Isolation of Novel Pelagic Bacteria from the German Bight and Their Seasonal Contributions to Surface Picoplankton (original) (raw)
Abstract
We tested new strategies for the isolation of abundant bacteria from coastal North Sea surface waters, which included reducing by several orders of magnitude the concentrations of inorganic N and P compounds in a synthetic seawater medium. Agar plates were resampled over 37 days, and slowly growing colonies were allowed to develop by repeatedly removing all newly formed colonies. A fivefold increase of colonies was observed on plates with reduced nutrient levels, and the phylogenetic composition of the culture collection changed over time, towards members of the Roseobacter lineage and other alpha-proteobacteria. Novel gamma-proteobacteria from a previously uncultured but cosmopolitan lineage (NOR5) formed colonies only after 12 days of plate incubation. A time series of German Bight surface waters (January to December 1998) was screened by fluorescence in situ hybridization (FISH) with isolate-specific and general probes. During spring and early summer, a prominent fraction of FISH-detectable bacteria (mean, 51%) were affiliated with the_Cytophaga-Flavobacterium_ group (CF) of the_Bacteroidetes_. One Cytophaga sp. lineage with cultured representatives formed almost 20% of the CF group. Members of the Roseobacter cluster constituted approximately 50% of alpha-proteobacteria, but none of the _Roseobacter_-related isolates formed populations of >1% in the environment. Thus, the readily culturable members of this clade are probably not representative of Roseobacter species that are common in the water column. In contrast, members of NOR5 were found at high abundances (>105 cells ml−1) in the summer plankton. Some abundant pelagic bacteria are apparently able to form colonies on solid media, but appropriate isolation techniques for different species need to be developed.
Early marine microbiologists were convinced that a high percentage of abundant pelagic bacteria could be recovered on substrate-amended agar plates (49). This was supported by comparing the morphologies and Gram-staining reactions of isolates and free-living cells. Since then, epifluorescence microscopy (24) and molecular biological techniques have changed our paradigm about the culturability of common aquatic microbes. Frequently, less than 1% of all the cells from marine picoplankton form colonies on solid media (15). The reasons for this low recovery remain subject of debate (46).
Molecular biology has developed tools that help us to study the contributions of individual microbial lineages to the picoplankton (11, 17, 19, 36, 38, 47). This knowledge provides ecological criteria for choosing bacterial species as the focus of cultivation attempts and conversely can show whether isolated strains belong to lineages that are frequent in the environment (14). With the exception of the _Roseobacter_lineage (19), there are no isolates available from many of the abundant phylogenetic groups in the picoplankton (e.g., the SAR86 cluster or the marine crenarchaeota) (21, 27). In contrast, a large fraction of frequently isolated bacterial strains are related to opportunistic genera, such as Vibrio,Alteromonas, and Marinomonas, which probably inhabit niches with high substrate concentrations (32) but do not form large populations in the water column (14).
On the other hand, although the environmental parameters found at abundance maxima of particular phylogenetic groups may provide important information, a knowledge of the in situ densities does not readily result in specific cultivation strategies. The sizes of individual populations in the picoplankton are determined not only by substrate availability but also by food web structure and by viral mortality (48). Moreover, many heterotrophic pelagic bacteria and archaea probably overlap in their substrate spectra (26, 28, 35). At present, the appropriate conditions for the cultivation of common pelagic microbes can only be determined empirically.
It is, therefore, generally believed that isolation of model organisms for the oligotrophic pelagic microbiota will involve laborious and time-consuming strategies, e.g., dilution to extinction (2, 6,7). This view has recently been challenged by Pinhassi et al. (37, 38), who reported that a majority of the common bacterial species from the water column of the Baltic Sea could be grown on agar plates amended with ZoBell's original substrate mix (49). However, Eilers et al. have pointed out the practical limitations of such an approach (14). In their study, molecular biological techniques indicated the importance of uncultured gamma-proteobacteria related to SAR86 in coastal North Sea surface waters, but no representatives from this lineage were found in >140 bacterial strains collected over 2 years. The probability of retrieving any additional, novel gamma-proteobacterial isolates without a change of isolation strategy had decreased to less than 1 in every 50 new strains. So even if all pelagic marine bacteria and archaea were capable of forming colonies on agar plates, many strains from important marine lineages might still remain undetected, because of a predominance of isolates from genera that are rare in the picoplankton. For example, some oligotrophic species might exhibit a prolonged growth delay upon transfer to a solid medium. Such bacteria would be rapidly overgrown by other strains, such as those that are able to actively disperse over plate surfaces by swarming (12).
We attempted to obtain novel alpha- and gamma-proteobacteria from coastal North Sea surface waters by modified isolation strategies. The concentrations of the inorganic nutrients N and P in the growth medium were reduced, and plate cultivation techniques were adapted to allow the colony formation of microbes with longer growth delays. The abundances of those phylogenetic lineages that had been the target of cultivation were determined in environmental samples collected during the previous year. In addition, we determined the in situ densities of members from one culturable lineage of the_Cytophaga-Flavobacterium_ (CF) group (14).
MATERIALS AND METHODS
Sampling site and fixation.
Surface water was collected weekly (total bacterial counts and phytoplankton) or biweekly (specific bacterial groups) for the analysis of picoplankton dynamics between January and December 1998. Samples were obtained by pumping from a 1-m depth at the Helgoland Roads station (54°09′N, 7°52′E) near the island of Helgoland, which is situated approximately 50 km offshore in the German Bight of the North Sea. For cultivation, seawater was sampled once on 25 August 1999 at the same site. Water was stored at 4°C and processed within approximately 1 h. For fluorescence in situ hybridization (FISH), portions of 100 ml of unfiltered seawater were processed as described by Glöckner et al. (16).
Colony formation and enrichment cultures.
A synthetic seawater medium (MPM) was prepared for cultivation on agar plates as described by Schut et al. (44). In a modified version of this salt mix (MPM-m), the pH was adjusted to 7.5 and the concentrations of NH4Cl and KH2PO4were reduced to 50 and 1.5 μM, respectively. Both media were amended with a mix of monomers (5.7 mg of C/liter) (14), and for plate cultivation, 1% (wt/vol) agar (Difco Laboratories, Detroit, Mich.) was added. Subsamples (100 μl) of unfiltered seawater were directly spread on triplicate petri dishes containing either MPM or MPM-m. Over a period of 37 days the numbers of visible colonies were determined with a binocular microscope at a ×4 magnification. After 2, 7, 12, 15, 19, 29, and 36 days, all visible colonies were removed from the plate by excising the entire colony and the underlying agar with a sterile spatula. One plate containing MPM-m was selected for further characterization of isolates. The collected strains were subcultured in liquid medium before replating.
For the enrichment of marine alpha-proteobacteria in liquid cultures, MPM-m without additional substrates was inoculated with unfiltered or prefiltered (pore size, 1.2 and 0.45 μm) seawater (1:100 to 1:1,000). The dilutions were incubated at 16°C in the dark. The enrichments were continously screened by FISH with the oligonucleotide probe ALF968, specific for the alpha-proteobacteria (33), over a period of 9 weeks. If the alpha-proteobacterial abundances in the enrichments significantly increased (>20% of total counts), aliquots were plated on solid medium containing the MPM-m salt mix without additional carbon source. Representatives of all discernible colony morphotypes were selected from agar plates and subcultured under the same conditions. Only isolates that hybridized with probe ALF968 were included in the subsequent analysis.
Phylogenetic analysis.
Bacterial 16S rRNA primers 8_f_ (5′-AGAGTTTGATCMTGGC-3′) and 1542r (5′-AAAGGAGGTGATCCA-3′) were used to amplify almost full-length 16S ribosomal DNAs (rDNAs) from isolates (5) by PCR (42). Amplified 16S rRNA genes from isolates were sequenced by Taq cycle sequencing and universal 16S rRNA-specific primers using an ABI377 (Applied Biosystems, Inc.) sequencer. Sequence data were analyzed with the ARB software package (http://www.mikro.biologie.tu-muenchen.de). Phylogenetic trees were reconstructed using neighbor-joining, maximum-parsimony, and maximum-likelihood analyses. Only sequences that were at least 90% complete were used for tree construction. Alignment positions at which less then 50% of sequences of the entire set of data had the same residues were excluded from the calculations to prevent uncertain alignments within highly variable positions of the 16S rDNA, which may cause mistakes in the tree topology.
Cell counts, FISH, and probe design.
Total picoplankton counts were determined by epifluorescence microscopy (24). Isolates were screened by FISH with the oligonucleotide probes EUB338, targeted to Bacteria (1), ALF968, GAM42a, targeted to the gamma proteobacteria (31), and CF319a, targeted to the CF group of the Bacteroidetes(30). The specific oligonucleotide probes NOR5-730, NOR5-130, CYT1448, CYT1438, ROS537, KT13-231, KT09a, KT09b, RC1031, RC1239, ROS7-1029, ERYTH69, and RHIZO218 (Table1) were designed using the ARB software package. Probe specificity was evaluated with ARB against the rRNA database of the Technical University Munich (release 12/98), by the BLAST queueing system (http://www.ncbi.nlm.nih.gov/blast/blast.cgi), and by FISH to selected strains with base pair mismatches at the probe target sites. Probes labeled with the cyanine dye CY3 were synthesized by Interactiva (Ulm, Germany). Stringent hybridization conditions for the newly designed probes were determined for isolates by varying concentrations of formamide (34).
TABLE 1.
Oligonucleotide probes developed for FISH screening of environmental samples
| Probe | Specificity | Probe sequence (5′-3′) | Target sitea (16S rRNA positions) | % FAb |
|---|---|---|---|---|
| CYT1448 | _Cytophaga_spp. | CTAGGCCGCTCCTTACGG | 1448–1465 | 30 |
| CYT1438 | _Cytophaga_spp. | CCGCTCCTTACGGTGACG | 1438–1455 | 30 |
| NOR5-730 | NOR5 cluster | TCGAGCCAGGAGGCCGCC | 730–747 | 30 |
| NOR5-130 | NOR5 isolate KT71 | CCCCACTACTGGATAGAT | 130–147 | 20 |
| ROS537 | Marine alpha cluster (20) | CAACGCTAACCCCCTCC | 537–553 | 35 |
| KT13-231 | KT1117 | ATCTAATCAAACGCGGGCC | 231–249 | 30 |
| KT09b | KT0917 | GCTATTCCGTAGATCTGG | 136–153 | 20 |
| KT09c | KT0917 | ACTGTGTCCCCTAAAGGA | 1025–1042 | 20 |
| RC1031 | KT0202a | ACCTGTCACTATGTCCCG | 1031–1049 | 20 |
| RC1239 | KT0202a | TAACTCACTGTAGTTGCCAT | 1239–1279 | 20 |
| Ros7-1029 | JP7.1 group | CTGTCACTTGGTCTCTTG | 1029–1046 | 35 |
| Erythro69 | JP13.1 group | GCCACTCACCCCGAGGGT | 69–106 | 20 |
| Rhizo218 | JP66.1 group | GGGCCCATCCTACTCCGA | 218–235 | 20 |
Samples from North Sea surface waters were analyzed by FISH with all of the above probes. Hybridizations, counterstaining with 4,6-diamidino-2-phenylindole (DAPI; 1 μg/ml), mounting, and microscopic evaluations were performed as described previously (16). The fraction of FISH-stained bacteria in at least 1,000 DAPI-stained cells per sample was quantified.
Nucleotide sequence accession numbers.
The 16S rDNA sequences from isolates generated in this study were deposited in GenBank under the accession numbers AY007676 to AY007684 and AF305498.
RESULTS
Colony-forming bacteria obtained by time series.
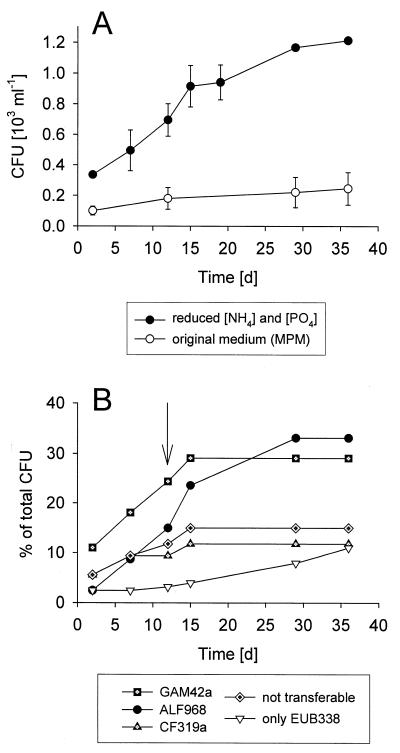
Over 37 days of incubation, only 0.25 × 103 ± 0.11 × 103 (mean ± 1 standard deviation) CFU ml−1 appeared on plates containing the original MPM medium, while 1.21 × 103 ± 0.01 × 103 CFU ml−1 were obtained on MPM-m plates (Fig. 1A). Of the isolates that appeared during the first 2 days of incubation on MPM-m, 41% were affiliated with the gamma-proteobacteria (14 out of 34), 20% were affiliated with the CF branch of the Bacteroidetes, and only 9% were affiliated with alpha-proteobacteria. Within the next 13 days the number of additional alpha-proteobacteria increased by a factor of 10, whereas the number of additional isolates from the gamma-proteobacteria and the CF group increased less than three-fold (Fig. 1B). During the second half of the incubation period the majority of newly retrieved isolates were affiliated with the alpha-proteobacteria. Altogether there were 19 colonies that did not grow after reinoculation in liquid medium (“not transferable” in Fig. 1B). At the end of incubation the gamma-proteobacteria constituted 29%, members of the CF group constituted 12%, and the alpha-proteobacteria constituted 33% of the culture collection (Fig. 1B). The number of alpha-proteobacteria affiliated with the “marine alpha” lineage (i.e., that hybridized with probe ROS537) (Fig. 2A) increased from a single colony during the first 2 days to a total of 11 strains (9%).
FIG. 1.
(A) Cumulative abundances of colony-forming bacteria from coastal North Sea surface waters on artificial seawater medium (MPM) during 37 days of incubation and on MPM with reduced concentrations of NH4 and PO4 (MPM-m). (B) Cumulative percentages of colonies from different phylogenetic groups on MPM-m. The arrow indicates the time of colony formation of isolate KT71, affiliated with the gamma-proteobacterial clade NOR5. “not transferable,” isolates that could not be subcultured.
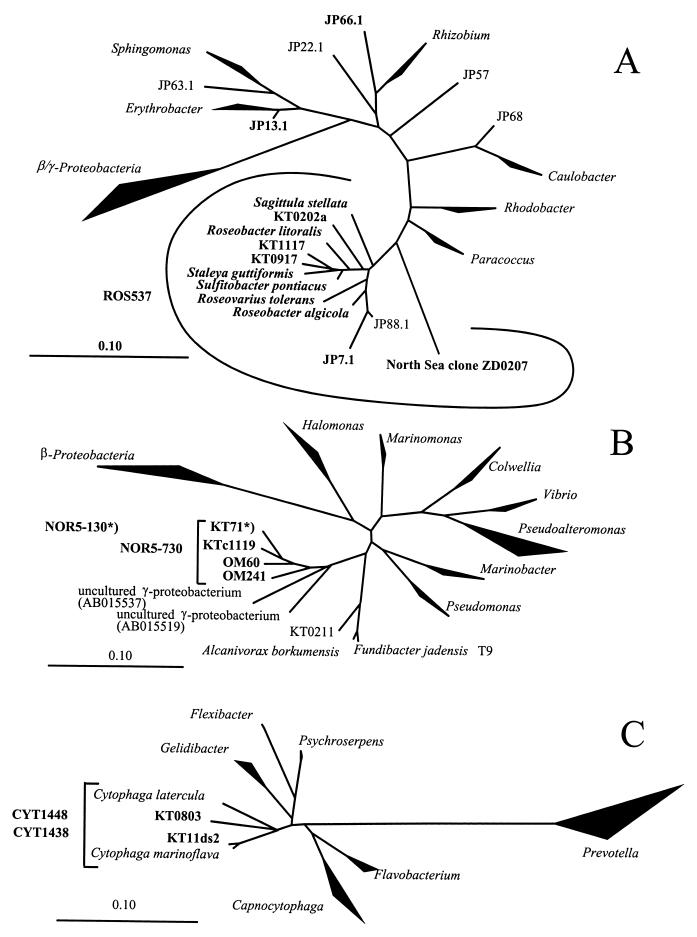
FIG. 2.
Phylogenetic trees based on comparative 16S rDNA sequence analysis of selected isolates of alpha-proteobacteria (A), gamma-proteobacteria (B), and the CF cluster (C). Brackets indicate specificity of the probes ROS537, NOR5-730, CYT1438, and CYT1448. Scale bars depict 10% sequence divergence. Names in bold indicate sequences targeted by newly designed specific probes. The asterisk indicates the probe that specifically targets the depicted phylotype KT71.
During the first 2 days of plate incubation, we obtained well-known alpha-proteobacteria such as_Roseobacter_ spp., gamma-proteobacteria affiliated with the genera Pseudoalteromonas, Alteromonas,Colwellia, and Photobacterium, and members of the CF cluster from the genera Cytophaga, Polaribacter, and Flavobacterium. Between days 3 and 11, bacteria from numerous additional genera were isolated, e.g.,Erythromicrobium, Agrobacterium,Paracoccus, and Sulfitobacter (alpha proteobacteria); Marinomonas,Shewanella, Sedimentobacter, and_Alcanivorax_ (gamma proteobacteria); and_Flexibacter_ (Bacteroidetes). One isolate (strain KT71) that was affiliated with a newly defined lineage of the gamma-proteobacteria (NOR5) (Fig. 2B) appeared only after 12 days of incubation (Fig. 1). This was also the case for several strains related to NOR1 (14). 16S rDNA sequence analysis of some isolates that did not hybridize with the three group-specific probes (ALF968, GAM42a, and CF319a) revealed that they belonged to other lineages, such as the actinobacterial genus_Microbacterium_ or the gamma-proteobacterial genus_Colwellia_.
During a period of several weeks the liquid enrichment cultures were regularly screened by FISH with probe ALF968. A total of 77 strains were isolated from those treatments, consisting of at least 20% alpha-proteobacteria. Twenty-three strains (30%) positively hybridized with probe ALF968. For initial phylogenetic analysis, partial 16S rDNA sequences of these isolates were determined. Sequences fell into eight distinct groups within the alpha-proteobacteria, and at least one nearly complete 16S rDNA sequence from each group was obtained (Table2; Fig. 2A). The most frequent isolates were related to the genus Erythrobacter (11 out of 23). Three sequences affiliated with two lineages within the_Roseobacter_ cluster (Fig. 2A). The phylogenetic affiliation of representative alpha-proteobacterial isolates from different lineages, and of other strains that were screened for their in situ occurrence in environmental samples, is shown in Table 2.
TABLE 2.
Phylogenetic affiliations of representative alpha-proteobacterial isolates and of isolates from other groups that were screened for their in situ abundances by FISH
| Phylogenetic group | Representative strain | Next relative to isolate or clonea | % 16S rRNA similarity | Specific probe(s) |
|---|---|---|---|---|
| CF | ||||
| Cytophaga | KT11ds2 (AF235111)b | Cytophaga marinoflavaM58770 | 98.0 | CYT1448, CYT1348 |
| KT0803 (AF235117)b | Cytophaga uliginosaM28238 | 93.6 | ||
| Alpha-proteobacteria | ||||
| Marine alpha cluster | ROS537 | |||
| Group A | JP88.1 (AY007684) | Alpha proteobacterium strain 303 (AF022392) | 99.7 | |
| Group B | JP7.1 (AY007679) | Marine isolate (L15345) | 93.3 | ROS7-1029 |
| Sulfitobacter | KT0917 (AF173972)b | Sulfitobacter pontiacus (Y13155) | 94–96 | KT09a, KT09b |
| KT1117 (AF173971)b | KT13-231 | |||
| KT0202a (AF305498)b | RC1031, RC1239 | |||
| Unknown affiliation | JP22.1 (AY007682) | Marine isolate (Y10914) | 98.9 | |
| Rhodospirillum | JP57 (AY007683) | Rhodospirillum salexigens (D14431) | 88.9 | |
| Sphingomonas | JP63.1 (AY007681) | Marine isolate (U85838) | 97.9 | |
| Caulobacter | JP68 (AY007678) | Caulobacter maris (AB008850) | 96.3 | |
| Erythrobacter | JP13.1 (AY007680) | Erythrobacter litoralis (ABO13354) | 97.3 | ERYTHRO69 |
| Phyllobacterium | JP66.1 (AY007677) | Phyllobacterium myrsinacearum (D12789) | 93.2 | RHIZO218 |
| Gamma-proteobacteria | ||||
| NOR5 | KT71 (AY007676) | Gamma-proteobacterium KTc1119 (AF235120) | 94.7 | NOR5-730, NOR5-130 |
Seasonal development of the plankton community.
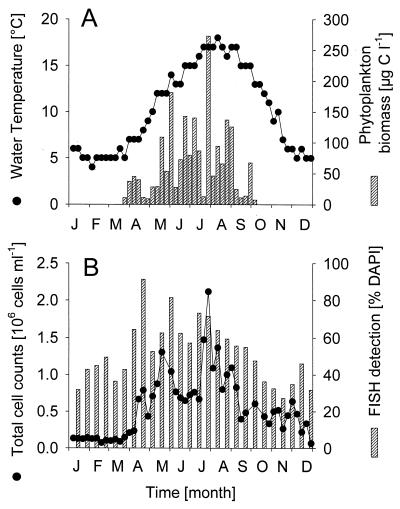
The annual temperature of surface waters of the German Bight in 1998 ranged between 3.2 and 17.9°C. It increased between April and August and declined thereafter (Fig. 3A). Between April and October, several short-lived maxima of phytoplankton biomass were observed (Fig. 3A) (www.pangaea.de/projects/). Interestingly, the spring phytoplankton bloom typically observed in the German Bight during April (40) was comparatively low. Bacterial cell densities remained more or less constant until late April (1.2 × 105 ml−1) (Fig. 3B), and cell numbers increased more than 10-fold over the next 3 months. Several peaks of total abundances were clearly separated by periods with lower cell densities. Bacterial numbers decreased between July and September and remained low thereafter.
FIG. 3.
(A) Seasonal fluctuations of water temperature and of phytoplankton biomass in German Bight surface waters in 1998. (B) Seasonal fluctuations of total picoplankton abundances and of FISH detection rates by the bacterial probe EUB338. Phytoplankton data are from www.pangaea.de/projects/.
Detection rates with probe EUB338, targeted to most_Bacteria_, roughly followed the patterns of total abundances (Fig. 3B) (Spearman rank correlation; n = 26, R = 0.7,P < 0.001). From January to March and from October to December, usually <50% of all DAPI-stained objects were detected by FISH. Between 52 and 92% of total picoplankton counts were visualized by FISH from April to September (mean, 66% of DAPI counts). The large majority of FISH-detectable bacteria (mean, 93%) could be identified as members of the CF group or of the alpha- and gamma-proteobacteria by the oligonucleotide probes CF319a, ALF968, and GAM42a. On average, 37% of FISH-detectable cells were affiliated with the CF group, 29% were affiliated with the alpha-proteobacteria, and 19% were affiliated with the gamma-proteobacteria (i.e., 21, 15, and 10% of DAPI counts, respectively). On several occasions, the sum of the group-specific probes even exceeded the total counts with probe EUB338, but with one exception (week 31) this always ranged within the expected cumulative counting error.
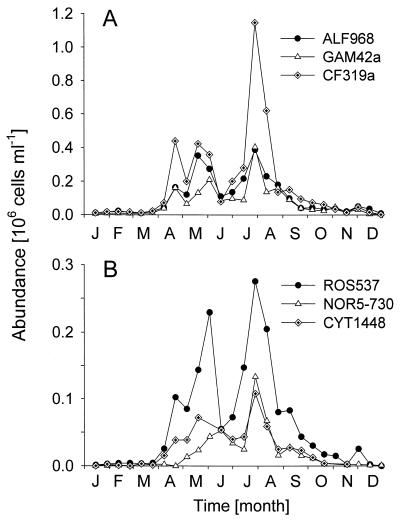
Between April and July, there were several peaks of abundance of members of the CF group, with an annual maximum of 1.2 × 106 cells ml−1 in July (Fig.4A). During this period they constituted up to 55% of total picoplankton counts (i.e., DAPI; mean, 35%) and up to 76% of FISH-detectable bacteria (mean, 51%). Bacteria affiliated with the _Cytophaga marinoflava-Cytophaga latercula_lineage (Fig. 2C and 4B) were detectable between April and September and formed a constant fraction of 6% ± 2% of the DAPI count. This corresponded to 0.75 × 105 cells ml−1 in May and 1.1 × 105 cells ml−1 in July and August.
FIG. 4.
(A) Seasonal fluctuations of alpha-proteobacteria (ALF968) and gamma-proteobacteria (GAM42a) and of members of the CF lineage (CF319a) detected by FISH in German Bight surface waters in 1998. (B) Abundances of Cytophaga spp. (CYT1448), members of the Roseobacter lineage (ROS537), and members of the NOR5 cluster (NOR5-730). Note the different _y_-axis scales.
The relative abundances of bacteria detected by ALF968 ranged from 12 to 33% of total DAPI counts between April and September. Alpha-proteobacteria formed local maxima in mid-May and July (Fig. 4A). In contrast to the CF group, the abundances of alpha-proteobacteria were not higher in July than in May. Roseobacter spp. and other members of the “marine alpha” group (19), as detected by probe ROS537 (Fig. 2A), formed the majority of the alpha proteobacteria between April and September (mean, 67%; range, 44 to 100%, corresponding to 12% of DAPI counts) (Fig. 4B). During that period, the counts with a published probe for the_Roseobacter_ group, MALF1 (19), were significantly lower than with probe ROS537 (mean, 69% of detection with ROS537; Wilcoxon matched-pair test, n = 9, P < 0.05). Culturable representatives from various phylogenetic lineages within the Roseobacter cluster (isolates KT0202a, KT0917, KT1117, and JP7.1) (Table 2) and from other alpha-proteobacterial lineages (isolates JP13.1 and JP66.1) (Table 2) never constituted more than 1% of DAPI counts in German Bight surface waters, as determined by FISH with specific probes (Table 1).
A first maximum of bacteria hybridizing with probe GAM42a was observed at the end of June (0.2 × 106 cells ml−1, 20% of DAPI) (Fig. 4A). Between April and September, the relative abundances of gamma-proteobacteria fluctuated between 10 and >20% of DAPI counts. Bacteria detected by GAM42a had densities similar to those of the alpha-proteobacteria during July and August (0.4 × 106 cells ml−1). For a period of approximately 3 months (May to August), members of the NOR5 cluster, as detected by probe NOR5-730, formed a large fraction (up to 61%) of gamma-proteobacteria. Two abundance maxima of NOR5 could be discerned, in early June and late July (Fig. 4B), when members of this group constituted 6 to 8% of total picoplankton (DAPI) counts.
DISCUSSION
Abundant culturable gamma-proteobacteria affiliated with NOR5.
So far, strain KT71 is the only isolated representative of a probably cosmopolitan gamma-proteobacterial lineage (NOR5) (Fig. 2B). Other members of this cluster are known exclusively from cloned 16S rDNA sequences, which have been amplified from the coastal North Sea, the Mediterranean Sea, and the Atlantic Ocean (14, 41,43). Almost all bacteria affiliated with NOR5 in coastal North Sea surface waters (Fig. 4B) were also detected by a second probe, NOR5-130 (Table 2), which was targeted exclusively to strain KT71.
Thus, within the limits of our rRNA-based identification, KT71 is among the first heterotrophic bacterial isolates that can be detected in the marine picoplankton in high abundances by direct microscopic counts. Bacteria related to KT71 formed populations of up to 1.5 × 105 cells ml−1 in the surface waters of the German Bight (8% of DAPI counts) (Fig. 4B). They constituted a major fraction of FISH-detectable gamma proteobacteria in the picoplankton during May and June (33%) and July and August 1998 (66%). At present we do not know whether NOR5 also plays a role in offshore microbial communities or in deeper water layers. The two observed abundance maxima coincided with biomass peaks of a single diatom species,Lauderia sp. (23). However, it would be too early to speculate about potential relationships between the two populations, since our environmental data are limited to a 1-year period and our sampling frequency probably did not provide sufficient resolution for an adequate description of plankton bloom situations (21).
Strain KT71 could be isolated on our mix of monomers and amino acids only after substantially reduction of the originally reported concentrations of N and P, and the strain formed a visible colony after 12 days of incubation. Eilers et al. (14) have shown that the probability of obtaining additional diversity of gamma-proteobacteria from the German Bight by plating on MPM medium was marginal, irrespective of a variety of different carbon sources. Therefore, the successful isolation of a member from the previously unculturable NOR5 lineage was most likely the consequence of our modifications of the original technique.
In a previous study, we discovered that frequently isolated opportunistic gamma-proteobacteria affiliated with_Alteromonas_ and Vibrio were quickly enriched in filtrates of coastal North Sea surface water (13). During earlier cultivation attempts (14), we furthermore noticed a rapid dispersal of some isolates over the agar surface, which resulted in a fusion of different colonies after 2 to 3 days of plate incubation. These observations inspired us to continuously remove newly appearing colonies from the petri dishes and, thus, to extend the potential period of colony formation to several weeks (Fig. 1A).
The isolation of KT71 after 12 days of incubation suggests that some common pelagic bacteria may grow on agar plates only after an extended lag phase. However, significant increases in the total number of isolates, and in their phylogenetic diversity, were observed only if the inorganic N and P concentrations in the medium were reduced to match the maximal ambient levels of coastal North Sea waters in 1998 (1.3 μmol of P liter−1, 29 μmol of N liter−1) (Fig. 1A). Cultivation of marine bacterioplankton on common complex media (2, 38) might, therefore, prevent the growth of autochthonous pelagic species because of excessively high nutrient concentrations. Preliminary observations indicate that strain KT71 does not grow on media containing 0.2% peptone and 0.1% yeast extract but can be maintained in liquid culture on our modified synthetic medium (H. Eilers, unpublished results).
Cultured members of the Roseobacter lineage are rare in the environment.
During recent years, the so-called “marine alpha” cluster (19), a phylogenetic lineage harboring genera such as Roseobacter, Sulfitobacter, and_Sagittula_ (Fig. 2A), has been shown to contribute significantly to coastal picoplankton (19, 21). Concomitantly, a number of isolates from this cluster could be readily obtained on substrates such as lignin or low-nutrient complex media (19, 22), and this has been regarded as evidence for the culturability of some abundant pelagic marine bacteria (19). Strains from the Roseobacter lineage serve as model organisms for the degradation abilities of “ecologically relevant” bacteria (4) and for the study of microbial sulfur cycling in the pelagic zone (18). This group, furthermore, plays a key role in controlling the turnover of the algal osmolyte dimethylsulfoniopropionate (50), and bacteria from the Roseobacter cluster form prominent populations during coccolithophore blooms (21, 50).
Altogether we obtained 14 strains that were affiliated with this phylogenetic group. A reduction of inorganic N and P in the medium, and an extended sampling of the agar plates, appeared to positively select for such bacteria. More than 10% of all isolates that formed colonies between day 2 and day 37 of plate incubation (10 of 87) were members of the Roseobacter lineage, as determined by group-specific FISH. In contrast, <6% of strains (8 of 142) were affiliated with this group in a previous collection of isolates that was established on the original MPM medium, and without plate resampling (14). Some strains from the marine alpha cluster were also isolated by plating on MPM-m medium after liquid enrichment on MPM-m without additional substrates. However, the latter approach was not particularly selective for this lineage but rather yielded numerous strains that were affiliated with_Erythrobacter_ (Fig. 2A).
We designed several probes targeting different groups of isolates from the Roseobacter cluster (Table 1) and a new group-specific probe, ROS537, with more complete coverage than a previously described probe (19). Cells detected by ROS537 (Fig. 4B) constituted a prominent fraction of all alpha-proteobacteria in German Bight surface waters (annual mean, 49%, corresponding to 8% of total DAPI counts). Yet the specific probes revealed that bacteria related to our isolates (Table 2; Fig. 2A) did not form large populations in the picoplankton, even when the FISH detection rates were generally high. Interestingly, the majority of_Roseobacter_ cells in samples from July to October 1998 (mean, 60%) could be visualized by a recently described probe, RSB67 (50), that is targeted to the 16S rDNA sequence of uncultured bacteria from this lineage (clone ZD0207) (Fig. 2A). Bacteria detected by this probe constituted the majority of the DNA- and protein-rich subpopulation in a picoplankton community during an algal bloom (50) in the northern part of the North Sea.
We present evidence that several culturable members of the marine_Roseobacter_ group are rare in the environment, as has been documented for the marine gamma-proteobacteria (13). The_Roseobacter_ lineage is physiologically rather versatile, including even phototrophic members (29). All previously described species are phylogenetically more closely related to our strains than to the uncultured phylotype ZD0207 (Fig. 2A). Culturable members of the Roseobacter lineage might, therefore, in general not be representative of the species that are abundant in coastal waters (Fig. 4B) (19) or during phytoplankton blooms (21, 50). Some readily culturable marine gamma-proteobacteria (Pseudoalteromonas and_Vibrio_) are known to be associated with animal and plant surfaces (25, 32). We speculate that the culturable species of the Roseobacter group may preferentially inhabit the surfaces of particular marine eucaryotes, e.g., algae (39). This could account for their superior ability to form colonies on solid media at reduced nutrient concentrations.
A lineage of culturable marine Cytophaga spp.
During an earlier study, only a few strains isolated on MPM medium were affiliated with the CF cluster of the Bacteroidetes(14). However, this group formed an important component of the surface picoplankton in the German Bight in 1998 and occasionally even numerically dominated the total bacterial community (Fig. 4A). High relative abundances of members of the CF group have also been found in the surface plankton of the coastal Atlantic Ocean during summer (8), as well as in Antarctic waters, co-occurring with a Phaeocystis bloom (45). Marine CF group members are apparently not limited to the attached bacterial fraction (10) but are common in the water column.
We therefore designed probes targeted to isolates from the culture collection of Eilers et al. (14) that were affiliated with the C. marinoflava-C. latercula lineage (Fig. 2C). This “marine cytophaga” cluster formed a small but constant fraction of the picoplankton during spring and summer. Between April and September, 5% of all DAPI-stained objects (19% of CF) were detected by the specific probes on average (Fig. 4B). Little is known about the physiology of the CF group members that are common in the aquatic environment. Preliminary results indicate that isolated strains from this C. marinoflava-C. latercula cluster do not grow on hydrolyzed chitin (Eilers, unpublished), which is preferentially consumed by some marine CF group members (9). Further studies are required to adequately cover the phylogenetic diversity within the CF lineage, and new isolation strategies that are more selective for abundant pelagic members of the CF group need to be developed.
Study of seasonal population changes by FISH.
We did not attempt an exhaustive analysis of the microbial community composition in German Bight surface picoplankton, and important groups such as SAR86, SAR11 (21), and the marine archaea have not been studied (27). Our investigation focused on the dynamics of a few lineages that were also targets of isolation attempts or from which isolates had been obtained earlier (Fig. 2) (14). Although a majority of the picoplankton community could be detected by FISH between April and September, at elevated water temperature and phytoplankton biomass (Fig. 1A), detection rates were low during winter. For the latter period it is thus impossible to evaluate whether and to what extent the per-cell activities of individual groups influenced our abundance estimates. Other microscopic approaches, e.g., FISH with polyribonucleotide probes (27), might help to overcome this limitation in the future.
Nevertheless, even a partial elucidation of the seasonal dynamics of picoplankton community composition provided us with targets for a directed screening and led to the investigation of isolates affiliated with Roseobacter spp. and Cytophaga spp. (Fig. 2A and C). The time series over the whole productive season (Fig. 4B) allowed a more integrated evaluation of the presence or absence of a particular phylotype in the pelagic zone than a single sampling. Bacteria isolated at any particular season could be extremely rare at that time point but could still form substantial populations during other periods of the year. Our biweekly sampling scheme might actually be too coarse to detect all abundance maxima of individual phylotypes, since, e.g., phytoplankton blooms in the North Sea occur on a scale of weeks (Fig. 1A). It is possible that some of the isolates affiliated with the alpha-proteobacteria formed similarly short-lived peaks in the German Bight. However, the probability of accidently missing all indication of six different populations (Fig. 2A) by our sampling is marginal. Nevertheless, investigations of single phylotypes at higher temporal resolution will be necessary to elucidate the actual dynamics and stability of bacterial populations in the plankton.
Conclusions.
Our results only partially support a hypothesis by Pinhassi et al. (38) and Gonzalez and Moran (19), who proposed that typical members of the marine picoplankton can form colonies on substrate-amended agar plates. Both the original and the modified synthetic medium yielded a few isolates that were closely related to abundant bacteria from coastal North Sea surface waters. However, most isolates were “laboratory weeds” with good culturability but low in situ abundance, e.g., all strains from the Roseobacter lineage. There is an obvious discrepancy between the rapid advancement of cultivation-independent approaches for the analysis of marine pelagic bacteria and the simplicity of typical isolation strategies, which have changed little during the last 50 years. In this study, the successful isolation of abundant bacteria from North Sea surface waters (Fig. 4B) was guided by information about bacterial in situ growth conditions and by data about microbial community composition. With this knowledge in hand, it is possible to isolate truly pelagic bacteria and to detect important members of the picoplankton hidden in culture collections.
ACKNOWLEDGMENTS
We thank Karl-Walter Klings for help during sampling and Birgit Rattunde for assistance in maintaining the culture collection. The BAH Helgoland is acknowledged for providing guest research facilities.
This study was supported by the Max Planck Society.
REFERENCES
- 1.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernard L, Schäfer H, Joux F, Courties C, Muyzer G. Genetic diversity of total, active and culturable marine bacteria in coastal seawater. Aquat Microb Ecol. 2000;23:1–11. [Google Scholar]
- 3.Brosius J, Dull T J, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 4.Buchan A, Collier L S, Neidle E L, Moran M A. Key aromatic-ring-cleaving enzyme, protocatechuate 3,4-dioxygenase, in the ecologically important marine Roseobacterlineage. Appl Environ Microbiol. 2000;66:4662–4672. doi: 10.1128/aem.66.11.4662-4672.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchholz-Cleven B, Rattunde B, Straub K. Screening for genetic diversity of isolates of anaerobic Fe(II)-oxidizing bacteria using DGGE and whole-cell hybridization. Syst Appl Microbiol. 1997;20:301–309. [Google Scholar]
- 6.Button D K, Robertson B R, Lepp P W, Schmidt T M. A small, dilute-cytoplasm, high-affinity, novel bacterium isolated by extinction culture and having kinetic constants compatible with growth at ambient concentrations of dissolved nutrients in seawater. Appl Environ Microbiol. 1998;64:4467–4476. doi: 10.1128/aem.64.11.4467-4476.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Button D K, Schut F, Quang P, Martin R, Robertson B. Viability and isolation of marine bacteria by dilution culture: theory, procedures, and initial results. Appl Environ Microbiol. 1993;59:881–891. doi: 10.1128/aem.59.3.881-891.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cottrell M T, Kirchman D L. Community composition of marine bacterioplankton determined by 16S rRNA gene clone libraries and fluorescence in situ hybridization. Appl Environ Microbiol. 2000;66:5116–5122. doi: 10.1128/aem.66.12.5116-5122.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cottrell M T, Kirchman D L. Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobactercluster consuming low- and high-molecular-weight dissolved organic matter. Appl Environ Microbiol. 2000;66:1692–1697. doi: 10.1128/aem.66.4.1692-1697.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeLong E F, Franks D G, Alldredge A L. Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol Oceanogr. 1993;38:924–934. [Google Scholar]
- 11.DeLong E F, Taylor L T, Marsh T L, Preston C M. Visualization and enumeration of marine planktonic archaea and bacteria by using polyribonucleotide probes and fluorescent in situ hybridization. Appl Environ Microbiol. 1999;65:5554–5563. doi: 10.1128/aem.65.12.5554-5563.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eberl L, Molin S, Givskov M. Surface motility of Serratia liquefaciensMG1. J Bacteriol. 1999;181:1703–1712. doi: 10.1128/jb.181.6.1703-1712.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eilers H, Pernthaler J, Amann R. Succession of pelagic marine bacteria during enrichment: a close look at cultivation-induced shifts. Appl Environ Microbiol. 2000;66:4634–4640. doi: 10.1128/aem.66.11.4634-4640.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eilers H, Pernthaler J, Glöckner F O, Amann R. Culturability and in situ abundance of pelagic bacteria from the North Sea. Appl Environ Microbiol. 2000;66:3044–3051. doi: 10.1128/aem.66.7.3044-3051.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferguson R L, Buckley E N, Palumbo A V. Response of marine bacterioplankton to differential filtration and confinement. Appl Environ Microbiol. 1984;47:49–55. doi: 10.1128/aem.47.1.49-55.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glöckner F O, Amann R, Alfreider A, Pernthaler J, Psenner R, Trebeslus K, Schleifer K-H. An in situhybridization protocol for detection and identification of planktonic bacteria. Syst Appl Microbiol. 1996;19:403–406. [Google Scholar]
- 17.Glöckner F O, Fuchs B M, Amann R. Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl Environ Microbiol. 1999;65:3721–3726. doi: 10.1128/aem.65.8.3721-3726.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonzalez J M, Kiene R P, Moran M A. Transformation of sulfur compounds by an abundant lineage of marine bacteria in the α-subclass of the class Proteobacteria. Appl Environ Microbiol. 1999;65:3810–3819. doi: 10.1128/aem.65.9.3810-3819.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzalez J M, Moran M A. Numerical dominance of a group of marine bacteria in the α-subclass of the class Proteobacteriain coastal seawater. Appl Environ Microbiol. 1997;63:4237–4242. doi: 10.1128/aem.63.11.4237-4242.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.González J M, Moran M A. Numerical dominance of a group of marine bacteria in the α-subclass of the class Proteobacteriain coastal seawater. Appl Environ Microbiol. 1997;63:4237–4242. doi: 10.1128/aem.63.11.4237-4242.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez J M, Simo R, Massana R, Covert J S, Casamayor E O, Pedros-Alio C, Moran M A. Bacterial community structure associated with a dimethylsulfoniopropionate-producing North Atlantic algal bloom. Appl Environ Microbiol. 2000;66:4237–4246. doi: 10.1128/aem.66.10.4237-4246.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzalez J M, Whitman W B, Hodson R E, Moran M A. Identifying numerically abundant culturable bacteria from complex communities: an example from a lignin enrichment culture. Appl Environ Microbiol. 1996;62:4433–4440. doi: 10.1128/aem.62.12.4433-4440.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hagmeier E. Phytoplankton der Helgoland-Reede. Landesamt für Natur und Umwelt des Landes Schleswig-Holstein. Germany: Flintbek; 1998. Nordseebericht 1998: [Google Scholar]
- 24.Hobbie J E, Daley R J, Jasper S. Use of Nuclepore filters for counting bacteria by fluorescence microscopy. Appl Environ Microbiol. 1977;33:1225–1228. doi: 10.1128/aem.33.5.1225-1228.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holmström C, Kjelleberg S. Marine Pseudoalteromonasspecies are associated with higher organisms and produce biologically active extracellular agents. FEMS Microbiol Ecol. 1999;30:285–293. doi: 10.1111/j.1574-6941.1999.tb00656.x. [DOI] [PubMed] [Google Scholar]
- 26.Jannasch H. Enrichments of aquatic bacteria in continuous culture. Arch Mikrobiol. 1967;59:165–173. doi: 10.1007/BF00406328. [DOI] [PubMed] [Google Scholar]
- 27.Karner M, DeLong E F, Karl D M. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature. 2001;409:507–509. doi: 10.1038/35054051. [DOI] [PubMed] [Google Scholar]
- 28.Karner M, Fuhrman J A. Determination of active marine bacterioplankton: a comparison of universal 16S rRNA probes, autoradiography, and nucleoid staining. Appl Environ Microbiol. 1997;63:1208–1213. doi: 10.1128/aem.63.4.1208-1213.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Labrenz M, Collins M D, Lawson P A, Tindall B J, Schumann P, Hirsch P. Roseovarius toleransgen. nov., sp. nov., a budding bacterium with variable bacteriochlorophyll a production from hypersaline Ekho Lake. Int J Syst Bacteriol. 1999;49:137–147. doi: 10.1099/00207713-49-1-137. [DOI] [PubMed] [Google Scholar]
- 30.Manz W, Amann R, Ludwig W, Vancanneyt M, Schleifer K-H. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum Cytophaga-Flavobacter-Bacteroidesin the natural environment. Microbiology. 1996;142:1097–1106. doi: 10.1099/13500872-142-5-1097. [DOI] [PubMed] [Google Scholar]
- 31.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 32.Montanari M P, Pruzzo C, Pane L, Colwell R R. Vibrios associated with plankton in a coastal zone of the Adriatic Sea (Italy) FEMS Microbiol Ecol. 1999;29:241–247. [Google Scholar]
- 33.Neef A. Ph.D. thesis. Munich, Germany: Technical University; 1997. [Google Scholar]
- 34.Neef A, Zaglauer A, Meier H, Amann R, Lemmer H, Schleifer K H. Population analysis in a denitrifying sand filter: conventional and in situ identification of Paracoccusspp. in methanol-fed biofilms. Appl Environ Microbiol. 1996;62:4329–4339. doi: 10.1128/aem.62.12.4329-4339.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ouverney C C, Fuhrman J A. Marine planktonic Archaea take up amino acids. Appl Environ Microbiol. 2000;66:4829–4833. doi: 10.1128/aem.66.11.4829-4833.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pernthaler J, Glöckner F O, Unterholzner S, Alfreider A, Psenner R, Amann R. Seasonal community and population dynamics of pelagic Bacteria and Archaeain a high mountain lake. Appl Environ Microbiol. 1998;64:4299–4306. doi: 10.1128/aem.64.11.4299-4306.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pinhassi J, Hagström A. Seasonal succession in marine bacterioplankton. Aquat Microb Ecol. 2000;21:245–256. [Google Scholar]
- 38.Pinhassi J, Zweifel U L, Hagström A. Dominant marine bacterioplankton species found among colony-forming bacteria. Appl Environ Microbiol. 1997;63:3359–3366. doi: 10.1128/aem.63.9.3359-3366.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prokic I, Brummer F, Brigge T, Gortz H D, Gerdts G, Schutt C, Elbrachter M, Muller W E G. Bacteria of the genus Roseobacter associated with the toxic dinoflagellate Prorocentrum lima. Protist. 1998;149:347–357. doi: 10.1016/S1434-4610(98)70041-0. [DOI] [PubMed] [Google Scholar]
- 40.Radach G, Berg J, Hagmeier E. Long-term changes of the annual cycles of meteorological, hydrographic, nutrient, and phytoplankton times series at Helgoland and at LV Elbe 1 in the German Bight. Continental Shelf Res. 1990;10:305–328. [Google Scholar]
- 41.Rappé M S, Kemp P F, Giovannoni S J. Phylogenetic diversity of marine coastal picoplankton 16S rRNA genes cloned from the continental shelf off Cape Hatteras, North Carolina. Limnol Oceanogr. 1997;42:811–826. [Google Scholar]
- 42.Saiki R K, Gelfand D H, Stoffel S, Scharf S J, Higuchi R, Horn G T, Mullins K B, Erlich H A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 43.Schäfer H, Bernard L, Courties C, Lebaron P, Servais P, Pukall R, Stackebrandt E, Troussellier M, Guindulain T, Vives-Rego J, Muyzer G. Microbial community dynamics in Mediterranean nutrient-enriched seawater. FEMS Microbiol Ecol. 2001;34:243–253. doi: 10.1111/j.1574-6941.2001.tb00775.x. [DOI] [PubMed] [Google Scholar]
- 44.Schut F, De Vries E J, Gottschal J C, Robertson B R, Harder W, Prins R A, Button D K. Isolation of typical marine bacteria by dilution culture growth maintenance and characteristics of isolates under laboratory conditions. Appl Environ Microbiol. 1993;59:2150–2160. doi: 10.1128/aem.59.7.2150-2160.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simon M, Glöckner F O, Amann R. Different community structure and temperature optima of heterotrophic picoplankton in various regions of the Southern Ocean. Aquat Microb Ecol. 1999;18:275–284. [Google Scholar]
- 46.Straskrabová V. The effect of substrate shock on populations of starving aquatic bacteria. J Appl Bacteriol. 1983;54:217–224. [Google Scholar]
- 47.Suzuki M T, Taylor L T, DeLong E F. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl Environ Microbiol. 2000;66:4605–4614. doi: 10.1128/aem.66.11.4605-4614.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thingstad T F, Lignell R. Theoretical models for the control of bacterial growth rate, abundance, diversity and carbon demand. Aquat Microb Ecol. 1997;13:19–27. [Google Scholar]
- 49.ZoBell C E. Marine microbiology. A monograph on hydrobacteriology. Waltham, Mass: Chronica Botanica Company; 1946. [Google Scholar]
- 50.Zubkov M V, Fuchs B M, Archer S D, Kiene R P, Amann R, Burkill P A. Linking the composition of bacterioplankton to rapid turnover of dissolved dimethylsulfoniopropionate in an algal bloom in the North Sea. Environ Microbiol. 2001;3:304–311. doi: 10.1046/j.1462-2920.2001.00196.x. [DOI] [PubMed] [Google Scholar]