In Vivo Transcription of the Escherichia coli oxyR Regulon as a Function of Growth Phase and in Response to Oxidative Stress (original) (raw)
Abstract
Simultaneous expression of seven genes in Escherichia coli was measured by a reverse transcription-multiplex PCR fluorescence procedure. Genes studied were (i) oxyR (transcriptional regulator); (ii) katG, dps, gorA, and ahpCF (controlled by OxyR); (iii) sodA (controlled by SoxRS); and (iv) trxA (not related to OxyR or SoxRS). Except for trxA, transcription of all genes was activated during the course of growth of wild-type bacteria, though notable variations were observed with respect to both the time and extent of activation. Whereas oxyR, katG, dps, and gorA were activated during exponential growth, ahpCF and sodA were stimulated in stationary phase. Maximal induction ranged from 4.6- to 86.5-fold, for gorA and dps, respectively. Treatment with H2O2 stimulated expression of the genes (katG, dps, ahpCF, and gorA) previously identified as members of the OxyR regulon, except for oxyR itself. Induction by H2O2 was a remarkably rapid and reversible process that took place in an OxyR-dependent and ςS-independent manner. NaCl induced expression of the genes controlled by OxyR, including the oxyR locus. This transcriptional up-regulation was preserved in a strain with the Δ_oxyR_::kan mutation, but it was abolished (ahpCF) or significantly reduced (oxyR and dps) in a strain with the rpoS::Tn_10_ mutation, potentially reflecting positive transcriptional regulation of the oxyR regulon by ςS. Expression of trxA was not increased either by H2O2 stress or by a shift to high-osmolarity conditions.
Different inducible responses are critical for survival of Escherichia coli after oxidative stress (20). Key regulators of adaptive responses to hydrogen peroxide (6) and superoxide anion (9) are OxyR (5) and SoxR together with SoxS (15, 30, 31), respectively.
The oxyR regulon contains at least eight genes, including those encoding hydroperoxidase I (katG), glutathione reductase (gorA), alkyl hydroperoxide reductase (ahpCF), and a nonspecific DNA-binding protein (dps) (2, 5). OxyR behaves as a transcriptional autorepressor under both inducing and noninducing conditions but activates the different regulon promoters only after H2O2 treatment (26). Direct oxidation of OxyR is the mechanism whereby cells sense hydrogen peroxide and induce the OxyR regulon (28). Recent studies have revealed that OxyR is activated by the formation of an intramolecular disulfide bond and is deactivated by enzymatic reduction with glutaredoxin 1 at the expense of reduced glutathione (32).
The soxRS regulon controls the expression of at least 10 genes, among them the gene encoding manganese-containing superoxide dismutase (sodA) (15). Regulation of the soxRS regulon occurs by a two-stage process. The constitutively expressed SoxR protein is first converted to an active form, which enhances soxS transcription. The increased levels of SoxS in turn activate expression of the various regulon genes. The mechanism of SoxR activation and the nature of the signaling molecule are still under debate (17, 18, 20); current mechanisms involve one-electron oxidation and assembly of the iron-sulfur centers of the molecule (7, 18).
An additional regulator for survival against oxidative stress in E. coli is the _rpoS_-encoded ςS subunit of RNA polymerase (20). The ςS regulon comprises a large number of genes, including katG (19), gorA (3), and dps (2), which are also controlled by OxyR. ςS expression is tightly regulated at the transcriptional, translational, and posttranslational levels (21). The cellular concentration of ςS increases during entry into stationary phase. Additionally, ςS and a rather large subset of ςS-controlled genes exhibit hyperosmotic induction during exponential growth (16).
The in vivo transcriptional activities of E. coli genes are regularly investigated by assaying for β-galactosidase activity in bacterial strains with fusions of the lac operon that contain the ribosome-binding site for lacZ (22). Data obtained by this procedure are not always consistent. Discrepancies are partly due to problems associated with differences among fusion constructions. We have recently designed and optimized a semiquantitative reverse transcription-multiplex PCR (RT-MPCR) procedure to examine simultaneously the in vivo expression of up to seven different genes (10). The assay is based on primer extension reactions with specific fluorophor-labeled primers and subsequent DNA sequencer (GeneScan) analysis of PCR products. This work investigates the applicability of the method to the study of the expression of genes involved in E. coli adaptive responses to oxidative stress.
MATERIALS AND METHODS
Chemicals.
Saturated phenol II and acrylamide-bis19:1 mixture were from Amresco (Solon, Ohio). The GeneAmp RNA PCR kit, Prism GeneScan-350 TAMRA (carboxytetramethylrhodamine-N_-hydroxysuccinimide) ladder, loading buffer, fluorescence-labeled primers, deoxynucleoside triphosphates (dNTPs), MgCl2 and Ampli_Taq were from Perkin-Elmer (Norwalk, Conn.). DNase (RNase free) was from Boehringer (Mannheim, Germany). MPCR buffer 3 was from Maxim Biotech (San Francisco, Calif.). Hydrogen peroxide (H2O2) and other chemicals were purchased from Sigma (St. Louis, Mo.). Reactive protein assay was from Bio-Rad (Hercules, Calif.).
Bacterial strains and growth conditions.
Bacterial strains used in this work are derived from E. coli K-12. Strain UC574 (arg56 nad113 araD81) (1) was considered the parental wild type. Strains UC1247 (Δ_oxyR_::kan) and UC1311 (rpoS::Tn_10_) were constructed by P1 transduction of the Δ_oxyR_::kan (obtained from G. Storz) or rpoS::Tn_10_ (obtained from P. Loewen) mutant allele into strain UC574. Transductants were first selected on Luria-Bertani (LB) medium with kanamycin or tetracycline and then scored for induction of hydroperoxidase I (HPI) (coded for by the katG gene) or hydroperoxidase II (coded for by the katE gene) activity upon exposure to 10 μM H2O2 (UC1247) or 500 mM NaCl (UC1311). Bacteria were grown in LB nutrient broth or M9 minimal medium (11). Minimal medium was supplemented with 2 g of glucose per liter, 40 μg of arginine per ml, 5 μg of d-biotin per ml, 5 μg of thiamine per ml, and 10 g of Casamino Acids per liter. Growth was monitored by measuring the optical density at 600 nm (OD600). Bacteria were inoculated into LB broth or M9 minimal medium and incubated for 15 h at 37°C with shaking at 150 rpm. The overnight cultures were then diluted into fresh LB or M9 medium (OD600 = 0.03) and incubated at 37°C and 150 rpm for the times indicated in Table 2 and the figure legends.
TABLE 2.
Gene expression during growth of wild-type bacteria in LB nutrient mediuma
| Sampling time (h) | OD600 | Mean fluorescence signal ± SEM of PCR productb | ||||||
|---|---|---|---|---|---|---|---|---|
| oxyR | katG | dps | gorA | ahpCF | sodA | trxA | ||
| 1.5 | 0.31 | 0.31 ± 0.07 (1.5) | 0.79 ± 0.11 (1.0) | 0.10 ± 0.02 (1.0) | 1.92 ± 0.18 (1.3) | 0.43 ± 0.07 (1.0) | 0.18 ± 0.06 (2.0) | 1.19 ± 0.12 (1.5) |
| 2 | 0.58 | 0.21 ± 0.02 (1.0) | 0.99 ± 0.08 (1.3) | 0.18 ± 0.02 (1.8) | 1.48 ± 0.28 (1.0) | 0.47 ± 0.10 (1.1) | 0.17 ± 0.06 (1.9) | 0.82 ± 0.14 (1.0) |
| 3 | 1.54 | 0.60 ± 0.15 (2.9) | 3.42 ± 0.55 (4.3) | 0.41 ± 0.02 (4.1) | 2.48 ± 0.33 (1.7) | 0.49 ± 0.08 (1.1) | 0.09 ± 0.02 (1.0) | 1.02 ± 0.13 (1.2) |
| 5 | 3.17 | 0.66 ± 0.07 (3.1) | 5.81 ± 0.21 (7.4) | 6.22 ± 0.38 (62.2) | 4.28 ± 0.43 (2.9) | 0.65 ± 0.09 (1.5) | 0.11 ± 0.02 (1.2) | 1.72 ± 0.25 (2.1) |
| 7 | 4.40 | 1.09 ± 0.30 (5.2) | 4.07 ± 0.40 (5.2) | 8.65 ± 1.50 (86.5) | 6.24 ± 1.57 (4.2) | 1.49 ± 0.14 (3.5) | 0.50 ± 0.12 (5.6) | 1.29 ± 0.30 (1.6) |
| 12 | 4.86 | 1.18 ± 0.22 (5.6) | 3.48 ± 0.43 (4.4) | 7.55 ± 1.37 (75.5) | 6.78 ± 0.91 (4.6) | 2.72 ± 0.55 (6.3) | 0.69 ± 0.13 (7.7) | 1.45 ± 0.27 (1.8) |
RNA purification.
Total RNA was extracted by the hot-phenol method, as previously described (8). The quality of the samples was checked electrophoretically, and quantification was done spectrophotometrically. At least two independent RNA preparations were isolated for each experimental condition.
RT-MPCR.
Synthesis of cDNA was carried out with the GeneAmp RNA PCR kit, as previously described (10). Each RNA sample was retrotranscribed at least twice. PCR amplification of cDNA was carried out with the primer pair sets listed in Table 1. Primers were designed with the Primer Select 3.03/96 (DNA Star, Madison, Wis.) and Oligo 5.0/96 (Molecular Biology Insights, Plymouth, Minn.) software programs, in order to obtain the highest specificity and performance. Conditions for MPCR were optimized as detailed by Gallardo-Madueño et al. (10) to produce fluorescence intensities of the desired products in the range of linearity. Thirty-five cycles were performed, each consisting of 1 min of denaturation at 94°C, 15 s of annealing at 68°C, and 30 s of enzymatic primer extension at 72°C. The PCR amplification mixture contained Ampli_Taq_ (1.25 U), MPCR buffer 3 (2.5 μl), MgCl2 (25 nmol), dNTP (at 1 mM each), and primers (3.1 pmol, oxyR; 1.5 pmol, katG; 1.4 pmol, dps; 1.125 pmol, gorA; 2.7 pmol, ahpCF; 3.1 pmol, sodA; 0.475 pmol, trxA; 0.7 pmol, gapA) in a final volume of 25 μl. PCR fragments of 129 bp (oxyR), 137 bp (katG), 142 bp (dps), 125 bp (gor), 128 bp (ahpCF), 121 bp (sodA), 148 bp (trxA) and 131 bp (gapA) were obtained. At least two MPCRs were performed for each cDNA.
TABLE 1.
PCR primer characteristics
| Primera | _n_-mer | Sequence | PCR fragment size (bp) |
|---|---|---|---|
| oxyR HEX labeled (forward) | 16 | 5′-CGC GAT CAG GCA ATG G-3′ | 129 |
| oxyR (reverse) | 21 | 5′-CAG CGC TGG CAG TAA AGT GAT-3′ | |
| katG (forward) | 21 | 5′-CTG CGT TTT GAT CCT GAG TTC-3′ | 137 |
| katG 6-FAM labeled (reverse) | 19 | 5′-GGC CCG ATG TAG CGA GAT T-3′ | |
| dps 6-FAM labeled (forward) | 22 | 5′-CAA AAC CCC GCT GAA AAG TTA C-3′ | 142 |
| dps (reverse) | 22 | 5′-GAT ATC TGC GGT GTC GTC ATC T-3′ | |
| gorA (forward) | 20 | 5′-GAT GTA TAC CGC CGT CAC CA-3′ | 125 |
| gorA HEX labeled (reverse) | 19 | 5′-AGC CCT GCA ACA TTT CGT C-3′ | |
| ahpCF 6-FAM labeled (forward) | 18 | 5′-CCG CAG GGT ATC ATC CAG-3′ | 128 |
| ahpCF (reverse) | 18 | 5′-TTA GCC GGG CAA ACT TCA-3′ | |
| sodA (forward) | 18 | 5′-CCG ATT ATG GGC CTG GAT-3′ | 121 |
| sodA 6-FAM labeled (reverse) | 17 | 5′-CAA AAC GTG CCG CTG CT-3′ | |
| trxA HEX labeled (forward) | 20 | 5′-TGC GGT CCG TGC AAA ATG AT-3′ | 148 |
| trxA (reverse) | 21 | 5′-AGC AGC AGA GTC GGG ATA CCA-3′ | |
| gapA 6-FAM labeled (forward) | 20 | 5′-GTC GCT GAA GCA ACT GGT CT-3′ | 131 |
| gapA (reverse) | 23 | 5′-AAG TTA GCG CCT TTA ACG AAC AT-3′ |
MPCR product quantification.
Following amplification, 0.5 μl of the MPCR product was mixed with 0.2 μl of Prism GeneScan-350 TAMRA ladder, 1.4 μl of deionized formamide, and 0.2 μl of loading buffer. Samples were denatured at 95°C for 2 min. Samples were run on a denaturing 4.24% polyacrylamide gel at 750 V in an ABI Prism 377 DNA sequencer/GeneScan from the Applied Biosystems Division of Perkin-Elmer (Foster City, Calif.). Data were collected and analyzed with the ABI Prism 377 Collection 2.1/97 and GeneScan Analysis 2.0.2/95 software programs, respectively (Perkin-Elmer/Applied Biosystems Division). Differences in amplification efficiencies among samples were normalized by comparing the fluorescence intensity of each band to that resulting from gapA amplification, which was used as reference gene. Samples for comparison of different experimental conditions were handled in parallel. Data are presented as means ± standard errors of the means (SEM) from n MPCR amplifications. Comparison between groups was done by Student’s t test. Significances at a P level of <0.05 are indicated in the text.
Enzymatic assays.
Ten milliliters of bacterial cultures were centrifuged at 16,000 × g at 4°C for 5 min. The cell pellet was washed and resuspended in 0.5 ml of 20 mM potassium phosphate buffer (pH 7.5) with 0.1 mM EDTA. Cells were disrupted at 4°C by ultrasonic disintegration (three 12-s pulses, 25 W). The peroxidase activity of HPI was assayed in dialyzed extracts by monitoring H2O2 decomposition at 460 nm with _o_-dianisidine as the hydrogen donor (24). One unit of peroxidase activity is defined as the amount of enzyme that decomposes 1 μmol of H2O2 per min at 25°C. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) activity was determined by using a coupled system in a reaction mixture containing 100 mM triethanolamine buffer (pH 7.6), 0.1 mM EDTA, 0.7 mM MgSO4, 0.1 mM ATP, 6 mM 3-phosphoglycerate, 1.5 U of phosphoglycerate kinase from chicken muscle per ml, and 0.2 mM NADH. A GAPDH unit of activity is defined as the amount of enzyme catalyzing the utilization of 1 μmol of substrate per minute at 30°C. Protein concentration was estimated by the method of Bradford (4). Specific activity values were expressed as means ± SEM for at least three independent determinations. Comparison between groups was done by Student’s t test. Significances at a P level of <0.05 are indicated in the text.
RESULTS AND DISCUSSION
Growth-phase dependent variation in gene expression.
Eight primer pairs were designed (Table 1) to study by RT-MPCR the simultaneous expression of genes related to E. coli adaptive responses to oxidative stress. The procedure was basically as previously described for a different group of genes (10). The fluorescent PCR products were separated on acrylamide gels with an ABI Prism 377 DNA sequencer and analyzed with GeneScan software. For purposes of semiquantitative analysis, data are expressed as the ratio of the signal obtained for each individual gene divided by the signal obtained from the reference mRNA of the corresponding sample.
As described previously (10), the gapA gene, which codes for GAPDH, was used as reference. Bacteria displayed similar basal GAPDH enzymatic activities through the growth curve (average of 790 ± 150 mU/mg of protein from three independent determinations). Genes under study were (i) oxyR (transcriptional regulator); (ii) katG, dps, gorA, and ahpCF (members of the H2O2-inducible regulon controlled by OxyR); (iii) sodA (member of the O2·−-induced regulon controlled by SoxRS); and (iv) trxA. The last gene, coding for thioredoxin, was included because its expression has not been linked to OxyR or SoxRS.
Expression of genes throughout the culture time of wild-type bacteria in rich LB medium (for which most data are available) is shown in Table 2. With the possible exception of trxA, all genes under study were clearly activated over the 12-h period of cultivation. In agreement with the oxidant resistance of stationary-phase E. coli (14), minimal expression levels were observed during the first 2 h of growth; then, gene expression increased with the time of culture, giving maximal induction levels at the late-exponential or stationary growth phase (5 to 12 h). Notable variations with respect to both the time and the extent of activation, however, were observed. Thus, whereas oxyR, katG, dps, and gorA were activated at exponential phase, ahpCF and sodA were stimulated somewhat later, at stationary phase. Maximal values ranged from 4.6- to 86.5-fold increases, for gorA and dps, respectively.
González-Flecha and Demple (13) reported a 3.5-fold increase in the steady-state level of oxyR mRNA during exponential growth (2 to 4 h) of E. coli in rich LB medium, followed by a rapid decline to yield initial values of expression. While we detected a similar rise in oxyR mRNA during exponential growth, we did not observe any decrease at stationary phase (Table 2). To the contrary, a maximal induction factor of 5.6-fold was quantified at 12 h of growth.
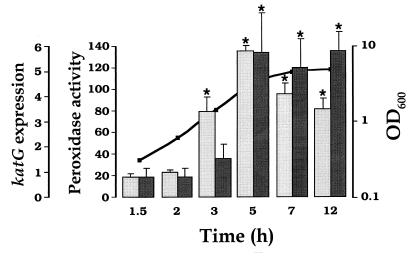
Activation of katG expression, however, exhibited a biphasic profile, with a minimal value at 1.5 h of outgrowth and a maximal induction factor of 7.4-fold at 5 h. After reaching this peak, the amount of katG transcript declined during stationary phase, and this decrease was statistically significant with respect to the 5-h value. This result is basically in agreement with a previous study (12) in which transcription was monitored by using a katG::lacZ operon fusion. However, the induction and maximum for katG mRNA reported in Table 2 preceded by ∼2 h that seen for β-galactosidase expression with the fusion gene. A delay for protein synthesis and the effect of the stability of the protein can explain differences between the pattern of enzymatic activity and the transcriptional behavior of a gene. These circumstances are investigated in Fig. 1 with respect to the peroxidase activity of HPI, coded for by katG. Low levels of peroxidase activity were observed during the first 3 h of outgrowth. Induction occurred at late exponential phase and was maintained during stationary phase. The maximal increase in peroxidase activity (7.3-fold) was identical to that observed in katG mRNA. As in the case of peroxidase, glutathione reductase (GRase) activity presented also an ∼2-h delay with respect to gorA expression (data not shown).
FIG. 1.
Peroxidase activity during course of growth of wild-type bacteria in nutrient LB medium. Cells were grown as described in Table 2, footnote a. Peroxidase activity (mU/mg of protein) was determined as specified in Materials and Methods. Activity data (darkly shaded bars) are from an average of four independent determinations. Expression data (lightly shaded bars) are those given in Table 2. Error bars represent SEM. Statistical significance (P < 0.05), for comparisons with minimal values at 1.5 h of outgrowth, is marked with asterisks. Bacterial growth, monitored as OD600, is indicated by the line.
Transcriptional regulation as a function of bacterial growth has been reported for two other genes controlled by OxyR, namely dps (2) and gorA (3). Our data agree with both previous reports in showing maximal transcriptional levels after the onset of stationary phase. Furthermore, they indicate the need for a much higher level of induction for dps than for gor transcripts (86.5- versus 4.6-fold increments). The physiological sense of such a difference might be in the ability of Dps to directly protect DNA from oxidative damage (23), compared with proteins which are either enzymes (HPI, GRase, AHP [alkyl hydroperoxide reductase], and Mn-SOD [manganese-containing superoxide dismutase]) or regulators (OxyR). To our knowledge, we present here the first examples of variations in the levels of ahpCF and sodA transcripts during aerobic growth in E. coli. It is noteworthy that transcriptional induction of ahpCF genes occurs much later than that of other OxyR-regulated genes (7 versus 3 h of outgrowth). This observation seems to be in disagreement with the idea of an H2O2-mediated change in OxyR regulon expression during growth (12).
Gene expression induction by hydrogen peroxide.
While the oxyR locus is central to the adaptive response of exponentially growing cells to H2O2, the constitutive increased resistance to oxidants of stationary phase cells is linked to rpoS (20). To examine the role of these two regulators on expression of the genes under study, strains carrying the Δ_oxyR_::kan or rpoS::Tn_10_ mutant allele were constructed and subjected to different stress conditions in conjunction with wild-type bacteria.
Optimal conditions for induction of gene expression by H2O2 were first established in experiments with the wild-type strain. Induction of transcripts was readily seen with 10 to 100 μM H2O2 during exponential growth (data not shown). Treatments in M9 minimal medium resulted in the induction of higher levels of gene expression than those in nutrient LB (data not shown), in agreement with previous data on the induction level of enzymes controlled by OxyR (5). The kinetics of induction following 10 μM H2O2 addition to wild-type cells at early exponential growth in M9 is shown in Fig. 2. The influence of the regulatory Δ_oxyR_::kan and rpoS::Tn_10_ mutations on H2O2 induction is investigated in Fig. 3.
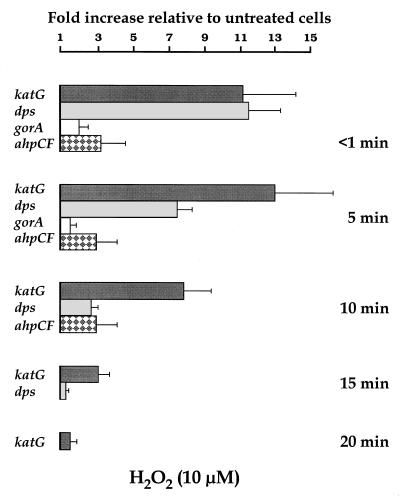
FIG. 2.
Wild-type cells from an overnight culture in M9 minimal medium were diluted in fresh medium and incubated at 37°C with shaking at 150 rpm. At an OD600 of 0.2, H2O2 was added to half of the culture, to make a final 10 μM solution, and the rest was used as a control. Samples were collected immediately (<1 min) after the addition of H2O2 and at 5, 10, 15, and 20 min of exposure. Samples were frozen with liquid nitrogen, and total RNA was purified as described in Materials and Methods. The fluorescence signal of each PCR product was compared to that of gapA. Data are from an average of eight MPCR amplifications. Values from treated samples were divided by those from the corresponding control. All genes were analyzed, but only those genes for which statistically significant (P < 0.05) increases were observed at a given time are represented. Error bars were estimated from the corresponding SEM.
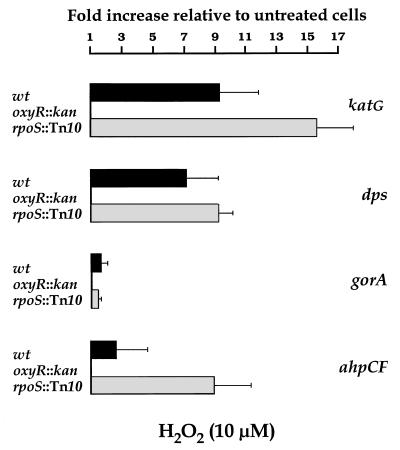
FIG. 3.
Wild-type (wt), Δ_oxyR_::kan, and rpoS::Tn_10_ bacteria from overnight cultures in M9 minimal medium were diluted in fresh medium and incubated at 37°C with shaking at 150 rpm. At an OD600 of 0.2, H2O2 was added to half of each culture, to make a final 10 μM solution, and the rest was used as a control. Samples were collected at 5 min after the addition of H2O2 and frozen with liquid nitrogen. Total RNA was purified as described in Materials and Methods. The fluorescence signal of each PCR product was compared to that of gapA. Data are from an average of eight MPCR amplifications. Values from treated samples were divided by those from the corresponding control. All genes were analyzed, but only those genes for which statistically significant (P < 0.05) increases were observed are represented. Error bars were estimated from the corresponding SEM.
Treatment with H2O2 stimulated expression of the four genes identified as being under oxyR control (2, 5). Therefore, maximal induction levels of 12.9-, 11.3-, 3.3-, and 2.0-fold were observed for katG, dps, ahpCF, and gorA, respectively (Fig. 2). Induction of katG and dps transcription in response to H2O2 was previously reported (2, 27). However, though _oxyR_-regulated promoters have been mapped upstream from the Salmonella typhimurium ahp genes and the E. coli gorA gene (27, 28), and a constitutive oxyR1 mutant of S. typhimurium contains higher levels of GRase and AHP activities than do wild-type extracts (5), the putative increments in ahpCF and gorA transcription by H2O2 stress have not been reported so far for E. coli.
The induction of katG, dps, gorA, and ahpCF expression produced by H2O2 was abolished by the introduction of the Δ_oxyR_::kan mutation (Fig. 3), indicating that the induction is OxyR dependent, in agreement with published reports on regulation of katG and dps expression (2, 27). In contrast, this transcriptional up-regulation was preserved in the strain with the rpoS::Tn_10_ mutation, indicating that ςS is not required in the OxyR-mediated response to H2O2 of exponentially growing cells. In fact, the factors of induction by H2O2 were somewhat higher in the rpoS::Tn_10_ mutant than in the wild-type strain, which might suggest that more ς70-containing RNA polymerase remains in the rpoS mutant than in wild-type cells to support OxyR-dependent transcription.
The four OxyR-dependent genes exhibited remarkably rapid induction in response to 10 μM H2O2 (Fig. 2). Therefore, transcription increased to maximal (or near maximal) induction levels immediately after addition of the oxidant and then fell back to basal levels within 10 to 20 min of treatment. Longer periods for this transient phenomenon were reported for oxyS expression upon exposure of wild-type cells to a much higher (200 μM) H2O2 concentration (32). Since expression of oxyR was not induced after oxidative stress with 10 μM H2O2, in agreement with previous results of OxyR protein synthesis (26), our data indicate that activation by oxidation of the OxyR transcription factor is an extremely rapid event in vivo, the oxidized OxyR being then quickly converted to the reduced and inactive form in the presence of cellular reductants such as glutaredoxins and thioredoxins (10, 32).
While expression of the OxyR-dependent genes was readily induced by H2O2, induction (fivefold) of sodA transcription (under the control of soxRS) was specific to paraquat (a redox-cycling compound) treatment (data not shown), in agreement with previous results (29). Expression of trxA was not induced after oxidative stress by either H2O2 or paraquat.
Gene expression induction by sodium chloride.
To obtain evidence of ςS-dependent regulation, we exploited the observation that ςS expression is induced posttranscriptionally in response to osmotic upshift in the growth medium (21). Osmotic shock by increasing the NaCl concentration to 500 mM resulted in the induction of gene expression in wild-type bacteria at both the exponential and stationary phases when grown in either nutrient LB or minimal M9 medium (data not shown). The kinetics of induction of transcription in response to increased medium osmolarity and comparisons of the wild-type strain and isogenic derivatives defective in OxyR or ςS are examined in Fig. 4 and 5, respectively.
FIG. 4.
Wild-type cells from an overnight culture in M9 minimal medium were diluted in fresh medium and incubated at 37°C with shaking at 150 rpm. At an OD600 of 0.2, NaCl was added to half of the culture, to make a final 500 mM solution, and the rest was used as a control. Samples were collected immediately (<1 min) after the addition of NaCl and at 5, 10, and 15 min of exposure. Samples were frozen with liquid nitrogen, and total RNA was purified as described in Materials and Methods. The fluorescence signal of each PCR product was compared to that of gapA. Data are from an average of eight MPCR amplifications. Values from treated samples were divided by those from the corresponding control. All genes were analyzed, but only those genes for which statistically significant (P < 0.05) increases were observed at a given time are represented. Error bars were estimated from the corresponding SEM.
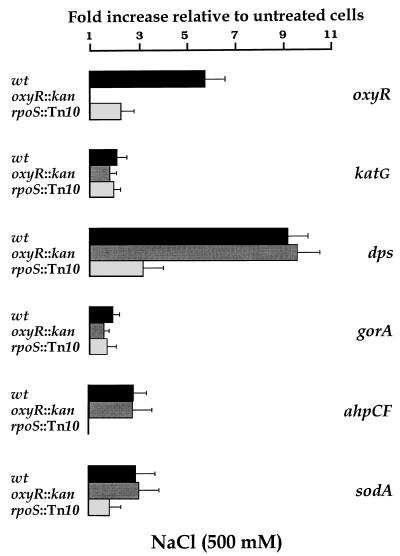
FIG. 5.
Wild-type (wt), Δ_oxyR_::kan, and rpoS::Tn_10_ bacteria from overnight cultures in M9 minimal medium were diluted in fresh medium and incubated at 37°C with shaking at 150 rpm. At an OD600 of 0.2, NaCl was added to half of each culture, to make a final 500 mM solution, and the rest was used as a control. Samples were collected at 15 min after the addition of NaCl and frozen with liquid nitrogen. Total RNA was purified as described in Materials and Methods. The fluorescence signal of each PCR product was compared to that of gapA. Data are from an average of eight MPCR amplifications. Values from treated samples were divided by those from the corresponding control. All genes were analyzed, but only those genes for which statistically significant (P < 0.05) increases were observed are represented. Error bars were estimated from the corresponding SEM. Bacteria carrying the Δ_oxyR_::kan mutation had undetectable levels of the corresponding mRNA.
Osmotic induction of transcription was a rapid process (Fig. 4), as previously reported for several _rpoS_-dependent genes (16). Nevertheless, induction by increased osmolarity was not as fast as induction in response to H2O2 (Fig. 2), considering that substantial increases in oxyR, katG, dps, gorA, ahpCF, and sodA expression were not observed until after 10 or 15 min of osmotic upshift. Osmotic induction of oxyR corresponded more or less with the maximal values found during the transition into stationary phase (Table 2). In contrast, stationary-phase induction greatly exceeded osmotic induction for dps and katG. Expression of gorA, ahpCF, and sodA was only weakly induced (two- to threefold) by NaCl under our experimental conditions. These results agree with the observation that for a given _rpoS_-dependent gene, the extents of osmotic induction and of stationary-phase induction do not necessarily correlate (16).
Induction by elevated osmolarity was unaffected in the Δ_oxyR_::kan mutant, but it was abolished (ahpCF) or significantly reduced (oxyR, dps, and sodA) in the rpoS::Tn_10_ mutant (Fig. 5), in agreement with published data on osmotic regulation of _rpoS_-dependent loci (16). Only katG and gorA expression did not follow this regulatory pattern, since neither a mutation in oxyR nor one in rpoS were able to prevent twofold induction by NaCl, which might correspond to a mild stimulation of transcription by ς70-containing RNA polymerase as a consequence of high-osmolarity stress.
Whereas positive transcriptional regulation by ςS has been previously reported for three of the genes of the oxyR regulon—dps (2), katG (19), and gorA (3)—negative regulation by ςS has been described for the oxyR regulatory locus (13). Therefore, the level of β-galactosidase expression from a single-copy _oxyR_′::lacZ fusion in a ςS-defective strain was higher (not lower) than in its wild-type parent strain as the cells entered and remained in stationary phase. Moreover, elevated expression of ςS prevented (not induced) normal expression of oxyR (13). In contrast, our data in Fig. 4 and 5 suggest that ςS is a direct or indirect positive regulator for oxyR transcription as well as for other OxyR-regulated genes, in accordance with the increased expression of oxyR at the onset of stationary phase (Table 2). It is not clear at present if differences in genetic backgrounds or methods for measurement of gene expression account for the apparent contradiction between our results and those of González-Flecha and Demple (13) on the role of ςS in transcriptional regulation of the oxyR gene.
Expression of trxA was not substantially induced by osmotic upshift and did not show an evident growth-phase-dependent variation (Table 2). Little is known about the control of the trxA gene in bacteria except for a recent report suggesting that expression of trxA (monitored with a trxA-lac translational fusion) in E. coli is negatively regulated by cyclic AMP (25). This regulation would adjust trxA expression to the growth rate of the bacteria, in accordance with the role of thioredoxin as a cofactor in the synthesis of deoxyribonucleotides. Nevertheless, by RT-MPCR, we have not found significant variations in trxA transcription under conditions in which a large increment, >30-fold, was observed in expression of grxA (which codes for a second cofactor for ribonucleotide reduction) (10).
In brief, this work monitors for the first time the simultaneous in vivo expression of multiple genes related to protection of bacteria against oxidative stress. The data presented contain new valuable information on gene expression during different stages of growth and in response to osmotic stress and confirm previous regulatory relationships. We propose that the RT-MPCR method applied in this work is a powerful tool for monitoring gene expression, particularly when the genes under study present a complex pattern of regulation, with outstanding advantages over alternative experimental approaches such as the use of lacZ fusions.
ACKNOWLEDGMENTS
C.M. and M.M. contributed equally to this work, and both should be considered first authors.
We are grateful to J. F. M. Leal, J. López-Barea, and R. Gallardo-Madueño for helpful discussions and to N. Abril for help in UC1247 construction.
M.M. was a recipient of a predoctoral fellowship from the Spanish Ministry of Education and Culture (MEC). This work was supported by grant P95-0557-CO2-01 (DGES) and by Junta de Andalucía (group CVI 0187).
REFERENCES
- 1.Abril N, Roldán-Arjona T, Prieto-Álamo M J, van Zeeland A A, Pueyo C. Mutagenesis and DNA repair for alkylation damages in Escherichia coli K-12. Environ Mol Mutagen. 1992;19:288–296. doi: 10.1002/em.2850190405. [DOI] [PubMed] [Google Scholar]
- 2.Altuvia S, Almirón M, Huisman G, Kolter R, Storz G. The dps promoter is activated by OxyR during growth and by IHF and ςS in stationary phase. Mol Microbiol. 1994;13:265–272. doi: 10.1111/j.1365-2958.1994.tb00421.x. [DOI] [PubMed] [Google Scholar]
- 3.Becker-Hapak M, Eisenstark A. Role of rpoS in the regulation of glutathione oxidoreductase (gor) in Escherichia coli. FEMS Microbiol Lett. 1995;134:39–44. doi: 10.1111/j.1574-6968.1995.tb07911.x. [DOI] [PubMed] [Google Scholar]
- 4.Bradford M M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 5.Christman M F, Morgan R W, Jacobson F S, Ames B N. Positive control of a regulon for defenses against oxidative stress and some heat-shock proteins in Salmonella typhimurium. Cell. 1985;41:753–762. doi: 10.1016/s0092-8674(85)80056-8. [DOI] [PubMed] [Google Scholar]
- 6.Demple B, Halbrook J. Inducible repair of oxidative DNA damage in Escherichia coli. Nature. 1983;304:466–468. doi: 10.1038/304466a0. [DOI] [PubMed] [Google Scholar]
- 7.Ding H, Demple B. Glutathione-mediated destabilization in vitro of [2Fe-2S] centers in the SoxR regulatory protein. Proc Natl Acad Sci USA. 1996;93:9449–9453. doi: 10.1073/pnas.93.18.9449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emory S A, Belasco J G. The ompA 5′ untranslated RNA segment functions in Escherichia coli as a growth-rate-regulated mRNA stabilizer whose activity is unrelated to translational efficiency. J Bacteriol. 1990;172:4472–4481. doi: 10.1128/jb.172.8.4472-4481.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farr S B, Natvig D O, Kogoma T. Toxicity and mutagenicity of plumbagin and the induction of a possible new DNA repair pathway in Escherichia coli. J Bacteriol. 1985;164:1309–1316. doi: 10.1128/jb.164.3.1309-1316.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallardo-Madueño R, Leal J F M, Dorado G, Holmgren A, López-Barea J, Pueyo C. In vivo transcription of nrdAB operon and of grxA and fpg genes is triggered in E. coli lacking both thioredoxin and glutaredoxin 1, or thioredoxin and GSH, respectively. J Biol Chem. 1998;273:18382–18388. doi: 10.1074/jbc.273.29.18382. [DOI] [PubMed] [Google Scholar]
- 11.Gerhardt P, Murray R G E, Wood W A, Krieg N R, editors. Methods for general and molecular bacteriology. Washington, D.C: American Society for Microbiology; 1994. [Google Scholar]
- 12.González-Flecha B, Demple B. Homeostatic regulation of intracellular hydrogen peroxide concentration in aerobically growing Escherichia coli. J Bacteriol. 1997;179:382–388. doi: 10.1128/jb.179.2.382-388.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.González-Flecha B, Demple B. Transcriptional regulation of the Escherichia coli oxyR gene as a function of cell growth. J Bacteriol. 1997;179:6181–6186. doi: 10.1128/jb.179.19.6181-6186.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodrich-Blair H, Uría-Nickelsen M, Kolter R. Regulation of gene expression in stationary phase. In: Lin E C C, Lynch A S, editors. Regulation of gene expression in Escherichia coli. New York, N.Y: Chapman and Hall; 1996. pp. 571–583. [Google Scholar]
- 15.Greenberg J T, Monach P, Chou J H, Josephy P D, Demple B. Positive control of a global antioxidant defense regulon activated by superoxide-generating agents in Escherichia coli. Proc Natl Acad Sci USA. 1990;87:6181–6185. doi: 10.1073/pnas.87.16.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hengge-Aronis R, Lange R, Henneberg N, Fisher D. Osmotic regulation of rpoS-dependent genes in Escherichia coli. J Bacteriol. 1993;175:259–265. doi: 10.1128/jb.175.1.259-265.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hidalgo E, Demple B. Adaptive responses to oxidative stress: the soxRS and oxyR regulons. In: Lin E C C, Lynch A S, editors. Regulation of gene expression in Escherichia coli. New York, N.Y: Chapman and Hall; 1996. pp. 435–452. [Google Scholar]
- 18.Hidalgo E, Ding H, Demple B. Redox signal transduction: mutations shifting [2Fe-2S] centers of the SoxR sensor-regulator to the oxidized form. Cell. 1997;88:121–129. doi: 10.1016/s0092-8674(00)81864-4. [DOI] [PubMed] [Google Scholar]
- 19.Ivanova A, Miller C, Glinsky G, Eisenstark A. Role of rpoS (katF) in oxyR-independent regulation of hydroperoxidase I in Escherichia coli. Mol Microbiol. 1994;12:571–578. doi: 10.1111/j.1365-2958.1994.tb01043.x. [DOI] [PubMed] [Google Scholar]
- 20.Jamieson D J, Storz G. Transcriptional regulators of oxidative stress response. In: Scandalios J G, editor. Oxidative stress and the molecular biology of antioxidant defenses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 91–115. [Google Scholar]
- 21.Lange R, Hengge-Aronis R. The cellular concentration of the ςS subunit of RNA polymerase in Escherichia coli is controlled at the levels of transcription, translation, and protein stability. Genes Dev. 1994;8:1600–1612. doi: 10.1101/gad.8.13.1600. [DOI] [PubMed] [Google Scholar]
- 22.Lin E C C, Lynch A S, editors. Regulation of gene expression in Escherichia coli. New York, N.Y: Chapman and Hall; 1996. [Google Scholar]
- 23.Martinez A, Kolter R. Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. J Bacteriol. 1997;179:5188–5194. doi: 10.1128/jb.179.16.5188-5194.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mukhopadhyay S, Schellhorn H. Induction of Escherichia coli hydroperoxidase I by acetate and other weak acids. J Bacteriol. 1994;176:2300–2307. doi: 10.1128/jb.176.8.2300-2307.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sa J-H, Namgung M A, Lim C-J, Fuchs J A. Expression of the Escherichia coli thioredoxin gene is negatively regulated by cyclic AMP. Biochem Biophys Res Commun. 1997;234:564–567. doi: 10.1006/bbrc.1997.6687. [DOI] [PubMed] [Google Scholar]
- 26.Storz G, Tartaglia L A, Ames B N. Transcriptional regulator of oxidative stress-inducible genes: direct activation by oxidation. Science. 1990;248:189–248. doi: 10.1126/science.2183352. [DOI] [PubMed] [Google Scholar]
- 27.Tartaglia L A, Storz G, Ames B N. Identification and molecular analysis of oxyR-regulated promoters important for the bacterial adaptation to oxidative stress. J Mol Biol. 1989;210:709–719. doi: 10.1016/0022-2836(89)90104-6. [DOI] [PubMed] [Google Scholar]
- 28.Toledano M B, Kullik I, Trinh F, Baird P T, Schneider T D, Storz G. Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: a mechanism for differential promoter selection. Cell. 1994;78:897–909. doi: 10.1016/s0092-8674(94)90702-1. [DOI] [PubMed] [Google Scholar]
- 29.Touati D. Transcriptional and posttranscriptional regulation of manganese superoxide dismutase biosynthesis in Escherichia coli, studied with operon and protein fusions. J Bacteriol. 1988;170:2511–2520. doi: 10.1128/jb.170.6.2511-2520.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsaneva I R, Weiss B. soxR, a locus governing a superoxide response regulon in Escherichia coli K-12. J Bacteriol. 1990;172:4197–4205. doi: 10.1128/jb.172.8.4197-4205.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu J, Weiss B. Two divergently transcribed genes, soxR and soxS, control a superoxide response regulon of Escherichia coli. J Bacteriol. 1991;173:2864–2871. doi: 10.1128/jb.173.9.2864-2871.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng M, Åslund F, Storz G. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science. 1998;279:1718–1721. doi: 10.1126/science.279.5357.1718. [DOI] [PubMed] [Google Scholar]