Formation of Chromosomal Tandem Arrays of the SXT Element and R391, Two Conjugative Chromosomally Integrating Elements That Share an Attachment Site (original) (raw)
Abstract
The SXT element, a conjugative, self-transmissible, integrating element (a constin) originally derived from a Vibrio cholerae O139 isolate from India, and IncJ element R391, originally derived from a South African Providencia rettgeri isolate, were found to be genetically and functionally related. Both of these constins integrate site specifically into the Escherichia coli chromosome at an identical attachment site within the 5′ end of prfC. They encode nearly identical integrases, which are required for chromosomal integration, excision, and extrachromosomal circularization of these elements, and they have similar tra genes. Therefore, these closely related constins have virtually identical mechanisms for chromosomal integration and dissemination. The presence of either element in a recipient cell did not significantly reduce its ability to acquire the other element, indicating that R391 and SXT do not encode surface exclusion determinants. In cells harboring both elements, SXT and R391 were integrated in tandem fashion on the chromosome, and homologous recombination appeared to play little or no role in the formation of these arrays. Interference between R391 and SXT was detected by measuring the frequency of loss of an unselected resident element upon introduction of a second selected element. In these assays, R391 was found to have a stronger effect on SXT stability than vice versa. The level of expression and/or activity of the donor and recipient integrases may play a role in the interference between these two related constins.
Similar genetic elements tend not to coexist within the same host cell. Instead, related elements of the same class usually “repel” one another in some fashion. For different types of genetic elements, the molecular bases of incompatibility differ. Plasmid incompatibility, for example, is generally mediated by competition for replication and/or partitioning systems (19). Conjugative plasmids also frequently inhibit host cell entry of related plasmids by altering the host cell's surface (1). Similarly, some bacteriophages alter the surface of host cells to exclude other phages. Phages can also prevent the replication of similar phages through immunity mechanisms (25). Thus, both plasmid and phage incompatibility can depend either on preventing entry of new DNA into a potential host or on inhibiting the replication of new DNA after it has breached the host cell barrier.
Chromosomally integrating mobile genetic elements that are transferred between cells via conjugation—often referred to as conjugative transposons—have been found with increasing frequency in both gram-negative and gram-positive bacteria. Unlike the case for phages and plasmids, relatively little is known about whether similar conjugative transposons can coexist within the same host cell. The well-studied conjugative transposon Tn_916_, which does not integrate site specifically, can be present in more than one copy at different sites in the host cell chromosome, and the presence of Tn_916_ in a recipient cell does not inhibit acquisition of a second copy (18, 26).
We previously identified a conjugative transposon-like element, the SXT element, encoding resistance to sulfamethoxazole (Sur), trimethoprim (Tmr), chloramphenicol (Cmr), and streptomycin (Smr) in the recently emerged O139 serogroup of Vibrio cholerae (30). The SXT element (henceforth referred to as SXT), or very closely related elements, have subsequently been detected in all O1 and O139 V. cholerae clinical isolates from the Indian subcontinent (11). In the laboratory, this element can be transferred between V. cholerae strains as well as to a number of other gram-negative bacterial species. An autonomously replicating extrachromosomal form of SXT has not been detected; however, an extrachromosomal circular form of the element has been observed and is thought to be an intermediate in its transfer (11). Formation of this extrachromosomal circular form of SXT requires the SXT-encoded site-specific recombinase (Int), which is closely related to the integrases found in lambdoid bacteriophages. Similar to these phages, SXT integrates site specifically into the chromosome in an _int_-dependent, _recA_-independent fashion via recombination between element (attP) and chromosomal (attB) sequences that are nearly identical. The SXT chromosomal attachment site is within the 5′ end of prfC, which encodes protein chain release factor 3 (11). Since integration of SXT resembles phage integration more than transposition, we thought that the term “conjugative transposon” would not be an informative classification for this element. Instead, we proposed a new term, constin, an acronym for SXT's properties (conjugative, self-transmissible, and integrating) to classify this and other elements with similar features (11).
The conjugative element R391, which mediates resistance to kanamycin (Knr) and mercury (Hgr), was isolated in Pretoria, South Africa, in 1967 from a strain originally classified as Proteus rettgeri (6) but subsequently reclassified as Providencia rettgeri (23). R391 was initially described as an R plasmid, because its antibiotic resistance genes could be transferred to other strains by conjugation (6). Since R391 could coexist with plasmids of all known incompatibility (Inc) groups, it was assigned to a new Inc group, IncJ (6). Since its first description in the early 1970s, only a few other members of the IncJ group have been described, although these elements have been derived from diverse species and geographic locations. Examples include R997, derived from an Indian isolate of Proteus mirabilis and carrying Apr, Sur, and Smr determinants (14), pMERPH (Hgr), isolated in England from Shewanella putrefaciens (24), and pJY1, isolated in the Philippines from V. cholerae El Tor (32). Like SXT, pJY1 mediates Sur, Smr, and Cmr. Although these elements were originally classified as plasmids, all attempts to isolate extrachromosomal DNA from strains containing these elements have failed, with one notable exception, in which small amounts of extrachromosomal DNA were detected (9). Instead, R391 was detected within the Escherichia coli chromosome (16, 20) and the integration site was localized by classical Hfr mapping techniques between uxuA (98 min on the E. coli map) and serB (99.5 min) (15). Based on these findings, Murphy and Pembroke reclassified R391 as a conjugative transposon (16).
Although IncJ elements were collectively assigned to a plasmid incompatibility group based on their compatibility with other plasmids, these elements do not exhibit classic plasmid incompatibility toward each other. That is, the presence of an IncJ element in a recipient cell does not reduce the frequency of transfer of an additional IncJ element in conjugation assays. Instead, incompatibility in the IncJ group has been assessed by measuring the frequency of loss of an unselected resident element upon introduction of a second selected element in conjugation assays (8, 24). In a recent investigation (22), Pembroke and Murphy studied the incompatibility of the two IncJ elements R391 and R997. They observed that loss of markers of the nonselected element was highly reduced in a recA mutant strain and concluded that R391 and R997 recombine to form hybrid elements carrying either the resistance genes of both original elements or only those which were selected for. Also, in recA recipient cells already harboring either R391 or R997, they were able to isolate an extrachromosomal form of the introduced element. Their observations suggested that when the chromosomal attachment site is occupied by an IncJ element, a newly introduced element either integrates into the chromosome by homologous recombination with the resident element or remains extrachromosomal in recA mutant strains. In this report, we show that R391 is closely related to SXT and that the interactions between R391 and SXT fundamentally differ from those described for R391 and R997. Cells harboring both SXT and R391 contain tandem arrays of these elements, and incompatibility between these two constins seems to be mediated by the activity of their int genes.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are described in Table 1. RW96 harboring R391 (9) was used as a donor for R391. Bacterial strains were routinely grown in Luria-Bertani (LB) broth (3). Bacterial strains were maintained at −70°C in LB broth containing 20% (vol/vol) glycerol. Antibiotics were used at the following concentrations: ampicillin, 100 mg liter−1; kanamycin, 50 mg liter−1; sulfamethoxazole, 160 mg liter−1; trimethoprim, 32 mg liter−1; tetracycline, 10 mg liter−1; and nalidixic acid, 40 mg liter−1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or phenotype | Reference or construction |
|---|---|---|
| E. coli K-12 | ||
| CAG18439 | MG1655 lacI42::Tn_10_ | 27 |
| HW220 | CAG18439 prfC::SXT | 11 |
| BI554 | HW220 Δ_int_ | 10 |
| JO99 | CAG18439 prfC::R391 | This study |
| JO113 | CAG18439 prfC::R391-SXT | Knr SXTr exconjugant from JO100 × HW220 |
| JO115 | CAG18439 prfC::R391-SXT | Knr SXTr exconjugant from BI537 × JO99 |
| JO116 | CAG18439 prfC::SXT-R391 | Knr SXTr exconjugant from BI537 × JO99 |
| BI533 | MG1655 Nalr | 10 |
| BI537 | BI533 prfC::SXT | This study |
| JO100 | BI533 prfC::R391 | This study |
| JO162 | JO100 Δ_int_ | This study |
| KB1 | MG1655 recA56 gutA52 gutR::Tn_10_ | K. Bettenbrock, unpublished data |
| BI616 | KB1 prfC::SXT | This study |
| JO139 | KB1 prfC::R391 | This study |
| BI742 | KB1 prfC::SXT-R391 | Knr SXTr exconjugant from BI537 × JO139 |
| BI743 | KB1 prfC::R391-SXT | Knr SXTr exconjugant from BI537 × JO139 |
| BI844 | KB1 prfC::R391-SXT | Knr SXTr exconjugant from JO100 × BI616 |
| BI845 | KB1 prfC::SXT-R391 | Knr SXTr exconjugant from JO100 × BI616 |
| RW96-R391 | Δ(umuDC)595::cat prfC::R391 | 9 |
| Plasmids | ||
| pGB2 | Spcr pSC101 derivative | 5 |
| pRLH421 | 22.5-kbp extrachromosomal R391 DNA cloned into pGB2, Spcr Knr | 9 |
| pCRII | Cloning vector, Apr Knr | Invitrogen |
| pINT6 | pCRII _int_R391+ | This study |
| pINT391-19 | pUC19 _int_R391+ | This study |
| pINT18 | pUC19 _int_SXT+ | This study |
To construct pINT6, a 1.8-kbp fragment was amplified with primers XIS5 (5′-TGCGTGACGAAAGCATCAATG-3′) and INT4 (5′-CTTCGCCAGAGCGTCGTATA-3′), with chromosomal DNA isolated from JO99 as template. The product was then cloned into the vector pCRII by using the Topo TA cloning kit (Invitrogen, Carlsbad, Calif.) as specified by the manufacturer. pINT391-19 is a pUC19 derivative with _int_R391 under the control of lacZP, and pINT18 is a pUC18 derivative with _int_SXT under the control of the same promoter. An int derivative of R391 in JO100 was constructed by a similar procedure to that described previously for SXT (10). In the resulting strain, JO162, bp 305 to 1025 of int are deleted.
Bacterial conjugations and incompatibility tests.
Conjugation experiments were routinely carried out as described previously (30). To compare the transfer frequency of matings performed on solid surfaces or in broth, donor and recipient cells were initially grown to mid-log phase in LB medium. Donor and recipient cells were mixed in a 1:1 ratio and incubated for 2 h in either 2 ml of LB broth or on LB agar plates. Subsequently, dilutions were spread on selective media to determine the number of exconjugants and donor and recipient cells. The frequency of transfer was calculated by dividing the number of SXTr or Knr exconjugants, respectively, by the number of donor cells. To assess the incompatibility between R391 and SXT, each element was transferred into a recipient strain that already carried the other element. In these conjugation assays, only the incoming element was selected; the resident element was not subject to selection. Finally, 100 exconjugants from each mating were tested for the presence of markers characteristic of the original element. Incompatibility was calculated as the percent loss of the nonselected element.
Molecular biology procedures.
Plasmid DNA was prepared using the Qiaprep Spin Miniprep kit (Qiagen, Valencia, Calif.), and chromosomal DNA was isolated with the Genome DNA kit (Bio 101, Vista, Calif.) as described by the manufacturer. Recombinant DNA manipulations were carried out by standard procedures (3). Southern blotting was performed with probes conjugated to horseradish peroxidase to enable hybridization to be detected with a chemiluminescent substrate (Amersham) as described previously (28). Automated DNA sequencing was carried out as described previously (29) at the Tufts Medical School DNA Sequencing Core Facility. Computer analysis of DNA sequences was performed with the MacVector and AssemblyLIGN programs (Oxford Molecular Group, Campbell, Calif.) and the BLAST programs (2), available on the website of the National Center for Biotechnology Information, Bethesda, Md.
DNA sequence analysis of the extrachromosomal form of R391.
Ho et al. (9) isolated small amounts of extrachromosomal R391 DNA from a recA718 strain, RW96. This extrachromosomal R391 DNA was partially digested with _Eco_RI and cloned into the low-copy-number vector pGB2 (5) by selecting for the R391-encoded kanamycin resistance gene. Several isolates were obtained that contained one or more _Eco_RI fragments. One of these, called pRLH421, contained an insert of ∼22.5 kb comprising four R391 _Eco_RI fragments of ∼11, 8, 3, and 0.5 kb and was chosen for further study. The DNA sequence 3′ to the rumAB operon (13) was obtained using standard “primer-walking” sequencing protocols by Lark Technologies (Houston, Tex). The sequence has been deposited in GenBank under accession number U13633.
PCR assays for detection of R391 integration into prfC in E. coli K-12 and for detection of the circular extrachromosomal form of R391.
Amplification of the chromosome-R391 junction fragments was carried out as described previously for SXT using primers 3, 6, 7, and 8 (11). Primers which are oriented toward the left and right SXT-chromosome junctions (primers 4 and 5 in reference 11) were used for detection of a circularized, extrachromosomal form of R391. These PCR assays were carried out using overnight cultures as template DNA as described previously (11).
Determination of the order of tandemly arranged R391 and SXT.
The leftmost element-chromosome junction in tandemly arranged R391 and SXT elements was assayed by PCR using the previously described primers 6 and 4 (11). Using these primers, integration of SXT into attL resulted in a 0.8-kbp product while integration of R391 into attL yielded a 2.8-kbp product. DNA corresponding to the rightmost element-chromosome junction was amplified using primer 8 (11) and primer 9 (5′-ATAGACTGAAGTGCTTGCGG-3′). The resulting 3.0-kbp product was subsequently cut with _Cla_I and electrophoresed in a 1% agarose gel. Since R391 carries an additional _Cla_I recognition site in the corresponding amplified region, integration of R391 and SXT into attR could be distinguished based on the sizes of the fragments obtained.
RESULTS
R391 integrates into prfC.
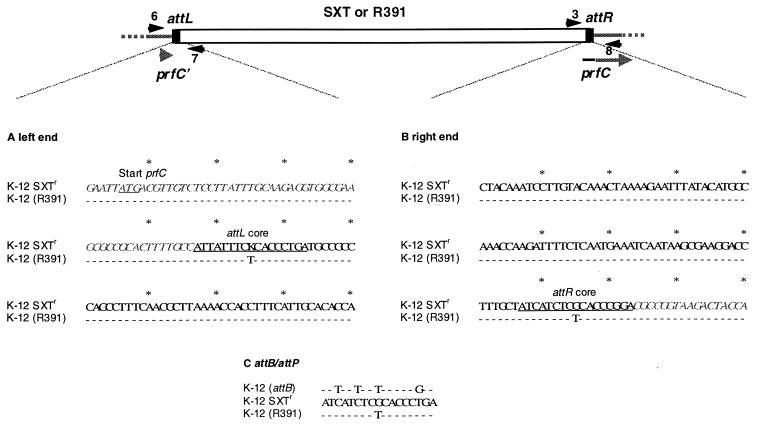
Murphy and Pembroke (15) found that R391 integrates into the E. coli chromosome in the region between 98.0 and 99.5 min. Since this area includes the SXT integration site, prfC, at 99.3 min (11), we speculated that R391 might also integrate into prfC and encode a similar integration system. To test this possibility, we transferred R391 into E. coli K-12 CAG18439, the strain we used to map the SXT integration site. With one of the resulting Knr Tcr exconjugants, designated JO99, PCR was performed using primer pairs that were previously found to amplify the left and right junctions of SXT and the CAG18439 chromosome (primers 6 and 7 and primers 3 and 8 in Fig 1). Strikingly, the PCR products were identical in size to those derived using HW220 (CAG18439 prfC::SXT) DNA as the template (396 and 393 bp, respectively). Therefore, like SXT, R391 integrates into the 5′ end of prfC. The DNA sequences of the PCR products were determined and compared to the corresponding sequences of SXT-HW220 junction fragments published previously (11) (Fig. 1). This comparison revealed that the left (attL) and right (attR) attachment sites of R391 and SXT are virtually identical, differing only in 2 bp. Furthermore, like SXT, R391 restores the reading frame of prfC (Fig. 1). Integrated into the chromosome, R391 is flanked by a 17-bp repeat sequence that probably corresponds to the core of the att site where strand exchange occurs during integration. The 17-bp R391 attR differed from the SXT attR by only 1 bp (a T at position 9 compared to a G in SXT+ strains). The near identity of the R391 and SXT attL and attR sequences strongly suggests that the mechanism of chromosomal integration of R391 is very similar to that of SXT.
FIG. 1.
Alignment of the nucleotide sequences of attL(A), attR (B), and attP (C) of SXT and R391. The primers (primers 6 and 7 and primers 3 and 8 [11]) used to amplify the junction fragments between the E. coli K-12 chromosome and R391 or SXT, respectively, are shown as black arrowheads. (A) Alignment of the sequences of attL of SXT (top) and R391 (bottom) obtained by sequencing the PCR product specific to the left junction fragment. Identical base pairs are represented by dashes. The 17-bp sequence encompassing the core of the att sites is underlined. The K in attL of SXT indicates that either T or G was found at this position (11). DNA specific to the chromosome of E. coli K-12 is in gray italic letters. (B) Alignment of the sequence of the right junction fragment of SXT (top) and R391 (bottom). Symbols are as in panel A. (C) Comparison of attB in the chromosome with attP of SXT and R391.
Identification of the R391 int gene.
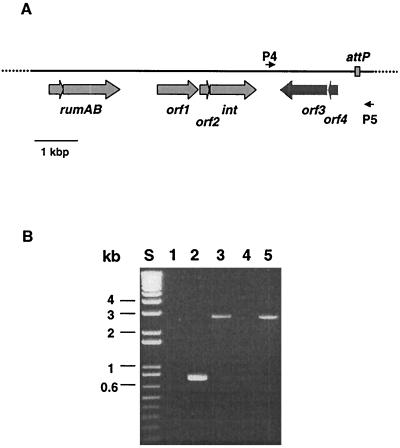
As part of an ongoing effort to determine the entire DNA sequence of SXT, we found that the DNA sequence of a region upstream of _int_SXT had 95% identity to the 3′ end of rumB from R391 (13). The rumAB operon encodes proteins that are phylogenetically related to a superfamily of novel error-prone DNA polymerases (31). This finding was a further indication of the similarity of R391 and SXT and suggested that an R391 int gene might be located downstream of R391 rumB. We have taken advantage of the fact that during the cloning of the rumAB operon, several recombinant plasmids were obtained that contained large fragments of extrachromosomal R391 DNA. Based on restriction analysis, it was determined that one of these plasmids, called pRLH421, contained approximately 7.5 kb 3′ to the rumAB operon. The nucleotide sequence of this fragment was determined using standard sequencing protocols and revealed that 2.17 kbp downstream of the stop codon of rumB there was a 1,242-bp open reading frame (ORF) which had 96% nucleotide sequence identity to _int_SXT. The deduced amino acid sequence of this ORF was 99.5% identical to the integrase of SXT, with only three amino acids that differed (Y123F, S198G, and S333G). In both R391 and SXT, int is preceded by two nearly identical ORFs (orf1 and orf2 in Fig. 2A). Neither of these ORFs shows similarities to any known protein, and their functions remain to be determined.
FIG. 2.
Organization of the int region of R391 and detection of a circular extrachromosomal form of R391. (A) Organization of the region downstream of the rumAB operon in the circular form of R391. Following rumAB, five ORFs were identified, including the int gene of R391. orf3 and orf4 (black arrows) are not present in SXT. Also shown is attP and primers P4 and P5 used in the experiment in panel B. Note that these primers are oriented toward the chromosomal junctions of the integrated form of R391. (B) Ethidium bromide-stained 1% agarose gel of PCR products amplified using primers P4 and P5. Lanes: S, molecular size marker; 1, CAG18439; 2, BI537 (SXT+); 3, JO100 (R391); 4, JO162 (R391 _int_−); 5, JO162/pINT391-19.
Detection of an excised circular form of R391.
_int_SXT is required for the site-specific recombination between the attL and attR sequences of SXT, leading to the generation of an extrachromosomal circular form of SXT (11). The identification of an _int_R391 that is nearly identical to _int_SXT suggested that R391 is also capable of excising from the chromosome and forming an extrachromosomal circle by a similar _int_-dependent mechanism. As no extrachromosomal DNA could be isolated by routine means from strains carrying R391, a more sensitive assay (11) based on PCR was used to identify an extrachromosomal circular form of R391. Since the primers in this assay are oriented toward the 5′ and 3′ ends of the integrated element, a product is only amplified if the element excises and circularizes (Fig. 2). Using this assay and control HW220 DNA (harboring SXT) as the template, the expected PCR product of 785 bp was obtained; with JO99 (harboring R391) as the template, the PCR amplification yielded a product of 2.5 kbp (Fig. 2). Thus, R391 also forms an extrachromosomal circle. However, the larger size of the PCR product from R391 than that from HW220 indicated that R391 possesses additional DNA at one of its ends. Indeed, sequence analysis of cloned extrachromosomal R391 DNA revealed that the R391 left end contains a region not present in SXT (Fig. 2). At 464 bp downstream of _int_R391, a 1,212-bp ORF (orf3) was identified. The deduced amino acid sequence of orf3 had 39% similarity and weak identity (∼25%) to HipA of E. coli K-12 as well as 55% identity (73% similarity) to a reported HipA orthologue (y4DM) in the Rhizobium symbiosis plasmid pNGR234 (7). HipA, together with HipB, controls tolerance to certain peptidoglycan and DNA synthesis inhibitors (4). orf3 seems to form an operon with orf4, since their likely start and stop codons are overlapping. Database comparisons with orf4 revealed partial identity to y4DL from plasmid pNGR234, the upstream gene of y4DM (7). Thus, this region appears to be arranged similary in pNGR234 and R391, except that R391 seems to have lost about 100 amino acids in its y4DL homologue. Whether the gene products of orf3 and orf4 play a role in integration or transfer of R391 remains to be studied. The sequence of the core region of the R391 attP differed by only 1 nucleotide from the SXT attP sequence (Fig. 1C), corresponding to the position where attR varied (Fig. 1B). These minor variations in the R391 attP and attR compared with the corresponding SXT sequences are consistent with the previously proposed cut sites for the SXT integrase (11).
The extrachromosomal circular form of SXT is apparently a requisite intermediate in its self-transfer (11). We expected this to be the case for transfer of R391 as well. To obtain data to support this hypothesis, we constructed a derivative of JO100 (MG1655 Nalr containing R391) with a deletion in int (strain JO162). We were unable to detect the extrachromosomal circular form of R391 in this strain (Fig. 2); however, circle formation could be complemented in trans with int expressed from plasmid pINT391-19 (Fig. 2). JO162 was unable to transfer R391 to recipient cells, and pINT391-19 had to be present in both donor and recipient cells to restore transfer, as previously observed for SXT (11). This suggests that excision and circularization of R391 precede its self-transfer. Furthermore, these data suggest that in recipient cells, R391 is unable to stably replicate as an extrachromosomal circle and instead integrates into the recipient's chromosome to be maintained.
R391 and SXT contain similar DNA sequences and have similar properties.
Our ongoing study of the SXT DNA sequence has revealed that this element has a modular organization, with clusters of genes with related functions. We designed oligonucleotides to amplify different areas of SXT by PCR in order to test whether R391 has sequence similarity to SXT outside of the int region. Three PCR primer pairs that amplify internal fragments of three genes in the putative SXT tra cluster—traC, traB, and traF orthologues—were successfully used to amplify identically sized fragments from R391. Similarly, an ORF (orfR) at the “right” end of SXT with similarity to the _c_I repressor of phage 434 (17) was found in R391, as was an intergenic region 5′ of the tra cluster. Additional Southern hybridization analyses using probes specific to the tra genes (data not shown) suggested that the tra genes detected by PCR in R391 are not only closely related in sequence to those of SXT but are also similarly organized. Although R391 and SXT share many genes, these elements also encode different properties, such as resistance to antimicrobial agents and heavy metals. As predicted from these phenotypic differences, primers used for PCR amplification of genes that encode the resistance of SXT to sulfonamide, trimethoprim, chloramphenicol, and streptomycin did not yield products when JO99 DNA was used as the template. Conversely, we detected merC, a gene of the mercury resistance operon of R391 (21), only in JO99 and not in HW220 (data not shown).
The similarity of R391 and SXT was not limited to DNA sequences and gene organization only; two defining properties of SXT were also found in R391. We previously found that transfer of SXT from donor cells is dependent on RecA (30). This was also the case for R391. When we introduced R391 into an E. coli MG1655 recA56 mutant strain (KB1), the resulting exconjugant, JO139, transferred R391 with a frequency 3 to 4 orders of magnitude lower than that for the wild type. In contrast to our findings, Murphy and Pembroke (16) observed efficient transfer of R391 from donor cells carrying various recA alleles including the recA56 allele used in our study. The reason for this difference is not known. The transfer frequency of SXT in liquid medium is greatly reduced compared to that on agar plates (30). We found a similar marked reduction in the transfer frequency of R391 in broth relative to plates (10−4 and 10−8, respectively). This result is again in contrast to previously published observations (6). Different mating conditions or strain variations might account for these differences.
SXT and R391 coexist in tandem arrays in the chromosome.
There have been relatively few studies concerning interactions between closely related constins like SXT and R391. Given the similarity of these two elements, including regions of DNA sequence identity, we explored whether they interfere with each other's transfer or maintenance. To assess this, we compared the transfer frequencies of R391 and SXT to a wild-type E. coli recipient strain with the transfer frequencies to isogenic strains harboring SXT or R391, respectively (Table 2). The presence of R391 in the recipient led to a minimal reduction in the measured transfer frequency of SXT from donor cells relative to recipients lacking R391 (Table 2). When SXT was present in the recipient cells, there was no detectable diminution in the R391 transfer frequency (Table 2). These data are consistent with previous observations (22, 32) suggesting that IncJ elements do not exclude each other.
TABLE 2.
The presence of SXT or R391 in recipient cells has a minimal effect on the transfer frequency of the other element
| Donor | Relevant genotype | Recipient | Relevant genotype | Transfer frequency (10−4)a with selectionb for: | ||
|---|---|---|---|---|---|---|
| SXT | Kanamycin | SXT + kanamycin | ||||
| BI537 | SXT+ | CAG18439 | 0.6 | NDc | ND | |
| BI537 | SXT+ | JO99 | R391+ | 0.1 | ND | 0.3 |
| JO100 | R391+ | CAG18439 | ND | 1 | ND | |
| JO100 | R391+ | HW220 | SXT+ | ND | 1 | 0.8 |
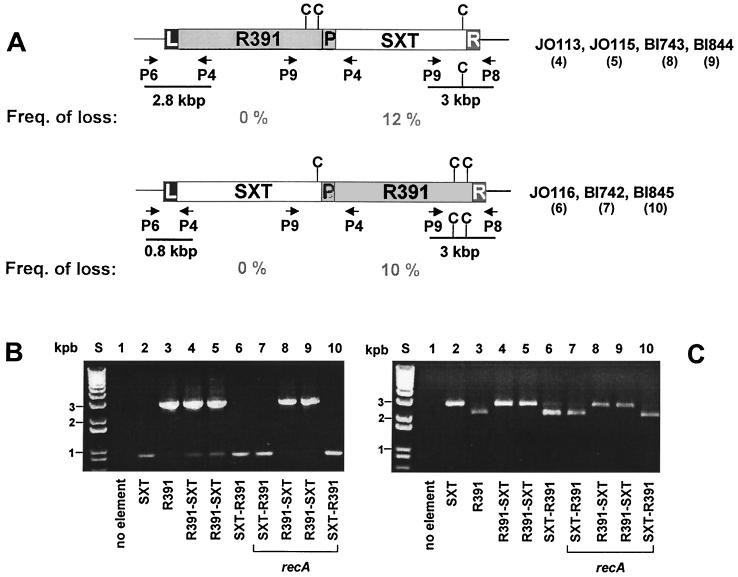
PCR assays (Fig. 3) and Southern hybridizations (data not shown) were used to analyze the organization of R391 and SXT in the exconjugants that contained both elements. The PCR assays were designed to reveal the right and left junctions of these elements within the chromosome. For these assays, we took advantage of two DNA sequence differences that allow the ends of R391 and SXT to be distinguished. These are the additional DNA found at the left end of R391 (Fig. 2) and a _Cla_I site, which is present only at the right end of R391. In every exconjugant that contained both elements, amplification of the right and left element-chromosome junctions never revealed junctions characteristic of only one element (either R391 or SXT). Instead, in all cases analyzed, one junction was derived from SXT and the other from R391. This result suggests that the two elements are arranged in a tandem fashion in the chromosome in these cells (Fig. 3). In several strains containing both elements (e.g., JO113 and JO115 [lanes 4 and 5 in Fig. 3B]) we could detect both the R391 and SXT right junction fragment, with the latter junction appearing less abundant. We believe that the presence of the right SXT-chromosome junction in these cases reflects excision of R391. In RecA− host cells containing both elements (strains BI742, BI743, BI844, and BI845 in Fig. 3), we also always observed junctions characteristic of both elements, suggesting that the process explaining the formation of Knr SXTr exconjugants does not involve homologous recombination between these two related elements. In fact, we could use any of our RecA+ Knr SXTr exconjugants (e.g., JO113, JO115, or JO116) as donors of either SXTr, Knr, or SXTr and Knr in subsequent conjugation experiments. In these experiments, SXTr or Knr exconjugants had chromosomal junctions characteristic of SXT or R391, respectively, strongly suggesting that in the Knr SXTr donors the two elements remain as independent units and have not recombined.
FIG. 3.
Tandem arrangements of SXT and R391. (A) Schematic depiction (not to scale) of the tandem arrangements of R391 and SXT with either R391 (top) or SXT (bottom) present as the 5′ element. The primers (P4, P6, P8, and P9) and the predicted sizes of products resulting from PCR assays in both arrangements are also shown. The additional _Cla_I restriction site found at the right end of R391 is indicated. Also shown are the average frequencies of loss of SXT or R391, respectively after ∼20 generations of growth. Abbreviations: C, _Cla_I; L, attL; P, attP; R, attR. (B and C) Amplification of the left (B) and right (C) chromosome-element junction fragments. Ethidium bromide-stained 1% agarose gels of PCR products amplified using primers P4 and P6 (B) and P8 and P9 (C) are shown. PCR products in the experiment in panel C were digested with _Cla_I. Only the largest of these fragments is shown. The deduced order of R391 and SXT is indicated at the bottom, with the element flanked by attL as the first followed by the element flanked by attR. Lanes: S, molecular size marker; 1, CAG18439; 2, HW220; 3, JO99; 4, JO113; 5, JO115; 6, JO116; 7, BI742; 8, BI743; 9, BI844; 10, BI845.
Our results indicate that when either R391 or SXT integrates into a chromosome of a cell where the element's attB sequence is already occupied by the other element, the second element uses the resident element's attL or attR for integration rather than integrating into a secondary att site elsewhere on the chromosome. SXT and R391 exhibited different preferences for each other's attL and attR sequences. R391 preferentially integrated into the SXT attL (Table 3), whereas SXT integrated into the R391 attL or attR with similar frequencies. The mechanism underlying this differential specificity of att site selection is not clear. Apparently, the minor differences of their integrases are not sufficient to explain this differential specificity, since expression of either _int_R391 or _int_SXT in trans in a recipient cell did not confer this specificity to an incoming Int− element (data not shown).
TABLE 3.
Integration specificity of R391 and SXTa
| Donor | Relevant genotype | Recipient | Relevant genotype | Frequencyb of tandem arrangement (%) | |
|---|---|---|---|---|---|
| R391-SXT | SXT-R391 | ||||
| BI537 | SXT+ | JO139 | recA R391+ | 53 (20) | 46 (18) |
| JO100 | R391+ | BI616 | recA SXT+ | 91 (32) | 9 (3) |
Tandem arrangements of SXT and R391 were relatively stable in both RecA+ and RecA− cells. After ∼20 generations of growth in LB broth without selection for either element, only ∼10% of the SXTr Knr exconjugants lost one of the two elements. Interestingly, there was a preferential loss of the element integrated into attR (Fig. 3). One potential explanation for this preference could be that the integrases of R391 and SXT bind to attR and attP (which is formed at the junction of the tandem elements) with greater affinity than to attL and attP. Subsequently, recombination between attP and attR results in excision and loss of the element located between these att sites.
Incompatibility between R391 and SXT.
Previously, “incompatibility” between IncJ elements has been assessed in conjugation assays by measuring the frequency of loss of an unselected resident element on introduction of a second selected element (24). We adopted this approach to assess incompatibility between R391 and SXT. To our surprise, selection for R391 resulted in a higher frequency of loss of a resident SXT (26%) than the frequency of loss of a resident R391 (8%) when selection was applied for an incoming SXT (Table 4). Since we have never observed spontaneous loss of either R391 or SXT, these results reveal some type of incompatibility between these two elements. The magnitude of these frequencies are fairly similar to those previously reported as evidence of incompatibility for IncJ elements (14, 24, 32). Nonreciprocal incompatibility has also been reported earlier for R997 and R391 (14).
TABLE 4.
Incompatibility between R391 and SXT
| Donor | Relevant genotype | Recipient | Relevant genotype | % Lossa of: | |
|---|---|---|---|---|---|
| SXT | R391 | ||||
| HW220 | SXT+ | JO100 | R391+ | NAb | 8 (1–15) |
| BI537 | SXT+ | JO139 | R391+recA | NA | 4 (0–10) |
| HW220 | SXT+ | JO162 | R391+int | NA | 1.5 (0–3) |
| BI554/pINT6 | SXT+int/int+ | JO100 | R391+ | NA | 11 (5–16) |
| JO100 | R391+ | HW220 | SXT+ | 26 (7–52) | NA |
| JO100 | R391+ | BI616 | SXT+recA | 58 (39–77) | NA |
| JO100 | R391+ | BI554 | SXT+int | 39 (18–69) | NA |
| JO162/pINT6 | R391+int/int+ | HW220 | SXT+ | 3 (0–8) | NA |
| JO162/pINT6 | R391+int/int+ | HW220/pINT391-19 | SXT+int+/int+R391 | 35 (12–58) | NA |
| JO162/pINT6 | R391+int/int+ | HW220/pINT18 | SXT+int+/int+SXT | 4 (2–6) | NA |
The molecular bases of plasmid incompatibility are rooted in the factors that mediate either the replication or partitioning of these autonomously replicating genetic elements (19). Since constins like SXT and R391 are part of the chromosome, it seemed unlikely that these factors could mediate incompatibility between these elements. The considerable degree of homology between R391 and SXT suggested that homologous recombination between the two elements might explain the incompatibility we observed. However, this was clearly not the case; a similar frequency of loss of the unselected element occurred in recA recipients (Table 4). The inequality in the frequencies of loss of R391 and SXT suggests that some property that distinguishes the two elements plays a role in their incompatibility.
Since an integrase is required for integration and excision of these elements—key steps in either their gain or loss—we speculated that int may be important for the observed incompatibility. Consistent with this idea, an R391+ int recipient had a lower frequency of loss of R391 than did an int+ recipient in the incompatibility assay (Table 4). In contrast, an SXT+ int recipient did not lose SXT at lower frequencies. A potential explanation for the disparate effects of the int deletions in the SXT and R391 int recipients is that _int_SXT is less active than _int_R391. In support of this idea, we found that deletion of _int_R391 from the donor strain JO162 resulted in a significant reduction in the frequency of loss of SXT from the recipient HW220 (Table 4). Furthermore, introduction of a plasmid containing _int_R391 into HW220 restored the higher frequency of loss of SXT from this strain, whereas introduction of the same vector containing _int_SXT did not have this effect (Table 4). Thus, expression of _int_R391 in the recipient seems to facilitate the loss of one constin when the second is introduced. The effect of _int_R391 is most notable when it is expressed from an extrachromosomal element; it is less pronounced when _int_R391 is integrated within the chromosome (e.g., in JO100), perhaps because the chromosomal gene is expressed at lower levels. The imbalance between the loss of SXT and the loss of R391 may also reflect the fact that SXT is more frequently found on the right end of an array, the position from which elements are usually lost.
DISCUSSION
We found that the conjugative, self-transmissible, integrating SXT element, originally derived from an Indian V. cholerae O139 isolate, is closely related to R391, a conjugative, self-transmissible, integrating “IncJ” element, originally derived from a South African P. rettgeri isolate. Both elements integrate into the same chromosomal attachment site (within the 5′ end of prfC) in a _recA_-independent process that results from site-specific recombination between nearly identical element and chromosomal sequences. Chromosomal integration, as well as excision of these two elements, is dependent on their almost identical int genes. Both R391 and SXT can form extrachromosomal circular elements by recombination between their respective attL and attR sequences in an _int_-dependent manner. In addition to sharing DNA sequences and genes required for integration and excision, these elements contain related tra genes. Taken together, our data suggest that R391 and SXT are highly related constins with virtually identical mechanisms of integration into the chromosome and for dissemination. Despite their significant similarities, both elements also encode unique properties, such as resistance to specific antibiotics and heavy metals. These elements seem to consist of similar basic building blocks (modules encoding integration and transfer functions) to which have been added genes encoding unique features. It will be interesting to learn whether other previously defined IncJ elements share integration and transfer functions with SXT and R391. At least for R997 this seems likely, since this element is known to integrate between uxuA and serB on the E. coli chromosome (15), a region that includes prfC. The great similarity between SXT and R391, even though they are derived from different bacterial species and their highly disparate sites and dates of isolation, suggests that this type of constin may be considerably more pervasive than was previously suspected.
The interactions between related conjugative integrating elements have not been thoroughly investigated. Previous studies with the conjugative transposon Tn_916_ and IncJ elements (18, 22, 32) have revealed that these elements do not prevent the transfer of a similar or identical element into a host already harboring such an element. Similar to this, our work showed that the presence of either R391 or SXT in a recipient cell had minimal to no detectable effect on the capability of the cell to serve as a recipient for the other element. This finding suggests that unlike plasmids such as F, these two elements do not encode surface exclusion mechanisms. Similarly, these elements do not seem to encode a repressor-mediated immunity function as described for bacteriophages such as λ or CTXφ (12, 25).
In cells carrying both R391 and SXT, the elements were arranged in tandem fashion on the chromosome. Homologous recombination played little or no role in the formation of these arrays. The arrays formed in recA recipients and either R391 or SXT could be transferred independently from a donor harboring both elements. Our data suggest that these tandem arrays were generated by site-specific recombination between an incoming element's attP sequence and the resident element's attL or attR sequence. The apparent lack of a role for homologous recombination as a form of interaction between R391 and SXT is in marked contrast to a recent report by Pembroke and Murphy, who studied the interaction between the two IncJ elements R997 and R391 (22). These authors concluded that homologous recombination is the predominant event which occurs following the introduction of R391 into a RecA+ host harboring R997 or vice versa. They concluded that in recA recipients the incoming element did not integrate but was maintained extrachromosomally as an autonomously replicating plasmid (22). This could be the case, although it is equally plausible that the extrachromosomal DNA they detected resulted from a shift in a equilibrium between the excised and integrated forms of R391 and R997 in cells harboring both of these IncJ elements. The differences between our observations and those of Pembroke and Murphy are most probably explained by the fact that we studied different IncJ elements. SXT and R391 may be less similar to each other than are R997 and R391. If this is the case, site-specific recombination mediated by the int genes of the elements may predominate over homologous recombination.
Incompatibility between R391 and SXT was determined by measuring the frequency of loss of an unselected resident element upon introduction of a second selected element, the established assay used to detect incompatibility of IncJ elements. While there was variability in our data regarding incompatibility between R391 and SXT (Table 4), there were always reproducible trends in these experiments that hint at a potential mechanism of incompatibility between R391 and SXT. Our data are consistent with a model where the level and/or specificity of activity of the donor and recipient integrases determines incompatibility. The factors regulating the amount or activity of these integrases are unknown; it is possible that there are differences in the production of integrase depending on whether an element is resident on the chromosome or present as an extrachromosomal circle. The three amino acid changes between the R391 and SXT integrases might also mediate subtle differences in the activities of these two enzymes, thereby explaining their different effects. A related factor that may play a role in the incompatibility of R391 and SXT is the apparently greater stability of the element in the 5′-most position in a tandem array. Since R391 had a preference for this site, this might in part account for the enhanced stability of this element.
Our findings demonstrate that the incompatibility between two constins that utilize the same attB is far less stringent than that observed for plasmids. Although the instability of R391 and SXT increased when both elements were present in a cell, these two elements can coexist relatively stably. Incompatibility between these constins is clearly a different process from that observed for plasmids. In fact, the term “incompatibility” has been previously applied only to replicating plasmids, and R391 and SXT in our assays do not appear to replicate as plasmids but ensure their vertical transmission by integrating into the chromosome. Assuming that our observations apply to other IncJ elements, the molecular bases of IncJ incompatibility are fundamentally different from the mechanisms known for other Inc groups. In this regard, it seems reasonable to abandon the term “IncJ incompatibility group” to describe this related group of constins and to apply a different term, perhaps “interference,” to describe the interactions between similar IncJ elements.
ACKNOWLEDGMENTS
We thank A. Camilli, A. Kane, B. Davis, and D. RayChaudhuri for critical reading of the manuscript. Strain KB1 was kindly provided by K. Bettenbrock.
This work was supported in part by funds from the NIH Intramural Research Program (R.W.), by the DFG (B.H.), NIH grant AI42347, the PEW Foundation, the Howard Hughes Medical Institute (M.K.W.), and a pilot project grant from the NEMC GRASP Center (P30DK-34928).
REFERENCES
- 1.Achtman M, Kennedy N, Skurray R. Cell-cell interactions in conjugating Escherichia coli: role of traT protein in surface exclusion. Proc Natl Acad Sci USA. 1977;74:5104–5108. doi: 10.1073/pnas.74.11.5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database research programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidmann J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene Publishing and Wiley Interscience; 1990. [Google Scholar]
- 4.Black D S, Kelly A J, Mardis M J, Moyed H S. Structure and organization of hip, and operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J Bacteriol. 1991;176:4081–4091. doi: 10.1128/jb.173.18.5732-5739.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Churchward G, Belin G, Nagamine Y. A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene. 1984;31:165–171. doi: 10.1016/0378-1119(84)90207-5. [DOI] [PubMed] [Google Scholar]
- 6.Coetzee J N, Datta N, Hedges R W. R factors from Proteus retgerri. J Gen Microbiol. 1972;72:543–552. doi: 10.1099/00221287-72-3-543. [DOI] [PubMed] [Google Scholar]
- 7.Freiberg C, Fellay R, Bairoch A, Broughton W J, Rosenthal A, Perret X. Molecular basis of symbiosis between Rhizobium and legumes. Nature. 1997;387:394–401. doi: 10.1038/387394a0. [DOI] [PubMed] [Google Scholar]
- 8.Hedges R W, Jacob A E, Datta N, Coetzee J N. Properties of plasmids produced by recombination between factors of group J and FII. Mol Gen Genet. 1975;140:289–302. doi: 10.1007/BF00267320. [DOI] [PubMed] [Google Scholar]
- 9.Ho C, Kulaeva O I, Levine A S, Woodgate R. A rapid method for cloning mutagenic DNA repair genes: Isolation of umu-complementing genes from multidrug resistance plasmids R391, R446b, and R471a. J Bacteriol. 1993;175:5411–5419. doi: 10.1128/jb.175.17.5411-5419.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hochhut B, Marrero J, Waldor M K. Mobilization of plasmids and chromosomal DNA mediated by the SXT element, a constin found in Vibrio cholerae O139. J Bacteriol. 2000;182:2043–2047. doi: 10.1128/jb.182.7.2043-2047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hochhut B, Waldor M K. Site-specific integration of the conjugal Vibrio cholerae SXT element into prfC. Mol Microbiol. 1999;32:99–110. doi: 10.1046/j.1365-2958.1999.01330.x. [DOI] [PubMed] [Google Scholar]
- 12.Kimsey H H, Waldor M K. CTXφ immunity: application in the development of cholera vaccines. Proc Natl Acad Sci USA. 1998;95:7035–7039. doi: 10.1073/pnas.95.12.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kulaeva O I, Wootton J C, Levine A S, Woodgate R. Characterization of the umu-complementing operon from R391. J Bacteriol. 1995;177:2737–2743. doi: 10.1128/jb.177.10.2737-2743.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matthew M, Hedges R W, Smith J T. Types of β-lactamase determined by plasmids in gram-negative bacteria. J Bacteriol. 1979;138:657–662. doi: 10.1128/jb.138.3.657-662.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murphy D B, Pembroke J T. Monitoring of chromosomal insertions of the IncJ elements R391 and R997 in Escherichia coli K-12. FEMS Microbiol Lett. 1999;174:355–361. doi: 10.1111/j.1574-6968.1999.tb13590.x. [DOI] [PubMed] [Google Scholar]
- 16.Murphy D B, Pembroke J T. Transfer of the IncJ plasmid R391 to recombination deficient Escherichia coli K-12: evidence that R391 behaves as a conjugal transposon. FEMS Microbiol Lett. 1995;134:153–158. doi: 10.1111/j.1574-6968.1995.tb07930.x. [DOI] [PubMed] [Google Scholar]
- 17.Nikolnikov S, Posfai G, Sain B. The construction of a versatile plasmid vector that allows direct selection of fragments cloned into six unique sites of the cl gene of coliphage 434. Gene. 1984;30:261–265. doi: 10.1016/0378-1119(84)90131-8. [DOI] [PubMed] [Google Scholar]
- 18.Norgren M G, Scott J R. Presence of the conjugative transposon Tn916 in the recipient strain does not impede transfer of a second copy of the element. J Bacteriol. 1991;173:319–324. doi: 10.1128/jb.173.1.319-324.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Novick R P. Plasmid incompatibility. Microbiol Rev. 1987;51:381–395. doi: 10.1128/mr.51.4.381-395.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nugent M E. A conjugative ‘plasmid’ lacking autonomous replication. J Gen Microbiol. 1981;126:305–310. doi: 10.1099/00221287-126-2-305. [DOI] [PubMed] [Google Scholar]
- 21.Osborn A M, Bruce K D, Ritchie D A, Strike P. The mercury resistance operon of the IncJ plasmid pMERH exhibits structural divergence from other Gram-negative mer operons. Microbiology. 1996;142:337–345. doi: 10.1099/13500872-142-2-337. [DOI] [PubMed] [Google Scholar]
- 22.Pembroke J T, Murphy D B. Isolation and analysis of a circular form of the IncJ conjugative transposon-like elements, R391 and R997: implications for IncJ incompatibility. FEMS Microbiol Lett. 2000;187:133–138. doi: 10.1111/j.1574-6968.2000.tb09149.x. [DOI] [PubMed] [Google Scholar]
- 23.Penner J L. The genera Proteus, Providencia, and Morganella. In: Balows A, Truper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. Vol. 3. New York, N.Y: Springer Verlag; 1992. pp. 2849–2862. [Google Scholar]
- 24.Peters S E, Hobman J L, Strike P, Ritchie D A. Novel mercury resistance determinants carried by IncJ plasmids pMERPH and R391. Mol Gen Genet. 1991;228:294–299. doi: 10.1007/BF00282479. [DOI] [PubMed] [Google Scholar]
- 25.Ptashne M. A genetic switch. Cambridge, United Kingdom: Cell Press and Blackwell Scientific Publications; 1992. [Google Scholar]
- 26.Scott J R, Churchward G G. Conjugative transposition. Annu Rev Microbiol. 1995;49:367–397. doi: 10.1146/annurev.mi.49.100195.002055. [DOI] [PubMed] [Google Scholar]
- 27.Singer M, Baker T A, Schnitzler G, Deischel S M, Goel M, Dove W, Jaacks K J, Grossman A D, Erickson J W, Gross C A. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989;53:6408–6411. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waldor M K, Mekalanos J J. Emergence of a new cholera pandemic: molecular analysis of virulence determinants in Vibrio cholerae O139 and development of a live vaccine prototype. J Infect Dis. 1994;170:278–283. doi: 10.1093/infdis/170.2.278. [DOI] [PubMed] [Google Scholar]
- 29.Waldor M K, Rubin E J, Pearson G D N, Kimsey H, Mekalanos J J. Regulation, replication, and integration functions of the Vibrio cholerae CTXφ are encoded by region RS2. Mol Microbiol. 1997;24:917–926. doi: 10.1046/j.1365-2958.1997.3911758.x. [DOI] [PubMed] [Google Scholar]
- 30.Waldor M K, Tschäpe H, Mekalanos J J. A new type of conjugative transposon encodes resistance to sulfamethoxazole, trimethoprim, and streptomycin in Vibrio cholerae O139. J Bacteriol. 1996;178:4157–4165. doi: 10.1128/jb.178.14.4157-4165.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woodgate R. A plethora of lesion-replicating DNA polymerases. Genes Dev. 1999;13:2191–2195. doi: 10.1101/gad.13.17.2191. [DOI] [PubMed] [Google Scholar]
- 32.Yokota T, Kuwahara S. Temperature-sensitive R plasmid obtained from naturally isolated drug-resistant Vibrio cholerae (biotype El Tor) Antimicrob Agents Chemother. 1977;11:13–20. doi: 10.1128/aac.11.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]