Analysis of Functional Domains of the Enterococcus faecalis Pheromone-Induced Surface Protein Aggregation Substance (original) (raw)
Abstract
Pheromone-inducible aggregation substance (AS) proteins of Enterococcus faecalis are essential for high-efficiency conjugation of the sex pheromone plasmids and also serve as virulence factors during host infection. A number of different functions have been attributed to AS in addition to bacterial cell aggregation, including adhesion to host cells, adhesion to fibrin, increased cell surface hydrophobicity, resistance to killing by polymorphonuclear leukocytes and macrophages, and increased vegetation size in an experimental endocarditis model. Relatively little information is available regarding the structure-activity relationship of AS. To identify functional domains, a library of 23 nonpolar 31-amino-acid insertions was constructed in Asc10, the AS encoded by the plasmid pCF10, using the transposons Tn_lacZ_/in and Tn_phoA_/in. Analysis of these insertions revealed a domain necessary for donor-recipient aggregation that extends further into the amino terminus of the protein than previously reported. In addition, insertions in the C terminus of the protein also reduced aggregation. As expected, the ability to aggregate correlates with efficient plasmid transfer. The results also indicated that an increase in cell surface hydrophobicity resulting from AS expression is not sufficient to mediate bacterial aggregation.
Enterococcus faecalis has become a growing health concern as a mediator of the spread of antibiotic resistance and a leading agent of nosocomial infections (for review, see reference 10). The surface protein aggregation substance (AS) appears to play a role in both antibiotic resistance spread and in the pathogenesis of enterococcal infections. Expression of AS, which is encoded on the sex pheromone plasmids of E. faecalis, is induced by small 7- to 8-amino-acid peptide pheromones (26). AS on the surface of the donor cell then binds its receptor, enterococcal binding substance, on the recipient cell, mediating close cell contact that leads to conjugative transfer of the plasmid. It is thought that AS has no role in forming the DNA channel machinery, as efficient conjugation can occur if AS is expressed on either the donor or recipient cells (26).
Over 20 different pheromone plasmids have been identified. Often, these pheromone plasmids express antibiotic resistance genes and other virulence factors, and many clinical isolates have multiple pheromone plasmids (36). The AS genes from the three most-studied plasmids, Asa1 from pAD1, Asp1 from pPD1, and Asc10 from pCF10 (encoded by the prgB gene), have been sequenced and show high identity (see below). The gene encoding Asa373, the AS protein of the pheromone plasmid pAM373, has also been sequenced but shows little homology with the other known AS proteins and appears to aggregate through a different mechanism (21). Expression of AS, which is normally tightly controlled in laboratory cultures, is induced in serum (13).
A number of functions of AS that may contribute to virulence have been identified. A major function of AS is host cell adhesion. Kreft et al. found that Asa1 increased adherence to cultured pig renal tubular cells (13). Increased uptake mediated by Asc10 into epithelial cells originating from the colon and duodenum but not from the ileum has also been observed (25, 30). Along these lines, Asa1 increases invasion in an ex vivo model of the colonic mucosa but does not increase translocation (11). Asc10 has been found to increase adherence to and uptake by polymorphonuclear leukocytes, possibly by binding the integrin CR3 (35). In other studies, Asc10-expressing enterococci had higher intracellular survival rates in polymorphonuclear leukocytes (27). Likewise, adherence to and survival inside macrophages were increased with the expression of Asa1 (33). In vivo examination of the role of AS has centered on the rabbit experimental endocarditis model. Infection of a rabbit with a damaged heart valve leads to development of a mass of bacteria, platelets, and fibrin known as a vegetation (17). Two studies have found more severe vegetation formation induced by AS-expressing enterococci (4, 31). Asc10 has also been shown to increase adherence to fibrin and cell surface hydrophobicity (8).
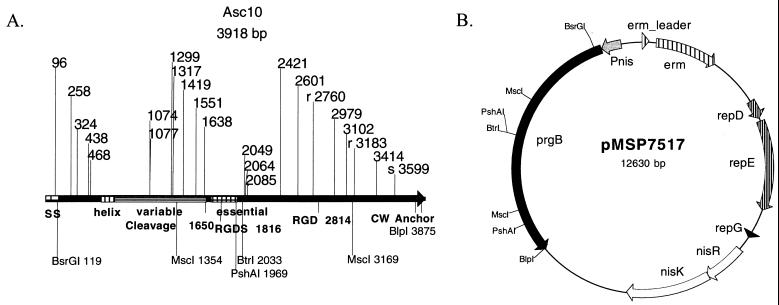
Although much study has focused on the functions of AS, it is unclear how the structure of the protein mediates these functions. Like most gram-positive surface proteins, Asc10 has an N-terminal signal sequence and C-terminal LPXTG cell wall anchor motif (see Fig. 1A). Analysis of the three sequenced genes encoding closely related proteins reveals striking conservation of >90% identity in the majority of the protein, excluding a variable region of 30 to 50% identity located between amino acids 266 and 559 in the N terminus of AS (36). All three proteins also have conserved Arg-Gly-Asp (RGD) motifs that have been implicated in binding to integrins (13, 29, 33, 35). Secondary structural analysis yields little information with the exception of a predicted alpha-helix domain from amino acids 200 to 280 (36). Isolation of AS yields both a full-length version of the protein (137 kDa) and a specific, 78-kDa, N-terminal cleavage product (9). Scanning electron microscopy of Asa1 on the cell surface suggests that the N terminus of the protein is more exposed than the C terminus (9). Finally, the only structural analysis of AS done to date found that an aggregation domain of Asa1 from amino acids 525 to 617 (of the mature protein with the signal sequence removed) exists and that the C terminus plays no essential role in aggregation (20).
FIG. 1.
(A) The positions of insertion mutations in prgB are shown on a linear map of the gene. Each mutation consists of an in-frame 31-amino-acid insertion. The insertions at r2760 and r3183 are in frame but in the reverse orientation, while the insertion at s3599 is out of frame and produces a stop codon. Structural map: SS, signal sequence; helix, predicted N-terminal helix domain; variable, unconserved AS region; essential, aggregation domain identified by Muscholl-Silberhorn; and cleavage, site of cleavage that produces the characteristic N-terminal 78-kDa fragment. (B) The mutant Asc10 proteins were expressed using the nisin-inducible Asc10 expression vector pMSP7517.
One dilemma with the use of conventional biochemical approaches to AS structure-function analysis is the high instability of purified protein. For this reason, we have taken a genetic approach to probe the protein for functional domains using the transposons Tn_lacZ_/in and Tn_phoA_/in (15, 16). In-frame insertions can be identified by functional fusions to the 5′ LacZ or PhoA reporter protein. Digestion of the insertion with _Bam_HI removes most of the transposon but leaves an in-frame 31-amino-acid insertion. These transposons have been successfully used to analyze the structure-function relationship of a number of membrane and cytosolic proteins (14, 15, 19, 22, 23), but this is the first attempt to use them in the analysis of a gram-positive surface protein.
A library of 23 insertional mutants distributed throughout the length of the prgB gene has been constructed. The stability of the AS protein expressed by these mutants was examined, and most proteins were found to be stable on the surface of E. faecalis. Phenotypic analysis of the insertion mutants in aggregation and conjugation revealed that both the N and C termini of the protein play significant roles in these processes. The ability of wild-type and mutant Asc10 proteins to increase cell surface hydrophobicity was also analyzed, and it was shown that increased hydrophobicity is not sufficient for aggregation.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
E. faecalis was grown at 37 or 30°C as indicated with gentle shaking in Todd-Hewitt broth (Difco). For DNA isolation and manipulation, Escherichia coli was grown at 37°C with shaking in Luria-Bertani (LB) medium or brain heart infusion broth (Difco) for erythromycin selection. Agar plates contained 1.5% agar. The antibiotic concentrations used for E. faecalis were erythromycin at 10 μg/ml, tetracycline at 15 μg/ml, and rifampin at 200 μg/ml, while the concentrations used for E. coli were erythromycin at 50 μg/ml (in brain heart infusion broth) or 200 μg/ml (in LB) and chloramphenicol at 50 μg/ml. All antibiotics were obtained from Sigma.
DNA manipulation.
Plasmids were isolated with the Qiagen midi or mini kit as recommended by the manufacturer. Restriction enzymes were purchased from Promega, Gibco BRL, and New England BioLabs. PCR was performed with a Perkin-Elmer Gene Amp PCR system or a Eppendorf Mastercycler using either BioXact DNA polymerase (Bioline) or Vent polymerase (New England BioLabs). All sequencing and primer synthesis were done by the Microchemical Facility of the University of Minnesota.
Phage isolation, infection, and screening of transposon insertions.
Preparation of λTn_lacZ_/in and λTn_phoA_/in was performed as previously described (1) using the E. coli suppressor strain CC245. Phage stocks were calculated at titers of 107 phage/ml. Phage infection was performed as previously described (15) with some minor modifications. Cultures of E. coli strain CC160 containing the target plasmid were grown overnight to stationary phase in λ broth (10 g of tryptone and 2.5 g of NaCl per liter supplemented with 0.2% maltose and 10 mM MgSO4). One milliliter of the overnight culture was mixed with 1 ml of the phage stock and was incubated at 37°C for 10 min. Three milliliters of LB broth was added, and the phage/bacteria were incubated at 30°C with gentle shaking for 6 h to overnight. The entire mixture was plated (150- by 15-mm LB agar petri plates; Falcon) with erythromycin and chloramphenicol and was grown overnight. The colonies were collected by washing the plates with distilled water. Plasmids were isolated from these cells; electroporated into competent E. coli strain CC118; and plated on LB supplemented with erythromycin, chloramphenicol, 5% sucrose (to counterselect against right-end insertions of the transposon), and 40 μg of 5-bromo-4-chloro-β-d-galactopyranoside (X-Gal) or 5-bromo-4-chloro-3-indolylphosphate (X-Phos) (Sigma) per ml. Blue transformants were screened by restriction digestion or colony PCR using a primer complementary to the nisin promoter (5′-CGGCTCTGATTAAATTCTGAAGTTTGTTAGATACAATGA-3′) and to the insertion sequence (5′-CCTGGACGGAACCTTTCCCG-3′). Colony PCR was performed by mixing a sterile pipette tip touched to the side of a colony into the PCR mix. The bacterial cells were first lysed by a 10-min 94°C incubation before the standard PCR was performed. The latter primer in the insertion sequence was also used to sequence transposon inserts in prgB. The bulk of the transposon was removed with _Bam_HI (Promega) digestion and religation with T4 DNA ligase (Gibco BRL). Relevant insertions in prgB were electroporated into E. faecalis, and transformants were screened by plasmid isolation (2) and restriction digestion.
Construction of insertion mutants.
Relevant plasmids are listed in Table 1. The initial target for insertion mutagenesis was the vector pMSP3602. This is a derivative of pINY1801 with the _Bam_HI sites removed and was constructed in two steps. First, the blunt-ended _Kpn_I-_Eco_RI fragment from pTRKH2 (the blunt end was generated using Klenow DNA polymerase [Promega] with the manufacturer's instructions) containing the erythromycin gene was inserted into the _Ban_II site of pINY1801 to generate pMSP3601. The _Bam_HI fragment from pMSP3601 was removed with Bam_HI digestion, and the overhangs of the fragment and vector backbone were filled with Klenow polymerase. The blunt-ended fragment was then religated into the pMSP3601 backbone to create pMSP3602. The Tn_lacZ/in insertions Ω1077, Ω1638, Ω2049, Ω2064, Ω2085, Ω2421, Ω2601, Ω2979, Ω3102, Ω3414, and Ωs3599 (Fig. 1) were generated in pMSP3602. Ωs3599 is an out-of-frame insertion at the very C terminus of the gene, resulting in production of most of the gene product without a cell wall anchor. The amino acid insertion sequence generated by the in-frame insertion sequence is 5′-XDSYTQVASWTEPFPFSIQGDPRSDQETXXX-3′, where X depends on the duplicated target sequence. Due to a lack of prgB expression in pMSP3602, these insertions were moved into pMSP7517 by isolating the _prgB Bsr_GI-_Blp_I fragment containing the insertion from pMSP3602 and by ligating it into pMSP7517, replacing the corresponding wild-type prgB sequence. pMSP7517 was also used as a target for insertional mutagenesis generating Ωr2760 and Ωr3183. These insertions are in frame but are in the reverse orientation, generating the amino acid insertion sequence 5′-XXXCLLIRSWIPLDGKRERFRPGRYLCIRVS/R-3′, where X depends on the duplicated target sequence. Out-of-frame and reverse insertions were frequently observed, as the transcription machinery of E. coli was likely recognizing artifactual promoters in the AT-rich E. faecalis DNA. In an attempt to decrease the target size of the gene, pMSP7517Δ_Msc_I was constructed by removing the Msc_I fragment from pMSP7517. Two Tn_phoA/in insertions, Ω96 and Ω258, were generated in this construct. These insertions were restored to the context of full-length prgB by reinserting the _prgB Msc_I fragment into the Msc_I restriction site. Note that the insertions left after removal of the Bam_HI fragments are identical for both λTn_lacZ/in and λTn_phoA/in.
TABLE 1.
Strains and plasmids used in this study
| Strain, phage, or plasmid | Strain or description | Source or reference |
|---|---|---|
| Bacteria | ||
| E. faecalis | CG1SSp | 6 |
| CG1RF | 6 | |
| E. coli | CC160 | C. Manoil |
| CC245 | C. Manoil | |
| CC118 | C. Manoil | |
| Phage | λTn_lacZ_/in | C. Manoil |
| λTn_phoA_/in | C. Manoil | |
| Plasmids | ||
| pWM402 | E. coli, E. faecalis shuttle vector | 37 |
| pGEX-4T | Expression–glutathione _S_-transferase fusion vector from the tac promoter | Pharmacia Biotech |
| pTRKH2 | Shuttle vector | 24 |
| pINY1801 | pCF10 positive control region from prgX through prgC | 5 |
| pMSP3601 | _Kpn_I-_Eco_RI fragment of pTRKH2 blunt ended and inserted into _Ban_II site of pINY1801 | This study |
| pMSP3602 | _Bam_HI sites of pMSP3601 filled, first mutagenesis target | This study |
| pMSP3603.1 | _Bsr_GI-_Btr_I PCR cloned into pGEXT-Easy | This study |
| pMSP3604 | _Bsp_HI-_Nco_I fragment of pMSP3603.1 inserted into _Xho_I-_Nco_I sites of pGEX-4T, making signal sequence deficient target | This study |
| pMSP7517 | Nisin-inducible prgB | 8 |
| pMSP7517Δ_Msc_I | _Msc_I fragment removed from pMSP7517 | This study |
| pMSP3535 | Nisin-inducible expression vector | 3 |
| pCF175 | Tn_917_ insertion into prgB of pCF10 | 5 |
| prgB insertions in pMSP7517a | ||
| pCWΩ96 | Tn_phoA_/in insertion at base 96 | This study |
| pCWΩ258 | Tn_phoA_/in insertion at base 258 | This study |
| pCWΩ324 | Tn_lacZ_/in insertion at base 324 | This study |
| pCWΩ438 | Tn_lacZ_/in insertion at base 438 | This study |
| pCWΩ468 | Tn_lacZ_/in insertion at base 468 | This study |
| pCWΩ1074 | Tn_lacZ_/in insertion at base 1074 | This study |
| pCWΩ1077 | Tn_lacZ_/in insertion at base 1077 | This study |
| pCWΩ1299 | Tn_lacZ_/in insertion at base 1299 | This study |
| pCWΩ1317 | Tn_lacZ_/in insertion at base 1317 | This study |
| pCWΩ1419 | Tn_lacZ_/in insertion at base 1419 | This study |
| pCWΩ1551 | Tn_lacZ_/in insertion at base 1551 | This study |
| pCWΩ1638 | Tn_lacZ_/in insertion at base 1638 | This study |
| pCWΩ2049 | Tn_lacZ_/in insertion at base 2049 | This study |
| pCWΩ2064 | Tn_lacZ_/in insertion at base 2064 | This study |
| pCWΩ2085 | Tn_lacZ_/in insertion at base 2085 | This study |
| pCWΩ2421 | Tn_lacZ_/in insertion at base 2421 | This study |
| pCWΩ2601 | Tn_lacZ_/in insertion at base 2601 | This study |
| pCWΩ2760 | Reverse in-frame Tn_lacZ_/in insertion at base 2760 | This study |
| pCWΩ2979 | Tn_lacZ_/in insertion at base 2979 | This study |
| pCWΩ3102 | Tn_lacZ_/in insertion at base 3102 | This study |
| pCWΩr3183 | Reverse in-frame Tn_lacZ_/in insertion at base 3183 | This study |
| pCWΩ3414 | Tn_lacZ_/in insertion at base 3414 | This study |
| pCWΩ3599 | Out-of-frame Tn_lacZ_/in insertion at base 3599 resulting in a stop codon and no cell wall anchor | This study |
A lack of N-terminal insertions in prgB led to the hypothesis that these fusions were lethal for E. coli, possibly through a protein hybrid jamming mechanism (32). To overcome this problem, a signal sequence-deficient clone of prgB was generated and was inserted into the vector pGEX-4T to create pMSP3604. pMSP3604 was constructed in two steps. A PCR product from the _Bsr_GI (5′-AGAGATCTACTGATAATGTACAAGC-3′, with a _Bgl_II site)-to-_Btr_I (5′-TAGGCTTAAGAAGCAGTCACGTCTTTCGC-3′, with an _Eco_RI site) sites of prgB in pMSP7517 was generated with BioXact and was cloned into the pGEM-T Easy (Promega) vector to generate pMSP3603.1. No erroneous mutations were found in the PCR product as determined by sequencing. The prgB fragment was removed by _Bgl_II-_Eco_RI digestion and was ligated into the _Bam_HI-_Eco_RI sites of pGEX-4T, creating pMSP3604. The insertions Ω324, Ω438, Ω468, Ω1074, Ω1299, Ω1317, Ω1419, and Ω1551 were generated in pMSP3604. These insertions were moved back into pMSP7517 in the context of wild-type prgB using two approaches. Initially, the _Bsr_GI-_Btr_I fragment containing the insertion was isolated and was exchanged with pMSP7517. However, problems with the _Btr_I restriction enzyme forced some of the insertions to be moved by isolating the _Bsr_GI-_Psh_AI fragment containing the insertion from pMSP3604 and exchanging it with the same fragment of pMSP7517.
Nisin induction, surface extraction, and Western blotting.
For nisin induction, cultures inoculated 1% from an overnight culture were grown for 3 h in Todd-Hewitt broth plus the appropriate antibiotics with gentle shaking. Nisin was added to a final concentration of 25 ng/ml (a stock solution of 10 mg of nisin/ml was made from a 2.5% nisin preparation [Sigma] in distilled water [effective nisin concentration was 250 μg/ml]), and the cultures were incubated for an additional 1.5 h. To overcome the differences in growth rate, the cultures grown for the stability difference seen at 30°C were induced overnight with 25 ng of nisin/ml. A lysozyme surface extract of each induced mutant culture was performed as previously described (7). The lysozyme extraction buffer was slightly modified to include 12.5 mM EDTA and 25 mg of lysozyme/ml for the extraction of the four cultures grown at 30 or 37°C to measure differences in protein stability. The protein concentration of each sample was determined using the bicinchoninic acid Protein Assay Kit (Pierce). An equivalent amount of each sample was electrophoresed on a sodium dodecyl sulfate–7.5% polyacrylamide gel electrophoresis gel and was transferred to a BA 85 nitrocellulose membrane (Schleicher & Schuell). Western blot analysis was performed with an antibody constructed against an N-terminal domain of Asc10 (18) at a dilution of 1/2,500. Detection was performed with the enhanced chemiluminescence protocol (Pierce).
Quantification of aggregation using flow cytometry and spectrophotometry.
Aggregation was quantified by two methods. Nisin-induced cultures were directly diluted 1/5 in phosphate-buffered saline–0.1% Tween 20 and were analyzed on a Becton Dickinson FACScan, and the data were analyzed using CellQuest Version 3.3 software (Becton Dickinson). Identically placed quadrants were used to analyze the percentage of each sample in each quadrant. One milliliter of nisin-induced cultures was also poured into plastic cuvettes and was left stationary for 1 h. The optical density at 600 nm (OD600) of each sample was read on a Beckman DU-70 Spectrophotometer.
Plasmid transfer.
The insertional mutants were induced with nisin as previously described with the exception that no antibiotic was added to the medium. The donor strain, OG1SSp(pCF175), an Asc10− pCF10 derivative, was induced in the same manner except that 25 ng of cCF10/ml was added instead of nisin. The recipient strains expressed either wild-type Asc10 or one of the insertion mutants, as AS can increase plasmid transfer when expressed on either the donor or recipient cell. The induced donor and recipient cultures were mixed at a ratio of 1:10 respectively and were incubated at 37°C for 30 min. Transconjugants were enumerated by serial dilution on Todd-Hewitt broth with rifampin and tetracycline.
Hydrophobicity assay.
Cell surface hydrophobicity was measured using a hexadecane extraction of induced cultures as previously described (28). Hydrophobicity is expressed as the percentage of cells that are extracted to the hexadecane as measured by OD.
Statistical analysis.
Statistical significance was calculated by determining confidence intervals for the differences of two population means when population variances are known (12).
RESULTS
Construction of PrgB insertion mutants.
To scan Asc10 for potential functional domains, the transposons Tn_lacZ_/in and Tn_phoA_/in were used to generate in-frame, nonpolar 31-amino-acid insertion mutations throughout prgB (15, 16). Previous work with the well-defined LacI repressor protein using these transposons identified the important functional motifs, validating their use in structure-function analysis of less-well-defined proteins (22). Briefly, prgB, carried on a shuttle plasmid, was targeted by Tn_lacZ_/in or Tn_phoA_/in in E. coli strain CC160. Relevant insertions were isolated and sequenced. The bulk of the transposon was then removed from prgB by _Bam_HI restriction digestion and religation, leaving a 31-amino-acid in-frame insertion. The 31-amino-acid insertion consists of 84 bp provided by the transposon and 9 bp derived from the duplicated target sequence. The prgB insertions were electroporated into E. faecalis and were analyzed for surface expression, loss of function of aggregation and of conjugation, and increase in cell surface hydrophobicity. The designation of the insertion mutations generated in this study indicates the nucleotide residue of prgB that immediately precedes the insertion junction (Table 1). A number of different plasmids were used to construct the insertion mutants (see Materials and Methods for rationale), but all of the functional analysis was performed with the insertions in the prgB gene of the nisin-inducible construct pMSP7517, allowing for controlled expression of the mutations.
Analysis of surface localization of Asc10 mutants.
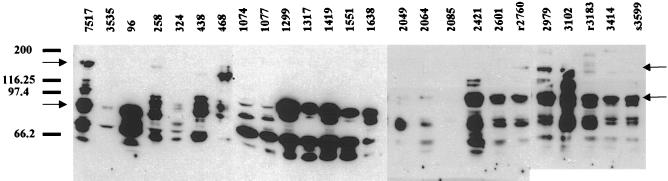
The stability of the mutant proteins was addressed by analyzing their surface expression in E. faecalis. Surface expression of the Asc10 insertion mutations in E. faecalis was analyzed by generating cell wall extracts of each mutant (7). An equivalent amount of total protein (measured by a bicinchoninic acid protein assay [Pierce Chemical] and silver staining) was electrophoresed on a Sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel and was Western blotted with a polyclonal antibody generated against the first 333 amino acids of Asc10 (18) (Fig. 2). When isolated from the cell wall, wild-type AS becomes very unstable and usually forms a laddering pattern on a Western blot. Most of the surface protein extracts from the insertion mutants reacted well in a Western blot, indicating normal cell wall localization. Only three mutants, Ω324, Ω2049, and Ω2064, had reduced levels of reactive protein. One mutant, Ω2085, had no reactive protein on the cell surface and was removed from the functional analysis. Surprisingly, Ωs3599, which has no cell wall anchor, also had reactive protein on the cell surface. Likely, anchorless Asc10 released from the cell immediately binds the AS receptor, enterococcal binding substance, on the cell surface. A Western blot of Ωs3599 expressed in the enterococcal strain INY3000 (negative for enterococcal binding substance) (34) had no reactive protein on the surface of the cell (data not shown). The mutant proteins also have different laddering patterns on the Western blot, suggesting differences in protein stability on the surface of the cell or during the extraction procedure. When different preparations of the same mutant protein were examined on different blots, they exhibited variable laddering patterns, making it difficult to draw conclusions about the mutant stability from the laddering patterns.
FIG. 2.
Western blot analysis of surface extracts from the Asc10 insertion mutants. The Western blot utilized a polyclonal antibody generated against the N terminus of Asc10. An equivalent amount of protein was added to each lane. The laddering pattern is typical of AS protein preparations, as they have high instability. Migration of molecular mass standard marker proteins is shown at the left of the blot, and the 137-kDa full-length Asc10 and 78-kDa fragment are indicated by arrows. 7517, Asc10+; 3535, vector control.
Identification of two domains that mediate aggregation.
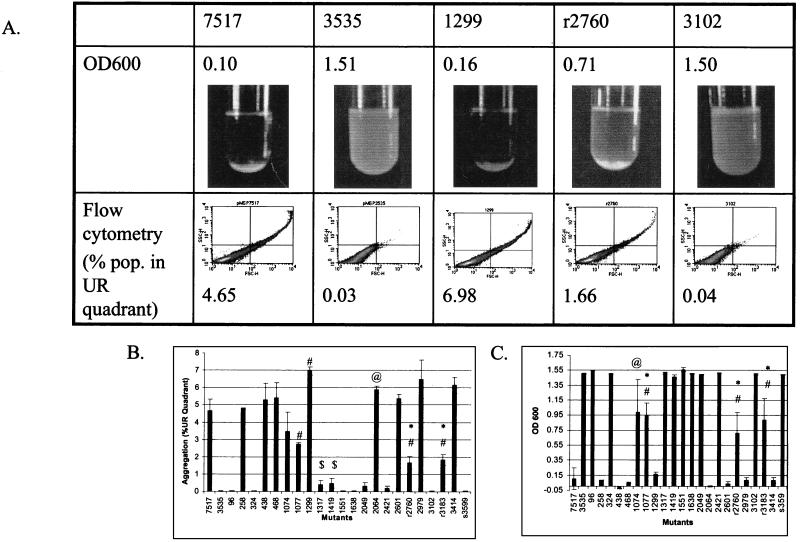
Aggregation of the insertion mutants was quantified by two methods. Figure 3A shows representative data from five mutants. First, the forward scatter and side scatter profiles of induced cultures were determined on a flow cytometer. Larger particles have larger forward scatter and side scatter profiles. Populations were separated into four quadrants (Fig. 3A, bottom). The vector control (3535) had very few events located in the upper right quadrant (0.03%), while a much larger percentage of the wild-type Asc10 nisin-induced (7517) population was located in the upper right quadrant (4.65%) (Fig. 3A and B). The profile of each mutants was determined two to three times (Fig. 3B). Many mutations throughout the gene resulted in complete inhibition of aggregation, while others maintained wild-type aggregation levels. Some mutants, Ω1077, Ωr2760, and Ωr3183, had intermediate levels of aggregation. Ω1317 and Ω1419 had very low but statistically significant levels of aggregation. Interestingly, Ω1299, Ω2064, and Ω3414 had statistically significant, increased levels of aggregation relative to wild-type Asc10. As expected, Ωs3599 was unable to aggregate.
FIG. 3.
Aggregation of the mutants was measured by spectrophotometry and flow cytometry. Representative data for Asc10+ (7517), the vector control (3535), a functional mutant (1299), intermediate mutant (r2760), and a nonaggregating mutant (3102) are shown. UR, upper right. (A). The aggregation of the mutants was measured by flow cytometry as a percentage of the induced populations in the (UR) quadrant (B). The OD600 of induced cultures after 1 h of settling was also determined (C). A decrease in OD600 indicates aggregation. #, P < 0.05 from 7517; @, P < 0.1 from 7517; ∗, P < 0.05 from 3535; $, P < 0.1 from 3535.
Aggregation was also quantified by determination of the OD600 of induced cultures after 1 h of settling (Fig. 3A, top, and C). For these data, increased aggregation is indicated by a decreased OD. Data obtained in this manner generally agreed with the results generated on the flow cytometer. In this method, no significant differences could be distinguished between cells expressing wild-type Asc10 and cells expressing mutant proteins that were functional aggregators. Likewise, no statistical difference was found between vector control 3535 and the nonaggregators Ω1317 and Ω1419. However, mutants displaying an intermediate level of aggregation when measured on the flow cytometer (Ω1074, Ω1077, Ωr2760, and Ωr3183) were also intermediate when measured by the spectrophotometer. Comparison of the two methods suggests that flow cytometry is more sensitive in quantifying small differences in aggregation levels.
The data measured by these two methods indicated a distinct aggregation domain expressed in the gene from nucleotide residues 1317 to 1638 (corresponding to amino acids 439 to 663 of the Asc10 protein). Also, many insertions in the C terminus (Ω2421, Ωr2760, Ω3102, and Ωr3183) had reduced or abolished aggregation, indicating regions in the C terminus that are involved in aggregation. However, two C-terminal functional aggregators (Ω2601 and Ω2979) were interspersed among the nonaggregators, suggesting that the entire region does not participate in aggregation.
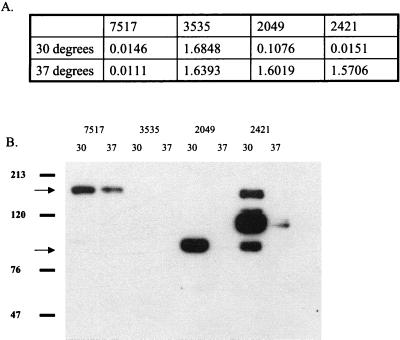
Temperature-sensitive stability of Asc10 from Ω2049 and Ω2421.
Growth of Ω2049 and Ω2421 at 30°C resulted in increased aggregation to near-wild-type levels (Fig. 4A). Analysis of surface extracts of these strains by Western blotting revealed that much higher levels of Asc10 were isolated from cells grown at 30°C. Note that increased exposure times would show reactive protein in the 37°C extract of 2421, confirming the presence of reactive protein seen in Fig. 2. Interestingly, more wild-type Asc10 can be seen from cultures grown at 30°C as well (Fig. 4B). These data suggest that the failure of Ω2049 and Ω2421 to aggregate at 37°C was likely due to instability of the mutant proteins rather than to disruption of an aggregation functional domain. The mechanism of this increased stability at 30°C is unknown. Growth at 30°C did not affect the aggregation phenotype of any of the other insertion mutants.
FIG. 4.
Insertion mutants 2049 and 2421 have near-wild-type aggregation levels when grown at 30°C as indicated by spectrophotometry (A). Western blot analysis of surface extracts indicated much more reactive protein on these cells when they were grown at 30°C (B). Migration of molecular mass standard marker proteins is shown to the left of the blot, and the 137-kDa full-length and 78-kDa Asc10 fragments are indicated by arrows.
Plasmid transfer of insertion mutants correlates with aggregation.
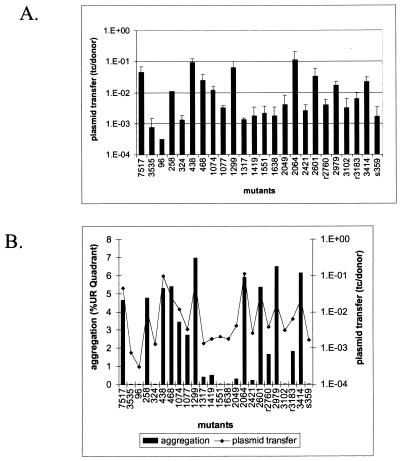
To determine the plasmid transfer capability of the insertion mutants, nisin-induced mutant cultures were used as recipients and were mixed with an E. faecalis OG1SSp(pCF175) donor strain. pCF175 is a pCF10 derivative that has a Tn_917_ insertion in the prgB gene, rendering it incapable of aggregation or efficient conjugation. Thus, aggregation could only be mediated by the recipient Asc10 mutants, as expression of AS can lead to increased conjugation when expressed on either the donor or recipient cell of the mating pair (26). Plasmid transfer is expressed as the number of transconjugant cells/donor cell. As expected, induced 7517 gave high transfer levels at 3.5 × 10−2, while the vector control transferred at 6.5 × 10−4. Various transfer levels were observed for the mutants (Fig. 5A), and as expected, transfer levels correlated with aggregation ability (Fig. 5B). Interestingly, the three mutants that had significantly higher aggregation levels as measured by flow cytometry, Ω1299, Ω2064, and Ω3414, did not have statistically significantly higher transfer levels.
FIG. 5.
The plasmid transfer levels of each mutant were determined and were expressed as transconjugants/donor (tc/donor) (A). Plasmid transfer levels are compared with the ability to aggregate (B). UR, upper right.
Hydrophobicity.
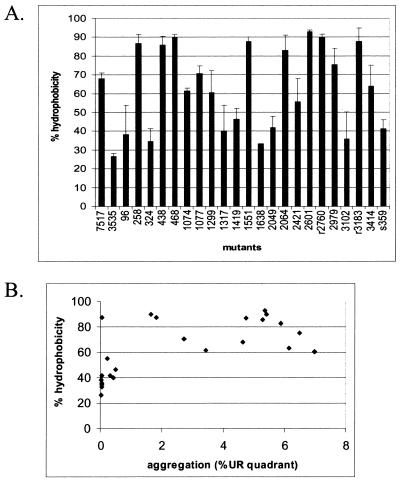
Cell surface hydrophobicity of the insertion mutants was measured by determining the percentage of cells that could be extracted from aqueous solution into hexadecane (Fig. 6A). A number of mutants maintained high levels of cell surface hydrophobicity, with some mutants having an even higher percentage of cell surface hydrophobicity than wild-type Asc10. Likewise, many mutants showed low levels of surface hydrophobicity comparable to those of non-Asc10-expressing strains. The amino acid sequences generated by both the forward and reverse insertions are relatively hydrophilic, with a percentage of polar amino acids at 63 and 50%, respectively. However, the actual amino acids of the insertion seemed to have little effect on the overall hydrophobicity of the mutants. The hydrophilicity, surface probability, and antigenicity indices were calculated for the amino acid sequence of Asc10 using the program PEPTIDESTRUCTURE (Wisconsin Package Version 10.1; Genetic Computer Group [GCG], Madison, Wis.). The values for a window of 10 amino acids adjacent to the insertion mutations were not predictive of the effect on cell surface hydrophobicity (data not shown).
FIG. 6.
Cell surface hydrophobicity of each mutant as a percentage of cells that were extracted with hexadecane (A). Comparison of hydrophobicity with aggregation for each mutant can be seen in the scatter plot (B). UR, upper right.
The effect of cell surface hydrophobicity on aggregation was examined by plotting aggregation ability on the x axis versus the percent hydrophobicity on the y axis (Fig. 6B). No strong correlation between cell surface hydrophobicity and aggregation levels was observed.
DISCUSSION
The functional analysis of the 23 prgB insertion mutants generated in this study led to four important conclusions: (i) the domain that mediates bacterial aggregation extends into the variable region of Asc10, farther into the amino terminus than previously reported (13), (ii) the C terminus of the protein does contribute to aggregation, (iii) efficient conjugation directly correlates with functional aggregation, and (iv) increased cell surface hydrophobicity caused by Asc10 is not sufficient to mediate aggregation.
The transposons Tn_lacZ_/in and Tn_phoA_/in have been successfully used to analyze the structure-function relationship of a number of gram-negative membrane and cytosolic proteins (14, 15, 22, 23) and even a mouse mammary tumor virus superantigen (19), but to our knowledge, this study is the first attempt to use these transposons for mutagenesis of a protein from a gram-positive organism. Although we had problems with E. coli transcription machinery recognizing artifactual promoters and inefficient export of the fusion proteins, 23 nonpolar, in-frame 31-amino-acid insertions were generated that were spaced throughout the prgB gene. The insertion mutants have good coverage of the protein, with the largest gap found from nucleotide residues 468 to 1074. This region was resistant to transposition, as multiple mutagenesis attempts yielded no insertions. Interestingly, previous Tn_5_ mutagenesis studies that targeted pCF10 DNA also had a large gap in the prgB gene that corresponds to the same transposition-resistant region identified in this study (26).
Western blotting of the surface extracts of induced mutant cultures revealed that most proteins were expressed on the surface of the cell. Full-length protein was difficult to observe in some mutants, but this result is not unexpected, as purified protein is very unstable. Three mutants, Ω324, Ω2049, and Ω2064, had reduced expression of Asc10 at 37°C, while Ω2085 showed no reactive protein. Consequently, Ω2085 was removed from the functional analysis. Interestingly, the insertion mutants Ω2049 and Ω2421 were found to aggregate to near-wild-type levels when grown at 30°C, suggesting that their lack of aggregation at 37°C is due to protein instability. The increased stability of these proteins at 30°C may be due to alterations in folding conformations or differences in activity of an enterococcal cell surface protease.
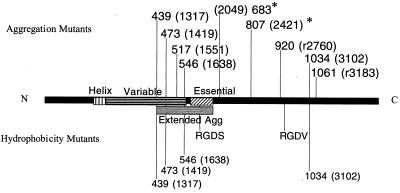
Insertions in two major regions of the protein inhibited or abolished aggregation. Insertions in the N terminus inhibited aggregation in agreement with Muscholl-Silberhorn's previous identification of an aggregation functional domain for Asa1 (20). The insertions generated in this study identify an aggregation functional domain in the gene from nucleotide residues 1317 to 1638, but it is not clear how far the functional domain extends beyond residue 1638. However, as Ω2049 aggregates at 30°C, the aggregation domain does not extend this far into the gene. The aggregation domain identified in Asa1 extends from base numbers 1704 to 1980. Combining the two identified regions would indicate an aggregation domain from 1317 to 1980 (Fig. 7). Also, a comparison of the two results indicates that the aggregation functional domain for Asc10 extends farther into the amino terminus of the protein than previously reported for Asa1. Interestingly, the domain identified in this study extends into the variable region, suggesting that it may play a role in aggregation. This result is supported by the observation that an Asa1-AspI variable region chimera fails to aggregate (20). Most of the insertions in the extreme N terminus of the protein, residues 258 to 1299, had no effect on aggregation levels with the exception of Ω96 and Ω324. Mutation Ω96 was in the signal sequence of the protein and likely disrupted correct migration through the cell membrane, while Ω324 had very low protein levels on the cell surface, suggesting decreased stability of the protein.
FIG. 7.
Functional domains identified in this study that disrupted aggregation (top) and hydrophobicity (bottom). All proteins generated from these insertions were expressed on the cell surface. The two mutants with increased stability at 30°C are indicated (∗). Amino acids preceding the insertions indicate the mutants (nucleotide residues are given in parentheses). The Extended Agg domain was identified by the mutations in this paper and by the previously identified aggregation functional domain (21). Note that the N-terminal aggregation domain extends into the variable region. C-terminal insertions that disrupt aggregation are hypothesized to play a structural role. Many mutants that are unable to aggregate still increase cell surface hydrophobicity.
Surprisingly, some C-terminal insertions had reduced or abolished aggregation. Specifically, the protein produced by insertions Ωr2760, Ω3102, and Ωr3183 were expressed well on the cell surface. These mutants also reacted with an anti-Asc10 monoclonal antibody as measured by fluorescence-activated cell sorter analysis (data not shown), further indicating reactive protein on the cell surface. Strikingly, the insertion at Ω3102 was completely deficient in aggregation. In contrast, Muscholl-Silberhorn (20) concluded that the C terminus of Asa1 is not essential in aggregation based on two observations: (i) a C-terminal deletion construct maintains aggregation, although at a third of wild-type levels, and (ii) an N-terminal fragment of the protein attached to a solid surface can mediate cell attachment. Based on (i) the data reported in this paper, (ii) the preceding observations with Asa1, and (iii) the fact that the N terminus is more surface exposed (9), it is likely that the C terminus plays a structural role in aggregation. Specifically, the regions identified by the insertions Ωr2760, Ω3102, and Ωr3183 serve to place the N-terminal functional domain in the correct conformation to mediate aggregation. This hypothesis could account for the decreased aggregation seen in the C-terminal deletion of Asa1. Moreover, the structural function of this C-terminal domain may not be necessary when the protein is attached to a solid surface, as observed by Muscholl-Silberhorn (20). Note that the fully functional insertion Ω2979 intersects this structural region, indicating that some sites are permissive. It should be noted that the use of these transposons in the analysis of structure-function relationships has been validated by previous work with the LacI repressor (22). In this study, all 18 31-codon insertions generated by random transposition of Tn_lacZ_/in yielded the predicted phenotypes based on previous structural analysis of the protein.
As expected, the ability to aggregate positively correlated with plasmid transfer levels, further supporting the notion that the only role of AS in conjugation is to initiate close contact between the members of the donor-recipient pair. Interestingly, the mutants that had statistically significant higher levels of aggregation as measured by flow cytometry did not have higher levels of plasmid transfer, suggesting that AS has evolved aggregation levels that maximize plasmid transfer.
The increased cell surface hydrophobicity elicited by Asc10 expression was also measured for each mutant. Various levels of hydrophobicity could be seen for the insertion mutants, but no one distinct domain necessary for increasing hydrophobicity was detected. Comparison of the hydrophobicity and aggregation levels reveals that increased cell surface hydrophobicity is not sufficient for aggregation. Specifically, Ω1551 is unable to aggregate but induces very high levels of cell surface hydrophobicity. Likewise, mutants that are intermediate aggregators have hydrophobicity levels equivalent to those of strong aggregators. Thus, AS mediates aggregation through a specific interaction, as opposed to a generalized alteration of cell surface hydrophobicity. It should be noted that no mutants that were completely deficient in increased cell surface hydrophobicity were able to aggregate, possibly indicating that hydrophobicity may be necessary but not sufficient for aggregation.
This study reports the construction of a library of 23 31-amino-acid insertions in Asc10, the AS of pCF10. By using Western blotting of surface extracts, it was shown that most of the insertion mutants were expressed on the surface of E. faecalis. Analysis of aggregation identified both N- and C-terminal domains important in aggregation and showed that the variable region may play a role in aggregation. Moreover, an increase in cell surface hydrophobicity is not sufficient for aggregation. This insertion library will be further analyzed for functional domains of Asc10 involved in the pathogenesis of E. faecalis infections.
ACKNOWLEDGMENTS
We thank Colin Manoil for supplying the transposons used in this study and for helpful advice. We thank Pat Cleary for reading the manuscript and Pat Schlievert, Helmut Hirt, and John McCormick for providing antibodies, other reagents, and helpful discussion.
This work was supported by NIH grants GM-49530 and HL-51987. C.M.W. was supported by NIH training grant 5 T32 AI07421-5.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1990. [Google Scholar]
- 2.Bae T, Clerc-Bardin S, Dunny G M. Analysis of expression of prgX, a key negative regulator of the transfer of the Enterococcus faecalis pheromone-inducible plasmid pCF10. J Mol Biol. 2000;297:861–875. doi: 10.1006/jmbi.2000.3628. [DOI] [PubMed] [Google Scholar]
- 3.Bryan E M, Bae T, Kleerebezem H, Dunny G M. Improved vectors for nisin-controlled expression in gram-positive bacteria. Plasmid. 2000;44:183–190. doi: 10.1006/plas.2000.1484. [DOI] [PubMed] [Google Scholar]
- 4.Chow J W, Thal L A, Perri M B, Vazquez J A, Donabedian S M, Clewell D B. Plasmid-associated hemolysin and aggregation substance production contribute to virulence in experimental enterococcal endocarditis. Antimicrob Agents Chemother. 1993;37:2474–2477. doi: 10.1128/aac.37.11.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christie P J, Dunny G M. Identification of regions of the Streptococcus faecalis plasmid pCF-10 that encode antibiotic resistance and pheromone response functions. Plasmid. 1986;15:230–241. doi: 10.1016/0147-619x(86)90041-7. [DOI] [PubMed] [Google Scholar]
- 6.Dunny G M, Brown B L, Clewell D B. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc Natl Acad Sci USA. 1978;75:3479–3483. doi: 10.1073/pnas.75.7.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galli D, Lottspeich F, Wirth R. Sequence analysis of Enterococcus faecalis aggregation substance encoded by the sex pheromone plasmid pAD1. Mol Microbiol. 1990;4:895–904. doi: 10.1111/j.1365-2958.1990.tb00662.x. [DOI] [PubMed] [Google Scholar]
- 8.Hirt H, Erlandsen S L, Dunny G M. Heterologous inducible expression of Enterococcus faecalis pCF10 aggregation substance Asc10 in Lactococcus lactis and Streptococcus gordonii contributes to cell hydrophobicity and adhesion to fibrin. J Bacteriol. 2000;182:2299–2306. doi: 10.1128/jb.182.8.2299-2306.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirt H, Wanner G, Galli D, Wirth R. Biochemical, immunological and ultrastructural characterization of aggregation substances encoded by Enterococcus faecalis sex-pheromone plasmids. Eur J Biochem. 1993;211:711–716. doi: 10.1111/j.1432-1033.1993.tb17600.x. [DOI] [PubMed] [Google Scholar]
- 10.Huycke M M, Sahm D F, Gilmore M S. Multiple-drug resistant enterococci: the nature of the problem and an agenda for the future. Emerg Infect Dis. 1998;4:239–249. doi: 10.3201/eid0402.980211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Isenmann R, Schwarz M, Rozdzinski E, Marre R, Beger H G. Aggregation substance promotes colonic mucosal invasion of Enterococcus faecalis in an ex vivo model. J Surg Res. 2000;89:132–138. doi: 10.1006/jsre.1999.5813. [DOI] [PubMed] [Google Scholar]
- 12.Khazanie R. Statistics in a world of applications, p 460. New York, N.Y: HarperCollins Publishers, Inc.; 1996. [Google Scholar]
- 13.Kreft B, Marre R, Schramm U, Wirth R. Aggregation substance of Enterococcus faecalis mediates adhesion to cultured renal tubular cells. Infect Immun. 1992;60:25–30. doi: 10.1128/iai.60.1.25-30.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee M H, Kosuk N, Bailey J, Traxler B, Manoil C. Analysis of F factor TraD membrane topology by use of gene fusions and trypsin-sensitive insertions. J Bacteriol. 1999;181:6108–6113. doi: 10.1128/jb.181.19.6108-6113.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manoil C, Bailey J. A simple screen for permissive sites in proteins: analysis of Escherichia coli lac permease. J Mol Biol. 1997;267:250–263. doi: 10.1006/jmbi.1996.0881. [DOI] [PubMed] [Google Scholar]
- 16.Manoil C, Traxler B. Insertion of in-frame sequence tags into proteins using transposons. Methods. 2001;20:55–61. doi: 10.1006/meth.1999.0905. [DOI] [PubMed] [Google Scholar]
- 17.McCormick J, Hirt H, Dunny G M, Schlievert P M. Pathogenic mechanisms of enterococcal endocarditis. Curr Infect Dis Rep. 2000;2:315–321. doi: 10.1007/s11908-000-0009-9. [DOI] [PubMed] [Google Scholar]
- 18.McCormick J K, Hirt H, Waters C M, Tripp T J, Dunny G M, Schlievert P M. Antibodies to a surface-exposed, N-terminal domain of aggregation substance are not protective in the rabbit model of Enterococcus faecalis infective endocarditis. Infect Immun. 2001;69:3305–3314. doi: 10.1128/IAI.69.5.3305-3314.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McMahon C W, Traxler B, Grigg M E, Pullen A M. Transposon-mediated random insertions and site-directed mutagenesis prevent the trafficking of a mouse mammary tumor virus superantigen. Virology. 1998;243:354–365. doi: 10.1006/viro.1998.9071. [DOI] [PubMed] [Google Scholar]
- 20.Muscholl-Silberhorn A. Analysis of the clumping-mediating domain(s) of sex pheromone plasmid pAD1-encoded aggregation substance. Eur J Biochem. 1998;258:515–520. doi: 10.1046/j.1432-1327.1998.2580515.x. [DOI] [PubMed] [Google Scholar]
- 21.Muscholl-Silberhorn A. Cloning and functional analysis of Asa373, a novel adhesin unrelated to the other sex pheromone plasmid-encoded aggregation substances of Enterococcus faecalis. Mol Microbiol. 1999;34:620–630. doi: 10.1046/j.1365-2958.1999.01631.x. [DOI] [PubMed] [Google Scholar]
- 22.Nelson B D, Manoil C, Traxler B. Insertion mutagenesis of the lac repressor and its implications for structure-function analysis. J Bacteriol. 1997;179:3721–3728. doi: 10.1128/jb.179.11.3721-3728.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nelson B D, Traxler B. Exploring the role of integral membrane proteins in ATP-binding cassette transporters: analysis of a collection of MalG insertion mutants. J Bacteriol. 1998;180:2507–2514. doi: 10.1128/jb.180.9.2507-2514.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Sullivan D J, Klaenhammer T R. High- and low-copy-number Lactococcus shuttle cloning vectors with features for clone screening. Gene. 1993;137:227–231. doi: 10.1016/0378-1119(93)90011-q. [DOI] [PubMed] [Google Scholar]
- 25.Olmsted S B, Dunny G M, Erlandsen S L, Wells C L. A plasmid-encoded surface protein on Enterococcus faecalis augments its internalization by cultured intestinal epithelial cells. J Infect Dis. 1994;170:1549–1556. doi: 10.1093/infdis/170.6.1549. [DOI] [PubMed] [Google Scholar]
- 26.Olmsted S B, Kao S M, van Putte L J, Gallo J C, Dunny G M. Role of the pheromone-inducible surface protein Asc10 in mating aggregate formation and conjugal transfer of the Enterococcus faecalis plasmid pCF10. J Bacteriol. 1991;173:7665–7672. doi: 10.1128/jb.173.23.7665-7672.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rakita R M, Vanek N N, Jacques-Palaz K, Mee M, Mariscalco M M, Dunny G M S, Van Winkle W B, Simon S I. Enterococcus faecalis bearing aggregation substance is resistant to killing by human neutrophils despite phagocytosis and neutrophil activation. Infect Immun. 1999;67:6067–6075. doi: 10.1128/iai.67.11.6067-6075.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosenberg M, Gutnick D, Rosenberg E. Adherence of bacteria to hydrocarbons: a Simple method for measuring cell-surface hydrophobicity. FEMS Microbiol Lett. 1980;9:29–33. [Google Scholar]
- 29.Ruoslahti E. RGD and other recognition sequences for integrins. Annu Rev Cell Dev Biol. 1996;12:697–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- 30.Sartingen S, Rozdzinski E, Muscholl-Silberhorn A, Marre R. Aggregation substance increases adherence and internalization, but not translocation, of Enterococcus faecalis through different intestinal epithelial cells in vitro. Infect Immun. 2001;68:6044–6047. doi: 10.1128/iai.68.10.6044-6047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlievert P M, Gahr P J, Assimacopoulos A P, Dinges M M, Stoehr J A, Hirt H, Dunny G M. Aggregation and binding substances enhance pathogenicity in rabbit models of Enterococcus faecalis endocarditis. Infect Immun. 1998;66:218–223. doi: 10.1128/iai.66.1.218-223.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Snyder W B, Silhavy T J. β-Galactosidase is inactivated by intermolecular disulfide bonds and is toxic when secreted to the periplasm of Escherichia coli. J Bacteriol. 1995;177:953–963. doi: 10.1128/jb.177.4.953-963.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sussmuth S D, Muscholl-Silberhorn A, Wirth R, Susa M, Marre R. Aggregation substance promotes adherence, phagocytosis, and intracellular survival of Enterococcus faecalis within human macrophages and suppresses respiratory burst. Infect Immun. 2000;68:4900–4906. doi: 10.1128/iai.68.9.4900-4906.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trotter K M, Dunny G M. Mutants of Enterococcus faecalis deficient as recipients in mating with donors carrying pheromone-inducible plasmids. Plasmid. 1990;24:57–67. doi: 10.1016/0147-619x(90)90025-8. [DOI] [PubMed] [Google Scholar]
- 35.Vanek N N, Simon S I, Jacques-Palaz K, Mariscalco M M, Dunny G M, Rakita R M. Enterococcus faecalis aggregation substance promotes opsonin-independent binding to human neutrophils via a complement receptor type 3-mediated mechanism. FEMS Immunol Med Microbiol. 1999;26:49–60. doi: 10.1111/j.1574-695X.1999.tb01371.x. [DOI] [PubMed] [Google Scholar]
- 36.Wirth R. The sex pheromone system of Enterococcus faecalis. More than just a plasmid-collection mechanism? Eur J Biochem. 1994;222:235–246. doi: 10.1111/j.1432-1033.1994.tb18862.x. [DOI] [PubMed] [Google Scholar]
- 37.Wirth R, An F Y, Clewell D B. Highly efficient protoplast transformation system for Streptococcus faecalis and a new Escherichia coli-S. faecalis shuttle vector. J Bacteriol. 1986;165:831–836. doi: 10.1128/jb.165.3.831-836.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]