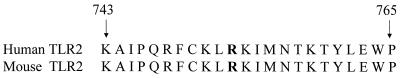A Novel Polymorphism in the Toll-Like Receptor 2 Gene and Its Potential Association with Staphylococcal Infection (original) (raw)
Abstract
The toll-like receptor 2 (TLR2) has gained importance as a major mammalian receptor for lipoproteins derived from the cell wall of a variety of bacteria, such as Borrelia burgdorferi, Treponema pallidum, and Mycoplasma fermentans. We were interested in identifying mutations in the TLR2 gene that might prove to be associated with altered susceptibility to septic shock. We performed a mutation screen of the TLR2 gene using single-stranded conformational polymorphism in 110 normal, healthy study subjects and detected an Arg753Gln mutation in three individuals. No other missense mutations were detected in the TLR2 open reading frame. Functional studies demonstrate that the Arg753Gln polymorphism, in comparison to the wild-type TLR2 gene, is significantly less responsive to bacterial peptides derived from B. burgdorferi and T. pallidum. In a septic shock population, the Arg753Gln TLR2 polymorphism occurred in 2 out of 91 septic patients. More importantly, both of the subjects with the TLR2 Arg753Gln polymorphism had staphylococcal infections. These findings suggest that a mutation in the TLR2 gene may predispose individuals to life-threatening bacterial infections.
Microbial pathogens are a major health concern worldwide. Invasion of the host by microbial pathogens causes the activation of the innate immune response, which acts as a first line of defense against pathogenic bacteria. The innate immune response triggers a sequence of events that results in the production and secretion of cytokines and chemokines, activation of macrophages and monocytes, and in some cases initiation of adaptive immunity (10).
The toll receptors appear to be very important in initiating the innate immune response. C3H/HeJ mice which have a mutation in the toll-like receptor 4 (TLR4) gene (Pro712His) and C57BL6/10ScCr mice which have a deletion of TLR4 are both hyporesponsive to lipopolysaccharide (LPS) (12). The TLR4 mutation in C3H/HeJ mice was the first direct proof for identifying TLR4 as a major LPS receptor in mammals. Subsequently TLR4−/− mice were shown to be LPS hyporesponsive as well (6). Binding of LPS to TLR4, via a complex involving CD14 as well as MD-2, is thought to initiate a signaling cascade that involves several kinases and accessory molecules such as MyD88 and IRAK, leading eventually to a nuclear translocation of transcription factors NF-κB and AP-1 (14). The binding of these transcription factors is responsible for initiating cytokine and chemokine expression subsequent to LPS exposure. Recently, mutations in the TLR4 receptor at residues 299 and 399 were shown to be associated with hyporesponsiveness to inhaled endotoxin in humans (2). Furthermore, the Asp299Gly TLR4 mutation may increase the risk to carriers of developing gram-negative septic shock (E. Lorenz, J. Mira, K. Frees, and D. Schwartz, submitted for publication). However, TLR4 does not seem to be involved in mediating the cellular response to gram-positive pathogens. TLR2, which is highly homologous to TLR4, has been shown to mediate a response to LPS, albeit to a lesser degree than TLR4. In fact, while TLR4−/− mice are unable to respond to LPS (6), TLR2−/− mice respond normally to LPS (16), indicating that TLR4 is the main LPS receptor in mammals. More importantly, evidence suggests that TLR2 is involved in the recognition of various other bacterial lipoproteins, such as peptides derived from Borrelia, Mycoplasma, and Treponema (9). Still more importantly, TLR2 has also been shown to be involved in staphylococcus signaling (1, 3).
This line of reasoning led us to hypothesize that mutations in TLR2 could be associated with a diminished response to gram-positive lipoproteins and place individuals at higher risk of gram-positive infections. We have identified a missense mutation that occurs in about 3% of people tested and results in an arginine to glutamine substitution at residue 753 of the human TLR2 gene. Transfection of 293T cells with either wild-type or mutant (Arg753Gln) TLR2 shows that the response to LPS 0111.B4 is unaffected by the mutation, while the response to different bacterial lipoproteins is reduced in the TLR2 Arg753Gln mutant compared to wild-type TLR2 cells. When a population of septic shock patients and healthy blood donor controls was analyzed, it was found that the TLR2 mutation occurred at the same frequency in both groups. However, two patients carrying the TLR2 Arg753Gln mutation both suffered from gram-positive infections. These results suggest that the TLR2 mutation Arg753Gln could be a risk factor for developing septic shock after infection by gram-positive bacteria.
MATERIALS AND METHODS
Study subjects.
The patient samples were collected as part of a multicenter study conducted in seven academic adult intensive care units (ICUs) in France between March 1996 and November 1997. The protocol was approved by the Institutional Review Board of Cochin Hospital, Paris, France. Informed consents were obtained from control subjects and patients or their relatives.
The control group comprised 73 healthy, unrelated, white blood donors at the Cochin Hospital blood bank; 42 of the controls had been previously genotyped for a study on tumor necrosis factor alpha (TNF-α) polymorphisms (11). An additional 31 healthy blood donors were included as control subjects for this study. The septic shock group, defined by the criteria of the consensus conference, included 91 ICU patients. Of these patients, 88 had been included in a previously published study on the effect of TNF-α on septic shock (11), and 3 patients were included after June 1997.
To be eligible for enrollment, the patients with septic shock had to be Caucasian and had to have the following six inclusion criteria of septic shock within a 12-h period: (i) clinical evidence of infection; (ii) hyperthermia (temperature of >38°C) or hypothermia (temperature of <35.6°C); (iii) tachycardia (>90 beats/min); (iv) tachypnea (120 breaths/min) or need for mechanical ventilation; (v) use of a vasopressor to maintain systolic blood pressure higher than 90 mm Hg or hypotension defined as systolic blood pressure less than 90 mm Hg for more than 30 min or a decrease in systolic blood pressure of more than 40 mm Hg from previously established values for more than 30 min (hypotension had to be present at enrollment and refractory to an intravascular volume challenge of at least 500 ml); and (vi) evidence of inadequate organ function or perfusion within 12 h of enrollment, as manifested by at least one of the following syndromes (previously described)—acute deterioration of the patient's mental status, arterial hypoxemia (PaO2/FiO2 < 280), plasma lactate concentration above the normal range or metabolic acidosis, oliguria, and disseminated intravascular coagulation. The exclusion criteria were the following: (i) older than 80 years, (ii) cardiac failure (class III or IV), (iii) liver insufficiency (Child C), (iv) bone marrow aplasia (white blood cell count of <0.50 × 109/liter), or (v) immunosuppression (positive human immunodeficiency virus serologic result, current immunosuppressive therapy including corticosteroids [equivalent prednisone, >0.5 mg/kg per day], or cancer).
Patients were monitored throughout their stays in the ICU. Information on age, sex, primary site of infection, infection-related organisms, and severity indexes (including the Simplified Acute Physiologic Score [SAPS II] [8], which uses a total of 15 variables to assess the severity of septic disease, and the Organ System Failure score (OSF) [7]) was collected at each patient's entry into the ICU. All the samples and information used in the study were coded, and patient confidentiality was preserved according to the guidelines for studies of human subjects.
Mutation detection.
Using single-stranded conformational polymorphism (SSCP), we initially screened the open reading frame of TLR2 in a population of 110 healthy volunteers at the University of Iowa. Genomic DNA was isolated from whole blood obtained from the study subjects using a rapid salt isolation procedure. Overlapping primer sets, less than 200 bp apart, were designed across the TLR2 coding sequence. Primers were derived from flanking intronic sequences to include all splice sites. Standard PCRs were prepared except that 10 to 20 ng of genomic DNA was used as a template. Amplification products were separated on nondenaturing, fan-cooled gels containing 5% acrylamide-bisacrylamide (19:1), 0.5× Tris-borate-EDTA (TBE), and 2.5% glycerol for 3 h at 20 W or 15 h at 5 W using standard procedures (17). The gels were subjected to silver staining, and aberrant bands were extracted from the gel, reamplified, and sequenced in both directions. To verify the sequence of the aberrant band, the same primers were used to amplify and sequence genomic DNA from each subject. TLR2 from at least one individual for which there was no aberrant band was also sequenced for comparison. The DNA sequence was determined with a Model 377 automated DNA sequencer (Perkin-Elmer, Norwalk, Conn.).
Sequencing of a human TLR2 cDNA plasmid to confirm the GenBank-submitted sequence showed the occurrence of an additional polymorphism at position 2251 of the open reading frame. This polymorphism results in an Arg (CGG) to a Gln (CAG) substitution. Subsequent sequencing of the control population showed that the G to A polymorphism, even though not detected by SSCP, occurred in 3% of the population. To determine the TLR2 genotype of the samples in both the Iowa test population and the French septic shock group with respect to the arginine to glutamine polymorphisms, the genomic DNA was amplified using forward primer 5′-TATGGTCCAGGAGCTGGAGA-3′ and reverse primer 5′-TGACATAAAGATCCCAACTAGACAA-3′, which span the region containing the polymorphism. PCR conditions were as follows: 5 min of initial denaturation at 95°C, followed by 30 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min. Following PCR amplification, fragments were purified using the Qiagen gel purification kit and were sequenced. The sequence of the genomic DNA from each subject was determined using a Model 377 automated DNA sequencer (Perkin-Elmer). The sequence analysis was performed blinded to the phenotype of the study subjects.
Mammalian cell culture and transfections.
293T cells were plated at a density of 300,000 in a 35-mm2 dish the day prior to the transfection. Transfections were performed using the CalPhos optimizer kit from Clontech according to the manufacturer's recommendation for 35-mm2 plates with 4 μg of DNA per dish. Each DNA mix consisted of 2 μg of reporter plasmid and 1 μg of each expression plasmid, replaced by an empty vector if only one expression plasmid was being used. In addition, 0.1 μg of β-galactosidase reporter plasmid was added to the transfection mix as an internal control for transfection efficiency. The transfection mix was added dropwise to the cells and was incubated for 4 h at 5% CO2. After the incubation, the transfection medium was removed and fresh Dulbecco modified Eagle medium supplemented with 1% fetal calf serum, 2 mM l-glutamine, and 10,000 U of PenStrep/ml was added overnight. Cells were lysed the following day using reporter lysis buffer (Promega, Madison, Wis.), and after overnight incubation at 4°C, luciferase and protein assays were done.
Protein assay.
Protein contents of the lysates were measured using the Bio-Rad protein assay reagent. In short, cells were lysed in 1× lysis buffer (Promega) overnight at 4°C. The following day, 1.2 μl of lysate was added to 200 μl of diluted Bio-Rad protein reagent, and the absorbance was read at 595 nm in a microplate reader from BioTek Instruments (Scotts Valley, Calif.). Each condition was assayed in triplicate, and the sample protein concentration was calculated based on known bovine serum albumin standards.
Luciferase assay.
Luciferase assay reagents were purchased from Promega. Briefly, cells were lysed in 1× lysis buffer overnight at 4°C. For each assay, 8 μl of lysate was added to 100 μl of luciferase assay reagent and read in a Luminometer Model Monolight 2010 (Analytical Luminescence Laboratories, San Diego, Calif.).
RESULTS
A novel polymorphism in the human TLR2 gene causes an arginine to glutamine substitution at the C terminus.
We used SSCP to screen the open reading frame of the human TLR2 gene in the original Iowa test population for possible point mutations. In each of three individuals, we detected two silent polymorphisms (Table 1). No missense mutations were detected. When sequencing a TLR2 cDNA from a human cDNA library (gift of F. Gusovsky), we identified a sequence change at the 3′ terminus of the TLR2 gene. This sequencing change causes an arginine to glutamine substitution at residue 753. Since the arginine is conserved between mice and humans and is part of a highly conserved stretch of amino acids at the C terminus of TLR2 (Fig. 1), we screened a population of healthy blood donors. The TLR2 Arg753Gln polymorphism occurred in 3% of the subjects tested.
TABLE 1.
Summary of SSCP screen of human TLR2 in a population of healthy blood donors (Iowa)a
| Nucleotide change | Amino acid | Conservedb | Frequency (out of 110) |
|---|---|---|---|
| C597T | Silent | NA | 3 |
| T1350C | Silent | NA | 3 |
| G2258A | Arg–Gln | Yes | 3 |
FIG. 1.
Alignment of mouse and human TLR2 C termini: the arginine residue is conserved across species. The arginine residue at position 753 is conserved between species. The sequence surrounding the TLR2 mutation was aligned for humans and mice (5). The arginine at position 753 is indicated in bold.
TLR2 Arg753Gln polymorphism does not affect the LPS response in vitro.
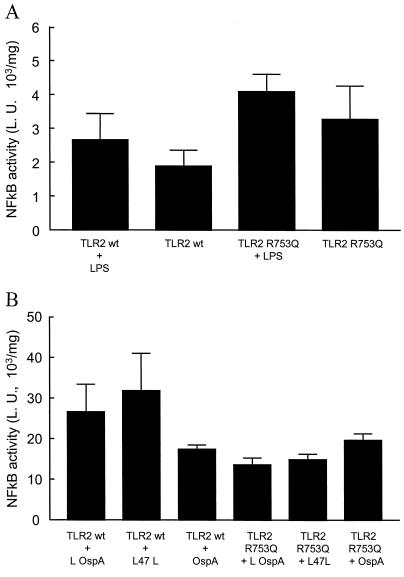
TLR2 wild-type (gift of S. Akira) and TLR2 Arg753Gln mutant cDNAs were transfected into LPS unresponsive 293T cells, and NF-κB activity was measured by luciferase assay. While at the same concentration of LPS (100 ng/ml) TLR4 shows a more than twofold stimulation by LPS 0111.B4, neither the wild-type TLR2 nor the TLR2 Arg753Gln mutant showed a significant stimulation of NF-κB in response to LPS 0111.B4 (Fig. 2). This result suggests that the TLR2 Arg753Gln mutation does not affect the ability of TLR2 to respond to LPS.
FIG. 2.
Functional significance of the R753Q mutation in 293T cells. 293T cells were transfected with TLR2 expression plasmids (wild type [wt] or Arg753Gln), and 24 h later the cells were stimulated with 100 ng of LPS/ml for 6 h (2A) or 33 ng of bacterial peptides/ml for 16 h (2B). In Fig. 2A, NF-κB activity was measured using a commercially available NF-κB reporter plasmid carrying the luciferase gene (Clontech, Palo Alto, Calif.). The NF-κB activity following LPS stimulation is unchanged in 293T cells transfected with the Arg753Gln plasmid compared to cells transfected with the wt TLR2 plasmid. In Fig. 2B, NF-κB activity was measured using a commercially available NF-κB reporter plasmid carrying the luciferase gene (Clontech). The NF-κB activity following stimulation with either _B. burgdorferi_-derived lipidated OspA (L OspA) or _T. pallidum_-derived lipidated 47L (L 47L) is absent in 293T cells transfected with the Arg753Gln plasmid compared to cells transfected with the wt TLR2 plasmid. The nonlipidated form of _B. burgdorferi_-derived OspA (OspA NL) did not activate either wild-type TLR2 or TLR2 Arg753Gln. L. U., luciferase units.
TLR2 Arg753Gln mutation affects the ability to respond to bacterial peptides in vitro.
We next measured the ability of the TLR2 Arg753Gln mutation to mediate the cellular response to bacterial peptides. We used two artificial peptides (kind gift of T. Sellati, Department of Pathology, University of Connecticut Health Center, Farmington, Conn.): _B. Burgdoferi_-derived OspA and _T. pallidum_-derived 47L (13). Transfection of 293T cells with the plasmids described above and subsequent luciferase assays showed that the wild-type TLR2 cDNA gave about a 1.5- to 2-fold increase in response to both active peptides compared to a nonlipidated control peptide. The TLR2 Arg753Gln mutation, on the other hand, did not show any activation in response to these peptides, similar to previous results in a TLR2-deficient strain (16). This indicates that mutation of a conserved arginine to glutamine reduces the ability of TLR2 to respond to bacterial peptides in vitro.
TLR2 Arg753Gln mutation occurs in two patients with staphylococcal infections.
Based on our in vitro data, we tested whether the TLR2 Arg753Gln mutation would occur in patients with gram-positive infection more commonly than in patients with gram-negative infections. In a well-characterized population of patients with septic shock, we found that the TLR2 Arg753Gln polymorphism occurred in 2 of the 91 septic shock patients. While only a minority of the septic shock patients had gram-positive infections (n = 22), both the patients heterozygous for the TLR2 Arg753Gln allele had staphylococcal infections. The presence of the TLR2 Arg753Gln mutation was higher in the patients with gram-positive septic shock (2 out of 22, or 9%) compared to all other patients with septic shock (0 out of 69, or 0%).
DISCUSSION
We report a novel polymorphism of the TLR2 gene that affects the response to bacterial peptides in vitro and may be associated with an increased risk for staphylococcal septic shock in patients with the mutation. The TLR2 Arg753Gln (G2251A) polymorphism has an allele frequency of about 3% in the populations tested and changes a highly conserved arginine residue to a glutamine. While the response to LPS is virtually unaffected, the response to peptides from B. burgdorferi and T. pallidum is lacking in cells carrying the TLR2 Arg753Gln polymorphism compared to that in wild type cells. Furthermore, screening of a septic shock population showed that the two patients with the TLR2 Arg753Gln polymorphism were diagnosed with staphylococcal infections. These results indicate that the TLR2 Arg753Gln polymorphism may place individuals at a higher risk for gram-positive septic shock. Because of the small sample size, we did not find a significant association between the TLR2 Arg753Gln polymorphism and an increased risk for gram-positive septic shock. While this may indicate that the association between the TLR2 polymorphism and gram-positive septic shock could be due to chance alone, previous in vitro results related to TLR2 function suggest that the lack of correlation is due to the small sample size and the rarity of the polymorphism.
TLR2 plays a unique role in innate immunity. While TLR4 seems to be highly specific for LPS-mediated signaling (16), TLR2 can respond to a variety of different bacterial cell wall components (9). Even though the homology between TLR2 and TLR4 is high, the proteins have different roles in the immune system. TLR2 has been shown to respond to bacterial lipoproteins. While it can be activated by lipoproteins derived from Borrelia, Treponema, and Staphylococcus (9). TLR2 is not universal for gram-positive cell wall components. It cannot be activated by group B type III streptococci (4). With respect to LPS, TLR4 seems to be a major mammalian LPS receptor. While TLR2 has been shown to respond to high doses of LPS in in vitro assays, it is not the main LPS receptor in vivo, as studies in both TLR4−/− and TLR2−/− mice have shown (6). The downstream signaling pathway is shared between TLR2 and TLR4, involving MyD88 and IRAK related proteins and resulting in activation of NF-κB and AP-1.
Since TLR2 can respond to a variety of peptides derived from gram-positive organisms, a mutation in TLR2 can affect the immune response to various bacterial stimuli and potentially place individuals at a significant risk for septic shock. Only 2 of the 22 patients with gram-positive septic shock had the TLR2 Arg753Gln mutation. Since the mutation is located at the very C terminus of TLR2, it likely affects the signaling function of the molecule, rather than ligand binding. So far no functional domains have been identified in human TLR2, but potential functions of the C terminus include dimerization to form TLR2 homodimers or heterodimers with downstream targets such as MyD88. Since the interleukin-1 (IL-1) receptor is highly homologous to the TLR2 protein, especially at the intracellular domain, evidence suggesting an involvement of the IL-1 receptor C terminus in IL-1-mediated signaling could indicate similar functions for the TLR2 C terminus (15). The results in this paper may indicate that the C terminus of TLR2 is involved in signaling and that Arg753, specifically, is important for this receptor function. More research is necessary to correlate mutations in TLR2 with functional consequences. The fact that only 2 out of 22 patients with gram-positive sepsis were carriers for the TLR2 mutation suggests that additional genes may be involved in determining the susceptibility to septic shock for gram-positive organisms. Mutations in additional genes, such as those for TNF-α (11) and TLR4 (E. Lorenz et al., submitted) have been linked to septic shock susceptibility. We did not find any evidence of linkage between the TLR2 Arg753Gln allele and these two markers in this population. Most likely, a larger sample size is needed because of the rarity of the TLR2 allele. It is therefore necessary to screen a variety of candidate genes, such as those for cytokines and members of the TLR signaling pathway, to characterize the genetics of septic shock susceptibility.
ACKNOWLEDGMENTS
We extend appreciation to Kathy Frees, Nicole Meyer, and Stephanie Swartz for their technical assistance.
This study was supported by grants from the Department of Veterans Affairs (Merit Review), the National Institute of Environmental Health Sciences (ES06537, ES07498, and ES09607), and the National Heart, Lung, and Blood Institute (HL62628).
REFERENCES
- 1.Aliprantis A, Yang R, Mark M, Suggett S, Devaux B, Radolf J, Klimpel G, Godowski P, Zychlinsky A. Cell activation and apoptosis by bacterial lipoproteins through toll-like receptor-2. Science. 1999;285:736–739. doi: 10.1126/science.285.5428.736. [DOI] [PubMed] [Google Scholar]
- 2.Arbour N C, Lorenz E, Schutte B, Zabner J, Kline J, Jones M, Frees K, Watt J L, Schwartz D A. TLR4 mutations are associated with endotoxin hyporesponsiveness in humans. Nat Genet. 2000;25:187–191. doi: 10.1038/76048. [DOI] [PubMed] [Google Scholar]
- 3.Brightbill H, Libraty D, Krutzik S, Yang R, Belisle J, Bleharski J, Maitland M, Norgard M, Plevy S, Smale S, Brennan P, Bloom B, Godowski P, Modlin R. Host defense mechanisms triggered by microbial lipoproteins through toll-like receptors. Science. 1999;285:732–736. doi: 10.1126/science.285.5428.732. [DOI] [PubMed] [Google Scholar]
- 4.Flo T, Halaas O, Lien E, Ryan L, Teti G, Golenbock D, Sundan A, Espevik T. Human toll-like receptor 2 mediates monocyte activation by Listeria monocytogenes, but not by group B streptococci or lipopolysaccharide. J Immunol. 2000;164:2064–2069. doi: 10.4049/jimmunol.164.4.2064. [DOI] [PubMed] [Google Scholar]
- 5.Heine H, Kirschning C, Lien E, Monks B, Rothe M, Golenbock D. Cutting edge: cells that carry a null allele for toll-like receptor 2 are capable of responding to endotoxin. J Immunol. 1999;162:6971–6975. [PubMed] [Google Scholar]
- 6.Hoshino K, Takeuchi O, Kawai T, Sanjo H, Ogawa T, Takeda Y, Takeda K, Akira S. Cutting edge: toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J Immunol. 1999;162:3749–3752. [PubMed] [Google Scholar]
- 7.Knaus W, Draper E, Wagner D. Multiple systems organ failures: epidemiology and prognosis. Crit Care Clin. 1989;5:221–232. [PubMed] [Google Scholar]
- 8.Le Gall J, Lemeshow S, Saulnier F. A new Simplified Acute Physiology Score (SAPS II) based on a European/North American multicenter study. JAMA. 1993;270:2957–2963. doi: 10.1001/jama.270.24.2957. [DOI] [PubMed] [Google Scholar]
- 9.Lien E, Sellati T, Yoshimura A, Flo T, Rawadi G, Finberg R, Carroll J, Espevik T, Ingalls R, Radolf J, Golenbock D. Toll-like receptor 2 functions as a pattern recognition receptor for diverse bacterial products. J Biol Chem. 1999;274:33419–33425. doi: 10.1074/jbc.274.47.33419. [DOI] [PubMed] [Google Scholar]
- 10.Medzhitov R, Janeway C A. Innate immunity: impact on the adaptive immune response. Curr Opin Immunol. 1997;9:4–9. doi: 10.1016/s0952-7915(97)80152-5. [DOI] [PubMed] [Google Scholar]
- 11.Mira J, Cariou A, Grall F, Delclaux C, Losser M, Heshmati F, Cheval C, Monchi M, Teboul J, Riche F, Leleu G, Arbibe L, Mignon A, Delpech M, Dhainaut J. Association of TNF2, a TNF-α promoter polymorphism, with septic shock susceptibility and mortality. JAMA. 1999;282:561–568. doi: 10.1001/jama.282.6.561. [DOI] [PubMed] [Google Scholar]
- 12.Poltorak A, He X, Smirnova I, Liu M-Y, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B. Defective LPS signaling in C3H/Hej and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 13.Sellati T, Bouis D, Caimano M, Feulner J, Ayers C, Lien E, Radolf J. Activation of human monocytic cells by Borrelia burgdorferi and Treponema pallidum is facilitated by CD14 and correlates with surface exposure of spirochetal lipoproteins. J Immunol. 1999;163:2049–2056. [PubMed] [Google Scholar]
- 14.Shimazu R, Akashi S, Ogata H, Nagai Y, Fukudome K, Miyake K, Kimoto M. MD-2, a molecule that confers lipopolysaccharide responsiveness on toll-like receptor 4. J Exp Med. 1999;189:1777–1782. doi: 10.1084/jem.189.11.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh R, Wang B, Shirvaikar A, Khan S, Kamat S, Schelling J, Konieczkowski M, Sedor J. The IL-1 receptor and Rho directly associate to drive cell activation in inflammation. J Clin Investig. 1999;103:1561–1570. doi: 10.1172/JCI5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, Ogawa T, Takeda K, Akira S. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity. 1999;11:443–451. doi: 10.1016/s1074-7613(00)80119-3. [DOI] [PubMed] [Google Scholar]
- 17.Warren W, Hovig E, Smith-Sorensen B, Borresen A, Fujimura F, Liu Q, Feng J, Sommer S. Detection of mutations by single-strand conformation polymorphism (SSCP) analysis and SSCP-hybrid methods. In: Dracopoli N C, Haines J L, Korf B R, Moir D T, Morton C C, Seidman C E, Seidman J G, Smith D R, editors. Current protocols in human genetics. New York, N.Y: John Wiley & Sons, Inc.; 1997. pp. 7.41–7.43. [DOI] [PubMed] [Google Scholar]