Genotypic Heterogeneity of Streptococcus oralis and Distinct Aciduric Subpopulations in Human Dental Plaque (original) (raw)
Abstract
The genotypic heterogeneity of Streptococcus oralis isolated from the oral cavity was investigated using repetitive extragenic palindromic PCR. Unrelated subjects harbored unique genotypes, with numerous genotypes being isolated from an individual. S. oralis is the predominant aciduric bacterium isolated from noncarious tooth sites. Genotypic comparison of the aciduric populations isolated at pH 5.2 with those isolated from mitis-salivarius agar (MSA) (pH 7.0) indicated that the aciduric populations were genotypically distinct in the majority of subjects (χ2 = 13.09; P = 0.0031). Neither the aciduric nor the MSA-isolated strains were stable, with no strains isolated at baseline being isolated 4 or 12 weeks later in the majority of subjects. The basis of this instability is unknown but is similar to that reported for Streptococcus mitis. Examination of S. oralis strains isolated from cohabiting couples demonstrated that in three of five couples, genotypically identical strains were isolated from both partners and this was confirmed by using Salmonella enteritidis repetitive element PCR and enterobacterial PCR typing. These data provide further evidence of the physiological and genotypic heterogeneity of non-mutans streptococci. The demonstration of distinct aciduric populations of S. oralis implies that the role of these and other non-mutans streptococci in the caries process requires reevaluation.
The initiation of dental caries is associated with the ability of dental plaque to produce acid from ingested foods on a caries-prone tooth surface. At caries-prone sites, smooth tooth surfaces, interproximal sites, pits and fissures where plaque accumulates (1, 3, 10), and certainly within carious lesions, the local pH is acidic, and the bacteria present in these sites must be aciduric, exhibiting an ability to replicate in the prevailing or transient acidic environment. Dental plaque contains many species of acidogenic and aciduric microorganisms. The acidogenic bacteria most closely associated with the dental caries process are mutans streptococci (Streptococcus mutans and Streptococcus sobrinus), lactobacilli, and perhaps Actinomyces spp. There has been considerable discussion as to the role of other bacteria, in particular the role of the non-mutans streptococci (NMS), in the initiation and progression of dental caries. In an attempt to obtain understanding of the potential pathogenic role of these, van Houte and colleagues studied the acidogenicity of NMS isolated from sound and carious tooth sites (29, 35, 36). In these studies, the NMS were heterogeneous with respect to acidogenicity. Thus, from infected dentine within carious lesions and from plaque on sound surfaces in the mouths of caries-active subjects, NMS which were more acidogenic than NMS were isolated from sound tooth surfaces in caries-free subjects. The clonality of these strains has not been reported nor was the aciduricity of the isolates investigated. However, this was the first detailed and focused report of heterogeneity amongst individual NMS species of a determinant expected to be a significant feature of any microorganism involved in the initiation of dental caries. The acidogenic NMS were also more numerous than mutans streptococci, and it was proposed that these organisms may play a significant role in the caries process.
The aciduricity of bacteria isolated from the dental plaque biofilm has been investigated in a number of studies with a variety of in vitro techniques, primarily by determining the ability of isolates to metabolize carbohydrates and survive at acidic pH levels (11, 12, 15, 18, 23, 31, 32). These studies have demonstrated that lactobacilli and mutans streptococci are the most aciduric dental plaque bacteria, while NMS and Actinomyces spp. are less aciduric. However, the strains tested in those experiments were all isolated from conventional selective and nonselective culture media, which may have influenced the phenotypes of the strains isolated and subsequently examined. The predominant aciduric component of dental plaque has not been extensively investigated. In a preliminary report, we indicated that the predominant aciduric bacteria isolated from dental plaque taken from noncarious surfaces were NMS, with Streptococcus oralis, Streptococcus parasanguinis, and Streptococcus intermedius being the most frequently isolated (8). In this paper, we extend these observations and report the genotypic characterization of aciduric S. oralis strains from saliva and interproximal dental plaque samples. Representatives of the aciduric isolates from each subject were genotyped by using repetitive extragenic palindromic PCR (REP-PCR [2]) and were compared to those isolated from conventional culture media (pH 7.0). The stability of the S. oralis populations was assessed over periods of up to 12 weeks, and the transmissibility of strains was determined by comparing the S. oralis genotypes in the plaque flora of cohabiting couples.
MATERIALS AND METHODS
Isolation of S. oralis from saliva and interproximal plaque by using MSA and aciduric media.
Paraffin wax (Ivoclar-Vivadent, Schaan, Liechtenstein)-stimulated saliva was collected from volunteers and decimally diluted in fastidious anaerobe broth (LabM; Salford, Lancs, England), and 100-μl aliquots were plated onto mitis salivarius agar (MSA) (Becton-Dickinson, Oxford, England) supplemented with 0.1% potassium tellurite (Becton-Dickinson) for the isolation of viridans streptococci. Interproximal dental plaque was collected from a single caries-free site in each subject. The plaque samples were collected by using sterile wooden toothpicks, and each sample was placed in 1 ml of sterile PBSTC (1.58 g of K2HPO4 · 3H2O, 0.34 g of KH2PO4, 8 g of NaCl, 1.0 g of sodium thioglycollate, and 0.001 g of cetyltrimethylammonium bromide per liter of distilled water). The samples were dispersed by vortexing with sterile glass beads (BDH), decimally diluted in PBSTC, and plated onto MSA. The MSA plates were incubated anaerobically at 37°C for 3 days.
In order to isolate sufficient S. oralis strains from each sample plated onto the MSA, 75 colonies not exhibiting extracellular polysaccharide production were picked from each sample and inoculated into 200 μl of Todd-Hewitt broth (Oxoid, Basingstoke, Hants, England) in flat-bottomed microtiter trays (Griffiths and Neilson, Billinghurst, Kent, England) and were grown anaerobically for 48 h. Each isolate was tested for sialidase activity (6) by transferring 50 μl of the Todd-Hewitt broth suspension to 20 μl of sialidase substrate [200 μg of 2′-(4-methylumbelliferyl)-α-d-_N_-acetylneuraminic acid (Sigma, Poole, Dorset, England) in 50 mM _N_-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid (TES) buffer, pH 7.5] in a flat-bottomed microtiter tray. The enzyme assays were incubated aerobically at 37°C for 3 h, and an increase in fluorescence, demonstrating sialidase activity, was measured by using a fluorimeter (Perkin-Elmer, Beaconsfield, Hants, England) at emission and excitation wavelengths of 380 and 460 nm, respectively. The remainder of the cultures were supplemented with glycerol to final concentrations of approximately 50% (vol/vol), and the microtiter plates were stored at −80°C.
The preliminary screening for sialidase-positive streptococci identified those isolates which were S. oralis, Streptococcus mitis, S. intermedius, or Abiotrophia adjaciens (17, 28, 39). In order to identify the S. oralis strains from amongst the collection of sialidase-positive strains, all of these isolates were tested for the production of β-fucosidase, β-_N_-acetylgalactosaminidase, and β-_N_-acetylglucosaminidase by using the appropriate fluorogenic substrate as previously described (4). Those strains which were negative for β-fucosidase, positive for the two hexosaminidase activities, and positive for sialidase activity were identified as S. oralis, and the identification was confirmed by using further physiological tests, including fermentation of a range of carbohydrates and hydrolysis of arginine and esculin (4).
Aciduric S. oralis strains were also isolated from each interproximal plaque sample. This isolation procedure employed was the most probable number (MPN) technique. The predominant aciduric bacteria in each sample capable of growth and proliferation in acidic media were isolated by using brain heart infusion (BHI) (Oxoid) adjusted to pH 5.2 by using citric acid and Na2HPO4 at final concentrations of 13 and 26 mM, respectively. Each plaque sample was decimally diluted in PBSTC to give a dilution series from 10−1 to 10−8, and 12 replicates of each dilution, 15 μl of plaque dilution in PBSTC, and 135 μl of acidic BHI were set up in sterile flat-bottomed microtiter trays and incubated anaerobically for 5 days. The terminal pH of wells exhibiting growth was measured and found to be ±0.05 pH units of the original pH of the medium. Terminal wells, the lowest dilutions exhibiting bacterial growth were subcultured onto Columbia agar (Oxoid) supplemented with 5% (vol/vol) horse blood and were incubated anaerobically for 48 h. The colonies were examined by gram-staining, and those that appeared as gram-positive cocci were identified. Aciduric S. oralis strains were isolated from each subject, and these were stored in glycerol broth at −80°C until required for subsequent genotypic analysis (2). For the majority of the investigations, the MPN method was used as a convenient multiwell procedure for the isolation of aciduric S. oralis strains from samples, but the MPN method was used to quantitate the proportion of S. oralis in 18 interproximal samples. From these samples, the number of aciduric S. oralis strains as a percentage of the total bacterial count determined using the MPN method with BHI at pH 7.0 was calculated, and the proportion of S. oralis strains isolated from MSA was expressed as a percentage of the total bacterial count on nonselective media (9).
Genotyping of S. oralis.
Individual S. oralis strains were genotyped by using repetitive extragenic palindromic-PCR (REP-PCR) which, as previously described, results in amplicon patterns which are unique for each independent isolate of S. oralis and all other species of viridans streptococci (2). For confirmatory purposes, enterobacterial PCR (ERIC-PCR) and Salmonella enteritidis repetitive element PCR (SERE-PCR) were also used as previously described (2).
Visualization of amplicons.
The amplification products of REP-PCR and ERIC-PCR were analyzed by using 2% Metaphor agarose (Flowgen, Staffordshire, England) containing 0.5 μg of ethidium bromide per ml and were separated electrophoretically on 20- by 25-cm gels at 140 V for 3 h in Tris-borate-EDTA buffer. SERE-PCR products (15 μl) were resolved on 0.8% agarose (Sigma) containing 0.5 μg of ethidium bromide per ml by electrophoretic separation at 140 V for 3 h. To all samples, 3 μl of tracking dye (0.25% bromophenol blue, 0.25% xylene cyanol FF, 30% glycerol) was added, and molecular size marker (pGEM DNA Markers; Promega, Southampton, England) were included on all gels, in three to four separate lanes, to facilitate comparison of tracks between gels. Gels were examined on a transilluminator and were photographed by using Polaroid type 665 positive-negative film (Sigma).
Computer-assisted analysis of the DNA patterns.
All of the patterns of the isolates from an individual were compared by using GelCompar version 4.0 (Applied Maths, Kortrijk, Belgium). The individual bands in each of the patterns produced by the different PCR methods were analyzed by applying the Dice coefficient to the peaks. For clustering, the unweighted pair group method using mathematical averages (UPGMA) was used, and a band position tolerance of 1.5% was used for comparison of the DNA patterns. The analysis of the patterns was undertaken in accordance with the instructions of the manufacturer. Differences in the frequency of recovery of genotypes were compared statistically using the χ2 test (Fisher's exact test).
Genotypic heterogeneity of S. oralis isolated from individuals.
Interproximal plaque samples were taken from 15 subjects, and S. oralis strains were isolated from each sample by using MSA and the multiwell method as described above. The genotypes present in each subject, isolated on each medium, were compared by cluster analysis, and the similarity of the strains from the two culture conditions in each subject was determined.
Stability of S. oralis genotypes.
The stability of the S. oralis populations in individual mouths was determined in two experiments. In the first, an oral rinse was obtained from each of five unrelated adults on three separate occasions: at baseline and 4 and 12 weeks later. The S. oralis component of the flora was isolated from each of the oral rinses by using MSA only. S. oralis isolates present in each sample were identified, and individual strains were genotyped by using REP-PCR. The genotypes of these populations were compared by cluster analysis, and the stability of the population present at baseline was determined over the 12-week period.
In the second experiment, to assess the stability of the aciduric S. oralis population in interproximal plaque, samples were taken from 10 adult subjects on two separate occasions 4 weeks apart. The predominant aciduric bacteria in each sample were isolated using the multiwell method as described above. The strains, primarily viridans streptococci, isolated in this way were identified. Those isolates identified as S. oralis were genotyped by using the REP-PCR method described above. The stability of the aciduric S. oralis populations in each subject was evaluated by cluster analysis.
Transmission of S. oralis between subjects.
Interproximal plaque samples were taken from five cohabiting couples at the same time, and the S. oralis populations were isolated from each sample by using both the multiwell and MSA methods. The genotypes of S. oralis in each sample were determined, and the genotypes of each couple were compared by using cluster analysis in order to ascertain the presence of strains, which were apparently indistinguishable from each other, in the interproximal samples of each couple. The presence of such strains would indicate transmission of S. oralis between couples.
When strains were isolated from couples which were indistinguishable by REP-PCR, these strains were subjected to additional strain genotyping by using SERE-PCR and ERIC-PCR methods. This was undertaken since strains which are different by REP-PCR are clearly different, but strains which exhibit the same pattern by only one typing method may be shown to be different when subjected to other typing systems. Therefore, indistinguishable strains should yield similar, but different, band patterns when examined with other typing methods.
RESULTS
Genotypic heterogeneity of S. oralis within individuals.
In the majority of the 15 subjects, the aciduric S. oralis populations were distinct from the S. oralis population isolated by using MSA. Representative examples of REP-PCR patterns of multiwell- and MSA-isolated S. oralis strains for individual subjects are shown in Fig. 1. In only 4 of the 15 subjects were S. oralis strains isolated by the MSA and multiwell methods indistinguishable from each other by REP-PCR typing. The data are summarized in Table 1, which shows the number of strains examined per subject, the number of genotypes per subject amongst those strains examined, and those genotypes recovered from each subject on the MSA and from the aciduric medium. The null hypothesis that each subject harbored only one S. oralis population, i.e., that the same genotypes should be recovered from both media, was tested, and it was found that the genotypes recovered from the two media were significantly different (χ2 = 13.09; P = 0.00031). Therefore, it may be concluded that the S. oralis populations isolated from the aciduric media were distinct from those recovered from MSA.
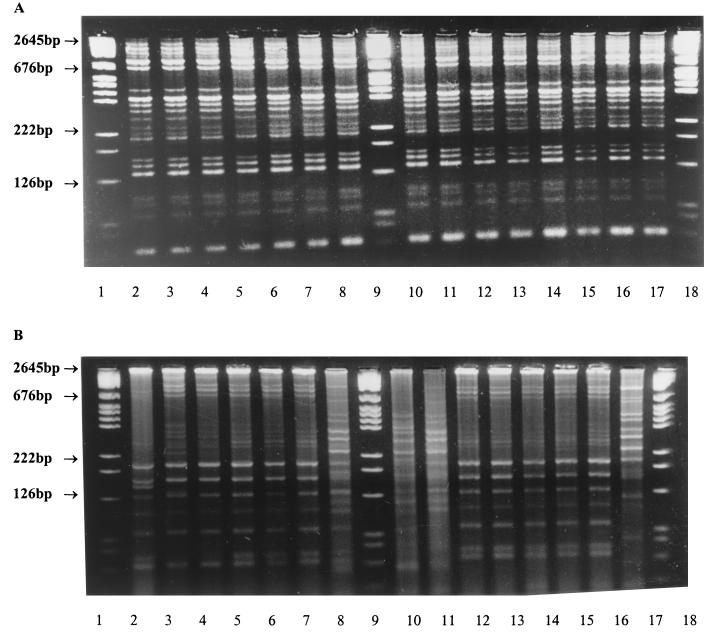
FIG. 1.
REP-PCR patterns of S. oralis strains isolated from interproximal plaque samples using (a) MSA and (b) MPN methods from two subjects. Lanes 1, 9, and 18 contain molecular size markers.
TABLE 1.
Genotypic heterogeneity of S. oralis strains isolated from interproximal plaque using MSA and aciduric media (MPN) of 15 adult subjects
| Subject | Medium | No. of isolates | Subject-specific S. oralis genotype no.a | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 |
| 1 | MSA | 15 | 14 | 1 | ||||||||||
| MPN | 15 | 10 | 2 | 1 | 1 | 1 | ||||||||
| 2 | MSA | 15 | 6 | 1 | 5 | 2 | 1 | |||||||
| MPN | 5 | 1 | 1 | 1 | 1 | 1 | ||||||||
| 3 | MSA | 15 | 8 | 2 | 1 | 1 | 1 | 1 | 1 | |||||
| MPN | 15 | 14 | 1 | |||||||||||
| 4 | MSA | 15 | 15 | |||||||||||
| MPN | 15 | 10 | 2 | 1 | 1 | 1 | ||||||||
| 5 | MSA | 14 | 8 | 2 | 1 | 1 | 1 | 1 | ||||||
| MPN | 3 | 1 | 1 | 1 | ||||||||||
| 6 | MSA | 16 | 4 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 |
| MPN | 20 | 15 | 4 | 1 | ||||||||||
| 7 | MSA | 15 | 6 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | ||||
| MPN | 7 | 2 | 1 | 1 | 1 | 2 | ||||||||
| 8 | MSA | 10 | 10 | |||||||||||
| MPN | 11 | 4 | 6 | 1 | ||||||||||
| 9 | MSA | 10 | 7 | 2 | 1 | |||||||||
| MPN | 9 | 6 | 1 | 1 | 1 | |||||||||
| 10 | MSA | 5 | 1 | 1 | 1 | 1 | 1 | |||||||
| MPN | 11 | 2 | 6 | 1 | 1 | 1 | ||||||||
| 11 | MSA | 20 | 20 | |||||||||||
| MPN | 20 | 19 | 1 | |||||||||||
| 12 | MSA | 17 | 8 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | ||||
| MPN | 16 | 10 | 4 | 1 | 1 | |||||||||
| 13 | MSA | 13 | 5 | 3 | 2 | 1 | 1 | 1 | ||||||
| MPN | 16 | 3 | 2 | 11 | ||||||||||
| 14 | MSA | 12 | 10 | 2 | ||||||||||
| MPN | 10 | 4 | 2 | 2 | 1 | 1 | ||||||||
| 15 | MSA | 11 | 5 | 2 | 2 | 1 | 1 | |||||||
| MPN | 6 | 1 | 1 | 1 | 1 | 1 | 1 |
The proportion of aciduric S. oralis in interproximal plaque samples.
The aciduric S. oralis isolates represented 1.9% (± 0.7%) of the MPN of bacteria in the samples isolated in BHI at pH 7.0. The MPN of bacteria at pH 5.2 represented 3.8% (± 1.2%) of the total MPN of bacteria at pH 7.0. Thus, on average, the S. oralis strains isolated in the media at pH 5.2 represented approximately 50% of the bacterial count. The S. oralis isolates recovered from the MSA plates formed 11.2% (± 3.6%) of the total count from the nonselective media.
Stability of S. oralis genotypes.
To determine the stability of the S. oralis populations, strains were recovered by using MSA from the saliva of five subjects sampled over a 12-week interval. There were considerable differences in the number of genotypes of S. oralis present in the S. oralis population of each subject. When the genotypes of each subject were compared, it was found that in only two subjects were any strains exhibiting the same REP-PCR pattern isolated at baseline and 4 weeks later. In no subject were strains which were identical to those isolated in the 12-week sample isolated in the baseline or 4-week samples (Table 2). A typical dendrogram of these relationships is shown in Fig. 2.
TABLE 2.
Number of different genotypes recovered from saliva samples of subjects at baseline, 1 month, and 6 months
| Subject | Time (h) | Subject-specific genotype no.a | ||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 |
| 1 | 0 | 1 | 1 | 3 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 4 | ||||||||||||||||
| 1 | 5 | 1 | 1 | 1 | 1 | 1 | 1 | |||||||||||||||||||||
| 6 | 1 | 1 | 1 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||||||||||
| 2 | 0 | 2 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 5 | 1 | 1 | ||||||||||||||||
| 1 | 1 | 1 | 2 | 4 | 1 | 1 | 1 | |||||||||||||||||||||
| 6 | 2 | 1 | 1 | 1 | 4 | 1 | 1 | 1 | 1 | 1 | 1 | |||||||||||||||||
| 3 | 0 | 1 | 1 | 10 | 1 | 1 | 1 | 1 | 1 | |||||||||||||||||||
| 1 | 1 | 2 | ||||||||||||||||||||||||||
| 6 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||||||
| 4 | 0 | 1 | 1 | 1 | 1 | |||||||||||||||||||||||
| 1 | 1 | 1 | 1 | 1 | 3 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | |||||||||||||||
| 6 | 6 | 1 | 1 | 1 | 1 | 1 | 1 | |||||||||||||||||||||
| 5 | 0 | 1 | 1 | 1 | 1 | 4 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||||||||
| 1 | 1 | 1 | 1 | |||||||||||||||||||||||||
| 6 | 1 | 1 | 1 | 1 |
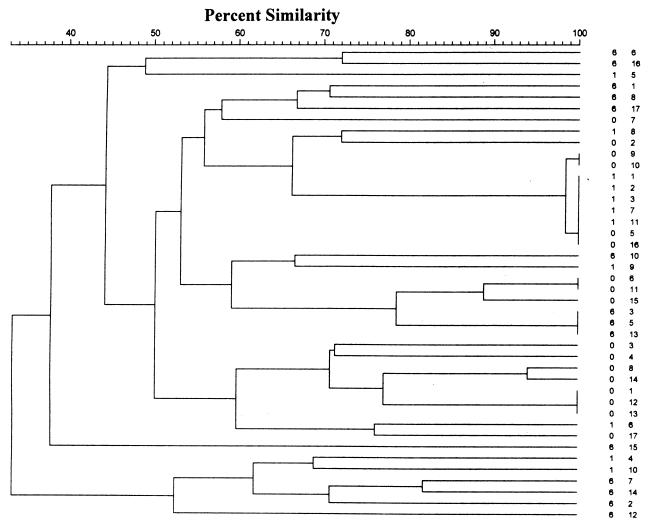
FIG. 2.
Dendrogram illustrating genotypic relationships between S. oralis strains isolated from the saliva of a single subject. Strain identification is rendered time (0, 1, or 6 months) and strain number. Isolates 1, 2, 3, 7, and 11 from the 1-month sample were indistinguishable from isolates from the 6-month sample. Individual REP-PCR amplicons were marked, and the individual bands were analyzed by using the Dice coefficient and clustering using the UPGMA method.
The stability of the aciduric S. oralis populations was determined in 10 subjects from whom interproximal plaque was sampled at baseline and after 4 weeks. In five subjects, identical genotypes were recovered on each of the two sampling times. However, in each subject, additional genotypes were recovered from the plaque samples at each sampling time. These data are summarized in Table 3 and illustrate the overall heterogeneity of aciduric S. oralis genotypes isolated from the interproximal plaque samples and the instability of the S. oralis populations in these subjects.
TABLE 3.
Numbers of genotypes identified amongst the aciduric S. oralis strains recovered from the interproximal plaque of 10 subjects sampled at baseline and at 1 month later
| Subject | Time (h) | Subject-specific genotype no.a | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 |
| 1 | 0 | 7 | 2 | 1 | 1 | 1 | 1 | 1 | ||||||||
| 1 | 8 | 1 | 1 | |||||||||||||
| 2 | 0 | 2 | 1 | 1 | 1 | 2 | 3 | 1 | 1 | 1 | 1 | 1 | ||||
| 1 | 8 | 1 | 6 | 1 | 1 | |||||||||||
| 3 | 0 | 8 | 3 | 2 | 1 | 1 | 1 | 1 | ||||||||
| 1 | 12 | 2 | 1 | 1 | 1 | |||||||||||
| 4 | 0 | 9 | 3 | 2 | 1 | 1 | ||||||||||
| 1 | 13 | 1 | 1 | |||||||||||||
| 5 | 0 | 4 | 1 | 1 | 1 | |||||||||||
| 1 | 5 | |||||||||||||||
| 6 | 0 | 8 | 2 | 1 | 1 | 1 | 1 | 1 | ||||||||
| 1 | 3 | 3 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||||
| 7 | 0 | 2 | 3 | 1 | 1 | 1 | ||||||||||
| 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||||||
| 8 | 0 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||
| 1 | 14 | 1 | 1 | 1 | ||||||||||||
| 9 | 0 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | |||||||
| 1 | 6 | 1 | 2 | 1 | 3 | 1 | ||||||||||
| 10 | 0 | 9 | 4 | 1 | 1 | 1 | 1 | |||||||||
| 1 | 6 | 2 | 2 | 1 | 1 | 1 | 1 | 1 |
Transmission of S. oralis.
S. oralis strains were isolated from five couples by using both MSA and the multiwell method. In all, 40 to 62 isolates per couple were genotyped; a total of 258 isolates. On examination of the dendrograms of all strains from each couple, it was apparent that in three couples S. oralis strains which were indistinguishable by REP-PCR were isolated from each member of the couple (Fig. 3). In one couple, 3 of 16 strains from the acidic media from one partner were identical to 10 of 16 strains from the other. In the second couple, 15 of 20 strains from the acidic medium were identical to 1 of 7 strains from the acidic medium from the other partner, and in the third couple, 19 of 20 strains from the acidic medium were identical to 1 of 5 strains from the acidic medium from the other partner. When the identical strains from each couple were further examined by SERE-PCR and ERIC-PCR, it was apparent that the strains were indistinguishable by both methods (data not shown).
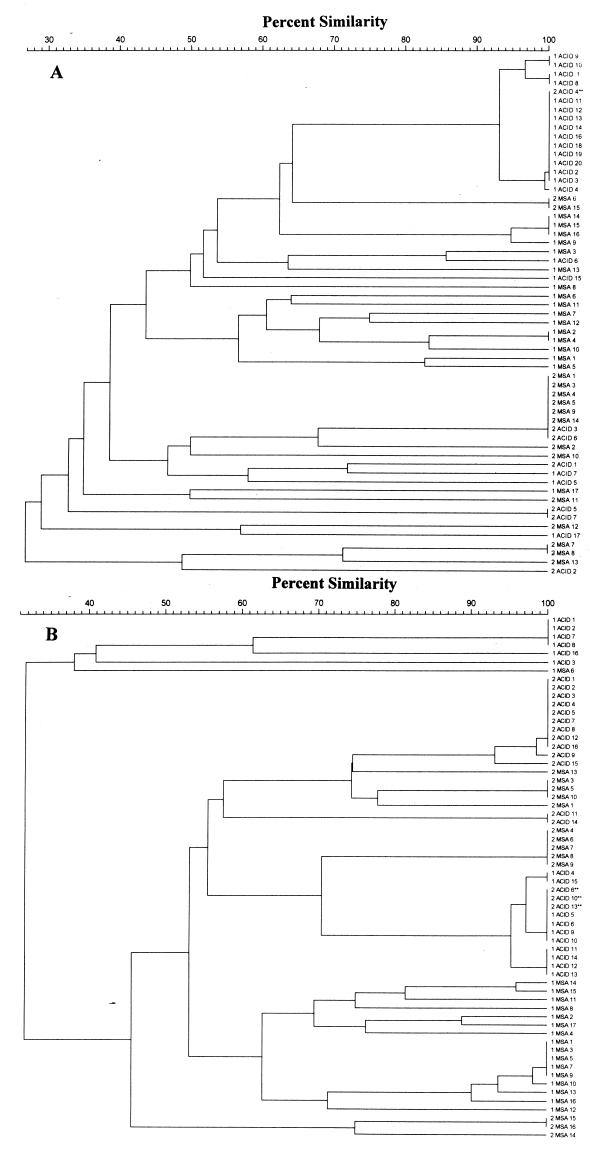
FIG. 3.
Representative dendrograms illustrating genotypic relationships between S. oralis strains isolated from cohabiting couples A and B with partners identified as 1 and 2. In each couple, the first number indicates the individual partner, MSA and ACID indicate the culture method used to isolate the strains, and the last number indicates the isolate number. In couple A, strain 2 ACID 4 was identical to strains from the other partner, and in couple B, strains 2 ACID 6, 10, and 13 were identical to strains from the other partner. For comparison, individual REP-PCR amplicons were marked, and the individual bands were analyzed by using the Dice coefficient and clustering using the UPGMA method.
DISCUSSION
This study has demonstrated that the S. oralis population in an individual is heterogeneous, that the aciduric S. oralis strains may be genotypically distinct from those isolated from the same plaque sample using MSA, and that the S. oralis populations are unstable but are apparently readily transmissible between cohabiting couples. In previous studies, heterogeneity in populations of other dental plaque bacteria has been reported (9, 13, 14, 16, 20–22, 33, 37). Studies of the clonality of the phylogenetically related species S. mitis biovar 1 have reported diversity similar to that found in the present study for S. oralis. Howhy and Kilian (16) identified 106 S. mitis biovar 1 isolates from amongst 250 streptococcal isolates from three members of the same family. The 106 isolates represented 24 different genotypes by restriction endonuclease analysis with 6 to 13 types being isolated from each individual. Similarly, Fitzsimmons et al. (13) isolated 101 S. mitis biovar 1 strains from 40 neonates, and when examined by ribotyping, these isolates represented 93 unique types, again demonstrating the high degree of diversity within this species. It is apparent from the data presented in this report that S. oralis populations behave in a similar manner. This high degree of genotypic diversity is not restricted to these two species of viridans streptococci, as we demonstrated in a previous study that unrelated members of each of the human species of viridans streptococci exhibited considerable diversity with either REP-PCR, ERIC-PCR, or SERE-PCR (2).
The instability of the S. oralis populations reported here is in contrast to the apparent population stability of S. mutans (19) and Prevotella intermedia and Prevotella nigrescens (22) populations in human dental plaque but is more like the reported instability of S. mitis biovar 1 in infants (13). The rate of genotypic change must be rapid and may be reflected in the antigenic composition of the organism, providing a mechanism with which to avoid the host's immune system and consequent elimination. It may be expected that all oral bacteria, especially those on mucosal surfaces, including those colonizing the periodontal pocket, would behave in a similar fashion; however, they do not behave in this manner, as evidenced by the reported stability of P. nigrescens and P. intermedia populations (22). Other mechanisms may underlie the rapid changes in population structure. We found that after 500 in vitro divisions there was no change in the REP-PCR pattern of recently isolated individual S. oralis strains (unpublished observations). However, more extensive growth periods, involving 10,000 replications of Escherichia coli, resulted in significant changes in genotype associated with movement of transposons within the genome (24). It would seem unlikely that this mechanism for modifying the genotypes of these streptococci is responsible for the changes observed, given the slow doubling time of bacteria in dental plaque (3) and the relatively short interval between sampling times. Both S. oralis and S. mitis are highly competent and may undergo transformation in vivo by horizontal gene transfer (25), which would be expected to produce the changes in REP-PCR patterns seen here in S. oralis. It has also been suggested that the rapid genotypic change may be related to the generation and loss of REP-PCR priming sites during DNA replication (34).
It was demonstrated in this study that S. oralis strains with the same genotype were isolated from the dental plaque of cohabiting couples. This is not unexpected, since other viridans streptococci have been shown to be transmitted between couples and between mothers and their infants (7, 13, 16, 21, 27). Transmission is presumably mediated by salivary transfer between partners. It is of note that in each of the three couples where transmission was demonstrable, the strain which appeared to have been transmitted was predominant in one of the partners. The use of the other PCR-based genotyping methods to confirm the similarity of the common genotypes provides unequivocal support for the isolation of the same strains of S. oralis from each member of these three couples, since the PCR priming sites are different for each reaction (26, 38).
We have previously reported that S. oralis is amongst the most numerous aciduric bacterial species in dental plaque (8) and that mutans streptococci are rarely isolated from among the predominant aciduric bacteria in plaque associated with caries-free tooth surfaces. In this report, we have subjected the S. oralis populations to extensive genotypic analysis, and these results considerably extend these previous observations. We have shown that aciduric S. oralis populations which are genotypically distinct from those isolated from MSA (initial pH 7.0) exist in plaque. The presence of this distinct population is unexpected, since in other studies it has been demonstrated that although NMS can survive in acidic conditions, their survival was regarded as phenotypic adaptation (32). The present data support the presence of genetically distinct aciduric S. oralis in which there may be a genetic basis to their survival and proliferation under the acidic conditions.
NMS are heterogeneous with respect to acidogenicity and, by virtue of their numerical superiority, these acidogenic NMS may play a significant role in the caries formation process (29, 35, 36). The present data may also be viewed in the same manner. The predominant aciduric bacteria isolated from caries-free tooth surfaces are not mutans streptococci but are rather S. oralis and other viridans streptococci including S. intermedius, S. parasanguinis, and Streptococcus anginosus. The role of these bacteria in the initiation of caries is not yet known, although many NMS, including S. oralis, are reported to induce caries in laboratory rats (40). The data presented here clearly indicate that S. oralis is genotypically heterogeneous with respect to aciduricity, and our previous study indicated that this species was predominant amongst the aciduric dental plaque microflora.
This is the first report of genotypic heterogeneity amongst NMS relating to aciduricity. The significance of the aciduric S. oralis and other aciduric NMS in the caries process warrants further consideration, and it may be that the central role assigned previously to S. mutans will require reevaluation.
ACKNOWLEDGMENTS
We thank the subjects who gave consent to be involved in the study.
This study was supported by Guy's, King's, and St. Thomas' Dental Institute and in part by Unilever Research, United Kingdom.
REFERENCES
- 1.Alaluusua S, Malmivirta R. Early plaque accumulation: a sign of caries risk in young children. Community Dent Oral Epidemiol. 1994;22:273–276. doi: 10.1111/j.1600-0528.1994.tb02049.x. [DOI] [PubMed] [Google Scholar]
- 2.Alam S, Brailsford S R, Whiley R A, Beighton D. PCR-based methods for genotyping viridans group streptococci. J Clin Microbiol. 1999;37:2772–2776. doi: 10.1128/jcm.37.9.2772-2776.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beighton D, Hayday H, Walker J. The relationship between the number of the bacterium Streptococcus mutans at discrete sites on the dentition of macaque monkeys (Macaca fascicularis) and the subsequent development of dental caries. Arch Oral Biol. 1985;30:85–88. doi: 10.1016/0003-9969(85)90029-9. [DOI] [PubMed] [Google Scholar]
- 4.Beighton D, Hardie J M, Whiley R A. A scheme for the identification of viridans streptococci. J Med Microbiol. 1991;35:367–372. doi: 10.1099/00222615-35-6-367. [DOI] [PubMed] [Google Scholar]
- 5.Beighton D, Smith K, Hayday H. The growth of bacteria and the production of exoglycosidic enzymes in the dental plaque of macaque monkeys. Arch Oral Biol. 1986;31:829–835. doi: 10.1016/0003-9969(86)90137-8. [DOI] [PubMed] [Google Scholar]
- 6.Beighton D, Whiley R A. Sialidase activity of the “Streptococcus milleri group” and other viridans group streptococci. J Clin Microbiol. 1990;28:1431–1433. doi: 10.1128/jcm.28.6.1431-1433.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berkowitz R J. Streptococcus mutans—establishment and transmission in infants. ASDC (Am Soc Dent Child) J Dent Child. 1976;43:192–195. [PubMed] [Google Scholar]
- 8.Brailsford S R, Byrne R W, Adams S, Zoitopoulos L, Allison C, Beighton D. Investigation of the aciduric microflora of plaque. Caries Res. 1999;33:290. [Google Scholar]
- 9.Brailsford S R, Leftwich H, Tregaskis R, Beighton D. The predominant Actinomyces spp. isolated from infected dentine of active root caries lesions. J Dent Res. 1999;78:1525–1534. doi: 10.1177/00220345990780090701. [DOI] [PubMed] [Google Scholar]
- 10.Carvalho J C, Ekstrand K R, Thylstrup A. Dental plaque and caries on occlusal surfaces of first permanent molars in relation to stage of eruption. J Dent Res. 1989;68:773–779. doi: 10.1177/00220345890680050401. [DOI] [PubMed] [Google Scholar]
- 11.Chestnutt I G, MacFarlane T W, Stephen K W. An in vitro investigation of the cariogenic potential of oral streptococci. Arch Oral Biol. 1994;39:589–593. doi: 10.1016/0003-9969(94)90134-1. [DOI] [PubMed] [Google Scholar]
- 12.de Soet J J, van Loveren C, Lammens A J, Pavicic M J, Homburg C H, ten Cate J M, de Graaff J. Differences in cariogenicity between fresh isolates of Streptococcus sobrinus and Streptococcus mutans. Caries Res. 1991;25:116–122. doi: 10.1159/000261353. [DOI] [PubMed] [Google Scholar]
- 13.Fitzsimmons S, Evans M, Pearce C, Sheridan M J, Wientzen R, Bowden G, Cole M F. Clonal diversity of Streptococcus mitis biovar 1 isolates from the oral cavity of human neonates. Clin Diagn Lab Immunol. 1996;3:517–522. doi: 10.1128/cdli.3.5.517-522.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.George K S, Reynolds M A, Falkler W A., Jr Arbitrarily primed polymerase chain reaction fingerprinting and clonal analysis of oral Fusobacterium nucleatum isolates. Oral Microbiol Immunol. 1997;12:219–226. doi: 10.1111/j.1399-302x.1997.tb00382.x. [DOI] [PubMed] [Google Scholar]
- 15.Hamilton I R, Svensater G. Acid-regulated proteins induced by Streptococcus mutans and other oral bacteria during acid shock. Oral Microbiol Immunol. 1998;13:292–300. doi: 10.1111/j.1399-302x.1998.tb00710.x. [DOI] [PubMed] [Google Scholar]
- 16.Hohwy J, Kilian M. Clonal diversity of the Streptococcus mitis biovar 1 population in the human oral cavity and pharynx. Oral Microbiol Immunol. 1995;10:19–25. doi: 10.1111/j.1399-302x.1995.tb00113.x. [DOI] [PubMed] [Google Scholar]
- 17.Kawamura Y, Hou X G, Sultana F, Liu S, Yamamoto H, Ezaki T. Transfer of Streptococcus adjacens and Streptococcus defectivus to Abiotrophia gen. nov. as Abiotrophia adiacens comb. nov. and Abiotrophia defectiva comb. nov., respectively. Int J Syst Bacteriol. 1995;45:798–803. doi: 10.1099/00207713-45-4-798. [DOI] [PubMed] [Google Scholar]
- 18.Kohler B, Birkhed D, Olsson S. Acid production by human strains of Streptococcus mutans and Streptococcus sobrinus. Caries Res. 1995;29:402–406. doi: 10.1159/000262099. [DOI] [PubMed] [Google Scholar]
- 19.Kozai K, Wang D S, Sandham H J, Phillips H I. Changes in strains of mutans streptococci induced by treatment with chlorhexidine varnish. J Dent Res. 1991;70:1252–1257. doi: 10.1177/00220345910700090401. [DOI] [PubMed] [Google Scholar]
- 20.Kulkarni G V, Chan K H, Sandham H J. An investigation into the use of restriction endonuclease analysis for the study of transmission of mutans streptococci. J Dent Res. 1989;68:1155–1161. doi: 10.1177/00220345890680070401. [DOI] [PubMed] [Google Scholar]
- 21.Li Y, Caufield P W. The fidelity of initial acquisition of mutans streptococci by infants from their mothers. J Dent Res. 1995;74:681–685. doi: 10.1177/00220345950740020901. [DOI] [PubMed] [Google Scholar]
- 22.Matto J, Saarela M, von Troil-Linden B, Kononen E, Jousimies-Somer H, Torkko H, Alaluusua S, Asikainen S. Distribution and genetic analysis of oral Prevotella intermedia and Prevotella nigrescens. Oral Microbiol Immunol. 1996;11:96–102. doi: 10.1111/j.1399-302x.1996.tb00342.x. [DOI] [PubMed] [Google Scholar]
- 23.Milnes A R, Bowden G H, Hamilton I R. Effect of NaF and pH on the growth and glycolytic rate of recently isolated strains of oral Lactobacillus species. J Dent Res. 1985;64:401–404. doi: 10.1177/00220345850640030101. [DOI] [PubMed] [Google Scholar]
- 24.Papadopoulos D, Schneider D, Meier-Eiss J, Arber W, Lenski R E, Blot M. Genomic evolution during a 10,000-generation experiment with bacteria. Proc Natl Acad Sci USA. 1999;96:3807–3812. doi: 10.1073/pnas.96.7.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poulsen K, Reinholdt J, Jespersgaard C, Boye K, Brown T A, Hauge M, Kilian M. A comprehensive genetic study of streptococcal immunoglobulin A1 proteases: evidence for recombination within and between species. Infect Immun. 1998;66:181–190. doi: 10.1128/iai.66.1.181-190.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rajashekara G, Koeuth T, Nevile S, Back A, Nagaraja K V, Lupski J R, Kapur V. SERE, a widely dispersed bacterial repetitive DNA element. J Med Microbiol. 1998;47:489–498. doi: 10.1099/00222615-47-6-489. [DOI] [PubMed] [Google Scholar]
- 27.Rogers A H. Evidence for the transmissibility of human dental caries. Aust Dent J. 1977;22:53–66. doi: 10.1111/j.1834-7819.1977.tb04444.x. [DOI] [PubMed] [Google Scholar]
- 28.Ruoff K L, Whiley R A, Beighton D. Streptococcus. In: Murray P M, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. Washington, D.C.: American Society for Microbiology; 1999. pp. 297–305. [Google Scholar]
- 29.Sansone C, Van Houte J, Joshipura K, Kent R, Margolis H C. The association of mutans streptococci and non mutans streptococci capable of acidogenesis at a low pH with dental caries on enamel and root surfaces. J Dent Res. 1993;72:508–516. doi: 10.1177/00220345930720020701. [DOI] [PubMed] [Google Scholar]
- 30.Suchett-Kaye G, Decoret D, Barsotti O. Intra-familial distribution of Fusobacterium nucleatum strains in healthy families with optimal plaque control. J Clin Periodontol. 1999;26:401–404. doi: 10.1034/j.1600-051x.1999.260611.x. [DOI] [PubMed] [Google Scholar]
- 31.Svensater G, Larsson U B, Greif E C, Cvitkovitch D G, Hamilton I R. Acid tolerance response and survival by oral bacteria. Oral Microbiol Immunol. 1997;12:266–273. doi: 10.1111/j.1399-302x.1997.tb00390.x. [DOI] [PubMed] [Google Scholar]
- 32.Takahashi N, Yamada T. Acid-induced acid tolerance and acidogenicity of non-mutans streptococci. Oral Microbiol Immunol. 1999;14:43–48. doi: 10.1034/j.1399-302x.1999.140105.x. [DOI] [PubMed] [Google Scholar]
- 33.Teanpaisan R, Douglas C W, Eley A R, Walsh T F. Clonality of Porphyromonas gingivalis, Prevotella intermedia and Prevotella nigrescens isolated from periodontally diseased and healthy sites. J Periodontal Res. 1996;31:423–432. doi: 10.1111/j.1600-0765.1996.tb00511.x. [DOI] [PubMed] [Google Scholar]
- 34.van Belkum A. The role of short sequence repeats in epidemiologic typing. Curr Opin Microbiol. 1999;2:306–311. doi: 10.1016/S1369-5274(99)80053-8. [DOI] [PubMed] [Google Scholar]
- 35.van Houte J, Lopman J, Kent R. The final pH of bacteria comprising the predominant flora on sound and carious human root and enamel surfaces. J Dent Res. 1996;75:1008–1014. doi: 10.1177/00220345960750040201. [DOI] [PubMed] [Google Scholar]
- 36.van Houte J, Sansone C, Joshipura K, Kent R. Mutans streptococci and non mutans streptococci acidogenic at low pH, and in vitro acidogenic potential of dental plaque in two different areas of the human dentition. J Dent Res. 1991;70:1503–1507. doi: 10.1177/00220345910700120601. [DOI] [PubMed] [Google Scholar]
- 37.van Steenbergen T J, Bosch-Tijhof C J, Petit M D, Van der Velden U. Intra-familial transmission and distribution of Prevotella intermedia and Prevotella nigrescens. J Periodontal Res. 1997;32:345–350. doi: 10.1111/j.1600-0765.1997.tb00543.x. [DOI] [PubMed] [Google Scholar]
- 38.Versalovic J, Koeuth T, Lupski J R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whiley R A, Beighton D. Current classification of the oral streptococci. Oral Microbiol Immunol. 1998;13:195–216. doi: 10.1111/j.1399-302x.1998.tb00698.x. [DOI] [PubMed] [Google Scholar]
- 40.Willcox M D, Drucker D B, Green R M. Relative cariogenicity and in-vivo plaque-forming ability of the bacterium Streptococcus oralis in gnotobiotic WAG/RIJ rats. Arch Oral Biol. 1987;32:455–457. doi: 10.1016/0003-9969(87)90083-5. [DOI] [PubMed] [Google Scholar]