Use of Recombinant Envelope Proteins for Serological Diagnosis of Dengue Virus Infection in an Immunochromatographic Assay (original) (raw)
Abstract
An immunochromatographic test that incorporates recombinant antigens (Dengue Duo Rapid Strip Test; PanBio, Brisbane, Australia) has recently become commercially available. This assay is performed in 15 min and detects both immunoglobulin M (IgM) and IgG in a capture format. The four recombinant proteins used represent the N-terminal 80% of the viral envelope glycoproteins of dengue viruses 1, 2, 3, and 4, respectively. The sensitivity and specificity of the recombinant-antigen-based assay were 90 and 86%, respectively. The similar diagnostic performance of these antigens to that of enzyme-linked immunosorbent assays using whole dengue virus suggests that they mimic whole dengue viruses in primary structure and epitope conformation. These results suggest that recombinant proteins can be used in diagnostic assays for dengue to overcome safety issues associated with the use of whole virus.
Dengue is one of the most important human arboviral infections in terms of morbidity and mortality (14). Effective and rapid diagnosis can contribute to the control of dengue and dengue hemorrhagic fever (DHF) through more accurate public health notification. Rapid laboratory diagnosis can confirm clinical suspicion of dengue to guide further treatment and prognosis. These outcomes help to mitigate the health care costs and allow early public health intervention to prevent the spread of dengue virus infection.
Numerous commercial assays are available for the diagnosis of dengue virus infection utilizing several different formats. The most common format is the enzyme-linked immunosorbent assay (ELISA), which offers a relatively quick and easy method for diagnosing dengue. Some of these assays have the additional ability to differentiate between primary and secondary infections (22, 27, 31, 38). Other formats available include the dot blot (4), the dipstick (41), and the immune fluorescent antibody assay (Progen Biotechnika GmbH, Heidelberg, Germany). Recently, lateral flow tests for the diagnosis of dengue have also become available. These offer the advantage of speed and have the potential to be used in field settings (21, 22, 32, 37, 41).
Reported methods for diagnosis of dengue virus infection commonly use whole virus (3, 26) or crude extracts (1, 19, 35) for the target antigen. The use of such materials presents a potential health hazard through exposure to infectious virus particles. In addition, production costs associated with the cultivation of virus for live or inactivated viral antigen are generally high. As a result, utilization of recombinant proteins, which can be produced more cheaply and present little or no health hazard, are an attractive alternative. This paper will discuss a new immunochromatographic test (PanBio Dengue Duo Rapid Strip Test; catalog no. DEN-25S) that utilizes recombinant dengue envelope proteins for the laboratory diagnosis and differentiation of primary and secondary dengue virus infections.
The PanBio rapid test relies on both dengue immunoglobulin M (IgM) and IgG detection to diagnose active dengue virus infection. The IgG test line is set to detect the high levels of IgG characteristic of secondary virus infection and hence is able to distinguish secondary from primary dengue and past infections. The IgM test line is set to detect IgM levels characteristically present in primary dengue virus infections and in the majority of secondary dengue virus infections. In this study, the performance of the PanBio rapid test was compared to that of the U.S. Armed Forces Research Institute of Medical Sciences (AFRIMS; Bangkok, Thailand) reference ELISA using sera taken from patients with primary and secondary dengue virus infections, Japanese encephalitis (JE) virus infection, and nonflavivirus infections.
MATERIALS AND METHODS
Case definitions for dengue.
In children experiencing a febrile illness consistent with dengue fever or DHF, dengue virus infections were defined as the isolation or identification of a dengue virus, the detection of IgM to dengue virus, or a sustained elevation (≥1:2,560) or a fourfold rise in dengue virus antibody between acute- and convalescent-phase sera as detected by hemagglutination inhibition (HAI) antibody titer. Dengue virus infection was categorized as primary or secondary according to the World Health Organization criteria (a titer of ≥1:2,560 in a single serum was taken as secondary dengue) (40) and the standard operating procedure for the reference ELISA (1).
Serological definitions.
The following serological definitions were used for the diagnosis of dengue virus infection across all age groups represented in this study. All acute- and convalescent-phase specimens were defined as sera collected over time and separated by more than 3 days.
(i) No evidence of recent dengue virus infection.
No evidence of recent dengue virus infection was defined as no detectable dengue virus antibody in acute- and convalescent-phase sera collected 5 to 7 days apart by ELISA or HAI assay or as stable antibody titers by HAI assay with titers of less than 1:2,560 to all dengue virus antigens without a fourfold rise.
(ii) Recent dengue virus infection.
Recent dengue virus infection was defined as antibody against dengue virus antigens detected by HAI titers greater than or equal to 1:2,560 in at least one specimen without a fourfold rise between acute- and convalescent-phase specimens.
(iii) Acute dengue virus infection.
Acute dengue virus infection was defined as the isolation of dengue virus or identification of a dengue virus genome by reverse transcription-PCR from serum or plasma during an acute febrile illness consistent with dengue fever or DHF and a dengue virus-specific IgM level of 40 U or more by IgM capture ELISA or a fourfold rise in HAI antibody against any dengue virus serotype between the acute- and convalescent-phase specimens.
(iv) Acute primary or secondary dengue.
Primary dengue virus infection was defined as an acute dengue virus infection with an IgM-to-IgG ratio of 1.8 or greater by IgM capture ELISA in acute- or convalescent-phase specimens taken at least 7 days apart. A ratio of less than 1.8 was defined as an acute secondary dengue virus infection. Primary dengue by HAI assay was defined as an acute dengue virus infection with a fourfold or greater change in HAI titer to one or more dengue serotypes in acute- and convalescent-phase specimens (taken at least 7 days apart) with no titer greater than 1:1,280. Acute secondary dengue virus infection was defined as a fourfold or greater change in HAI titer to two or more dengue serotypes in acute- and convalescent-phase specimens with titers greater than 1:2,560.
(v) Acute JE virus infection.
Acute JE virus infection was defined by using an ELISA specific for JE IgM as described previously (2). A binding index is calculated as follows: (optical density [OD] of the test sample − OD of the negative control)/(OD of the weak positive control − OD of the negative control). The binding index multiplied by 100 is equal to the number of units, where units of ≥40 are considered a positive test result. A ratio of anti-dengue IgM to anti-JE IgM of ≥1.0 is typical of acute dengue, whereas a ratio of ≤1.0 is consistent with acute JE virus infection.
Serum samples.
In this study, the PanBio rapid test was evaluated using sera collected in Thailand from patients with and without dengue virus infections. The samples were characterized by AFRIMS using their in-house dengue IgM and IgG ELISA and HAI assays. Sera from patients with suspected dengue virus infections were collected at the time of admission to and discharge from the Queen Sirikit National Institute of Child Health (Bangkok Children's Hospital, Bangkok, Thailand) during 1999 and frozen at −70°C prior to being assayed.
Paired sera from 16 patients with secondary dengue and 20 patients with primary dengue virus infections were tested. Multiple samples were tested from one patient with a secondary infection (n = 4) and from two patients with a primary infection (n = 3). Paired sera from 17 patients with suspected dengue but no laboratory evidence of dengue virus infection (NEI) were also tested. A panel of sera from patients with nonflavivirus infections was also included, representing indirect immunoperoxidase-positive cases of rickettsial scrub typhus (paired sera; n = 20), microscopic agglutination test-positive cases of leptospirosis (paired sera; n = 20), and blood smear-positive cases of malaria (n = 15). Sera (n = 9) and cerebrospinal fluid (n = 9) from patients with laboratory-confirmed JE virus infections were also tested to determine cross-reactivity between these two flaviviruses.
Recombinant proteins.
Recombinant dengue proteins were incorporated into the rapid test, and its performance was determined through clinical evaluation. The recombinant proteins represent the N-terminal 80% of the viral envelope glycoproteins of dengue viruses 1, 2, 3, and 4. The following strains of dengue virus were the sources of the recombinant proteins: dengue virus 1, 258848; dengue virus 2, PR159/S1; dengue virus 3, CH53489; and dengue virus 4, H241. The recombinant 80% E proteins were expressed in Drosophila melanogaster strain Schneider 2 (S2) cells (33). Transfection with the appropriate expression vectors resulted in stable transfectants and the expression and secretion of the 80% E subunits into the culture medium. The recombinant subunits were purified by immunoaffinity chromatography using the monoclonal antibody 4G2 (17). Based on gel electrophoresis, the proteins were greater than 80% pure, had a molecular mass of approximately 43 kDa, and appeared to have no significant differences from whole dengue viruses in primary structure. The ability of the conformationally sensitive monoclonal antibodies, 4G2 and 9D12 (12), to bind to the recombinant 80% E proteins in both immunofluorescence assays and ELISA is an indication that the proteins are folded correctly and hence should mimic whole dengue virus.
PanBio Dengue Duo Rapid Strip Test.
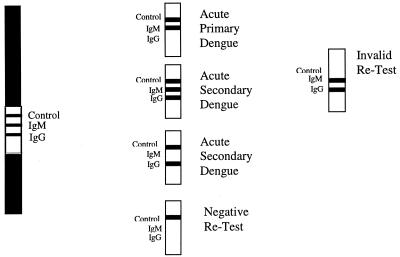
The PanBio strip test (catalog code DEN-25S) was performed according to the manufacturer's instructions. Three drops of the buffer was added to a glass tube. Using the loop provided, 1 μl of serum was added to the tube and gently mixed. A single test strip was then added to the tube, and the results were read after 15 min. Any trace of a pink or purple line in the test area was indicative of a positive result. A primary infection was characterized by a positive IgM and a negative IgG test line. A secondary infection was characterized as a positive IgG test line with or without a positive IgM test line. No visible lines in the test area represented a negative result. This test result could indicate no recent dengue virus infection or that a later convalescent specimen should be tested. A control line was also included to ensure the correct running and validation of the assay (refer to Fig. 1 for interpretation).
FIG. 1.
Interpretation of the Dengue Duo Rapid Strip Test.
HAI assay.
Acetone-extracted sera were tested for HAI antibodies as described previously, with the assay modified to a microtiter plate format (6). Dengue virus types 1 to 4 and JE virus (8 U each) were used. Antigens were produced by sucrose acetone extraction of the brains of suckling mice infected with the following prototype mouse-adapted virus strains: dengue virus 1 Hawaii, dengue virus 2 New Guinea C, dengue virus 3 H-87, dengue virus 4 H-241, and JE virus Nakayama. A fourfold increase was considered positive for acute flavivirus infection. The infection was diagnosed as primary if titers were less than 1:2,560 a week or more after the onset of illness or as secondary infection if antibody titers were greater than or equal to 1:2,560.
AFRIMS ELISA.
The in-house IgM and IgG capture ELISAs were performed as described previously (1). For single specimens, 40 U of IgM to dengue virus (with the amount of dengue IgM greater than that of JE IgM) was considered evidence of an acute dengue virus infection (or for paired samples, a rise from less than 15 U to more than 30 U). A dengue IgM/IgG ratio equal to or greater than 1.8:1 defined a primary dengue virus infection. A ratio less than 1.8:1 defined a secondary dengue virus infection. Using serial specimens, a twofold increase in IgG to dengue with an absolute value of 100 U or greater indicated a secondary flavivirus infection in the absence of 40 U or more of IgM to dengue.
Data analysis.
Fisher's exact test was performed to compare sensitivity and specificity. Spearman's correlation analysis was performed to compare ELISA ratios in individual sera. Analysis of variation was performed to compare the average ELISA ratios with the rapid test scores. Statistics were performed using Instat (Graphpad Software Inc., San Diego, Calif.).
RESULTS
The sensitivities obtained with the PanBio rapid test and the AFRIMS ELISA are summarized in Table 1. Of the 46 primary cases, 42 (91%) were detected via a positive IgM test line in the rapid test compared to 43 (93%) in the IgM ELISA. Thirty-one (86%) of the 36 secondary dengue cases were detected using the IgG rapid test line compared to 25 (69%) in the IgG ELISA. Of the secondary sera, 53% (19 of 36) were IgM positive in the rapid test compared to 50% (18 of 36) in the AFRIMS IgM ELISA. The overall sensitivity for dengue virus infections in the rapid test was not significantly different from that achieved using the ELISA (90 versus 87%, respectively; P = 0.4022).
TABLE 1.
Sensitivities of recombinant-antigen rapid test and AFRIMS in-house ELISA for primary and secondary dengue virus infections
| Assay | No. of positive samples/total no. tested (% sensitivity) | |
|---|---|---|
| Primary dengue | Secondary dengue | |
| IgM rapid test | 42/46 (91) | 19/36 (53) |
| IgM ELISA | 43/46 (93) | 18/36 (50) |
| IgG rapid test | 3/46 (6) | 31/36 (86) |
| IgG ELISA | 2/46 (4) | 25/36 (69) |
| IgM and IgG rapid test | 42/46 (91) | 32/36 (89) |
| IgM and IgG ELISA | 43/46 (93) | 28/36 (78) |
Paired serum analysis revealed that the rapid test detected 18 of 20 (90%) primary and 14 of 16 (88%) secondary infections using the acute sample (S1 [Table 2]), with the ELISA detecting 19 of 20 (95%) primary and 8 of 16 (50%) secondary infections. Analysis of the discharge (S2) sera revealed that the rapid test detected 20 of 20 primary infections and 15 of 16 (94%) secondary infections (Table 2), while the ELISA also detected all primary infections and 14 of 16 (88%) secondary infections. Of the two triplicate samples taken from patients with primary dengue virus infection, one showed seroconversion with the second and third samples, while the other patient had detectable levels of IgM in all three samples. All four samples taken from the one patient with secondary dengue virus infection produced positive IgG results in the rapid test. Consequently, the rapid test detected 38 of 40 (95%) primary infections and 29 of 32 (91%) secondary infections compared to the ELISA, which detected 39 of 40 (98%) primary infections and 18 of 32 (56%) secondary infections.
TABLE 2.
Detection of dengue virus infection in paired sera using the rapid test
| Diagnosis | No. of positive samples/total no. tested (% sensitivity)a | |||
|---|---|---|---|---|
| IgM rapid test S1 | IgM rapid test S2 | IgG rapid test S1 | IgG rapid test S2 | |
| Primary dengue | 18/20 (90) | 20/20 (100) | 0/20 (0) | 3/20 (15) |
| Secondary dengue | 4/16 (25) | 14/16 (88) | 14/16 (88) | 15/16 (94) |
The overall specificity of the rapid test (Table 3) in nonflavivirus infections was 85% (76 of 89), with false-positive results occurring with sera from patients with NEI (n = 5), malaria (n = 2), leptospirosis (n = 5), and scrub typhus (n = 1) infections. The majority (12 of 13) of the false-positive results occurred at the IgM test line. No cross-reactivity with JE occurred at the IgM test line, with only two samples giving a positive IgG test line (89% specificity). The specificity was 89% for the IgM test line and 97% for the IgG test line, resulting in an overall specificity of 86% for the rapid test.
TABLE 3.
Specificities of dengue rapid test for JE virus and nonflavivirus infections
| Test | No. of negative samples/total no. tested (% specificity) | ||||||
|---|---|---|---|---|---|---|---|
| JE | NEI | Malaria | Leptosporosis | Scrub typhus | Nonflavivirus total | Nondengue total | |
| IgM line | 18/18 (100) | 30/34 (88) | 13/15 (87) | 15/20 (75) | 19/20 (95) | 77/89 (87) | 95/107 (89) |
| IgG line | 16/18 (89) | 33/34 (97) | 15/15 (100) | 20/20 (100) | 20/20 (100) | 88/89 (99) | 104/107 (97) |
| IgM or IgG line | 16/18 (89) | 29/34 (85) | 13/15 (87) | 15/20 (75) | 19/20 (95) | 76/89 (85) | 92/107 (86) |
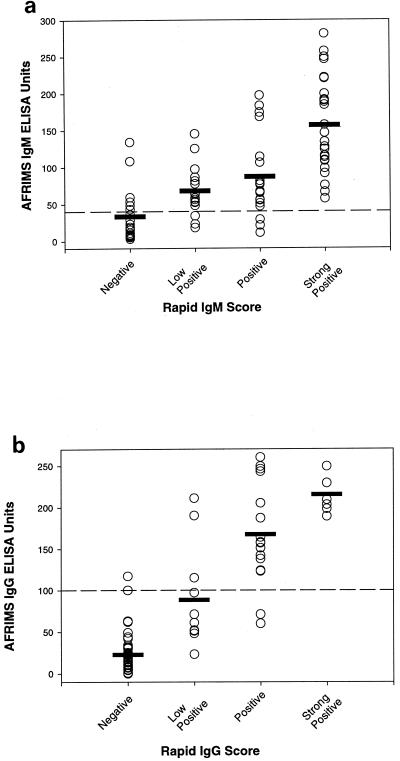
The individual results of the AFRIMS ELISA were well correlated using either the IgM or IgG tests (Fig. 2). There was a significant association between the mean ELISA ratio and the rapid score (analysis of variance, P < 0.0001 for IgM and P < 0.0001 for IgG). However, in the IgM and IgG assays it was apparent that a number of ELISA-negative samples were positive in the rapid test. The majority of discrepant sera gave low positive results in the rapid test, indicating that the cutoffs used in this assay are slightly lower than those used in the ELISAs.
FIG. 2.
Correlation between the ELISA and the rapid test for IgM (a) and IgG (b). The broken lines represent the cutoff units used in the ELISA. The horizontal bars represent the mean ELISA ratio for each rapid score.
DISCUSSION
Due to the higher mortality associated with secondary infections (15), it is important to use diagnostic assays that are able to differentiate between the two forms of dengue virus infection. In endemic areas, secondary infections are most common, as the majority of children have antibodies against dengue by the time they are 5 years of age (18, 30, 36). As primary and secondary dengue virus infections show markedly different immunological responses (13), the detection of antibodies is a valuable procedure to diagnose and differentiate dengue virus infections. Traditionally, the HAI assay has been used as the “gold standard” serological test (39), though ELISA has recently been adopted in most laboratories due to its convenience and performance (13).
To further simplify diagnosis, a number of commercial lateral flow tests for the serological diagnosis of dengue virus infections are now available (from PanBio Ltd.; Cortez Diagnostics, Calabasas, Calif.; Glysby, Arcore, Italy; and AmeriTek, Seattle, Wash.). This format offers the benefit of short incubation times (5 to 30 min) and the ability to be used in field settings or in laboratories without adequate equipment or electricity. The PanBio Dengue Duo Rapid Strip Test is the first commercial lateral flow test for the diagnosis of dengue that incorporates recombinant proteins. The four proteins used (from dengue viruses 1, 2, 3, and 4) each represent the N-terminal 80% of the respective envelope glycoprotein. The expression and secretion of these recombinant subunits in the S2 cells has resulted in molecules that have retained native-like characteristics.
In the PanBio Rapid Test, IgM and IgG are detected simultaneously using a single addition of diluted serum. Therefore, differentiation between primary and secondary infections can be made through a single application of diluted serum rather than a series of dilutions, as is needed in the HAI assay (6). The sensitivity of this rapid test has been set so that in patients with primary dengue, IgM is positive while IgG is negative. In contrast, patients with secondary infections will have a positive IgG result with or without a positive IgM result.
In-house comparison studies between the whole virus and recombinant antigens in an IgM and IgG capture ELISA show a high level of agreement (94% for both). The data obtained suggest that these proteins should be useful in diagnostic assays for the detection of IgM and IgG to dengue virus (data not shown). In this study, the rapid test performed similarly to the in-house reference ELISA. The rapid test detected 91% of primary and 86% of secondary dengue cases compared to 93 and 69%, respectively, using the reference ELISA. There was no significant difference between the sensitivities in primary dengue virus infections (P = 1.0000), though there was a significant difference in the sensitivities in secondary dengue virus infections (P = 0.0063).
No significant differences were reported between the abilities of the rapid test and the reference ELISA to detect primary infections using the acute sample only (90 versus 85%, respectively; P = 1.0000). Based on these results, the rapid test was able to diagnose 90% of all primary cases at hospital admission. Eighty-eight percent of secondary cases were also diagnosed using only the acute sample in the rapid test. Consequently, 89% of all dengue patients were diagnosed using the acute sample in the rapid test, indicating that a second sample was necessary in only 11% of cases. A laboratory diagnosis of acute dengue virus infection at the time of admission will refine the differential diagnosis. However, a negative result early in the course of illness requires repeat testing, as does a diagnosis of secondary dengue virus infection if it is important to know the antibody response pattern.
There was no significant difference between the sensitivities when either the acute- or convalescent-phase sample was used for the detection of IgM in primary dengue (P = 0.4872) or IgG in secondary dengue (P = 1.0000). The only significant difference observed between the first and second samples was the sensitivity of IgM in secondary dengue (P = 0.0010). A sensitivity of 25% was achieved using the first sample, and this rose to 88% using the second sample. This supports theories stating that the IgM in secondary dengue is generally produced at lower levels and can take longer to reach detectable levels than IgM in primary infections (18, 30, 36).
As the recombinant proteins include 80% of the envelope protein, it is likely that they have a level of cross-reactivity similar to that of the whole virus. In this study, we found there were false positives with malaria, leptospirosis, and scrub typhus sera at the IgM test line, with the IgG test line being very specific. This pattern is different from that generally observed with whole virus (24, 31, 37, 38).
The only cross-reactivity seen at the IgG test line in the rapid test was with sera from patients infected with JE virus. Consequently, this rapid test would be useful in areas where dengue and JE are endemic, as a positive IgM result in this assay is not likely to be due to an active JE virus infection. Cross-reactivity between flaviviruses at the IgG level is common and has been reported in diagnostic assays (7, 8, 20, 24).
The recombinant proteins were chosen because they represent the majority of the active section of the envelope protein, the target for neutralizing epitopes (16), and hence are more likely to mimic whole virus in serological assays. In addition, the envelope protein has been reported to elicit the first immune response in primary infections (5). As this response is also long lasting, the inclusion of recombinant glycoproteins in a dengue assay is a logical choice.
Several groups have expressed recombinant dengue envelope subunits as soluble secreted proteins using a variety of recombinant expression systems (9, 10, 23, 25, 28, 34). These are claimed to elicit neutralizing and HAI antibodies (11) and to induce a stronger and longer-lasting immune response in animals (29). These efforts to express soluble dengue envelope subunits have met with various degrees of success. In each case, efforts to establish that the expressed subunits have maintained native-like structure were made; however, it is difficult to assess the relative quality of one subunit versus another based on the in vitro assays used. The ability of the envelope subunits to stimulate the production of neutralizing antibodies and to provide a protective response when used to immunize mice or monkeys has been the key to assessing the quality of recombinant dengue envelope proteins. The _Escherichia coli-_expressed E proteins (25), while reactive to appropriate monoclonal antibodies, failed to provide a protective response in a mouse challenge assay. The baculovirus-expressed E proteins (9, 10, 28) have resulted in various levels of neutralizing antibodies and protection in both mice and monkeys, indicating that these insect cell-produced subunits have a more native-like conformation. In similar studies with the S2-expressed 80% E subunits, the induction of levels of neutralizing antibodies and protection against challenge has been achieved in both mice and monkeys (B.-A. G. Coller, D. E. Clements, G. S. Bignami, I. D. Peters, J. R. Putnak, M. McDonell, and T. Humphreys, unpublished data). These results indicate that the S2-expressed 80% E subunits have maintained a native-like conformation. This study also confirmed that the 80% E subunit proteins expressed in the S2 expression system have native-like characteristics based on the ability of these recombinant subunits to produce sensitivities similar to those of whole-virus antigens in the assays described.
We have presented our results on the performance of the PanBio Duo Rapid Test using these recombinant dengue proteins and demonstrated a high level of sensitivity and specificity in diagnosing acute primary and secondary dengue. This test offers a safe alternative to diagnostic assays using whole-virus antigens and will provide the clinician with a reliable and rapid assay to distinguish acute dengue virus infections and identify secondary dengue virus infections and thus patients at risk for severe dengue. Due to these advantages, this rapid test is likely to be the first of the next generation of diagnostic assays for dengue.
ACKNOWLEDGMENTS
This work was supported by the U.S. Army Medical Research and Material Command and PanBio Ltd.
REFERENCES
- 1.Bundo K, Igarashi A. Antibody capture ELISA for detection of immunoglobulin M antibodies in sera from Japanese encephalitis and dengue haemorrhagic fever patients. J Virol Methods. 1985;11:15–22. doi: 10.1016/0166-0934(85)90120-x. [DOI] [PubMed] [Google Scholar]
- 2.Burke D S, Nisalak A, Hoke C H., Jr Field trial of a Japanese encephalitis diagnostic kit. J Med Virol. 1986;18:41–49. doi: 10.1002/jmv.1890180106. [DOI] [PubMed] [Google Scholar]
- 3.Cardosa M J, Zuraini I. Comparison of an IgM capture ELISA with a dot enzyme immunoassay for laboratory diagnosis of dengue virus infections. Southeast Asian J Trop Med Public Health. 1991;22:337–340. [PubMed] [Google Scholar]
- 4.Cardosa M J, Bahavudin F, Hamid S, Hooi T P, Nimmanitya S. A nitrocellulose membrane based IgM capture enzyme immunoassay for etiological diagnosis of dengue virus infection. Clin Diagn Virol. 1995;3:343–350. doi: 10.1016/0928-0197(94)00049-z. [DOI] [PubMed] [Google Scholar]
- 5.Churdboonchart V, Bhamarapravati N, Peampramprecha S, Sirinavin S. Antibodies against dengue viral proteins in primary and secondary dengue hemorrhagic fever. Am J Trop Med Hyg. 1991;44:481–493. doi: 10.4269/ajtmh.1991.44.481. [DOI] [PubMed] [Google Scholar]
- 6.Clarke D H, Casals J. Techniques for haemagglutination and haemagglutination-inhibition with arthropod-borne viruses. Am J Trop Med Hyg. 1958;7:561–573. doi: 10.4269/ajtmh.1958.7.561. [DOI] [PubMed] [Google Scholar]
- 7.Cuzzubbo A J, Vaughn D W, Nisalak A, Solomon T, Kalayanarooj S, Aaskov J, Dung N M, Devine P L. Alternate strategies for the serological diagnosis of dengue infection: comparison of PanBio Dengue Duo ELISA and MRL dengue fever virus IgM capture ELISA. J Clin Diagn Lab Immunol. 1999;6:705–712. doi: 10.1128/cdli.6.5.705-712.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cuzzubbo A J, Vaughn D V, Nisalak A, Solomon T, Kalayanarooj S, Aaskov J, Dung N M, Devine P L. Comparison of PanBio Dengue Duo IgM and IgG capture ELISA and Venture Technologies dengue IgM and IgG dot blot. J Clin Virol. 1999;16:135–144. doi: 10.1016/s1386-6532(99)00076-1. [DOI] [PubMed] [Google Scholar]
- 9.Delenda C, Staropoli I, Frenkiel M P, Cabanie L, Deubel V. Analysis of C-terminally truncated dengue 2 and dengue 3 virus envelope glycoproteins: processing in insect cells and immunogenic properties in mice. J Gen Virol. 1994;75:1569–1578. doi: 10.1099/0022-1317-75-7-1569. [DOI] [PubMed] [Google Scholar]
- 10.Feighny R, Burrous J, Putnak R. Dengue type-2 virus envelope protein made using recombinant baculovirus protects mice against virus challenge. Am J Trop Med Hyg. 1994;50:322–328. doi: 10.4269/ajtmh.1994.50.322. [DOI] [PubMed] [Google Scholar]
- 11.Fonseca B A, Pincus S, Shope R E, Paoletta E, Manson P W. Recombinant vaccinia viruses co-expressing dengue-1 glycoproteins prM and E induce neutralising antibodies in mice. Vaccine. 1994;12:279–285. doi: 10.1016/0264-410x(94)90206-2. [DOI] [PubMed] [Google Scholar]
- 12.Gentry M K, Henchal E A, McCown J M, Brandt W E, Dalrymple J M. Identification of distinct antigenic determinants on dengue-2 virus using monoclonal antibodies. Am J Trop Med Hyg. 1982;33:548–555. doi: 10.4269/ajtmh.1982.31.548. [DOI] [PubMed] [Google Scholar]
- 13.Gubler D J. Serological diagnosis of dengue haemorrhagic fever. Dengue Bull. 1996;20:20–23. [Google Scholar]
- 14.Gubler D J, Meltzer M. Impact of dengue/dengue hemorrhagic fever on the developing world. Adv Virus Res. 1999;53:35–70. doi: 10.1016/s0065-3527(08)60342-5. [DOI] [PubMed] [Google Scholar]
- 15.Halstead S B. Pathogenesis of dengue: challenges to molecular biology. Science. 1988;239:476–481. doi: 10.1126/science.3277268. [DOI] [PubMed] [Google Scholar]
- 16.Heinz F X. Epitope mapping of flavivirus glycoproteins. Adv Virus Res. 1986;31:103–168. doi: 10.1016/s0065-3527(08)60263-8. [DOI] [PubMed] [Google Scholar]
- 17.Henchal E A, McCown J M, Burke D J, Seguin M C, Brandt W E. Epitopic analysis of antigenic determinants on the surface of dengue-2 virions using monoclonal antibodies. Am J Trop Med Hyg. 1985;34:162–167. doi: 10.4269/ajtmh.1985.34.162. [DOI] [PubMed] [Google Scholar]
- 18.Innis B. Antibody responses to dengue virus infection. In: Gubler D J, Kuno G, editors. Dengue and dengue haemorrhagic fever. New York, N.Y: CAB International; 1997. pp. 221–243. [Google Scholar]
- 19.Innis B L, Nisalak A, Nimmannitya S, Kusalerdchariya S, Chongswasdi V, Suntayakorn S, Puttisri P, Hoke C H. An enzyme-linked immunosorbent assay to characterize dengue infections where dengue and Japanese encephalitis co-circulate. Am J Trop Med Hyg. 1989;40:418–427. doi: 10.4269/ajtmh.1989.40.418. [DOI] [PubMed] [Google Scholar]
- 20.Kuno G, Cropp C B, Wong-Lee J, Gubler D J. Evaluation of an IgM immunoblot kit for dengue diagnosis. Am J Trop Med Hyg. 1998;59:757–762. doi: 10.4269/ajtmh.1998.59.757. [DOI] [PubMed] [Google Scholar]
- 21.Lam S K. Rapid dengue diagnosis and interpretation. Malay J Pathol. 1993;15:9–12. [PubMed] [Google Scholar]
- 22.Lam S K, Devine P L. Evaluation of capture ELISA and rapid immunochromatographic test for the determination of IgM and IgG antibodies produced during dengue infection. Clin Diagn Virol. 1998;10:75–81. doi: 10.1016/s0928-0197(98)00002-6. [DOI] [PubMed] [Google Scholar]
- 23.Makino Y, Tadano M, Arakaki S, Fukunaga T. Potential use of a baculovirus-expressed dengue-4 E protein as a diagnostic antigen in regions endemic for dengue and Japanese encephalitis. Am J Trop Med Hyg. 1991;45:636–643. doi: 10.4269/ajtmh.1991.45.636. [DOI] [PubMed] [Google Scholar]
- 24.Makino Y, Tadano M, Saito M, Maneekarn N, Sittisombut N, Sirisanthana V, Poneprasert B, Fukunaga T. Studies on serological cross-reaction in sequential flavivirus infections. Microbiol Immunol. 1994;38:951–955. doi: 10.1111/j.1348-0421.1994.tb02152.x. [DOI] [PubMed] [Google Scholar]
- 25.Mason P W, Zugel M U, Semproni A R, Fournier M J, Mason T L. The antigenic structure of dengue type 1 virus envelope and NS1 proteins expressed in Escherichia coli. J Gen Virol. 1990;71:2107–2114. doi: 10.1099/0022-1317-71-9-2107. [DOI] [PubMed] [Google Scholar]
- 26.Nawa M. An enzyme-linked immunosorbent assay using a chaotropic agent (sodium thiocyanate) for serotype specific reaction between crude dengue viral antigen and anti-dengue mouse antibody. Microbiol Immunol. 1992;36:721–730. doi: 10.1111/j.1348-0421.1992.tb02074.x. [DOI] [PubMed] [Google Scholar]
- 27.Palmer C J, King S D, Caudrado R R, Perez E, Baum M, Ager A L. Evaluation of the MRL diagnostics dengue fever virus IgM capture ELISA and the PanBio rapid immunochromatographic test for diagnosis of dengue fever in Jamaica. J Clin Microbiol. 1999;37:1600–1601. doi: 10.1128/jcm.37.5.1600-1601.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Putnak R, Feighny R, Burrous J, Cochran M, Hackett C, Smith G, Hoke C. Dengue-1 virus envelope glycoprotein gene expressed in recombinant baculovirus elicits virus-neutralizing antibody in mice and protects them from virus challenge. Am J Trop Med Hyg. 1991;45:159–167. doi: 10.4269/ajtmh.1991.45.159. [DOI] [PubMed] [Google Scholar]
- 29.Raviprakash K, Kochel T J, Ewing D, Simmons M, Phillips I, Hayes K R. Immunogenicity of dengue virus type1 DNA vaccines expressing truncated length envelope protein. Vaccine. 2000;18:2426–2434. doi: 10.1016/s0264-410x(99)00570-8. [DOI] [PubMed] [Google Scholar]
- 30.Ruechusatawat K, Morita K, Tanaka M, Vongcheree S, Rojanasuphot S, Warachit P, Kanai K, Thongtradol P, Nimnakorn P, Kanungkid S, Igarashi A. Daily observation of antibody levels among dengue patients detected by enzyme-linked immunosorbent assay (ELISA) Jpn J Trop Med Hyg. 1994;22:9–12. [Google Scholar]
- 31.Sang C T, Cuzzubbo A, Devine P L. Evaluation of commercial capture enzyme-linked immunosorbent assay for the detection of immunoglobulin M (IgM) and IgG antibodies produced during dengue infection. Clin Diagn Lab Immunol. 1998;5:7–10. doi: 10.1128/cdli.5.1.7-10.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sang C T, Lim S H, Cuzzubbo A, Devine P L. Clinical evaluation of rapid immunochromatographic test for the diagnosis of dengue infection. Clin Diagn Lab Immunol. 1998;5:407–409. doi: 10.1128/cdli.5.3.407-409.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schneider I. Cell lines derived from late embryonic stages of Drosophila melanogaster. J Embryol Exp Morphol. 1972;27:353–365. [PubMed] [Google Scholar]
- 34.Sugrue R J, Cui T, Xu Q, Fu J, Chan Y C. The production of recombinant dengue virus E protein using Escherichia coli and Pichia pastoris. J Virol Methods. 1997;69:159–169. doi: 10.1016/s0166-0934(97)00151-1. [DOI] [PubMed] [Google Scholar]
- 35.Talarmin A, Labeau B, Lelarge J, Sarthou J-L. Immunoglobulin A-specific capture enzyme-linked immunosorbent assay for diagnosis of dengue fever. J Clin Microbiol. 1998;36:1189–1192. doi: 10.1128/jcm.36.5.1189-1192.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vaughn D W, Green S, Kalayanarooj S, Innis B L, Nimmannitya S, Suntayakorn S, Rothman A L, Ennis F A, Nisalak A. Dengue in early febrile phase: viremia and antibody responses. J Infect Dis. 1997;176:322–330. doi: 10.1086/514048. [DOI] [PubMed] [Google Scholar]
- 37.Vaughn D W, Nisalak A, Kalayanarooj S, Solomon T, Dung N M, Cuzzubbo A, Devine P L. Evaluation of rapid immunochromatographic test for diagnosis of dengue virus infection. J Clin Microbiol. 1998;36:234–238. doi: 10.1128/jcm.36.1.234-238.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vaughn D W, Nisalak A, Solomon T, Kalayanarooj S, Nguyen M D, Kneen R, Cuzzubbo A, Devine P L. Rapid serologic diagnosis of dengue virus infection using a commercial capture ELISA that distinguishes primary and secondary infections. Am J Trop Med Hyg. 1999;60:693–698. doi: 10.4269/ajtmh.1999.60.693. [DOI] [PubMed] [Google Scholar]
- 39.World Health Organization. Dengue haemorrhagic fever: diagnosis, treatment and control. Geneva, Switzerland: World Health Organization; 1986. [Google Scholar]
- 40.World Health Organization. Dengue haemorrhagic fever: diagnosis, treatment, prevention and control. Geneva, Switzerland: World Health Organization; 1997. [Google Scholar]
- 41.Wu S J L, Hanson B, Paxton H, Nisalak A, Vaughn D W, Rossi C, Henchal E A, Porter K R, Watts D M, Hayes C G. Evaluation of dipstick enzyme-linked immunosorbent assay for detection of antibodies to dengue virus. Clin Diagn Lab Immunol. 1997;4:452–457. doi: 10.1128/cdli.4.4.452-457.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]