Achieving 100% Typeability of Pseudomonas aeruginosa by Pulsed-Field Gel Electrophoresis (original) (raw)
Typing bacterial isolates by macrorestriction analysis of DNA by pulsed-field gel electrophoresis (PFGE) has become a widespread technique because of the general applicability of this method to any species. Among DNA-based typing techniques, macrorestriction analysis has often been found to be the most powerful and discriminatory method for the classification of isolates (3, 4). For Pseudomonas aeruginosa macrorestriction analysis is the method of choice for typing of isolates (3, 8, 9). However, we and others have recently reported that the typeability, i.e., the percentage of strains that could be assigned a type, is <100% (1, 3, 8). DNA degradation in the gel, which yields random fragments most commonly ranging from 40 to 150 kbp, prevented the typing of a specific strain (Fig. 1A and B). However, typing by other DNA-based methods, such as ribotyping, was not affected, so that the typeability remained 100% (3).
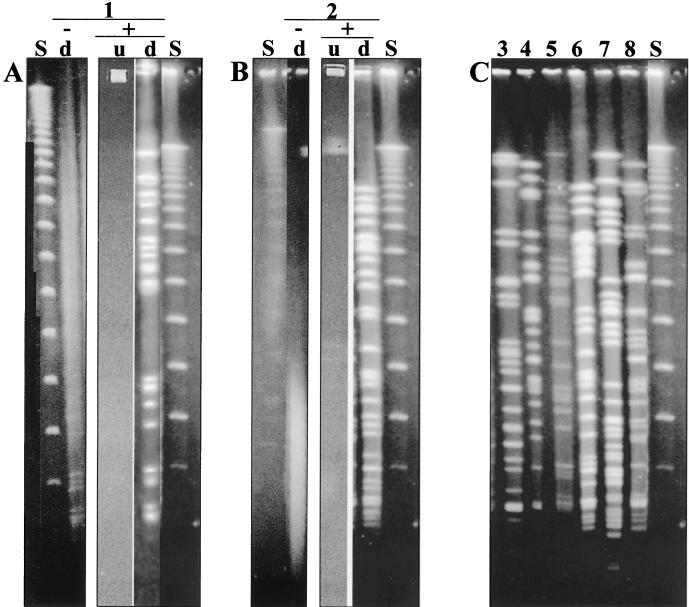
FIG. 1.
(A and B) PFGE results from strains sensitive to DNA degradation during electrophoresis. (A) Lane 1, cystic fibrosis patient P12 (9); (B) lane 2, reference hospital 1 (RH1). Symbols and abbreviations: −, conventional PFGE; +, addition of 50 μM thiourea to the buffer during the run; u, undigested chromosomal DNA; d, _Spe_I-digested P. aeruginosa isolate. (C) Examples of _Spe_I-digested P. aeruginosa genotypes from various sources with degradation-sensitive DNA after addition of 50 μM thiourea to the running buffer. Running conditions were set according to published protocol (9). Lanes: 3, P1 (9); 4, P59; 5, P20; 6, P4 (9); 7, clinical environment; 8, aquatic habitat; S, λ DNA concatemer size standard in 48.5-kbp increments.
For P. aeruginosa we achieved a typeability of 96% by PFGE when typing more than 1,200 isolates; this percentage is in the range reported by others (3). However, there were occasions when the lack of typeability hindered the answering of epidemiological questions. For example, 7 of 24 sequential isolates from one patient with cystic fibrosis and 4 of 10 isolates from a reference hospital were not typeable (Fig. 1A and B) (8, 9).
We reasoned that a nuclease must be responsible for the degradation of the DNA. This putative nuclease may have an infrequent recognition site and/or may retain a low activity despite the digestion of the DNA with proteinase K (8). Therefore, we modified our standard protocol for the isolation of intact chromosomal DNA in an agarose plug (9). We significantly increased the incubation time for the digestion with proteinase K, from 24 to 72 h, and introduced a fivefold increase in the concentration of proteinase K, changing the buffer solution daily. We also tried several other proteases. Others have reported that nucleases could be inhibited by the addition of dimethyl sulfoxide and aurintricarboxylic acid (6). However, in our hands none of these methods showed the desired effect (data not shown).
During our search for a cure for the problem, we learned of a report about Tris buffer-dependent cleavage by electrophoresis of two strains of the gram-positive Streptomyces spp. (2). Degradation of DNA was prevented by the usage of HEPES buffer instead of Tris or the addition of 50 μM thiourea in the gel buffer to scavenge reactive Tris radicals.
Using the latter method, we were able to gain macrorestriction patterns with no background for all our P. aeruginosa strains that were previously affected by DNA degradation (more than 50 strains) (Fig. 1). DNA preparation and digestion of DNA by restriction enzymes was done by our standard method without modification (9).
Strains affected by DNA degradation came from all sources examined, including isolates from patients as well as isolates from the clinical environment and aquatic habitats. Various genotypes were found among the isolates previously affected by DNA degradation (examples in Fig. 1C); however, in line with previous results isolates from one patient that were affected by DNA degradation belonged to one genotype (9; data not shown). DNA degradation by electrophoresis is not a stable trait. In a few cases we found one isolate from a group of clonal variants to be affected by the degradation process, indicating that DNA degradation is not a clonal trait (data not shown).
There are reports from studies of various bacteria, such as Clostridium difficile, Klebsiella spp., Ralstonia CDC group IV c-2, and Vibrio parahaemolyticus, which demonstrate that DNA degradation might be a problem in typing by PFGE (4, 5, 7). All eight clinical isolates of Ralstonia CDC group IV c-2 and a significant proportion of the V. parahaemolyticus isolates suffered from such degradation (5, 7). Based on our experience, in which the cure for DNA degradation was transferred from gram-positive Streptomyces (2) to gram-negative P. aeruginosa (this study), we conclude that the inexpensive and convenient addition of 50 μM thiourea to the gel buffer may also solve the problem of DNA degradation in these bacteria.
REFERENCES
- 1.Barth A L, Pitt T L. Auxotrophic variants of Pseudomonas aeruginosa are selected from prototrophic wild-type strains in respiratory infections in patients with cystic fibrosis. J Clin Microbiol. 1995;33:37–40. doi: 10.1128/jcm.33.1.37-40.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Evans M, Kaczmarek F S, Stutzman-Engwall K, Dyson P. Characterization of a Streptomyces-lividans-type site-specific DNA modification system in the avermectin-producer Streptomyces avermitilis permits investigation of two novel giant linear plasmids, pSA1 and pSA2. Microbiology. 1994;140:1367–1371. doi: 10.1099/00221287-140-6-1367. [DOI] [PubMed] [Google Scholar]
- 3.Grundmann H, Schneider C, Hartung D, Daschner F D, Pitt T L. Discriminatory power of three DNA-based typing techniques for Pseudomonas aeruginosa. J Clin Microbiol. 1995;33:528–534. doi: 10.1128/jcm.33.3.528-534.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kristjansson M, Samore M H, Gerding D N, DeGirolami P C, Bettin K M, Karchmer A W, Arbeit R D. Comparison of restriction endonuclease analysis, ribotyping, and pulsed-field gel electrophoresis for molecular differentiation of Clostridium difficile strains. J Clin Microbiol. 1994;32:1963–1969. doi: 10.1128/jcm.32.8.1963-1969.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marshall S, Clark C G, Wang G, Mulvey M, Kelly M T, Johnson W M. Comparison of molecular methods for typing Vibrio parahaemolyticus. J Clin Microbiol. 1999;37:2473–2478. doi: 10.1128/jcm.37.8.2473-2478.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matveyev A V, Rutgers E, Söderbäck E, Bergman B. A novel genome rearrangement involved in heterocyst differentiation of the cyanobacterium Anabaena sp. PCC 7120. FEMS Microbiol Lett. 1994;116:201–208. [Google Scholar]
- 7.Moissenet D, Goujon C P, Garbarg-Chenon A, Vu-Thien H. CDC group IV c-2: a new Ralstonia species close to Ralstonia eutropha. J Clin Microbiol. 1999;37:1777–1781. doi: 10.1128/jcm.37.6.1777-1781.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Römling U, Wingender J, Müller H, Tümmler B. A major Pseudomonas aeruginosa clone common to patients and aquatic habitats. Appl Environ Microbiol. 1994;60:1734–1738. doi: 10.1128/aem.60.6.1734-1738.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Römling U, Fiedler B, Bosshammer J, Grothues D, Greipel J, von der Hardt H, Tümmler B. Epidemiology of chronic Pseudomonas aeruginosa infections in cystic fibrosis. J Infect Dis. 1994;170:1616–1621. doi: 10.1093/infdis/170.6.1616. [DOI] [PubMed] [Google Scholar]