Automated High-Throughput Genotyping for Study of Global Epidemiology of Mycobacterium tuberculosis Based on Mycobacterial Interspersed Repetitive Units (original) (raw)
Abstract
Large-scale genotyping of Mycobacterium tuberculosis is especially challenging, as the current typing methods are labor-intensive and the results are difficult to compare among laboratories. Here, automated typing based on variable-number tandem repeats (VNTRs) of genetic elements named mycobacterial interspersed repetitive units (MIRUs) in 12 mammalian minisatellite-like loci of M. tuberculosis is presented. This system combines analysis of multiplex PCRs on a fluorescence-based DNA analyzer with computerized automation of the genotyping. Analysis of a blinded reference set of 90 strains from 38 countries (K. Kremer et al., J. Clin. Microbiol. 37:2607–2618, 1999) demonstrated that it is 100% reproducible, sensitive, and specific for M. tuberculosis complex isolates, a performance that has not been achieved by any other typing method tested in the same conditions. MIRU-VNTRs can be used for analysis of the global genetic diversity of M. tuberculosis complex strains at different levels of evolutionary divergence. To fully exploit the portability of this typing system, a website was set up for the analysis of M. tuberculosis MIRU-VNTR genotypes via the Internet. This opens the way for global epidemiological surveillance of tuberculosis and should lead to novel insights into the evolutionary and population genetics of this major pathogen.
Infectious diseases, in particular tuberculosis, represent perhaps more than ever a global threat for human health. Mycobacterium tuberculosis causes annually about 10 million new cases of active tuberculosis and 3 million deaths. Moreover, it is estimated that over one-third of the world population have latent tuberculosis infection, which represents a huge reservoir for the disease. Exponential development of travel extends this threat worldwide (8, 10). Because of these factors, efficient disease control can be achieved only by international epidemiological surveillance systems able to accurately monitor epidemic trends at a global level.
For this purpose, large-scale genotyping of M. tuberculosis is a highly appropriate tool. Genotyping allows one to identify outbreaks that remained undetected by traditional epidemiological surveillance alone and may thus be used as a basis, and not only as a help, for conventional epidemiological surveillance (1). Moreover, it can give unique insights into the international dissemination dynamics of M. tuberculosis by the comparison of isolates from widespread geographic areas and allows one to analyze evolutionary changes of pathogen populations. Several molecular-typing studies recently demonstrated the existence of clonally related M. tuberculosis families, which disseminated locally or globally (e.g., references 1, 2, 12, and 22 to 24). However, in many of these studies relatively limited numbers of strains were analyzed and they provided data that cannot be cross-compared due to the inherent drawbacks of the current “gold standard” typing method, IS_6110_ restriction fragment length polymorphism (RFLP). This method is labor-intensive; other drawbacks include the difficulty of reproducing results and comparing them among different laboratories. This renders the comparison of data from multicenter studies problematic, especially when large numbers of fingerprints are involved (12). Unfortunately, other methods, such as multilocus sequence typing, appropriate for large multicenter studies of many other bacteria (13), are not applicable to M. tuberculosis, since the gene sequence polymorphism among different strains of this species is very limited (12, 17).
Here, we propose a solution to these problems by the use of high-throughput PCR analysis of M. tuberculosis genomic loci containing variable-number tandem repeat (VNTR) sequences. Using this approach, strains can be typed by a numerical code corresponding to the numbers of VNTRs in 12 different loci containing novel genetic elements named mycobacterial interspersed repetitive units (MIRUs) (18, 19). These genetic markers provide nonambiguous data which are highly portable between different laboratories (7, 14, 19) and which are therefore highly suitable for global epidemiological studies. Analysis of M. tuberculosis isolates collected in France indicated that they have a discrimination power close to that of IS_6110_ RFLP and that their stability is adequate to track outbreak episodes. In addition, unlike IS_6110_ RFLP, they are well suited for population genetics analyses, for they unambiguously reveal the variability of independent genetic loci, a requisite for linkage disequilibrium analysis (14). In this study, we report the incorporation into the MIRU-VNTR approach of technologies used for high-throughput human VNTR-based genotyping. We analyzed the relevance and reliability of this high-capacity system for the analysis of the global genetic diversity of M. tuberculosis at different levels of evolutionary divergence by testing a blinded reference set of 90 strains of the M. tuberculosis complex from 38 countries.
MATERIALS AND METHODS
Strains and genomic DNA.
The automated MIRU-VNTR typing system was developed using heat-treated M. tuberculosis colonies and purified DNA from isolates from Paris hospitals described previously (9, 21) and from transmissions, relapses, and laboratory cross-contaminations from the collection of the Centre de Référence des Mycobactéries (Institut Pasteur, Paris, France) described previously (14). The mycobacterial genomic DNA samples used in the blinded typing experiment were described previously (12). Briefly, genomic DNA was isolated from 90 M. tuberculosis complex strains originating from 38 countries throughout the world and from 10 non-M. tuberculosis complex strains. A set of 31 duplicate M. tuberculosis complex samples was also included. These samples were provided as blinded dried aliquots by the National Institute of Public Health and the Environment (Bilthoven, The Netherlands) and decoded after analysis.
Multiplex PCRs.
The multiplex PCR mixtures were prepared as follows, using 96-well plates and the HotStartTaq DNA polymerase kit (Qiagen, Hilden, Germany). Five nanograms of DNA was added to a final volume of 50 μl containing 0.2 μl of DNA polymerase (1 U), 10 μl of Q solution (from the above-mentioned kit), 0.2 mM (each) dATP, dCTP, dGTP, and dTTP (Pharmacia, Uppsala, Sweden), 5 μl of PCR buffer, 0.4 μM (each) primer, and 3.0, 2.0, 2.5, and 1.5 mM MgCl2 for mixtures A to D, respectively (Table 1). The oligonucleotides used in the PCR corresponded to the flanking regions of the polymorphic MIRU-VNTR loci identified in the M. tuberculosis H37Rv genome (19). For each multiplex mixture, one primer of each oligonucleotide pair was tagged with a different fluorescent dye.
TABLE 1.
Conditions for multiplex PCRs of the 12 MIRU-VNTR loci
| Polymorphisma | Multiplex PCR mixture | MIRU locus | MIRU-VNTR length (bp)b | Primer sequence (5′–3′) (labeling) |
|---|---|---|---|---|
| High | A | 4 | 77 | GCGCGAGAGCCCGAACTGC (FAM) |
| GCGCAGCAGAAACGTCAGC | ||||
| 26 | 51 | TAGGTCTACCGTCGAAATCTGTGAC | ||
| CATAGGCGACCAGGCGAATAG (HEX) | ||||
| 40 | 54 | GGGTTGCTGGATGACAACGTGT (NED) | ||
| GGGTGATCTCGGCGAAATCAGATA | ||||
| High | B | 10 | 53 | GTTCTTGACCAACTGCAGTCGTCC |
| GCCACCTTGGTGATCAGCTACCT (FAM) | ||||
| 16 | 53 | TCGGTGATCGGGTCCAGTCCAAGTA | ||
| CCCGTCGTGCAGCCCTGGTAC (HEX) | ||||
| 31 | 53 | ACTGATTGGCTTCATACGGCTTTA | ||
| GTGCCGACGTGGTCTTGAT (NED) | ||||
| Low | C | 2 | 53 | TGGACTTGCAGCAATGGACCAACT |
| TACTCGGACGCCGGCTCAAAAT (FAM) | ||||
| 23 | 53 | CTGTCGATGGCCGCAACAAAACG (HEX) | ||
| AGCTCAACGGGTTCGCCCTTTTGTC | ||||
| 39 | 53 | CGCATCGACAAACTGGAGCCAAAC | ||
| CGGAAACGTCTACGCCCCACACAT (NED) | ||||
| Low | D | 20 | 77 | TCGGAGAGATGCCCTTCGAGTTAG (FAM) |
| GGAGACCGCGACCAGGTACTTGTA | ||||
| 24 | 54 | CGACCAAGATGTGCAGGAATACAT | ||
| GGGCGAGTTGAGCTCACAGAA (HEX) | ||||
| 27 | 53 | TCGAAAGCCTCTGCGTGCCAGTAA | ||
| GCGATGTGAGCGTGCCACTCAA (NED) |
The thermocycler programs for the four multiplex reactions were identical. The PCRs were carried out using a PCR Express cycler (Hybaid, Ashford, Great Britain) starting with a denaturing step of 15 min at 95°C, followed by 40 cycles of 1 min at 94°C, 1 min at 59°C, and 1 min 30 s at 72°C. The reactions were terminated by incubation for 10 min at 72°C. Negative controls consisted of the PCR performed on reaction mixtures lacking mycobacterial DNA.
Automated MIRU-VNTR analysis.
For each multiplex PCR, 1 μl of 20-fold-diluted PCR products was added to a loading buffer containing 1.25 μl of deionized formamide (Amresco, Solon, Ohio), 0.5 μl of blue dextran-EDTA (PE Applied Biosystems, Courtaboeuf, France), and 0.25 μl of Genescan 2500 Rox-labeled size standard (PE Applied Biosystems) using a Multimek automated 96-channel pipettor (Beckman, Gagny, France). Before being loaded, the samples were denatured at 95°C for 2 min and then kept on ice. The samples were subjected to electrophoresis using a 96-well ABI 377 automatic sequencer, Long Ranger Singel packs (Bio-Whittaker Molecular Applications, Rockland, Maine), and a 36-cm-long plate. After a 2-min prerun step, each lane was loaded with a 1-μl portion of sample using an eight-channel syringe. One run of 2 min 30 s was made between the loading of even- and odd-numbered lanes. The samples were run for 5.5 h at 3,000 V, 2,400 scans per h, and 51°C using the D filter. The sizes of the PCR fragments were estimated using the GeneScan and Genotyper software packages (PE Applied Biosystem). The sizes of the labeled amplicons were analyzed using the local Southern size calling method, with an analysis range from 1,500 to 12,000 scans and a detection threshold set to a peak height of 50. The peak sizes from the 94- to 1,115-bp fragments of the internal size standards of the different lanes were matched using a sample run at the center of the gel as a reference. Custom macro programs (available upon request) associated with Genotyper were used for automated conversion of these data into MIRU-VNTR alleles. The assignment of the various alleles was based on the corresponding expected sizes of the PCR products for the 12 MIRU-VNTR loci, calculated from the data of Supply et al. (19). Size tolerances of ±4 and ±6 bp were fixed for PCR fragments below and above 500 bp, respectively, with an offset of −1 bp. The genotype data were then exported (via the Microsoft Excell 98 software) to File Maker Pro, version 4.0 (Claris Corporation, Santa Clara, Calif.), database management software, which was used for automated retrieval of duplicate samples and repeated genotypes.
MIRU-VNTR genetic distance analysis.
Neighbor-joining analysis was performed with PAUP (D. Swofford, PAUP—phylogenetic analysis using parsimony (and other methods), 4.0 beta version, Sinauer Associates, Inc., Sunderland, Mass.). A consensus tree based on 10,000 bootstrap replicates was generated using the majority rule formula. The differences in MIRU-VNTR numbers were used to estimate the genetic distance. The data were treated as ordered character states, based on the assumption of a stepwise mutation mechanism for the MIRU-VNTR loci. This assumption is supported by the analysis of MIRU-VNTR changes in locus 4 in the Mycobacterium bovis BCG genealogy (19).
RESULTS
Automated fluorescence-based MIRU-VNTR genotyping.
We have recently identified 12 M. tuberculosis minisatellite-like loci that contain MIRU-VNTRs. In these loci, the sizes of the repeat units range from 51 to 77 bp (19). The numbers of variable units in each locus are calculated by measuring the sizes of the fragments amplified with specific primers hybridizing to the MIRU flanking regions (14, 19). First, we developed four different multiplex PCR assays in order to analyze four groups of three loci simultaneously. The groups were composed based on the compatibilities of the PCRs and on the ranking of polymorphisms of the different loci, as measured by the numbers and frequencies of their respective alleles (14), such that the loci with high to low polymorphism were grouped together (mixture A to mixture D; Table 1). For each multiplex PCR assay, one primer per oligonucleotide pair was tagged with a specific fluorescent dye (Fig. 1). The PCR cycles for the four multiplex reactions were identical, which allows them to be run simultaneously on a thermocycler.
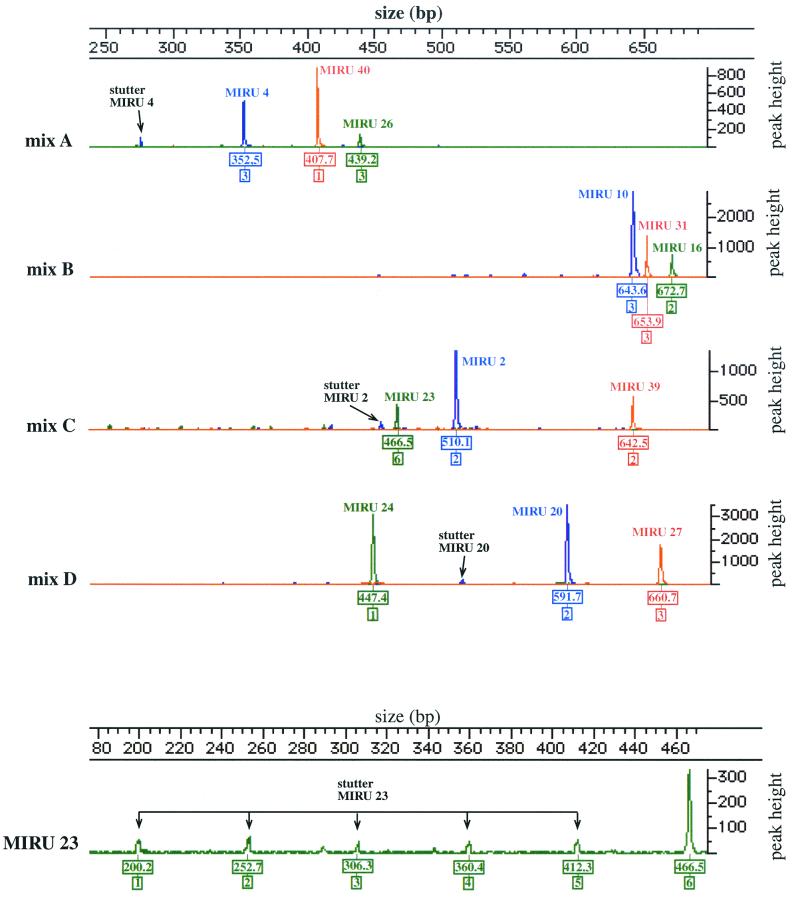
FIG. 1.
Multiplex PCR analysis of the 12 MIRU-VNTR loci (top) of M. tuberculosis H37Rv with a detailed view of locus 23 (bottom). Four multiplex PCRs were performed on genomic DNA from M. tuberculosis H37Rv, and the products were loaded onto four lanes of an automated sequencer. The GeneScan software was used for the collection of the raw sizing data. The results from the analysis of these data with the Genotyper software are shown. Boxed numbers below the signal peaks, calculated sizes (in base pairs) of the labeled PCR products and the corresponding identified MIRU-VNTR alleles. Positions of stutter peaks, easily diagnosed as low-intensity PCR artifacts, are shown.
The PCR fragments labeled with the three different fluorescent dyes from each multiplex PCR were combined with an internal size standard, labeled by a fourth dye, and then analyzed in single lanes on an automated DNA sequencer. The electrophoresis and analysis parameters were adapted in order to size amplicons from 200 to 1,000 bp, which covers the size ranges of all MIRU-VNTR alleles obtained for M. tuberculosis isolates studied to date (14, 19). The accuracy of the size determination was controlled by analyzing labeled PCR fragments amplified in multiplex PCR from M. tuberculosis H37Rv and other isolates, for which the exact sizes were determined by sequence analysis (19). Sizing was found to be reproducible, with within-run and between-run average precisions of ±0.5 and ±0.6 bp, respectively. Mean errors ± standard deviations for the sizing accuracy were 1.1 ± 1.0 bp for fragments below 500 bp and 0.8 ± 4.6 bp for fragments from 500 to up to 971 bp. This ensures the unambiguous identification of MIRU-VNTR alleles, which differ in size by 51 to 77 bp.
Stutter peaks are common during the genotyping of short tandem repeat sequences and mostly reflect artifactual strand slippage of the polymerase during PCR. Such stutter peaks were also quite frequently observed for the PCRs of the various MIRU-VNTR loci. However, they could be easily diagnosed because, as for short tandem repeat sequences, they appeared as a ladder of much-lower-intensity peaks, corresponding to sizes of PCR fragments that lack one or more repeats (Fig. 1). We noticed two exceptions: M. tuberculosis isolates with as many as 10 and 11 MIRU-VNTRs in locus 23 (isolates 67 and 93; Table 2). In these cases, the stutter peaks from the multiplex reaction had heights comparable to those of the fragments corresponding to these alleles. When these samples were reanalyzed by PCR with only the primer pair corresponding to this locus, single high-intensity peaks were obtained, allowing us to correctly assign the alleles.
TABLE 2.
MIRU-VNTR analysis of 90 strains of the M. tuberculosis complex
| Straina | Copy no. for MIRU locusb | Species | Origin | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 4 | 10 | 16 | 20 | 23 | 24 | 26 | 27 | 31 | 39 | 40 | |||
| 1 | 2 | 2 | 5 | 3 | 1 | 3 | 1 | 5 | 3 | 3 | 1 | 1 | M. tuberculosis | The Netherlands |
| 2c | 2 | 2 | 2 | 3 | 2 | 4 | 2 | 5 | 3 | 3 | 2 | 2 | M. bovis BCG | The Netherlands |
| 4c | 2 | 2 | 3 | 3 | 1 | 5 | 1 | 4 | 3 | 4 | 2 | 3 | M. tuberculosis | Rwanda |
| 6 | 2 | 2 | 5 | 3 | 2 | 4 | 2 | 4 | 3 | 5 | 2 | 2 | M. bovis | The Netherlands |
| 7c | 1 | 2 | 4 | 3 | 2 | 5 | 1 | 5 | 3 | 2 | 2 | 4 | M. tuberculosis | Mongolia |
| 8 | 2 | 2 | 4 | 3 | 2 | 5 | 1 | 5 | 3 | 3 | 2 | 3 | M. tuberculosis | Vietnam |
| 10c | 2 | 2 | 4 | 2 | 2 | 6 | 1 | 5 | 3 | 3 | 2 | 1 | M. tuberculosis | Ecuador |
| 11c | 2 | 6 | 2 | 2 | 1 | 1 | 1 | 5 | 3 | 3 | 2 | 3 | M. tuberculosis | Greenland |
| 12 | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 5 | 2 | 2 | 2 | 3 | M. tuberculosis | Tunisia |
| 13c | 2 | 2 | 5 | 3 | 2 | 5 | 1 | 5 | 3 | 3 | 2 | 3 | M. tuberculosis | Sri Lanka |
| 14 | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 7 | 3 | 5 | 4 | 3 | M. tuberculosis | South Africa |
| 15c | 2 | 2 | 2 | 3 | 2 | 5 | 1 | 2 | 3 | 3 | 1 | 3 | M. tuberculosis | Iran |
| 16 | 2 | 3 | 3 | 3 | 2 | 5 | 1 | 5 | 3 | 3 | 2 | 5 | M. tuberculosis | Canada |
| 17 | 3 | 6 | 2 | 2 | 1 | 1 | 1 | 5 | 3 | 3 | 2 | 4 | M. tuberculosis | Greenland |
| 18c | 2 | 2 | 4 | 2 | 2 | 6 | 1 | 5 | 3 | 3 | 2 | 4 | M. tuberculosis | United States |
| 19 | 2 | 2 | 6 | 4 | 2 | 5 | 1 | 5 | 3 | 5 | 3 | 2 | M. tuberculosis | India |
| 20c | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 7 | 3 | 6 | 3 | 3 | M. tuberculosis | Mongolia |
| 21 | 2 | 5 | 4 | 3 | 1 | 6 | 2 | 2 | 3 | 6 | 4 | 3 | M. tuberculosis | Zimbabwe |
| 23c | 1 | 2 | 4 | 2 | 2 | 3 | 1 | 5 | 3 | 2 | 2 | 4 | M. tuberculosis | Honduras |
| 24 | 2 | 3 | 7 | 4 | 2 | 4 | 1 | 4 | 3 | 4 | 2 | 2 | M. bovis | Saudi Arabia |
| 25 | 2 | 6 | 7 | 4 | 1 | 4 | 2 | 2 | 2 | 1 | 2 | 2 | M. microti | United Kingdom |
| 26 | 2 | 2 | 4 | 3 | 1 | 5 | 1 | 5 | 3 | 2 | 2 | 1 | M. tuberculosis | Zimbabwe |
| 27c | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 4 | 3 | 3 | 2 | 4 | M. tuberculosis | Ethiopia |
| 28c | 2 | 2 | 5 | 3 | 2 | 5 | 1 | 5 | 3 | 2 | 2 | 3 | M. tuberculosis | Sri Lanka |
| 29 | 2 | 2 | 5 | 3 | 2 | 5 | 1 | 5 | 3 | 3 | 2 | 3 | M. tuberculosis | India |
| 30 | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 8 | 3 | 5 | 3 | 3 | M. tuberculosis | South Africa |
| 31 | 2 | 2 | 2 | 3 | 2 | 5 | 1 | 5 | 3 | 3 | 2 | 3 | M. tuberculosis | Iran |
| 32c | 2 | 3 | 3 | 2 | 2 | 6 | 1 | 3 | 3 | 3 | 2 | 1 | M. tuberculosis | H37Rv |
| 33 | 2 | 2 | 5 | 3 | 2 | 3 | 1 | 5 | 3 | 3 | 2 | 3 | M. tuberculosis | The Netherlands |
| 34 | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 7 | 3 | 5 | 3 | 3 | M. tuberculosis | Malaysia |
| 35c | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 5 | 4 | 4 | 2 | 4 | M. tuberculosis | Rwanda |
| 36 | 2 | 4 | 4 | 3 | 2 | 6 | 2 | 2 | 3 | 5 | 2 | 2 | M. tuberculosis | India |
| 37c | 2 | 2 | 3 | 3 | 1 | 5 | 1 | 4 | 3 | 4 | 2 | 3 | M. tuberculosis | Uganda |
| 38c | 2 | 2 | 4 | 3 | 2 | 5 | 1 | 5 | 3 | 3 | 2 | 5 | M. tuberculosis | Tahiti |
| 40 | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 3 | 3 | 3 | 2 | 3 | M. tuberculosis | Burundi |
| 41c | 2 | 2 | 3 | 3 | 2 | 6 | 1 | 3 | 3 | 3 | 2 | 1 | M. tuberculosis | Chile |
| 42c | 1 | 2 | 4 | 3 | 2 | 6 | 1 | 5 | 3 | 2 | 2 | 6 | M. tuberculosis | Tahiti |
| 43 | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 7 | 3 | 5 | 3 | 3 | M. tuberculosis | China |
| 44 | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 5 | 3 | 5 | 3 | 3 | M. tuberculosis | Thailand |
| 45c | 2 | 2 | 3 | 3 | 1 | 5 | 1 | 5 | 3 | 5 | 4 | 4 | M. tuberculosis | Malaysia |
| 46 | 2 | 2 | 4 | 2 | 2 | 7 | 1 | 5 | 3 | 3 | 2 | 3 | M. tuberculosis | Chile |
| 47 | 2 | 5 | 3 | 3 | 2 | 6 | 2 | 2 | 3 | 5 | 1 | 3 | M. bovis | The Netherlands |
| 49 | 2 | 2 | 8 | 3 | 2 | 5 | 1 | 1 | 3 | 4 | 3 | 4 | M. tuberculosis | Tanzania |
| 50c | 2 | 2 | 5 | 3 | 2 | 5 | 1 | 5 | 3 | 3 | 2 | 3 | M. tuberculosis | Czech Republic |
| 51 | 2 | 2 | 5 | 3 | 2 | 5 | 1 | 5 | 3 | 3 | 2 | 3 | M. tuberculosis | The Netherlands |
| 53c | 2 | 2 | 5 | 3 | 1 | 3 | 1 | 5 | 3 | 3 | 2 | 1 | M. tuberculosis | Argentina |
| 54 | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 7 | 3 | 5 | 3 | 2 | M. tuberculosis | Thailand |
| 56 | 2 | 2 | 5 | 3 | 2 | 5 | 1 | 5 | 3 | 2 | 2 | 4 | M. tuberculosis | Curacao |
| 58 | 2 | 2 | 6 | 3 | 2 | 5 | 1 | 5 | 3 | 3 | 2 | 3 | M. tuberculosis | Argentina |
| 60 | 2 | 2 | 5 | 3 | 2 | 5 | 1 | 5 | 3 | 3 | 2 | 3 | M. tuberculosis | Bolivia |
| 62 | 2 | 6 | 7 | 3 | 1 | 4 | 2 | 2 | 2 | 1 | 2 | 2 | M. microti | United Kingdom |
| 63c | 2 | 2 | 5 | 3 | 1 | 3 | 1 | 5 | 3 | 3 | 2 | 4 | M. tuberculosis | Italy |
| 65c | 2 | 5 | 4 | 3 | 2 | 6 | 2 | 2 | 3 | 6 | 1 | 3 | M. tuberculosis | The Netherlands |
| 66c | 2 | 2 | 4 | 3 | 2 | 6 | 1 | 3 | 3 | 3 | 2 | 3 | M. tuberculosis | Spain |
| 67 | 2 | 5 | 4 | 3 | 2 | A | 2 | 2 | 3 | 6 | 3 | 3 | M. tuberculosis | Comoro Islands |
| 71 | 2 | 3 | 2 | 3 | 2 | 4 | 2 | 5 | 3 | 3 | 2 | 2 | M. bovis BCG | Japan |
| 72 | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 4 | 3 | 5 | 3 | 3 | M. tuberculosis | Central African Republic |
| 73 | 2 | 3 | 2 | 2 | 2 | 4 | 2 | 4 | 3 | 3 | 2 | 2 | M. bovis | The Netherlands |
| 74c | 2 | 5 | 4 | 3 | 2 | 6 | 2 | 2 | 3 | 5 | 3 | 4 | M. tuberculosis | India |
| 76 | 2 | 1 | 6 | 4 | 2 | 4 | 2 | 2 | 2 | 3 | 2 | 2 | M. bovis | Argentina |
| 77 | 2 | 2 | 5 | 3 | 2 | 3 | 1 | 5 | 3 | 3 | 2 | 3 | M. tuberculosis | Tunisia |
| 81 | 2 | 5 | 6 | 3 | 2 | 4 | 2 | 2 | 2 | 3 | 2 | 1 | M. bovis | Argentina |
| 82 | 2 | 3 | 3 | 3 | 2 | 5 | 1 | 4 | 3 | 3 | 2 | 4 | M. tuberculosis | Canada |
| 83 | 2 | 2 | 2 | 3 | 2 | 4 | 2 | 5 | 3 | 3 | 2 | 2 | M. bovis BCG | Russia |
| 85c | 2 | 1 | 5 | 4 | 2 | 4 | 2 | 5 | 3 | 5 | 2 | 2 | M. bovis | The Netherlands |
| 86 | 2 | 2 | 5 | 3 | 2 | 5 | 1 | 5 | 3 | 3 | 2 | 3 | M. tuberculosis | Bolivia |
| 87c | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 4 | 3 | 3 | 2 | 3 | M. tuberculosis | United States |
| 88 | 2 | 2 | 7 | 2 | 2 | 5 | 1 | 1 | 3 | 2 | 2 | 3 | M. tuberculosis | Russia |
| 90 | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 7 | 3 | 2 | 3 | 3 | M. tuberculosis | South Korea |
| 92 | 2 | 3 | 4 | 4 | 2 | 4 | 2 | 2 | 4 | 2 | 2 | 1 | M. africanum | The Netherlands |
| 93 | 2 | 5 | 4 | 3 | 2 | B | 2 | 2 | 3 | 6 | 3 | 3 | M. tuberculosis | Tanzania |
| 95 | 2 | 2 | 4 | 2 | 2 | 6 | 1 | 5 | 3 | 3 | 2 | 1 | M. tuberculosis | Spain |
| 96c | 2 | 2 | 3 | 1 | 2 | 5 | 1 | 5 | 3 | 3 | 2 | 2 | M. tuberculosis | The Netherlands |
| 97 | 2 | 1 | 3 | 3 | 1 | 5 | 1 | 2 | 3 | 4 | 2 | 3 | M. tuberculosis | Uganda |
| 98c | 2 | 2 | 4 | 2 | 2 | 6 | 1 | 6 | 3 | 3 | 2 | 1 | M. tuberculosis | Ecuador |
| 100 | 2 | 2 | 6 | 2 | 2 | 5 | 1 | 7 | 3 | 4 | 3 | 3 | M. africanum | The Netherlands |
| 101 | 2 | 5 | 6 | 4 | 2 | 4 | 2 | 2 | 2 | 3 | 2 | 1 | M. bovis | Argentina |
| 103c | 2 | 2 | 2 | 3 | 2 | 5 | 1 | 5 | 3 | 2 | 2 | 3 | M. tuberculosis | China |
| 109 | 2 | 3 | 3 | 2 | 2 | 6 | 1 | 3 | 3 | 3 | 2 | 1 | M. tuberculosis | H37Ra |
| 111 | 2 | 2 | 3 | 4 | 2 | 5 | 1 | 7 | 3 | 5 | 3 | 3 | M. tuberculosis | South Korea |
| 112 | 2 | 2 | 6 | 4 | 2 | 5 | 1 | 7 | 3 | 5 | 3 | 2 | M. tuberculosis | The Netherlands |
| 116c | 3 | 2 | 3 | 2 | 1 | 2 | 6 | 3 | 2 | 4 | 2 | 8 | M. canettii | Somalia |
| 117 | 2 | 3 | 2 | 2 | 2 | 4 | 2 | 6 | 3 | 3 | 2 | 2 | M. bovis | Argentina |
| 118 | 2 | 2 | 4 | 2 | 2 | 6 | 1 | 3 | 4 | 2 | 2 | 3 | M. tuberculosis | Honduras |
| 120 | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 5 | 3 | 5 | 2 | 2 | M. tuberculosis | Burundi |
| 121 | 2 | 2 | 3 | 3 | 2 | 5 | 1 | 5 | 3 | 4 | 2 | 3 | M. tuberculosis | Central African Republic |
| 123 | 2 | 3 | 5 | 3 | 2 | 6 | 1 | 4 | 3 | 3 | 2 | 4 | M. tuberculosis | Czech Republic |
| 126 | 2 | 3 | 2 | 2 | 2 | 4 | 2 | 5 | 3 | 3 | 2 | 2 | M. bovis | Argentina |
| 130 | 2 | 3 | 2 | 3 | 2 | 4 | 2 | 5 | 3 | 3 | 2 | 2 | M. bovis | The Netherlands |
Genotyping of a blinded set of M. tuberculosis complex isolates from 38 countries.
A synthetic interlaboratory study recently compared all currently known typing methods for M. tuberculosis complex isolates with respect to their reproducibility, sensitivity, specificity, and discrimination (12). The methods were tested by using a blinded set of 131 samples, including 90 isolates from 38 countries, as well as duplicate samples and non-M. tuberculosis complex strains. We analyzed the same isolate set similarly blinded to evaluate the performances of the automated MIRU-VNTR system.
As shown in Table 2, the MIRU-VNTR typing method was 100% sensitive, as all M. tuberculosis complex strains were fully typeable, with the 12 alleles amplified in all cases, and 100% reproducible, as all blinded duplicate samples analyzed were correctly assigned. The method was also 100% specific, as none of the non-M. tuberculosis complex strains gave the patterns expected for the M. tuberculosis complex. For all non-M. tuberculosis complex strains, either the PCRs yielded no amplification product or the amplified fragments had sizes that did not correspond to any of the M. tuberculosis complex MIRU-VNTR alleles (not shown). Thus, the MIRU-VNTR system can be used to identify the M. tuberculosis complex strains.
The MIRU-VNTR typing detected 78 different patterns among the 90 M. tuberculosis complex isolates (Table 2 and Fig. 2). Eight new alleles not found in isolates analyzed before were detected (14, 19), which raises the potential number of different MIRU-VNTR combinations to more than 2 × 108. In contrast to what is found for IS_6110_ RFLP typing, nearly all low-IS_6110_-copy-number (n ≤ 6) isolates were discriminated by MIRU-VNTR typing (20 distinct patterns out of 21 isolates). The only exceptions were the Russian and French M. bovis BCG vaccine sister strains (see below). In addition, most of the high-copy-number isolates (n > 6) were also discriminated (58 patterns out of 69 isolates), except for the cases discussed below.
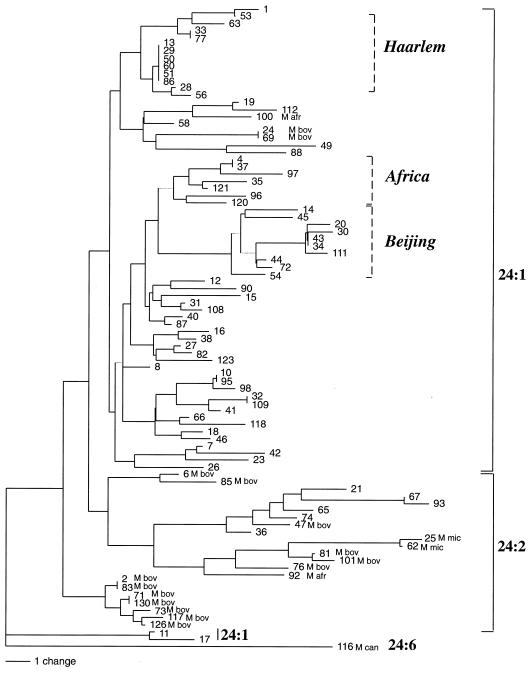
FIG. 2.
Dendrogram of genetic relationships among 90 isolates of the M. tuberculosis complex from 38 countries based on the 12 MIRU-VNTR loci. A majority rule consensus tree was built using the neighbor-joining distance algorithm as described in Materials and Methods. The linkage distance scale is indicated at the bottom. M. bovis isolate 130 is discriminated from M. bovis isolate 71 based on the presence of an additional 53-bp invariable MIRU unit in locus 4 in the first strain (this unit is not taken into account in the MIRU-VNTR dendrogram analysis). Dotted lines, groups including mainly Haarlem, Africa, and Beijing isolates identified by Kremer et al. (12). The numbers of MIRU-VNTRs in locus 24 (24:n) are indicated at the right. Isolates are M. tuberculosis, except for those indicated as M bov, M afr, M can, and M mic, corresponding to M. bovis, Mycobacterium africanum, M. canettii, and M. microti isolates, respectively.
The genetic relationships among the 90 isolates were analyzed on the basis of the MIRU-VNTR genotypes using the neighbor-joining distance algorithm (Fig. 2). Beijing, Africa, and Haarlem genotype families, including M. tuberculosis isolates originating from various parts of the world, were identified in this collection (12). The isolates within these families possess distinct, but at least 50% similar, IS_6110_ RFLP patterns. Many of them also appear highly similar and often identical based on other typing markers. This is especially the case for the Beijing family. Most of the members previously assigned to these families were readily distinguished from each other by MIRU-VNTRs. Seven distinct patterns were obtained for the 8 Beijing isolates, seven were obtained for the 8 Africa isolates, and nine were obtained for the 13 Haarlem isolates. In addition, most of them (all of the Beijing family) were recognized as closely related by analysis of MIRU-VNTR genetic distances. Exceptions were isolates 8, 58, 87, and 123 (Haarlem family) and 72 and 40 (Africa family). Furthermore, an additional isolate from South Korea (111) was included in the Beijing group by MIRU-VNTR analysis, which is consistent with its IS_6110_ RFLP and spoligotype, which display motifs similar to those typical of the Beijing group (12). Two additional isolates (29 and 77) were included in two clusters with identical MIRU-VNTRs containing five Haarlem strains and one Haarlem strain, respectively. This clustering was also consistent with their respective IS_6110_ RFLPs and spoligotypes, which also display motifs typical of the previously defined Haarlem strains (12). However, one or two M. tuberculosis isolates with more distantly related or unrelated IS_6110_ RFLPs were included in the three families by MIRU-VNTR genetic distance analysis (isolates 1 and 56 for the Haarlem family, 96 for the Africa family, and 72 for the Beijing family).
The remaining clinical isolates with identical MIRU-VNTR genotypes included a group of two M. tuberculosis strains from Ecuador (isolate 10) and Spain (isolate 95) and a group of two high-copy-number M. bovis strains from Saudi Arabia (isolate 24) and The Netherlands (isolate 69). These isolates also have highly similar, although not identical, IS_6110_ RFLP patterns (12), suggesting that they are clonal. M. tuberculosis laboratory strain H37Rv and its avirulent derivative H37Ra, which have been cultivated separately for several decades, also displayed identical MIRU-VNTR patterns, although they differ by 3 of 14 IS_6110_ bands. Moreover, the Russian and French M. bovis BCG vaccine sister strains displayed identical MIRU-VNTRs, whereas the Japanese sister strain differed from these by only a single unit change in locus 4, consistent with the data of Supply et al. (19).
DISCUSSION
The 12-locus MIRU-VNTR method described here represents a powerful approach to high-resolution genotyping of M. tuberculosis isolates. It uses multiplex PCR with fluorescence-labeled primers to analyze four sets of three MIRU-VNTR loci simultaneously and provides the automation that is necessary for quick and reliable production and management of the genotyping data. To our knowledge, up to now, only one comparable VNTR-based method has been reported for typing another bacterium, Bacillus anthracis (11). MIRU-VNTR typing was found to be 100% reproducible, sensitive, and specific for M. tuberculosis complex isolates, a performance that has not been achieved by any other resolution typing method, including IS_6110_ RFLP, tested in the same conditions (12). Moreover, the discrimination power of MIRU-VNTR typing was close to that of IS_6110_ RFLP (78 compared to 84 patterns, respectively), and MIRU-VNTR typing performed better than the other RFLP-based or IS_6110_-independent PCR-based typing methods (compare the present results with those in reference 12). In addition, the genetic-distance dendrogram built from MIRU-VNTRs correlated well with that based on IS_6110_ RFLP, with about 80% of the isolates similarly grouped by the two methods. These findings are similar to those of a more limited study of M. tuberculosis strains isolated in Paris (14). The results for 90 strains from 38 countries presented here thus extend these observations to the scale of global genetic diversity of the M. tuberculosis complex worldwide.
MIRU-VNTRs clearly distinguished the majority of the isolates of Beijing, Africa, and Haarlem families from each other. Most of them were also recognized as being closely related by MIRU-VNTRs (see also below). The Beijing family appeared the most homogenous family, which is consistent with the results obtained with IS_6110_ RFLP and other markers (12). Our analysis using the high-resolution typing system fully independent from IS_6110_ thus supports the assumption that these isolates have expanded clonally from common ancestors and subsequently disseminated globally (12). In contrast, almost all of the clinical isolates with high IS_6110_ copy numbers that had not been assigned to the Beijing, Africa, and Haarlem families had unique MIRU-VNTRs, except for two groups of two isolates each. However, these isolates might correspond to other genotype families, as they also had high similarities in their IS_6110_ RFLPs. In addition, MIRU-VNTRs distinguished all strains with low IS_6110_ copy numbers except for two M. bovis BCG vaccine sister strains, in contrast to IS_6110_ RFLP, for which two clusters of two and five strains with identical patterns were obtained. This class of isolates includes many M. tuberculosis isolates from several regions, especially Asia, and about 80% M. bovis isolates from all regions. Their typing is often problematic and requires combinations of several techniques (4). Thus, MIRU-VNTR typing is especially useful for analyzing the genetic diversity among these isolates.
The differences in genetic-relationship results between MIRU-VNTR typing and IS_6110_ RFLP analysis, observed for only a minority of isolates, may be due to several factors. Some of these differences correspond to strains or groups of strains which apparently share a few IS_6110_ bands with the other strains of the complex (for instance, Mycobacterium microti isolates 25 and 62). IS_6110_ RFLP analysis suggested the closest relationships between these strains and groups including M. tuberculosis isolates (12), while MIRU-VNTR analysis suggested the closest relationships to M. bovis strains (Fig. 2). The latter grouping is consistent with results from comparative genomics suggesting closer relationships between these strains, which are pathogenic for animals (3). This difference might be due to the fact that markers such as IS_6110_ RFLP may evolve too fast to be reliable for estimating higher levels of divergence (20). Moreover, the documented existence of IS_6110_ insertion hot spots (5, 6, 15) together with the intrinsic difficulties of interpreting RFLP patterns can conceivably mislead the analysis of some strains, especially those sharing only few bands. On the other hand, we cannot exclude the possibility that independent convergence to closely related MIRU-VNTR genotypes by unrelated isolates may occasionally occur. This might explain the grouping by MIRU-VNTR typing of a few isolates assigned to the Africa and Haarlem families, which, based on analysis with IS_6110_ RFLP and other markers, are classified as unrelated isolates (and vice versa). However, this convergence is necessarily limited by the huge number of possible MIRU-VNTR combinations (>2 × 108) provided by the 12 loci, which strongly reduces the probability of obtaining totally identical genotypes only by chance. Consistently, it was noted that the few isolates with identical MIRU-VNTR types in the different genotype families, as well as H37Rv and H37Ra strains, all had only slightly distinct IS_6110_ RFLP patterns.
The last observation supports the assumption that the combined molecular clock of the 12 MIRU-VNTR loci runs slightly slower than that of IS_6110_ RFLP. It may be useful to note that, like multilocus enzyme electrophoresis for instance, the genetic variation indexed by MIRU-VNTR typing relies on independent genomic loci that can be separately targeted by specific PCR primers. This is different from IS_6110_ RFLP, where all genomic loci containing IS_6110_ are probed simultaneously, without the possibility to assess individual loci. This provides the possibility to modulate the speed of the combined molecular clock of MIRU-VNTRs by selecting one or several targeted loci to analyze different scales of evolutionary divergence within the same system. Interestingly, the inclusion of two other VNTR loci identified by Frothingham and Meeker-O'Connell (7), namely, VNTR-A and VNTR-C, allowed us to distinguish three additional patterns among strains with identical MIRU-VNTRs, including two members of the Africa family (strains 4 and 37), BCG Pasteur and BCG Russia (strains 2 and 83), and two M. bovis isolates with high IS_6110_ copy numbers (strains 24 and 69) (12). Inclusion of other polymorphic tandem repeat loci in the M. tuberculosis complex genomes (16; Supply et al., unpublished results) may thus further improve the performance of MIRU-VNTR typing compared to that of IS_6110_ RFLP for tracing recent transmission of strains circulating within defined geographic areas.
Conversely, the selective use of some less-variable MIRU-VNTR loci, such as locus 24, may be especially informative for higher levels of evolutionary divergence. Locus 24 is among the least diverse loci, with only three alleles (Table 2). This locus is polymorphic between three groups, which appear genetically distant by both MIRU-VNTR and IS_6110_ RFLP typing (12), while it is monomorphic within each of these groups (Fig. 2). One group includes mainly M. tuberculosis and M. bovis isolates with high IS_6110_ copy numbers, M. microti isolates together with low-IS_6110_-copy-number M. bovis and M. tuberculosis isolates, and a single M. canettii isolate. This polymorphism, only observed among genetically distant groups, suggests a very slow evolution rate of locus 24. Therefore, we speculate that the hierarchy of allelic diversity among the MIRU-VNTR loci (14) may be correlated to the speed of their respective molecular clocks, in addition perhaps to selective constraints on the numbers of alleles.
Using the automated MIRU-VNTR typing reported here, several hundred isolates can be genotyped per week on a single sequencer, starting from genomic DNA or bacterial colonies. This is considerably faster than IS_6110_ RFLP or manual VNTR analysis. Adequate equipment and software are widely available, while specific files and macro software for standardized allele scoring will be provided by the authors upon request. Compared to IS_6110_ RFLP, the portability of MIRU-VNTR typing results will greatly facilitate the comparisons of results between different laboratories worldwide. To ensure consistency of results on a daily basis within and among laboratories, genotypic tests of standardized reference strains H37Rv or H37Ra (Table 2) should be run as validating controls. To generate a database allowing laboratories to compare their data, we have set up a website for the analysis of M. tuberculosis complex MIRU-VNTR genotypes via the Internet (http://www.ibl.fr/mirus/mirus.html). We hope that the use of this automated typing system for large-scale genotyping of M. tuberculosis will provide a new tool for M. tuberculosis global epidemiological surveillance and lead to novel comprehensive insights into the population genetics and evolution of this major human pathogen.
ACKNOWLEDGMENTS
Vincent Vatin and Karl Oulmi are gratefully acknowledged for their help in software utilization.
The work was supported by INSERM, Institut Pasteur de Lille, a grant from the Ministère de l'Education Nationale, de la Recherche et de la Technologie, and the European Community (grant QLK2-CT-2000-00630). P.S. is a Chercheur du Centre National de Recherche Scientifique. E.S. holds a Poste Vert from the INSERM.
REFERENCES
- 1.Bifani P J, Mathema B, Liu Z, Moghazeh S L, Shopsin B, Tempalski B, Driscol J, Frothingham R, Musser J M, Alcabes P, Kreiswirth B N. Identification of a W variant outbreak of Mycobacterium tuberculosis via population-based molecular epidemiology. JAMA. 1999;282:2321–2327. doi: 10.1001/jama.282.24.2321. [DOI] [PubMed] [Google Scholar]
- 2.Bifani P J, Plikaytis B B, Kapur V, Stockbauer K, Pan X, Lutfey M L, Moghazeh S L, Eisner W, Daniel T M, Kaplan M H, Crawford J T, Musser J M, Kreiswirth B N. Origin and interstate spread of a New York City multidrug-resistant Mycobacterium tuberculosis clone family. JAMA. 1996;275:452–457. [PubMed] [Google Scholar]
- 3.Brosch R, Gordon S V, Eiglmeier K, Garnier T, Tekaia F, Yeramanian E, Cole S T. Genomics, biology, and evolution of the Mycobacterium tuberculosis complex. In: Hatful G F, Jacobs W R Jr, editors. Molecular genetics of mycobacteria. Washington, D.C.: American Society for Microbiology; 1999. pp. 19–36. [Google Scholar]
- 4.Collins D M. Molecular epidemiology: Mycobacterium bovis. In: Ratledge C, Dale J, editors. Mycobacteria. Molecular biology and virulence. Oxford, United Kingdom: Blackwell Science Ltd.; 1999. pp. 123–135. [Google Scholar]
- 5.Fang Z, Forbes K J. A Mycobacterium tuberculosis IS6110 preferential locus (ipl) for insertion into the genome. J Clin Microbiol. 1997;35:479–481. doi: 10.1128/jcm.35.2.479-481.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang Z, Morrison N, Watt B, Doig C, Forbes K J. IS6110 transposition and evolutionary scenario of the direct repeat locus in a group of closely related Mycobacterium tuberculosis strains. J Bacteriol. 1998;180:2102–2109. doi: 10.1128/jb.180.8.2102-2109.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frothingham R, Meeker-O'Connell W A. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology. 1998;144:1189–1196. doi: 10.1099/00221287-144-5-1189. [DOI] [PubMed] [Google Scholar]
- 8.Gleissberg V. The threat of multidrug resistance: is tuberculosis ever untreatable or uncontrollable? Lancet. 1999;353:998–999. doi: 10.1016/S0140-6736(99)01439-7. [DOI] [PubMed] [Google Scholar]
- 9.Goguet de la Salmoniere Y O, Li H M, Torrea G, Bunschoten A, van Embden J, Gicquel B. Evaluation of spoligotyping in a study of the transmission of Mycobacterium tuberculosis. J Clin Microbiol. 1997;35:2210–2214. doi: 10.1128/jcm.35.9.2210-2214.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grange J M, Zumla A. Paradox of the global emergency of tuberculosis. Lancet. 1999;353:996. doi: 10.1016/S0140-6736(99)01196-4. [DOI] [PubMed] [Google Scholar]
- 11.Keim P, Price L B, Klevytska A M, Smith K L, Schupp J M, Okinaka R, Jackson P J, Hugh-Jones M E. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J Bacteriol. 2000;182:2928–2936. doi: 10.1128/jb.182.10.2928-2936.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kremer K, van Soolingen D, Frothingham R, Haas W H, Hermans P W, Martin C, Palittapongarnpim P, Plikaytis B B, Riley L W, Yakrus M A, Musser J M, van Embden J D. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. J Clin Microbiol. 1999;37:2607–2618. doi: 10.1128/jcm.37.8.2607-2618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maiden M C, Bygraves J A, Feil E, Morelli G, Russell J E, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant D A, Feavers I M, Achtman M, Spratt B G. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mazars E, Lesjean S, Bañuls A-L, Gilbert M, Vincent V, Gicquel B, Tibayrenc M, Locht C, Supply P. High-resolution minisatellite-based typing as a portable approach to global analysis of Mycobacterium tuberculosis molecular epidemiology. Proc Natl Acad Sci USA. 2001;98:1901–1906. doi: 10.1073/pnas.98.4.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McHugh T D, Gillespie S H. Nonrandom association of IS6110 and Mycobacterium tuberculosis: implications for molecular epidemiological studies. J Clin Microbiol. 1998;36:1410–1413. doi: 10.1128/jcm.36.5.1410-1413.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smittipat N, Palittapongarnpim P. Identification of possible loci of variable number of tandem repeats in Mycobacterium tuberculosis. Tuber Lung Dis. 2000;80:69–74. doi: 10.1054/tuld.2000.0236. [DOI] [PubMed] [Google Scholar]
- 17.Sreevatsan S, Pan X, Stockbauer K E, Connell N D, Kreiswirth B N, Whittam T S, Musser J M. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc Natl Acad Sci USA. 1997;94:9869–9874. doi: 10.1073/pnas.94.18.9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Supply P, Magdalena J, Himpens S, Locht C. Identification of novel intergenic repetitive units in a mycobacterial two-component system operon. Mol Microbiol. 1997;26:991–1003. doi: 10.1046/j.1365-2958.1997.6361999.x. [DOI] [PubMed] [Google Scholar]
- 19.Supply P, Mazars E, Lesjean S, Vincent V, Gicquel B, Locht C. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol Microbiol. 2000;36:762–771. doi: 10.1046/j.1365-2958.2000.01905.x. [DOI] [PubMed] [Google Scholar]
- 20.Tibayrenc M. Toward an integrated genetic epidemiology of parasitic protozoa and other pathogens. Annu Rev Genet. 1999;33:449–477. doi: 10.1146/annurev.genet.33.1.449. [DOI] [PubMed] [Google Scholar]
- 21.Torrea G, Offredo C, Simonet M, Gicquel B, Berche P, Pierre-Audigier C. Evaluation of tuberculosis transmission in a community by 1 year of systematic typing of Mycobacterium tuberculosis clinical isolates. J Clin Microbiol. 1996;34:1043–1049. doi: 10.1128/jcm.34.5.1043-1049.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Soolingen D, Borgdorff M W, de Haas P E, Sebek M M, Veen J, Dessens M, Kremer K, van Embden J D. Molecular epidemiology of tuberculosis in The Netherlands: a nationwide study from 1993 through 1997. J Infect Dis. 1999;180:726–736. doi: 10.1086/314930. [DOI] [PubMed] [Google Scholar]
- 23.van Soolingen D, Qian L, de Haas P E, Douglas J T, Traore H, Portaels F, Qing H Z, Enkhsaikan D, Nymadawa P, van Embden J D. Predominance of a single genotype of Mycobacterium tuberculosis in countries of east Asia. J Clin Microbiol. 1995;33:3234–3238. doi: 10.1128/jcm.33.12.3234-3238.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yaganehdoost A, Graviss E A, Ross M W, Adams G J, Ramaswamy S, Wanger A, Frothingham R, Soini H, Musser J M. Complex transmission dynamics of clonally related virulent Mycobacterium tuberculosis associated with barhopping by predominantly human immunodeficiency virus-positive gay men. J Infect Dis. 1999;180:1245–1251. doi: 10.1086/314991. [DOI] [PubMed] [Google Scholar]