Amplification of Mutator Cells in a Population as a Result of Horizontal Transfer (original) (raw)
Abstract
Mutator cells that lack the mismatch repair system (MMR−) occur at rates of 10−5 or less in laboratory populations started from wild-type cells. We show that after selection for recombinants in an interspecies mating between Salmonella enterica serovar Typhimurium and Escherichia coli, the percentage of MMR− cells rises to several percent of the recombinant population, and after a second successive mating and selection, greater than 95% of the recombinants are MMR−. Coupling a single cross and selection with either mutagenesis or selection for spontaneous mutants also results in a dramatic increase in MMR− cells. We discuss how horizontal transfer can result in mutator strains during adaptive evolution.
Mutators, cells with higher rates of mutation than normal cells, play a role in human disease and adaptive evolution (see reviews in references 7, 18, and 19). For instance, the human mismatch repair system (MMR), the counterpart of the bacterial and yeast mismatch repair systems (10, 21, 22, 27), is involved in inherited predispositions to colon (HNPCC), endometrial, and ovarian cancer (3, 6, 12, 24). Tumor lines from HNPCC patients are mutators with greatly increased repeat-tract or microsatellite instability (1, 9, 12, 25, 26). How mutator cells arise and proliferate in cell populations as a result of different processes and selective forces have been the object of recent studies (15, 20). Reports of these studies previously described how selection for mutants resulting from spontaneous mutations amplifies the mutator subpopulation, to the point where an entire population of cells becomes mutator (15, 20). Most of the mutators arising from this process are mismatch repair system deficient (MMR−) and make frequent mutations due to their inability to repair replication errors. However, the MMR not only protects against replication errors but also acts as a barrier to recombination between divergent chromosomes. Radman and coworkers showed that MMR− cells lacking either the MutS or MutL function carry out homeologous recombination resulting from interspecies crosses between Salmonella enterica serovar Typhimurium and Escherichia coli three orders of magnitude more frequently than MMR+ cells (28, 31). MutS binding to mismatches may limit the heteroduplex region (see reference 28 and references therein; see also references 2 and 34). Since the mutators in a wild-type population have such an elevated frequency of recombination in interspecies crosses, does the selection for recombination in such a mating enhance the mutator fraction among the surviving cells? We describe here how interspecies crosses and homeologous recombination does select for rare MMR− cells, amplifying them in the population. Two successive homeologous crosses can convert a population with as few as 10−5 mutators to greater than 95% mutator. This suggests that horizontal transfer can ultimately be a mutagenic process at the population level.
MATERIALS AND METHODS
Bacterial strains and strain construction.
Table 1 lists the strains used in this work. Strain PA101 was constructed by transducing strain CSH110 (17) to Tetr using a P1 vir lysate grown on strain CAG12185 (30) and scoring for the retention of the Met− marker. The Tn_10_ in strain CAG12185 is integrated into the argH gene, and transduction of CSH110 to Tetr crosses out the argE mutation in CSH110, which can be verified by selecting for Arg+ revertants that lose the Tetr marker. Strain PA210 was obtained by transducing PA101 to Camr with a P1_vir_ lysate grown on a strain carrying a miniTn_10cam_ inserted into mutS (J. H. Miller, P. Funchain, and A. Yeung, unpublished data). Strain PA102 was obtained by transducing PA101 to Arg+ using a P1_vir_ lysate grown on strain P90C and scoring for the retention of the Met− marker and the loss of the Tetr marker. PA103 was constructed by transferring the F′ factor from strain CC107 to PA102, selecting for Pro+ Nalr colonies after a short mating, and scoring for the other markers in PA102. Strain PA104 was constructed by transducing strain PA102 with a P1_vir_ lysate grown on strain CAG18442 (30) (thr::Tn_10_) and selecting for Tetr [Thr+] recombinants. Strain CAG18442 carries a Tn_10_ integrated into the thr operon. All crosses and transductions used in strain construction were performed as described by Miller (17).
TABLE 1.
Bacterial strains described in this report
| Strain | Genotype | Reference |
|---|---|---|
| CC107 | ara Δ_(gpt-lac)5_/F′128 lacIZ proA+B+ (carries a +1 frameshift in I portion of I-Z fusion) | 4 |
| CSH110 | ara Δ_(gpt-lac)5 supE gyrA metB argE_(Am) rpoB | 17 |
| PA101 | ara Δ_(gpt-lac)5 supE gyrA metB argE_::Tn_10_ | This work |
| PA201 | PA101 mutS::minTn_10cam_ | This work |
| PA102 | ara Δ_(gpt-lac)5 supE gyrA metB_ | This work |
| PA103 | ara Δ_(gpt-lac)5 supE gyrA metB_/F′lacpro (F′ from CC107) | This work |
| PA104 | ara Δ_(gpt-lac)5 supE gyrA metB thr-34_::Tn_10_ | This work |
| CAG18442 | MG1655 thr-34::Tn_10_ | 30 |
| CAG12185 | MG1655 argE86::Tn_10_ | 30 |
| CAG12173 | MG1655 cysC95::Tn_10_ | 30 |
| CAG18427 | MG1655 zje-2241::Tn_10_ | 30 |
| DPB267 | MG1655 recD1901::Tn_10_ | 30 |
| SA975 | HfrK13 thrA49 leuBCD39 ara-7 (Salmonella serovar Typhimurium) | 29 |
| SA534 | Hfr K4 serA13 rfa-3058 (Salmonella serovar Typhimurium) | 29 |
Mutagenesis.
Ethyl methanesulfonate (EMS) mutagenesis was carried out as described by Miller (17). EMS was used at a dose of 60 min of exposure to 0.03 ml of EMS added to 2 ml of resuspended washed cells in minimal phosphate buffer, pH 7.0. Cells were diluted 1:10 and grown overnight in Luria-Bertani (LB) broth.
Conjugational matings.
Overnight cultures of both donors and recipients grown in LB broth without aeration (except in the case of EMS mutagenized cells and successive selection; see below) were diluted 1:50 and grown in LB broth in a water bath at 37° for approximately 4 h and then mated by mixing 0.2 ml of donor with 0.2 ml of recipient in a test tube for 1 h. After the addition of 0.5 ml of LB broth, the mixture was placed on a rotor at 30 rpm for 1 h before being plated on selective medium. Nalidixic acid (17) was used to counterselect against the Hfr. The crosses were carried out at a 1:1 ratio of Hfr to F−, and typically both were at 1 × 108 to 2 × 108 cells/ml. Therefore, the number of recombinants per Hfr also equals the number of recombinants per F−.
Successive selections.
Cultures were prepared by inoculating different single colonies. After selection for Lac+, Lac+ colonies were scraped with a glass spreader and transferred to 50 ml of LB broth in a 250-ml flask and grown overnight. This mixture was then diluted 1:50 to prepare for conjugational matings as described above. Similarly, Thr+ colonies resulting from an Hfr cross were scraped and pooled into a flask and grown overnight before being diluted for a second mating.
Identification of mutators.
We tested for mutators by screening for increased numbers of spontaneous Rifr mutants. Initially, we gridded purified colonies onto LB plates and incubated them overnight before replicating onto a second LB plate, and after 8 h of incubation, this plate was replicated onto an LB plate with 100 μg of rifampin/ml and incubated for 16 to 24 h. Patches of wild-type cells showed either no colonies or an occasional colony, but MMR− cells showed numerous colonies growing out of the patch. (Even weaker mutators reveal themselves by this procedure.) All candidates for mutators were tested more quantitatively by growing in broth overnight and plating samples onto LB + rifampin, and in many cases we calibrated the efficiency of the gridding procedure by testing the entire set of 50 or 100 colonies by growing overnight cultures and plating on such selective medium. The difference between MMR+ and MMR− derivatives of PA101 is easy to detect since MMR+ strains routinely give 0 to 5 colonies on an LB plate with rifampin per 0.05 ml of a saturated overnight culture whereas the MMR− derivatives give between 200 and 1,000 colonies. We mapped the mutation causing the mutator phenotype in examples of the mutator colonies by P1 cotransduction. We transduced the mutator strains to Tetr using P1_vir_ lysates grown on strains carrying Tn_10_ inserts near mutS (CAG12173) (30; see Table 1), mutL (DPB267) (Table 1), or mutH (CAG18427) (Table 1). We tested 16 Tetr transductants to determine whether the mutator character was lost or retained, since the linkage of the mutator allele is close to 50% to each relevant Tn_10_. In several cases, including the one example of a mutH mutation we detected, we transduced the mutation into a wild-type strain using a linked Tn_10_ and demonstrated the mutator phenotype.
RESULTS
Homeologous crosses.
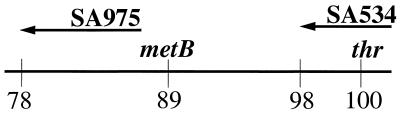
We carried out crosses between Salmonella serovar Typhimurium Hfr strains and E. coli F− recipients. Table 1 describes all of the strains used in this work. Figure 1 shows the position of the points of origin and the markers in the strains used for these crosses. The Salmonella Hfr SA975 (29) donates markers in a clockwise fashion from a point near 78 min on the circular Salmonella and E. coli maps. It donates the Met+ marker (metB; 89 min) after about 11 min of mating. We used as the E. coli recipient strain PA101, a Met− (metB) Rifr derivative of CSH110 (17). When SA975 is crossed with PA101, a very low level of Met+ recombinants are found, in contrast to a cross of SA975 with an MMR− derivative (mutS) of PA101 (PA201). As Table 2 shows, there is an approximately 1,000-fold increase in the number of Met+ recombinants detected in the mutS strain. This is the effect first described by Radman and coworkers (28, 31). Normally, MMR− cells are present on the order of 10−5 in a wild-type E. coli population started from a single cell and grown overnight (15). However, because some of the MMR− cells (mutS and mutL) (28) have such an enhanced efficiency of carrying out the homeologous recombination, their frequency should be amplified in the Met+ recombinant population relative to the starting Met− recipient (PA101) population. We therefore monitored the MMR− phenotype by determining the Rifr mutant frequency in Met+ recombinant colonies in the cross of SA975 with PA101. Using both replica plating and broth tests, we tested 200 Met+ recombinants from each of two independent crosses. We found 24 of the total 400 Met+ recombinants (6%) were mutators (Table 2), as judged by their high frequency of Rifr (see Materials and Methods). None of the 400 tested Met− colonies of PA101 (before the cross with SA975) were mutators.
FIG. 1.
Points of origin and direction of transfer of Salmonella Hfr strains used in this work.
TABLE 2.
Efficiency of recombination (Met+/Hfr)a or (Thr+/Hfr)b and % of MMR− recombinants in crosses between Salmonella Hfr and E. coli F− strains
| Recombinant type | Frequency in E. coli F− (WTe; PA101) | No. (%) of MMR− mutators | Frequency in E. coli F− (mutS; PA201) |
|---|---|---|---|
| Met+ | 1 × 10−6 to 2 × 10−6 | 24 of 400 (6) | 2 × 10−3 to 4 × 10−3 |
| After EMS: Met+ | 1 × 10−5 | 44 of 50, 147 of 200 (81) | |
| After Lac+ reversionc: Lac+ Met+ | 42 of 50, 35 of 48 (78 ) | ||
| After Thr+ selectiond: Thr+ Met+ | 96 of 99, 98 of 100 (97) |
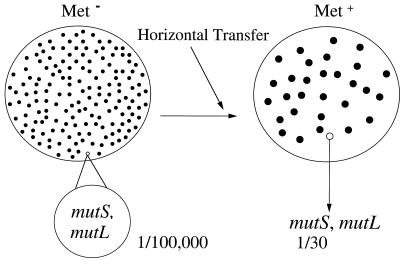
We mapped a random set of six of the mutators found in the above experiment and determined that five had mutations in mutS and one in mutL. These experiments (Fig. 2) demonstrate that the horizontal transfer from Salmonella to E. coli amplifies the mutator population in the recipient E. coli up to several percent (6% in these experiments, 1 to 3% in other experiments with different strains) in a single step of homeologous recombination, in this case selecting for acquisition of the Met+ character.
FIG. 2.
Increase in MMR− mutators among E. coli Met+ recombinants after an interspecies cross with a Salmonella Hfr.
Successive steps of mutator enhancement.
If a single step of horizontal transfer can amplify the mutator population close to 1,000-fold, then carrying out an interspecies mating in a population that already has an elevated frequency of mutators should enhance the mutator population even further, to the point of virtually overtaking the population. There are several ways a population can experience an elevated mutator population, for instance, exposure to mutagens, following selection for a mutant phenotype, and following horizontal transfer (see above). After treatment with a mutagen, MMR− cells can represent up to 1 per 1,000 of the population (see, for instance, reference 20). Mutators are also amplified in a population following each round of selection for a phenotype (15). For example, after selection for Lac+ resulting from the reversion of a frameshift, mutators are present in 0.5% of the revertants (15). Moreover, as shown in Table 2, horizontal transfer elevates the mutator population. We examined each of these effects as the first step in a two-step procedure, with a horizontal transfer being the second step. (In the last of the three methods, the horizontal transfer would represent a second, successive horizontal transfer.)
Exposure to mutagens followed by an interspecies cross.
We treated cells of PA101 with EMS and grew them for five to six generations to allow mutants generated by EMS to segregate out. We then mated a sample of this culture with SA975, as described above (see also Materials and Methods). Table 2 shows the results. EMS creates MMR− mutants on the order of 10−3 in the population, as measured by direct selection (20; data not shown). In fact, we detected one mutator among the 200 colonies examined from two separate experiments after EMS treatment. However, after mating with the Salmonella Hfr SA975, we noted a fourfold increase in the level of Met+ recombinants (column 2 in Table 2), even though the cultures had undergone five to six generations following the EMS treatment. An examination of the Met+ recombinants (Table 2) showed that 80% were strong mutators. Mapping a sample of these showed that they carried mutations in either mutS or mutL (see below).
Selection for new phenotypes followed by an interspecies cross.
We introduced the F′lacpro plasmid that carries a frameshift in lacZ that reverts by the addition of a -G- to a run of six -G-'s (CC107) (4) into an Arg+ derivative of PA101 (PA102). We used this lacZ marker to show that in strains carrying it, 0.5% of the Lac+ revertants were MMR− mutators (15). We plated four cultures of this new strain, PA103, on lactose minimal medium and pooled the Lac+ colonies into two flasks, each flask containing Lac+ cells from two different cultures. After overnight growth in rich medium, the cells were mated with the Salmonella Hfr SA975 (see above) and Met+ recombinants were selected. We tested 50 purified Met+ revertants from each of the two experiments. Whereas less than 1% of the Lac+ revertants (before the cross) were found to be mutators, close to 80% of the Met+ recombinants from the two experiments were shown to be MMR− mutators (Table 2).
Successive homeologous matings.
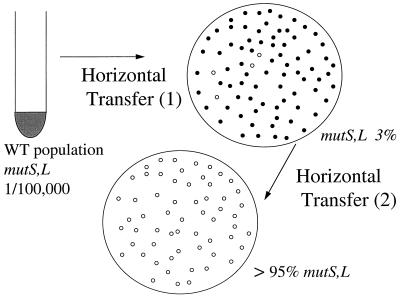
We prepared a Thr− derivative of PA102, termed PA104 (see Table 1 and Materials and Methods), and crossed it with the Salmonella Hfr SA534 (29) that donates clockwise from 98 min, bringing in thr (0 min) very early and metB (89 min) very late (Fig. 1). Thr+ colonies were pooled and grown up overnight in a single flask in rich medium and then mated with the Salmonella Hfr SA975 (described above; 29). Met+ recombinants were selected in this second mating. Purified Met+ colonies were then tested for mutator activity. Figure 3 depicts the experiment. Greater than 95% of the colonies proved to be strong mutators, as shown in Table 2.
FIG. 3.
Increase in MMR− mutators after successive interspecies crosses. WT, wild type.
Mapping mutations resulting in the MMR− phenotype.
We mapped 28 of the mutations resulting in the MMR− phenotype derived from the experiments described above by using P1 transduction (see Materials and Methods). We found that 22 of the mutations were in mutS, 5 in mutL, and 1 in mutH.
DISCUSSION
Cells acquire new traits not only by mutation, but also by horizontal transfer. In fact, it is now believed that horizontal transfer and recombinational reshuffling has played the major role in the generation of microbial diversity rather than stepwise mutations, as recently reviewed by Ochman and coworkers (23). For instance, Lawrence and Ochman (11) analyzed the sequenced genome of E. coli and determined that 755 of the 4,288 open reading frames in E. coli were introduced by lateral transfer events in the 100 million years since E. coli diverged from Salmonella. They also concluded that none of the phenotypic traits that distinguish E. coli from Salmonella arose by stepwise mutation. However, the MMR acts as a barrier to recombination between divergent chromosomes. Thus, MMR− cells lacking either the MutS or MutL function are 1,000 times more efficient in carrying out homeologous recombination between divergent sequences, such as the 18% divergence between Salmonella serovar Typhimurium and E. coli (28, 31). Denamur and coworkers have suggested that the fact that the MMR encoding genes are more mosaic than normal indicates that these genes were lost and reacquired by horizontal transfer several times (5). They have shown how the sequence divergence generated by continuous growth of a mutator strain after 20,000 generations can already act as a detectable recombination barrier in the presence of a functioning MMR system (33).
In this paper we demonstrate how recombination after an interspecies cross leads to an enhancement of mutators in populations of cells after horizontal transfer (Fig. 2; Table 2). After one mating and selecting for the acquisition of one character, the percentage of mutators in the recombinant population can be elevated as much as several percent. This is because the subpopulation of mutators has such a high rate of success relative to the main nonmutator population in an interspecies mating that the mutator fraction will be substantially enhanced among the successful recombinants in a homeologous cross. Thus, if the subpopulation of MMR− cells is present at a frequency of 1 × 10−5 to 2 × 10−5 but has a success rate of becoming Met+ in an interspecies cross 1,000 to 2,000 higher than the main population, the fraction of mutators goes from near 1/100,000 to near 1 per 30 or more.
Coupling the interspecies transfer and recombination to any other process that increases the mutator population leads to very high percentages of mutators. For instance, after EMS treatment, the MMR− mutators are elevated to between 0.01% and 0.1%. Subjecting the mutagenized recipient population to an interspecies mating followed by a single selection elevates the mutators to 80 to 90% (Table 2). Also, selecting for Lac+ revertants in a Lac− strain followed by selection for Met+ in an interspecies cross results in close to 80% of the Met+ recombinants being MMR− mutators (Table 2). The Lac+ selection elevates the mutators to between 0.1% and 1% (15), and the homeologous recombination provides a further enhancement. In a third experiment (Fig. 3; Table 2), two successive rounds of horizontal transfer and recombination lead to conversion of most (approximately 97%) of the population to mutators. This is provocative, since it argues that horizontal transfer itself is a mutagenic process, amplifying mutator phenotypes as a byproduct of crosses.
In laboratory populations started from single MMR+ cells, MMR− mutators occur as a result of random mutagenic events and, after growth of a typical overnight culture, represent only about 1 per 100,000 cells in E. coli (15, 20) and an even smaller fraction in Salmonella serovar Typhimurium (14). However, populations of E. coli and serovar Typhimurium in the wild have several percent mutators (13, 16). It is not certain whether this is due to (i) growing in a constantly changing environment that places these populations under frequent if not continuous selection, (ii) having been subject in the recent past to a more mutagenic environment than normal, or (iii) as yet undiscovered reasons. It has been suggested that higher mutator subpopulations may be associated with pathogenicity (13). In any case, based on the experiments depicted here, it is evident for populations in the wild that after only one horizontal transfer and accompanying homeologous recombination event, the selected recombinants would be 80 to 100% mutator. Yet, this poses a paradox, since continuous growth as a mutator has detrimental effects. For instance, we have shown (8) that continuous growth of MMR− mutator lineages generates a continued accumulation of mutations in unselected genes that leads to a multiple loss of function that can be very costly in other environments. When passaged through severe bottlenecks, mutator lineages also accumulate mutations that confer loss of fitness (8). The deleterious effects of growing as a mutator act as a counterbalancing force that selects for MMR+ cells once the selections for new phenotypes by mutation or horizontal transfer have receded. (See reference 32 for a discussion of these same points.)
ACKNOWLEDGMENTS
We thank Miroslav Radman for communicating unpublished results and for helpful discussions and Kenneth Sanderson for supplying the Salmonella Hfr strains.
This work was supported by grant GM32184 to J.H.M. from the National Institutes of Health.
REFERENCES
- 1.Aaltonen L A, Peltomaki P, Leach F, Sistonen P, Pylkkanen S M, Mecklin J-P, Jarvinen H, Powell S, Jen J, Hamilton S R, Petersen G M, Kinzler K W, Vogelstein B, de la Chapelle A. Clues to the pathogenesis of familial colorectal cancer. Science. 1993;260:812–816. doi: 10.1126/science.8484121. [DOI] [PubMed] [Google Scholar]
- 2.Alani E, Reenan R A G, Kolodner R D. Interaction between mismatch repair and genetic recombination in Saccharomyces cerevisiae. Genetics. 1994;137:19–39. doi: 10.1093/genetics/137.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhattacharyya N P, Skandalis A, Ganesh A, Groden J, Meuth M. Mutator phenotypes in human colorectal carcinoma cell lines. Proc Natl Acad Sci USA. 1994;91:6319–6323. doi: 10.1073/pnas.91.14.6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cupples C G, Cabrera M, Cruz C, Miller J H. A set of lacZ mutations in Escherichia coli that allow rapid detection of specific frameshift mutations. Genetics. 1990;125:275–280. doi: 10.1093/genetics/125.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Denamur E, Lecointre G, Darlu P, Tenaillon O, Acquaviva C, Sayada C, Sunjevaric I, Rothstein R, Elion J, Taddei F, Radman M, Matic I. Evolutionary implications of the frequent horizontal transfer of mismatch repair genes. Cell. 2000;103:711–721. doi: 10.1016/s0092-8674(00)00175-6. [DOI] [PubMed] [Google Scholar]
- 6.Fishel R, Lescoe M K, Rao M R, Copeland N G, Jenkins N A, Garber J, Kane M, Kolodner R. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 7.Friedberg E C, Walker G C, Seide W. DNA repair and mutagenesis. Washington, D.C.: American Society for Microbiology; 1995. [Google Scholar]
- 8.Funchain P, Yeung A, Stewart J L, Lin R, Slupska M M, Miller J H. The consequences of growth of a mutator strain of Escherichia coli as measured by loss of function among multiple gene targets and loss of fitness. Genetics. 2000;154:959–970. doi: 10.1093/genetics/154.3.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ionov Y, Peinado M A, Malkhosyan S, Shibata D, Peruco M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1995;363:558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 10.Kolodner R. Biochemistry and genetics of eukaryotic mismatch repair. Genes Dev. 1996;10:1433–1442. doi: 10.1101/gad.10.12.1433. [DOI] [PubMed] [Google Scholar]
- 11.Lawrence J G, Ochman H. Molecular archaeology of the Escherichia coli genome. Proc Natl Acad Sci USA. 1998;95:9413–9417. doi: 10.1073/pnas.95.16.9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leach F S, Nicolaides N C, Papadopoulos N, Liu B, Jen J, Parsons R, Peltomaki P, Sistonen P, Aaltonen L A, Nystrom-Lahti M, Guan X-Y, Zhang J, Meltzer P S, Yu J-W, Kao F-T, Chen D J, Cerosaletti K M, Fournier R E K, Todd S, Lewis T, Leach R J, Naylor S L, Weissenbach J, Meckin J-P, Jarvinen H, Petersen G M, Hamilton S R, Green J, Jass J, Watson P, Lynch H T, Trent J M, de la Chapelle A, Kinsler K W, Vogelstein B. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell. 1993;75:1215–1225. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- 13.LeClerc J E, Li B, Payne W L, Cebula T A. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science. 1996;274:1208–1211. doi: 10.1126/science.274.5290.1208. [DOI] [PubMed] [Google Scholar]
- 14.LeClerc J E, Payne W L, Kupchella E, Cebula T A. Detection of mutator subpopulations in Salmonella typhimurium LT2 by reversion of his alleles. Mutat Res. 1998;400:89–97. doi: 10.1016/s0027-5107(98)00069-4. [DOI] [PubMed] [Google Scholar]
- 15.Mao E F, Lane L, Lee J, Miller J H. Proliferation of mutators in a cell population. J Bacteriol. 1997;179:417–422. doi: 10.1128/jb.179.2.417-422.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matic I, Radman M, Taddei F, Picard B, Doit C, Bingen E, Denamur E, Elion J. Highly variable mutation rates in commensal and pathogenic Escherichia coli. Science. 1997;277:1833–1834. doi: 10.1126/science.277.5333.1833. [DOI] [PubMed] [Google Scholar]
- 17.Miller J H. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 18.Miller J H. Spontaneous mutators in bacteria: insights into pathways of mutagenesis and repair. Annu Rev Microbiol. 1996;50:625–643. doi: 10.1146/annurev.micro.50.1.625. [DOI] [PubMed] [Google Scholar]
- 19.Miller J H. Mutators in Escherichia coli. Mutat Res. 1998;409:99–106. doi: 10.1016/s0921-8777(98)00049-4. [DOI] [PubMed] [Google Scholar]
- 20.Miller J H, Suthar A, Tai J, Yeung A, Truong C, Stewart J L. Direct selection for mutators in Escherichia coli. J Bacteriol. 1999;181:1576–1584. doi: 10.1128/jb.181.5.1576-1584.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Modrich P. Mechanisms and biological effects of mismatch repair. Annu Rev Genet. 1991;25:229–253. doi: 10.1146/annurev.ge.25.120191.001305. [DOI] [PubMed] [Google Scholar]
- 22.Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 23.Ochman H, Lawrence J G, Groisman E A. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 24.Papadopoulos N, Nicolaides N C, Wei Y F, Ruben S M, Carter K C, Rosen C A, Haseltine W A, Fleischmann R D, Fraser C M, Adams M D, Venter J C, Hamilton S R, Petersen G M, Watson P, Lynch H T, Peltomaki P, Mecklin J-P, de la Chapelle A, Kinzler K W, Vogelstein B. Mutation of a mutL homolog in hereditary colon cancer. Science. 1994;263:1625–1629. doi: 10.1126/science.8128251. [DOI] [PubMed] [Google Scholar]
- 25.Parsons R, Myeroff L, Liu B, Willson J K V, Markowitz S D, Kinzler K W, Vogelstein B. Microsatellite instability and mutations of the transforming growth factor beta type II receptor gene in colorectal cancer. Cancer Res. 1995;55:5548–5550. [PubMed] [Google Scholar]
- 26.Peltomaki P, Lothe R A, Aaltonen L A, Pylkkanen L, Nystrom-Lahti M, Seruca R, David L, Holm R, Ryberg D, Gaugen A, Brogger A, Borresen A-L, de la Chapelle A. Miscrosatellite instability is associated with tumors that characterize the hereditary non-polyposis colorectal carcinoma syndrome. Cancer Res. 1993;53:5853–5855. [PubMed] [Google Scholar]
- 27.Radman M, Wagner R. Mismatch repair in Escherichia coli. Annu Rev Genet. 1986;20:523–538. doi: 10.1146/annurev.ge.20.120186.002515. [DOI] [PubMed] [Google Scholar]
- 28.Rayssiguier C, Thaler D S, Radman M. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature. 1989;342:396–401. doi: 10.1038/342396a0. [DOI] [PubMed] [Google Scholar]
- 29.Sanderson K E. F-mediated conjugation, F+ strains, and Hfr strains of Salmonella typhimurium and Salmonella abony. In: Neidhardt F C, editor. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C.: American Society for Microbiology; 1996. pp. 2406–2412. , [Google Scholar]
- 30.Singer M, Baker T A, Schnitzler G, Deischel S M, Goel M, Dove W, Jaacks K W, Grossman A D, Erickson J W, Gross C A. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989;53:1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stambuk S, Radman M. Mechanism and control of interspecies recombination in Escherichia coli. I. Mismatch repair, methylation, recombination, and replication functions. Genetics. 1998;150:533–542. doi: 10.1093/genetics/150.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taddei F, Vulic M, Radman M, Matic I. Genetic variability and adaptation to stress. In: Bijlsma R, Loeschcke V, editors. Environmental stress, adaptation and evolution. O. Basel, Switzerland: Birkhäuser Verlag; 1997. [DOI] [PubMed] [Google Scholar]
- 33.Vulic M, Lenski R E, Radman M. Mutation, recombination, and incipient speciation of bacteria in the laboratory. Proc Natl Acad Sci USA. 1999;96:7348–7351. doi: 10.1073/pnas.96.13.7348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Worth L, Clark S, Radman M, Modrich P. Mismatch repair proteins MutS and MutL inhibit RecA-catalyzed strand transfer between diverged DNAs. Proc Natl Acad Sci USA. 1994;91:3238–3241. doi: 10.1073/pnas.91.8.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]