Gene Transfer in the Gastrointestinal Tract (original) (raw)
Abstract
The maximum in vivo transfer rate of plasmid pAMβ1 in the gut was 0.03 transconjugant per recipient cell, and this rate could be simulated in vitro only by forced filter mating. Transfer was not detected in liquid culture matings. Our findings demonstrate that in vitro methods, such as forced filter mating and liquid mating, underestimate the in vivo rates of gene transfer.
Genetically modified organisms (GMOs) are being developed and approved for use in many areas of food production; these organisms include microbial GMOs that are used in cheese and bread making (1, 7) and a variety of GMO food crops (11). Safety studies have indicated that the use of GMOs presents no greater risk than the use of the original unmodified products (10). However, there are still public and scientific concerns about the safety of foods produced by gene manipulation technology. One of the main concerns is the possibility of gene transfer from GMOs to organisms in the environment, particularly transfer of genes to pathogenic bacteria that could increase their virulence. Gene transfer has been observed in the gastrointestinal (GI) tracts of mammals (2, 6). However, several studies have showed that gene transfer in the environment, such as in the GI tract of rats (14) and in wastewater treatment plants (4, 5), occurs at rates that are 101- to 105-fold lower than the rates measured under laboratory conditions. In this study we investigated the persistence of GMO probiotics and transgene transfer in the avian GI tract.
Persistence of a GMO in the GI tract.
For a GMO to have an impact in the gut, the probiotic strain must be stable and be able to transiently colonize the gut; however, ideally, it should not persist following removal of the probiotic from the diet. The GMO probiotic used in this study was a genetically manipulated Enterococcus faecium strain (NCIMB 11508) with chromosomally encoded rifampin resistance (Rifr), in which the Ruminococcus flavefaciens β-1,4-glucanase was present on a non-self-transmissible plasmid (pVACMC1, conferring erythromycin resistance [Eryr]) and was maintained as an extrachromosomal element (15). The construct was 98% stable for five generations in vitro and produced 0.125 U of cellulase/ml of culture, which was equivalent to 1.4 U of cellulase/mg of total protein.
The in vivo studies were carried out by using broiler chicks and probiotic added to feed at a concentration of 105 CFU/g; the method used is described in the accompanying paper (8). Probiotic bacteria were recovered from the GI tract by selecting for Rifr colonies on MRS medium (Oxoid Ltd.). This selection strategy was appropriate as the background level of Rifr bacteria in the GI microflora is <101 CFU/ml, 100,000-fold lower than the level of the probiotic in the GI tract. The number of generations of the GMO probiotic that occurred during transit of the GI tract is not known, but the plasmid was detected in 69% (percentage of Rifr enterococci which were also Eryr) of GMO probiotic isolates obtained from the GI tract following 4 weeks of probiotic treatment. Thus, while the GMO probiotic was maintained in the gut, a major proportion retained the recombinant genotype.
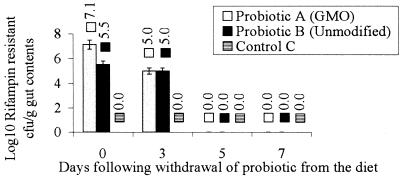
The carriage rate of the probiotics was initially determined to be 105 CFU/g of gut contents. After 28 days of probiotic feeding, the GMO carriage rate had increased to 107 CFU/g of gut contents, while the level of unmodified probiotic remained 105 CFU/g (Fig. 1). This may indicate that the GMO had a selective advantage over the unmodified strain, which may have been due to the production of cellulase which provided an accessible energy source for the microorganism and the host.
FIG. 1.
Persistence of probiotics in the cecum following removal from the feed: numbers of CFU (log10) of probiotics isolated on selective media containing rifampin/g of cecum contents, as determined at 2-day intervals after the probiotics were removed from the diet. The probiotics were removed from the diet following 28 days of treatment. The error bars indicate standard errors of the means. Three replicates were used in each experiment.
Neither the GMO nor unmodified probiotic strains persisted in the GI tract of chickens for more than 5 days following removal from the diet (Fig. 1). Therefore, the probiotic strains do not permanently colonize the avian GI tract when the rate of application of the probiotic in the feed is 105 CFU/g, the rate used in this study.
To investigate whether pVACMC1 was transferred to the endogenous avian microflora, 5,000 Eryr cellulase-producing bacteria recovered from the GI tracts of birds inoculated with the probiotic were found to be Rifr, suggesting that these organisms were probiotic strains. In addition, after the probiotic strain was washed out following removal of the strain from the diet, no Eryr cellulase-expressing bacteria were detected in the GI tract. Although it is not possible to conclusively prove that pVACMC1 was not transferred to the endogenous avian microflora, we detected no transfer events, indicating that if the plasmid was transferred, the transfer occurred at a very low rate.
Gene transfer in the avian GI tract.
One of the major concerns in using genetically modified organisms in food production is the risk of gene transfer to other organisms in the environment. Naturally occurring gene transfer in the environment takes place relatively infrequently (3) and is therefore difficult to measure experimentally with currently available techniques. To estimate the rate of transfer of transgenes from the GMO probiotic, in vitro and in vivo techniques were compared by using broad-host-range conjugative mobilizing plasmid pAMβ1 (26 kb, Eryr) to determine the relative sensitivity of each of the methods. The in vitro method used included the filter mating technique of Sasaki et al. (12) and liquid mating, which consisted of the same basic method involving mixing optimum concentrations of log-phase donors and recipients with and without a filter step.
The transfer rate of recombinant plasmid pVACMC1 (Eryr) was investigated initially in vitro under forced filter mating conditions by using 108 CFU of the donor strain (Rifr Eryr encoded by pVACMC1) per ml and 107 CFU of the Rifr recipient strain per ml and in the presence of the mobilizing Eryr plasmid pAMβ1 at a concentration of 108 CFU/ml. This was considered the worst-case scenario for detection of gene transfer from the recombinant probiotic. Gene transfer, as shown by the appearance of Rifr Eryr cellulose-expressing colonies, was not detected under these conditions but may have occurred at a rate of less than 10−8, which was the lowest level of detection.
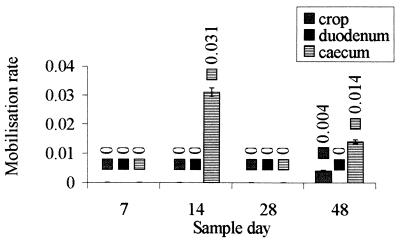
The gene transfer rate of conjugative plasmid pAMβ1 from a Rifs probiotic strain to a Rifr probiotic strain in the GI tract was found to be 0.03 transconjuant per donor organism at probiotic concentrations of 105 CFU of donor (Rifs probiotic strain)/ml of gut contents and 104 CFU of recipient (Rifr probiotic strain)/ml of gut contents (Fig. 2). This rate can be considered greater than the rate predicted by the in vitro study (also 0.03 transconjugant per donor), but it was obtained under optimal forced filter mating conditions at maximal probiotic concentrations (12).
FIG. 2.
Rates of mobilization of the probiotic in the chicken GI tract: distribution of gene transfer events in the crop, duodenum, and cecum on four different sampling dates. The rate of mobilization rate was the mean number of transconjugants per recipient cell. The error bars indicate standard errors. Three replicates were used in each experiment.
The gene transfer rate is most commonly expressed as the number of transconjugants per recipient cell, but this method of expression does not take into account the volume in which the organisms are placed and therefore the potential for intimate contact that is required for transfer to take place. In vitro, the volume in which mating takes place is infinitely small, and the donors and recipients are in intimate contact on a filter; thus, the potential for conjugal transfer should be expected to be much greater than the potential for conjugal transfer in the GI tract. Therefore, we also should consider the density of the bacteria in the mating mixture. Our in vitro transfer rate is similar to the maximal rates reported previously (13), and therefore the discrepancy between the in vitro and in vivo rates is not due to suboptimal application of in vitro techniques but truly reflects the inadequacy of the techniques used to represent in vivo conditions.
Gene transfer in vitro was not detected without filter mating. Liquid mating at a donor concentration of 108 CFU/ml and a recipient concentration of 107 CFU/ml was carried out, both with and without gut contents, but no transfer was observed.
Neither the filter mating technique nor the liquid mating technique accurately represent the natural conditions in the gut. The effects of bacterial binding to the gut wall membrane and solid matter in the lumen create conditions for gene transfer at estimated bacterial concentrations that are 1,000 fold lower than the concentrations used in vitro. Therefore, the in vivo conditions actually represent the worst-case scenario for gene transfer rather than the optimized in vitro conditions. However, this presents a problem in assessing the risk of the GMO probiotic, as the methods used to detect transfer in vivo have severe sensitivity limitations and tend to underestimate the true rate of transfer in vivo.
The in vivo measurements obtained for sample sites and individuals were not consistent (Fig. 2). Most of the transfer occurred in the cecum, but transfer was detected in only 5 of the 72 chicks tested (the detection rates were represented by counts of between 30 and 200 CFU/plate at rates of dilution of gut contents of 10−0.5 to 10−1). Transfer may have occurred at rates below the level of detection in the other individuals, but techniques with greater sensitivity are necessary to improve the accuracy of the estimated in vivo transfer rates. The current method produced large numbers of CFU when transfer did occur, but this seemed to occur in a minority of the chicks tested. Transfer was not detected in some chicks, which may have been due to individual variability of the chicks. For example, gene transfer may have occurred with a different bacterial species, unidentified factors in the GI tract may have inhibited transfer, or E. faecium may have entered an unculturable state. The relatively low concentrations of the donor and recipient probiotics in the gut (105 and 104 CFU/ml) could have contributed to the sporadic detection of gene transfer, as these concentrations are near the limit of sensitivity of the culture techniques.
Within the gut, a probiotic organism is not present as a synchronized culture, and only a small proportion of the cells may be at a stage of growth capable of conjugation. Thus, the in vivo culture technique probably underestimated the measurable rate of transfer in the avian GI tract, as only the recipient probiotic strain was detected and this strain represented a very minor population of the enterococci in the GI tract, all of which were potential recipients but were difficult to detect in the presence of a large number of background Eryr donor organisms.
The presence of other bacteria and solid matter may also reduce the likelihood that a donor and a recipient meet and therefore the likelihood of gene transfer. As the intestinal tract contains many naturally competent bacteria at concentrations greater than probiotic application rates, we cannot predict that transfer to other bacteria does not occur in the GI tract via transformation. The resulting transformants would not have been detected on the selective media which we used, and therefore the true rate of transfer would have been underestimated. The presence of DNases in the GI tract, however, reduces the likelihood of transfer by this mechanism.
In conclusion, our findings indicate that the true gene transfer rates in vivo are higher than the rates determined in vitro, which means that there are indigenous effects in the GI tract that promote a higher rate of transfer than the rate observed in vitro. Similar “hot spots” for gene transfer have been identified in the environment (for example, the bean phylloplane [9]) and are characterized by the availability of nutrients and high bacterial densities.
Acknowledgments
This work was supported by a grant from the United Kingdom Ministry of Agriculture, Fisheries and Food as part of the Novel Foods Programme.
We thank Harry Flint, Rowett Research Institute, Aberdeen, United Kingdom for donation of the pVACMC1 plasmid.
REFERENCES
- 1.Fitzgerald G, Coffey A, Daly C. Molecular manipulations of Lactococcus starter cultures for food fermentations. J Chem Technol Biotechnol. 1993;58:195–199. [Google Scholar]
- 2.Gruzza M, Langella P, Duval-Iflah Y, Ducluzeau R. Gene transfer from engineered Lactococcus lactis strains to Enterococcus faecalis in the digestive tract of gnotobiotic mice. Microb Releases. 1993;2:121–125. [PubMed] [Google Scholar]
- 3.Lorenz M G, Wackernagel W. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol Rev. 1994;58:563–602. doi: 10.1128/mr.58.3.563-602.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mach P A, Grimes D J. R-plasmid transfer in a wastewater treatment plant. Appl Environ Microbiol. 1982;44:1395–1403. doi: 10.1128/aem.44.6.1395-1403.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marcinek H, Wirth R, MuschollSilberhorn A, Gauer M. Enterococcus faecalis gene transfer under natural conditions in municipal sewage water treatment plants. Appl Environ Microbiol. 1998;64:626–632. doi: 10.1128/aem.64.2.626-632.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McConnell M A, Mercer A A, Tannock G W. Transfer of plasmid pAMβ1 between members of the normal microflora inhabiting the murine digestive tract and modification of the plasmid in a Lactococcus reuteri host. Microb Ecol Health Dis. 1991;4:343–355. [Google Scholar]
- 7.McGarry A, Law J, Coffey A, Daly C, Fox P F, Fitzgerald G F. Effect of genetically modifying the lactococcal proteolytic system on ripening and flavor development in cheddar cheese. Appl Environ Microbiol. 1994;60:4226–4233. doi: 10.1128/aem.60.12.4226-4233.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Netherwood T, Gilbert H J, Parker D S, O’Donnell A G. Probiotics shown to change bacterial community structure in the avian gastrointestinal tract. Appl Environ Microbiol. 1999;65:5134–5139. doi: 10.1128/aem.65.11.5134-5138.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Normander B, Christensen B B, Molin S, Kroer N. Effect of bacterial distribution and activity on conjugal gene transfer on the phylloplane of the bush bean (Phaseolus vulgaris) Appl Environ Microbiol. 1998;64:1902–1909. doi: 10.1128/aem.64.5.1902-1909.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Redenbaugh K, Hiatt W, Martineau B, Emlay D. Determination of the safety of genetically engineered crops. Am Chem Soc Symp Ser. 1995;605:72–87. [Google Scholar]
- 11.Redenbaugh K, Hiatt W, Martineau B, Lindemann J, Emlay D. Aminoglycoside 3′-phosphototransferase-Ii (Aph(3′)Ii)—review of its safety and use in the production of genetically engineered plants. Food Biotechnol. 1994;8:137–165. [Google Scholar]
- 12.Sasaki Y, Taketomo N, Sasaki T. Factors affecting transfer frequency of pAMβ1 from Streptococcus faecalis to Lactobacillus plantarum. J Bacteriol. 1988;170:5939–5942. doi: 10.1128/jb.170.12.5939-5942.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schaberg D R, Zervos M J. Intergeneric and interspecies gene exchange in gram-positive cocci. Antimicrob Agents Chemother. 1986;30:817–822. doi: 10.1128/aac.30.6.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schlundt J, Saadbye P, Lohmann B, Jacobsen B L, Nielsen E M. Conjugal transfer of plasmid DNA between Lactococcus lactis strains and distribution of transconjugants in the digestive tract of gnotobiotic rats. Microbiol Ecol Health Dis. 1994;7:59–69. [Google Scholar]
- 15.Whitehead T R, Flint H J. Heterologous expression of an endoglucanase gene (EndA) from the ruminal anaerobe Ruminococcus flavefaciens 17 in Streptococcus bovis and Streptococcus sanguis. FEMS Microbiol Lett. 1995;126:165–169. doi: 10.1111/j.1574-6968.1995.tb07411.x. [DOI] [PubMed] [Google Scholar]