique (nine protocols). Four different methods of PCR product detection were applied: agarose or polyacrylamide gel electrophoresis and ethidium bromide staining (eight protocols); hybridization techniques, such as Southern blotting with biotinylated probes (five protocols); detection by an enzyme-linked immunosorbent assay (two protocols); and real-time PCR (one protocol). None of the participants applied sequencing techniques. Table 1 displays and contrasts the 16 testing methods applied in this study.
Controls.
The results obtained from the blinded controls are shown in Table 2. All 16 testing methods identified the positive controls as positive to the level of one C. pneumoniae inclusion or 500 chlamydial particles/ml of tissue homogenate (100%). Furthermore, 11 testing methods (69%) allowed the detection of 0.1 chlamydial inclusion (50 particles) and 3 protocols (19%) allowed the detection of 0.01 chlamydial inclusion (5 particles) per ml of tissue homogenate. Taking into account the amount of tissue used for DNA preparation and finally the volume of DNA extract used for PCR when calculating the detection limit for C. pneumoniae in spiked tissue samples, the achieved sensitivities were in the range of 0.3 to 0.0001 inclusion, corresponding to 1.5 × 102 to 5 × 10−2 particles (Table 3). Three out of 16 negative controls (19%) were rated positive.
TABLE 2.
Detection of C. pneumoniae DNA in controls in nine laboratories using 16 testing methods
| Control | No. of: | Resultc reported by the following laboratory: | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Inclusionsa | Particlesb | A | B | C | Dd | E | F | Gd | H | Id | ||||||||
| 1a | 1b | 2a | 2b | 1 | 2 | 3 | 1 | 2 | 3 | |||||||||
| C1 | 10 | 5 × 103 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| C2 | 1 | 5 × 102 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| C3 | 0.1 | 5 × 101 | + | + | − | + | − | + | − | + | − | + | + | + | + | + | + | − |
| C4 | 0.01 | 5 | − | + | − | − | − | − | − | − | − | + | − | − | + | − | − | − |
| C5 | 0 | 0 | − | − | − | + | − | + | − | − | + | − | − | − | − | − | − | − |
TABLE 3.
Detection of C. pneumoniae DNA in endarterectomy specimens in nine laboratories using 16 testing methods
| Laboratory | Detection limita | No. (%) of 15 atheroma samples reported positive | |
|---|---|---|---|
| Inclusions | Elementary bodies | ||
| A | 0.001 | 0.5 | 9 (60) |
| B | 0.0001 | 0.05 | 5 (33) |
| C | 0.3 | 150 | 3 (20) |
| Db | |||
| 1a | 0.001 | 0.5 | 2 (13) |
| 1b | 0.01 | 5 | 0 (0) |
| 2a | 0.001 | 0.5 | 0 (0) |
| 2b | 0.01 | 5 | 0 (0) |
| E | 0.003 | 1.5 | 1 (7) |
| F | 0.1 | 50 | 1 (7) |
| Gb | |||
| 1 | 0.0005 | 0.25 | 0 (0) |
| 2 | 0.005 | 2.5 | 0 (0) |
| 3 | 0.005 | 2.5 | 0 (0) |
| H | 0.003 | 1.5 | 0 (0) |
| Ib | |||
| 1 | 0.002 | 1 | 0 (0) |
| 2 | 0.003 | 1.5 | 0 (0) |
| 3 | 0.03 | 15 | 0 (0) |
Endarterectomy samples.
In general, 240 analyses were performed on 15 atheroma samples by means of 16 testing methods. In summary, 21 out of the 240 analyses (9%) were considered positive, but none of them was rated positive by more than 4 out of 16 protocols, showing a maximum concordant result for positivity of only 25% for one carotid artery.
Out of 240 analyses performed on 15 samples, 219 (91%) were reported negative. Two laboratories (G [detection methods 1, 2, and 3] and H) spiked specimens with C. pneumoniae DNA and reported their DNA extracts to be free of PCR inhibitors. Of the 15 endarterectomy samples, 4 (27%) were rated negative by all 16 testing methods.
Statistical analysis.
Based on the two-parameter logistic model, the expected number of positive results for each method was calculated (Table 4). A priori, differences could be expected due to the differences in the sensitivities of the methods. Considering sensitivity as determined by the positive controls, a consistent variation of about 1:2 is to be expected for low positivity and less variation is to be expected for higher positivity. Apparently, there is no relationship between the numbers of specimens reported positive and the sensitivities of the methods.
TABLE 4.
Observed and expected numbers of and frequency distribution for _C. pneumoniae_-positive specimensa
| Result found by method | Frequency distribution | |||||||
|---|---|---|---|---|---|---|---|---|
| Testing method | Expected no. of positive results | Observed no. of positive results | 30% positives expected | 40% positives expected | No. of positives per testing method | Observed no. of testing methods | 30% positives expected | 40% positives expected |
| A | 1.34 | 9 | 0.0146222 | 0.0713237 | 0 | 10 | 0.1 | 0.0 |
| B | 1.88 | 5 | 0.1798045 | 0.0766298 | 1 | 2 | 0.4 | 0.0 |
| C | 0.93 | 3 | 0.2412578 | 0.1509848 | 2 | 1 | 1.2 | 0.2 |
| D | 3 | 1 | 2.3 | 0.7 | ||||
| 1a | 1.34 | 2 | 0.0799224 | 0.0176888 | 4 | 0 | 3.3 | 1.6 |
| 1b | 1.34 | 0 | 0.0203368 | 0.0033813 | 5 | 1 | 3.4 | 2.6 |
| 2a | 0.93 | 0 | 0.0037302 | 0.0003412 | 6 | 0 | 2.6 | 3.2 |
| 2b | 0.93 | 0 | 0.0203368 | 0.0033813 | 7 | 0 | 1.6 | 3.0 |
| E | 1.34 | 1 | 0.0252752 | 0.0035962 | 8 | 0 | 0.8 | 2.3 |
| F | 0.93 | 1 | 0.0904558 | 0.0233947 | 9 | 1 | 0.3 | 1.3 |
| G | >9 | 0 | 0.1 | 0.9 | ||||
| 1 | 1.88 | 0 | 0.0003880 | 0.0000182 | ||||
| 2 | 1.34 | 0 | 0.0037302 | 0.0003412 | ||||
| 3 | 1.34 | 0 | 0.0037302 | 0.0003412 | ||||
| H | 1.88 | 0 | 0.0003880 | 0.0000182 | ||||
| I | ||||||||
| 1 | 1.33 | 0 | 0.0037302 | 0.0003412 | ||||
| 2 | 1.33 | 0 | 0.0037302 | 0.0003412 | ||||
| 3 | 0.93 | 0 | 0.0033813 | 0.0033813 |
Because of an average of 30 to 40% _C. pneumoniae_-positive findings reported for atherosclerotic lesions in the literature, we calculated the probability of estimating the exact number of positive results for each method under the hypothesis that the average proportions of true positive specimens are in fact 30 and 40%. Under these assumptions, while method (laboratory) A had a low probability due to the high positivity rate, most methods, especially those with a high sensitivity, had a low probability due to the presence of few positive results (Table 4). Table 4 also compares the observed frequency distributions of positive results with the expected distributions under the assumption of true positivity rates of 30 and 40%. The difference between observed and expected distributions was highly significant (P < 0.000001).
DISCUSSION
Several studies evaluating PCR for the detection of C. pneumoniae in vessel wall specimens have been published in recent years (2). Only one study compared detection methods between different centers (17). In that study, coronary arteries from patients seeking heart transplantation were investigated. At least one testing method showed C. pneumoniae in coronary arteries in 7 of 10 patients with coronary atherosclerosis. The concordant positivity in that study ranged between 9 and 82%. Since different parts of the atherosclerotic vessels were analyzed in the participating laboratories, a random distribution of C. pneumoniae within the plaques may explain some of the discrepancies in the detection of this organism. However, lack of standardization of diagnostic methods also may be responsible for false-positive as well as false-negative results.
Our study was conducted to compare DNA extraction methods and PCR protocols—as are established in different centers—using aliquots of homogenized specimens. We are aware that such aliquots are similar but not absolutely identical. The number of targets in clinical samples might be very low and aliquots of the same sample may contain, as a result of a random distribution, various numbers of C. pneumoniae DNA targets.
In our study, there was no consistent pattern of positive results among the various laboratories, and the rate of detection of positive results (0 to 60%) in the various laboratories did not correlate with the sensitivity of their assays, as determined with a panel of spiked control specimens. Some laboratories reported positive controls to 0.01 inclusion or five chlamydial particles/ml as positive but reported no positive results for the atheroma specimens; however, others reported several atheroma specimens positive but some of the positive controls negative. Therefore, neither an extraordinarily high sensitivity nor an accumulation of aliquots with a relatively high copy number (which is, displayed in Table 4, statistically extremely unlikely) could explain the high percentage of positive findings in laboratories A and B.
Contamination of negative controls occurred in two laboratories, but the methods affected were not the methods that provided the largest number of positive results for the atheroma specimens (in fact, one of them reported none of the atheroma specimens positive). Therefore, the observed contamination seems to be related to the high copy number of the positive control preceding the negative control. As suggested by Boman et al. (2), the use of strongly positive controls, i.e., highly concentrated solutions of extracted C. pneumoniae DNA, should be avoided in future studies.
A possible drawback of this study was the inclusion of only one negative control, which might not be enough to determine cross contamination. However, “in-house” negative controls were used by the individual laboratories (range, 2 to 10 in each run per testing method), and only results of testing attempts with correctly identified negative as well as positive controls were considered for analyses. Furthermore, taking into account that more than 90% of the atheroma samples were rated negative, contamination was not a major problem in this study. Additionally, it should be emphasized that the primary goal of this study was the comparison of already established protocols but not the determination of sensitivities and specificities of each method, a strategy which makes the number of control samples less critical.
The overall detection rate (9%) and the maximum concordant positive rate (25% for one carotid artery) for the atheroma samples were lower than expected. Possible explanations for these rather unexpected findings are DNA degradation due to grinding of the samples and the release of endonucleases (13) and the presence of PCR inhibitors within clinical samples. Two laboratories evaluated samples for inhibition and reported the absence of detectable PCR inhibitors. Although only sections of the vessel walls without obvious calcifications, thrombi, or fatty tissue were processed, one cannot conclude that all samples prepared by the remaining protocols were free of PCR inhibitors, since PCR inhibition was not studied by all participants. We tried to minimize the presence of PCR inhibitors and therefore removed the calcified stony parts of the vessel walls, which are known PCR inhibitors. If it is assumed that this part of a specimen contains C. pneumoniae, it must be taken into account that the specimens used in this study were from patients suffering from extensive and advanced atherosclerotic lesions. Therefore, it is very likely that C. pneumoniae, if present, would have disrupted the endothelial barrier and been distributed into surrounding areas of the vessel walls at the time of surgical intervention.
The results of this study indicate that procedures suitable for extracting DNA from spiked control samples are not necessarily suitable for extracting DNA from clinical samples. Hence, DNA extraction by itself could be a determining factor. Different DNA extraction protocols with minimized variables and performed simultaneously by participating laboratories, followed by “DNA sharing” and PCR analysis, could help to answer this question in future studies.
Although this study was not designed to find out whether or not C. pneumoniae is present in atheromatous tissues, the large number of negative results obtained with the majority of protocols remains difficult to explain. Unfortunately, during the study period, no quantitative amplification technique had been established and validated against a conventional PCR protocol in any of the participating centers. Such a quantitative PCR might have provided a better definition of the actual chlamydial load and a consensus regarding true-positive results in this study.
It can be concluded that C. pneumoniae is present only at a very low target level in atheromatous specimens, if at all; that current detection methods are relatively reliable; and that discordant positivity rates are generated as a result of a problem with the distribution of C. pneumoniae targets (i.e., due to sampling error). However, the statistical calculation concerning the observed distribution of positive results in the atheroma panel does not support these assumptions. Rates of detection of between 15 and 60% have been reported in previous studies (1, 7, 8, 10, 11, 15, 16, 19, 22). Based on an average positivity rate of 30 to 40%, as reported in the literature, the expected and observed distributions of positive results differ significantly (Table 4). If the only difference between methods is sensitivity, the probability of observing the distribution found in this study just by chance is less than 1 in a million!
It is impossible to decide which methods are correct or whether any of the methods applied is correct. It is possible that the clinical specimens analyzed contained almost no C. pneumoniae DNA, that the occasional positive results resulted from incidental contamination, and that the few methods yielding high proportion of positive results had an inherent yet undetected error. However, it is also possible that C. pneumoniae DNA was present in the majority of samples and that the few methods yielding positive results were closer to the truth, while those failing to yield a positive reaction could not detect the target either due to the failure of DNA extraction from atheroma tissues or due to some undetected inhibitory mechanism. Since no reference method is available, a decision between these two explanations is impossible to make.
ACKNOWLEDGMENTS
We thank Peter Polterauer for providing surgical specimens. We are indebted to the staff of Operating Theater Group II at Vienna University Hospital, whose personal engagement made the transport of specimens possible. We also appreciate Hermelinde Steiner's assistance in preparing the manuscript. Furthermore, the excellent technical assistance of Marion Loidl is gratefully acknowledged.
REFERENCES
- 1.Blasi F, Denti F, Erba M, Cosentini R, Raccanelli R, Rinaldi A, Fagetti L, Esposito G, Roberti U, Allegra L. Detection of Chlamydia pneumoniae but not Helicobacter pylori in atherosclerotic plaques of aortic aneurysms. J Clin Microbiol. 1996;34:2677–2769. doi: 10.1128/jcm.34.11.2766-2769.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boman J, Gaydos C A, Quinn T C. Molecular diagnosis of Chlamydia pneumoniae infection. J Clin Microbiol. 1999;37:3791–3799. doi: 10.1128/jcm.37.12.3791-3799.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell L A, Perez Melgosa M, Hamilton D J, Kuo C C, Grayston J T. Detection of Chlamydia pneumoniae by polymerase chain reaction. J Clin Microbiol. 1992;30:434–439. doi: 10.1128/jcm.30.2.434-439.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fredricks D N, Relman D A. Application of polymerase chain reaction to the diagnosis of infectious diseases. Clin Infect Dis. 1999;29:475–488. doi: 10.1086/598618. [DOI] [PubMed] [Google Scholar]
- 5.Gaydos C A, Fowler C L, Gill V J, Eiden J J, Quinn T C. Detection of Chlamydia pneumoniae by polymerase chain reaction-enzyme immunoassay in an immunocompromised population. Clin Infect Dis. 1993;17:718–723. doi: 10.1093/clinids/17.4.718. [DOI] [PubMed] [Google Scholar]
- 6.Gaydos C A, Quinn T C, Eiden J J. Identification of Chlamydia pneumoniae by DNA amplification of the 16S rRNA gene. J Clin Microbiol. 1992;30:796–800. doi: 10.1128/jcm.30.4.796-800.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jantos C A, Nesseler A, Waas W, Baumgärtner W, Tillmanns H, Haberbosch W. Low prevalence of Chlamydia pneumoniae in atherectomy specimens from patients with coronary heart disease. Clin Infect Dis. 1999;28:988–992. doi: 10.1086/514763. [DOI] [PubMed] [Google Scholar]
- 8.Juvonen J, Juvonen T, Laurila A, Alakärppä H, Lounatmaa K, Surcel H M, Leinonen M, Kairaluoma M I, Saikku P. Demonstration of Chlamydia pneumoniae in the walls of abdominal aortic aneurysms. J Vasc Surg. 1997;25:499–505. doi: 10.1016/s0741-5214(97)70260-x. [DOI] [PubMed] [Google Scholar]
- 9.Kuo C C, Jackson L A, Campbell L A, Grayston J T. Chlamydia pneumoniae (TWAR) Clin Microbiol Rev. 1995;8:451–461. doi: 10.1128/cmr.8.4.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuo C C, Shor A, Campbell L A, Fukushi H, Patton D L, Grayston J T. Demonstration of Chlamydia pneumoniae in atherosclerotic lesions of coronary arteries. J Infect Dis. 1993;167:841–849. doi: 10.1093/infdis/167.4.841. [DOI] [PubMed] [Google Scholar]
- 11.Maass M, Krause E, Engel P M, Krüger S. Endovascular presence of Chlamydia pneumoniae in patients with hemodynamically effective carotid artery stenosis. Angiology. 1997;48:699–706. doi: 10.1177/000331979704800805. [DOI] [PubMed] [Google Scholar]
- 12.Madico G, Quinn T C, Boman J, Gaydos C A. Touchdown enzyme time release-PCR for detection and identification of Chlamydia trachomatis, C. pneumoniae, and C. psittaci using the 16S and 16S–23S spacer rRNA genes. J Clin Microbiol. 2000;38:1085–1093. doi: 10.1128/jcm.38.3.1085-1093.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meijer A, van der Vliet J A, Roholl P J M, Gielis-Proper S K, de Vries A, Ossewaarde J M. Chlamydia pneumoniae in abdominal aortic aneurysms—abundance of membrane components in the absence of heat shock protein 60 and DNA. Arterioscler Thromb Vasc Biol. 1999;19:2680–2686. doi: 10.1161/01.atv.19.11.2680. [DOI] [PubMed] [Google Scholar]
- 14.Meijer A, van der Vliet J A, Schouls L M, de Vries A, Roholl P J M, Ossewaarde J M. Detection of microorganisms in vessel wall specimens of the abdominal aorta: development of a PCR in the absence of a gold standard. Res Microbiol. 1998;149:577–583. doi: 10.1016/s0923-2508(99)80005-9. [DOI] [PubMed] [Google Scholar]
- 15.Nadrchal R, Makristathis A, Apfalter P, Rotter M, Trubel W, Huk I, Polterauer P, Hirschl A M. Detection of Chlamydia pneumoniae DNA in atheromatous tissues by polymerase chain reaction. Wien Klin Wochenschr. 1999;111:153–156. [PubMed] [Google Scholar]
- 16.Ong G, Thomas B J, Mansfield A O, Davidson B R, Taylor-Robinson D. Detection and widespread distribution of Chlamydia pneumoniae in the vascular system and its possible implications. J Clin Pathol. 1996;49:102–106. doi: 10.1136/jcp.49.2.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramirez J A the Chlamydia pneumoniae/Atherosclerosis Study Group. Isolation of Chlamydia pneumoniae from the coronary artery of a patient with coronary atherosclerosis. Ann Intern Med. 1996;125:979–982. doi: 10.7326/0003-4819-125-12-199612150-00008. [DOI] [PubMed] [Google Scholar]
- 18.Saikku P, Leinonen M, Tenkanen L, Linnanmäki E, Ekman M R, Manninen V, Mänttäri M, Frick M H, Huttunen J K. Chronic Chlamydia pneumoniae infection as a risk factor for coronary heart disease in the Helsinki Heart Study. Ann Intern Med. 1992;116:273–278. doi: 10.7326/0003-4819-116-4-273. [DOI] [PubMed] [Google Scholar]
- 19.Thomas M, Wong Y, Thomas D, Ajaz M, Tsang V, Gallaher P J, Ward M E. Relation between direct detection of Chlamydia pneumoniae DNA in human coronary arteries at postmortem examination and histological severity (Stary grading) of associated atherosclerotic plaque. Circulation. 1999;99:2733–2736. doi: 10.1161/01.cir.99.21.2733. [DOI] [PubMed] [Google Scholar]
- 20.Tong C Y W, Donnelly C, Harvey G, Sillis M. Multiplex polymerase chain reaction for the simultaneous detection of Mycoplasma pneumoniae, Chlamydia pneumoniae, and Chlamydia psittaci in respiratory samples. J Clin Pathol. 1999;52:257–263. doi: 10.1136/jcp.52.4.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tong C Y W, Sillis M. Detection of Chlamydia pneumoniae and Chlamydia psittaci in sputum samples by PCR. J Clin Pathol. 1993;46:313–317. doi: 10.1136/jcp.46.4.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weiss S M, Roblin P M, Gaydos C A, Cummings P, Patton D L, Schulhoff N, Shani J, Frankel R, Penney K, Quinn T C, Hammerschlag M R, Schachter J. Failure to detect Chlamydia pneumoniae in coronary atheromas of patients undergoing atherectomy. J Infect Dis. 1996;173:957–962. doi: 10.1093/infdis/173.4.957. [DOI] [PubMed] [Google Scholar]