Expression of ExsA in trans Confers Type III Secretion System-Dependent Cytotoxicity on Noncytotoxic Pseudomonas aeruginosa Cystic Fibrosis Isolates (original) (raw)
Abstract
Twelve Pseudomonas aeruginosa cystic fibrosis isolates that are not able to exert a type III secretion system (TTSS)-dependent cytotoxicity towards phagocytes have been further studied. The strains, although possessing TTSS genes and exsA, which encodes a positive regulator of the TTSS regulon, showed no transcriptional activation of the exsCBA regulatory operon. The expression of exsA in trans restored the in vitro secretion of TTSS proteins and ex vivo cytotoxicity.
Pseudomonas aeruginosa is an opportunistic pathogen which affects numerous compromised human hosts, most notably cystic fibrosis (CF) patients. Chronic respiratory infections and inflammatory responses cause progressive pulmonary tissue destruction, which is often fatal for CF patients (5). The pulmonary damage is attributed on the one hand to the uncontrolled release of toxic mediators from an excessive number of necrotic polymorphonuclear neutrophils (PMNs) at the site of infection (17, 19) and on the other to the synthesis and accumulation of bacterial products (4). A number of virulence proteins produced by P. aeruginosa that contribute to its pathogenesis have been characterized (11).
A novel virulence mechanism has been recently identified as the type III secretion system (TTSS), which is able to provoke eukaryotic cell intoxication (15, 27). TTSSs are conserved in many gram-negative pathogens and encode on the order of 20 proteins assembled into a complex to secrete and translocate effectors into eukaryotic cell (14). To date, four TTSS-secreted effectors have been identified in P. aeruginosa isolates: ExoS, ExoT, ExoU, and ExoY. ExoS and ExoT are closely related ADP-ribosyltransferases (9, 18, 20–22), ExoY is an adenylate cyclase (29), and ExoU (PepA) is a cytotoxin with unidentified activity (7, 12, 23). We have recently reported that a CF clinical isolate, CHA, is able to induce rapid oncosis of PMNs and macrophages. The cytotoxicity is TTSS dependent but is independent of the toxin ExoU (2, 3). A survey of 29 P. aeruginosa CF isolates showed that 6 strains were able to secrete type III proteins in vitro and to provoke ExoU-independent oncosis of phagocyte cells. However most of the strains found to be noncytotoxic in an ex vivo infection model possess the type III effector genes and the exsA gene, which encodes the transcriptional activator of the TTSS regulon (3, 13).
We showed in this study that expression of the activator of the TTSS regulon, ExsA, in trans was sufficient to activate in vitro secretion and ex vivo cytotoxicity toward phagocytes in 9 out of 12 noncytotoxic isolates tested.
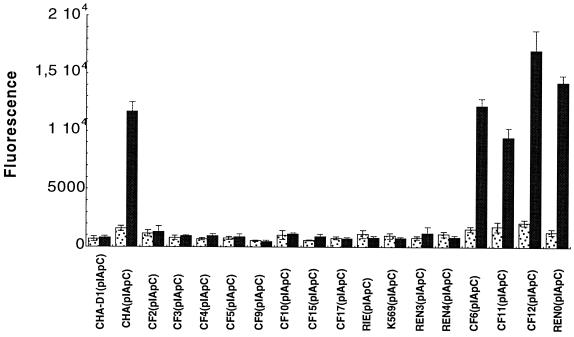
The relevant properties of all strains and plasmids used in this study are summarized in Table 1. To measure and visualize the expression of TTSS genes in living P. aeruginosa, the promoter of the exsCBA operon (8) isolated from the CHA strain was fused with gfp_mut3 (encoding green fluorescent protein [GFP]) (1) in the P. aeruginosa-Escherichia coli vector pUCP20 (25). First, the gfp_mut3 gene was cloned into Xba_I-Pst_I-digested pUCP20. P. aeruginosa strains transformed with pUCP20-gfp_mut3 showed high GFP fluorescence due to the activity of p_lac present just upstream from the cloned gfp_mut3 gene. To avoid interference with p_lac activity, the −10 region of p_lac was deleted with mutagenic oligonucleotide 5′GCTCACAATTCCACACACGAGCCGGAAG using the Altered Sites in vitro mutagenesis system (Promega, Madison, Wis.) according to the manufacturer's instructions. The sequence of mutagenized plasmid (named pIA101*) was confirmed by sequencing. No background GFP fluorescence could be detected in P. aeruginosa strains transformed with pIA101* when cultivated in Luria-Bertani (LB) medium. The p_exsC (8) region was amplified by PCR using genomic DNA of CHA as the template. Primers 5′TCGGATCCCCCATGAAGGACGTC and 5′AGGGATCCTGCGAACTCGGCAAGCAG were synthesized, with Bam_HI sites (underlined) incorporated at the 5′ ends to facilitate cloning. A 280-bp p_exsC PCR product was cloned into the Bam_HI site of pIA101*, giving pIApC, and introduced by electroporation into the CHA strain (6). To test transcriptional fusion, gfp expression under control of p_exsC was measured in the CHA(pIApC) strain grown either in LB or in LB supplemented with 5 mM EGTA and 20 mM MgCl2, a condition known to activate p_exsC transcription (26). GFP expression in individual bacteria was visualized by fluorescence microscopy (Zeiss) and quantified with a FluoroImager (Vistra Fluorescens). The expression of p_exsC-gfp in CHA(pIApC) increased about eightfold during growth under activating conditions (Fig. 1). When the fusion was introduced into the exsA_-mutated CHA strain CHA-D1 (2), no expression of GFP could be detected under activating growth conditions (Fig. 1). These results showed that the p_exsC-gfp fusion could be used as an efficient reporter for monitoring transcription of the exsCBA operon.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or phenotypea | Source and/or reference |
|---|---|---|
| Strains | ||
| E. coli DH5a | Gibco BRL | |
| P. aeruginosa | ||
| CHA | Mucoid CF isolate | 24 |
| CHA-D1 | exsA::Gm mutant of CHA | 2 |
| CF2, CF3, CF4, CF5, CF9, CF10, CF15, RIE, K569, REN3, REN4 | Noncytotoxic CF isolates possessing exsA, exoS, exoT, and exoY genes | 3 |
| CF17 | Noncytotoxic CF isolate possessing exsA, exoS, exoT, exoY, and exoU genes | 3 |
| CF6, CF11, CF12, REN0 | Cytotoxic CF isolates possessing exsA, exoS, exoT, and exoY genes | 3 |
| Plasmids | ||
| pUCP20 | Apr | 25 |
| pDD2 | pUCP20-derived plasmid containing the exsA gene in the sense orientation | 2 |
| pDD3 | pUCP20-derived plasmid containing the exsA gene in the inverse orientation | This work |
| pUCP20-_gfp_mut3 | pUCP20 containing the _gfp_mut3 gene | This work |
| pIA101* | pUCP20-gfp_mut3 with deletion of the p_lac −10 region | This work |
| pIApC | pIA101* containing p_exsC_ | This work |
| pIAX12 | pIA101*-derived plasmid containing the _gfp_mut3 gene under a constitutive promoter X12 | This work (2) |
FIG. 1.
GFP fluorescence in P. aeruginosa isolates transformed with reporter plasmid pIApC, grown in LB medium (stippled bars) or in LB medium supplemented with 5 mM EGTA and 20 mM MgCl2 (inducing conditions) (solid bars). Error bars represent the standard deviation. Data are from at least three independent experiments.
To determine the transcriptional level of the exsCBA operon in noncytotoxic P. aeruginosa isolates, the p_exsC-gfp_ fusion was introduced into strains by electroporation (6). The transformants were selected on Pseudomonas Isolation Agar (Difco) supplemented with 1,000 μg of carbenicillin per ml. For measurement of p_exsC_ activity, the transformed strains were cultivated overnight at 37°C and 300 rpm in LB liquid medium with 300 μg of carbenicillin per ml. Each culture was diluted to an optical density at 600 nm (OD600) of 0.1 and grown to an OD600 of 1 under two culture conditions: in LB medium, where p_exsC_ was not active (no GFP fluorescence could be detected), and in calcium-depleted LB medium, a condition that activated p_exsC_. GFP fluorescence was quantified in a 96-well plate with a FluoroImager using 2.5 × 108 bacteria per well, washed once with sterile water, and resuspended in 100 μl of sterile water.
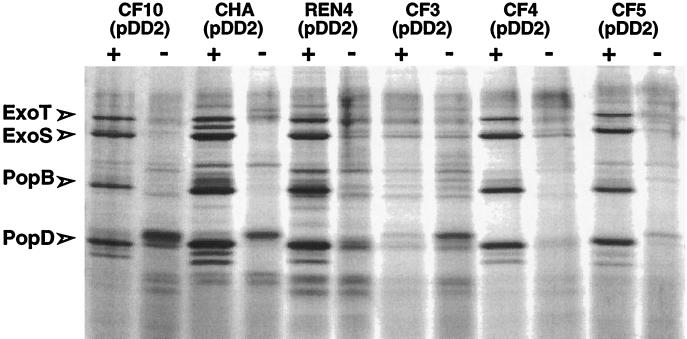
In four selected cytotoxic strains, CF6, CF11, CF12, and REN0, harboring the p_exsC-gfp_ fusion, the expression of p_exsC_ was increased six- to ninefold under inducing conditions, similar to the activation observed for CHA(pIApC) (Fig. 1). In contrast, strains that were found to be noncytotoxic toward PMNs and macrophages over a period of 6 h in the infection model showed no transcriptional activation of p_exsC_ under conditions of calcium depletion (Fig. 1). These results indicate that the absence of secretion of TTSS proteins in vitro and, as a consequence, the absence of a cytotoxic phenotype in the infection model may be due to inefficient transcriptional activation of the exsCBA operon (3). The third gene of the operon encodes ExsA, the transcriptional activator necessary for the expression of operons encoding type III secretion complex proteins, translocation proteins, and effectors (26, 28). In order to see whether the absence of functional ExsA is responsible for the noncytotoxic phenotype, we introduced the functional exsA gene, isolated from CHA, into all 12 noncytotoxic strains. The plasmid pDD2, which was previously used to complement the CHA exsA mutant strain, CHA-D1, contains the exsA gene under the control of a constitutive promoter. The ability of the transformants to secrete in vitro TTSS proteins was first tested. Bacteria were grown in calcium-depleted LB medium, and extracellular proteins were analyzed by 0.1% sodium dodecyl sulfate–12% polyacrylamide gel electrophoresis. The protein profiles were compared to the protein profile obtained with the CHA strain, in which secreted ExoS, ExoT, PopB, and PopD were previously identified by matrix-assisted laser desorption ionization–time-of-flight mass spectrometry (2). The results showed that 11 out of the 12 strains harboring pDD2 were able to secrete TTSS proteins (ExoS, ExoT, PopB, and PopD) in vitro (Fig. 2 and data not shown). To check that the in vitro reactivation of the type III secretion was due to the expression of exsA in trans, the same strains were electrotransformed with pDD3, in which exsA is cloned in the antisense orientation with respect to the promoter. As expected, the strains harboring pDD3 were unable to secrete type III proteins under inducing conditions (data not shown).
FIG. 2.
In vitro TTSS secretion profiles. Silver nitrate-stained 0.1% sodium dodecyl sulfate–12% polyacrylamide gel of supernatants of strains transformed with plasmid pDD2 cultured with (+) or without (−) induction conditions (5 mM EGTA and 20 mM MgCl2) compared to cytotoxic strain CHA(pDD2). The positions of ExoS, ExoT, PopB, and PopD are shown. The strains CF2, CF9, CF15, CF17, RIE, K569, and REN3 transformed with plasmid pDD2, which are able to secrete the four type III secreted proteins, are not shown.
Only one isolate, CF3(pDD2), although efficiently transformed, was unable to secrete any of the TTSS proteins in vitro. We considered the possibility that the constitutive promoter isolated from CHA and used to control exsA expression in pDD2 might not be recognized in the CF3 strain. To test this, plasmid pIAX12 containing _gfp_mut3 placed under the control of the same constitutive promoter as in pDD2 was introduced into the isolates CF3, CF5, and CHA. The fluorescences (in arbitrary units) of CF3(pIAX12), CHA(pIAX12), and CF5(pIAX12) were 26,361 ± 687, 31,722 ± 1588, and 21,620 ± 812, respectively. These similar levels, compared to the low level of fluorescence with transformed strains with pIA101* as a negative control (data not shown), indicated that the promoter is equally active in the three strains. Thus, the nonreactivation of secretion in CF3(pDD2) is not due to a dysfunction of the promoter but may be to a mutation acting further downstream in the activation process.
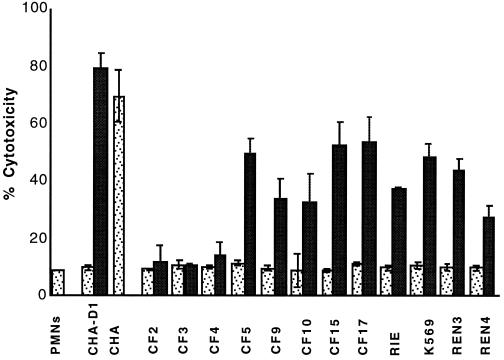
P. aeruginosa CF isolates which are cytotoxic to PMNs and macrophages ex vivo demonstrate in vitro secretion of TTSS proteins (3). In order to test whether the reactivation of in vitro secretion of TTSS proteins in the strains transformed with pDD2 was sufficient to reactivate cytotoxicity, infection experiments using PMNs and J774 macrophages were performed as previously described (2, 3). Briefly, PMNs were isolated from whole blood by Percoll gradient centrifugation, and the macrophage cell line J774 was grown in Dulbecco modified Eagle medium (Gibco) supplemented with 10% heat-inactivated fetal calf serum (Gibco). For infection, bacteria were grown to an OD600 of between 1 and 1.2 after dilution of overnight cultures at an OD600 of 0.1, washed once and resuspended in the appropriate medium. Infection was carried out in 96-well plates with a multiplicity of infection of 10 in a CO2 incubator at 37°C. Cytotoxicity was determined by measuring the release of the cytosolic enzyme lactate dehydrogenase into infection supernatant using a Cytotoxicity Detection Kit (Roche Molecular Biochemical, Meylan, France). The cytotoxic CF isolate CHA was able to induce type III secretion-dependent cell death of PMNs (Fig. 3) and macrophages (data not shown), with 80% cell lysis measured at 3 h postinfection. The same phenotype was previously described for cytotoxic CF clinical isolates (2, 3). Untransformed noncytotoxic isolates, similarly to the exsA mutant CHA-D1, were unable to provoke cell lysis, even after 6 h postinfection (Fig. 3). The isolates CF5, CF9, CF10, CF15, CF17, RIE, K569, REN3, and REN4 transformed with pDD2 were able to induce 30 to 55% PMN death (Fig. 3) and 30 to 90% macrophage death (data not shown) at 3 h postinfection. The strains harboring pDD3 (exsA cloned in the inverse orientation) were still noncytotoxic, with values similar to those obtained with untransformed strains (data not shown). The variations of cytotoxicity observed between strains in ex vivo infection model (Fig. 3) were not related to the quantity of secreted type III proteins in vitro (Fig. 2). We have previously shown that, although cytotoxic, different CF isolates may have different kinetics of cytotoxicity (3). This might be due to differences between strains in cell growth, adhesion, level of activation of the TTSS, and/or resistance to PMNs. Two pDD2-transformed strains (CF2 and CF4) which were capable of secreting TTSS proteins in vitro failed to induce cell death of PMNs and macrophages (Fig. 3 and data not shown). Although the secretion complex seemed to be intact, an inefficient translocation process may have caused the lack of cytotoxicity.
FIG. 3.
Cytotoxicity to PMNs in P. aeruginosa strains transformed with pDD2. The percentage of cytotoxicity was calculated according to the release of lactate dehydrogenase activity (multiplicity of infection of 10). Stippled bars, PMNs infected by untransformed strains; solid bars, PMNs infected by strains transformed with pDD2. Error bars indicate standard deviations.
In this study, we have shown that 9 out of 12 successfully transformed strains previously characterized as noncytotoxic in a cellular model of infection could be made cytotoxic just by expressing the exsA gene in trans from a constitutive promoter. ExsA is an activator of TTSS operons, and its expression is autoregulated (26). ExsA is encoded by the last gene in the exsCBA operon, which is expressed from the regulated p_exsC_ promoter. The exsC and exsB genes are involved in the processing effect on exsA mRNA (10). The cytotoxic phenotype conferred by exsA in trans using a constitutive promoter suggests that the ExsA protein, which is necessary for the expression of the TTSS, is not functional in noncytotoxic strains. This might result either from a mutation(s) in the exsCBA operon affecting the coding or regulatory regions or from inappropriate signaling in the regulatory cascade upstream from p_exsC_.
The bacterial TTSS is an efficient mechanism for eukaryotic cell intoxication and in many cases a requirement for pathogen survival in the host. However, the synthesis of the system is an energetically expensive process for the bacterial cell, involving more than 20 proteins that make up the secretion complex, translocation complex, effectors, and regulatory proteins. As is the case for the complex regulatory network controlling the synthesis of the P. aeruginosa exopolysaccharide alginate (16), the regulation of the TTSS might involve a signaling pathway upstream from ExsA, which would allow expression of TTSS genes only when appropriate conditions inside the host are met. The environmental stimuli and the signaling pathway upstream from p_exsC_ affecting expression of the regulatory loci remain to be determined. Our survey of CF clinical isolates showed that about 96% of noncytotoxic strains possess genes capable of encoding proteins of the TTSS (3). Our results suggest that these strains are all potentially cytotoxic.
Acknowledgments
This work was supported by grant 98033 from the Association Française de Lutte contre la Mucoviscidose (AFLM) and by grants from DGA (DSP/STTC).
We thank F. Morel (Laboratoire d'Enzymologie, CHU-Grenoble) for whole blood and A. Colbeau and W. Dischert for valuable discussion and critical reading of the manuscript.
REFERENCES
- 1.Cormack B P, Valdivia R H, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 2.Dacheux D, Attree I, Schneider C, Toussaint B. Cell death of human polymorphonuclear neutrophils induced by a Pseudomonas aeruginosa cystic fibrosis isolate requires a functional type III secretion system. Infect Immun. 1999;67:6164–6167. doi: 10.1128/iai.67.11.6164-6167.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dacheux D, Toussaint B, Richard M, Brochier G, Croize J, Attree I. Pseudomonas aeruginosa cystic fibrosis isolates induce rapid, type III secretion-dependent, but ExoU-independent, oncosis of macrophages and polymorphonuclear neutrophils. Infect Immun. 2000;68:2916–2924. doi: 10.1128/iai.68.5.2916-2924.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Döring G. Pseudomonas aeruginosa as an opportunistic pathogen. New York, N.Y: Plenum Press; 1993. [Google Scholar]
- 5.Elborn J S, Shale D J. Cystic fibrosis. 2. Lung injury in cystic fibrosis. Thorax. 1990;45:970–973. doi: 10.1136/thx.45.12.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enderle P J, Farwell M A. Electroporation of freshly plated Escherichia coli and Pseudomonas aeruginosa cells. Biotechniques. 1998;25:954–956. doi: 10.2144/98256bm05. , 958. (Erratum, 26:716, 1999.) [DOI] [PubMed] [Google Scholar]
- 7.Finck-Barbancon V, Goranson J, Zhu L, Sawa T, Wiener-Kronish J P, Fleiszig S M, Wu C, Mende-Mueller L, Frank D W. ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol Microbiol. 1997;25:547–557. doi: 10.1046/j.1365-2958.1997.4891851.x. [DOI] [PubMed] [Google Scholar]
- 8.Frank D W, Iglewski B H. Cloning and sequence analysis of a trans-regulatory locus required for exoenzyme S synthesis in Pseudomonas aeruginosa. J Bacteriol. 1991;173:6460–6468. doi: 10.1128/jb.173.20.6460-6468.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frithz-Lindsten E, Du Y, Rosqvist R, Forsberg A. Intracellular targeting of exoenzyme S of Pseudomonas aeruginosa via type III-dependent translocation induces phagocytosis resistance, cytotoxicity and disruption of actin microfilaments. Mol Microbiol. 1997;25:1125–1139. doi: 10.1046/j.1365-2958.1997.5411905.x. [DOI] [PubMed] [Google Scholar]
- 10.Goranson J, Hovey A K, Frank D W. Functional analysis of ExsC and ExsB in regulation of exoenzyme S production by Pseudomonas aeruginosa. J Bacteriol. 1997;179:1646–1654. doi: 10.1128/jb.179.5.1646-1654.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Govan J R, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hauser A R, Kang P J, Engel J N. PepA, a secreted protein of Pseudomonas aeruginosa, is necessary for cytotoxicity and virulence. Mol Microbiol. 1998;27:807–818. doi: 10.1046/j.1365-2958.1998.00727.x. [DOI] [PubMed] [Google Scholar]
- 13.Hovey A K, Frank D W. Analyses of the DNA-binding and transcriptional activation properties of ExsA, the transcriptional activator of the Pseudomonas aeruginosa exoenzyme S regulon. J Bacteriol. 1995;177:4427–4436. doi: 10.1128/jb.177.15.4427-4436.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hueck C J. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee C A. Type III secretion systems: machines to deliver bacterial proteins into eukaryotic cells? Trends Microbiol. 1997;5:148–156. doi: 10.1016/S0966-842X(97)01029-9. [DOI] [PubMed] [Google Scholar]
- 16.Martin D W, Schurr M J, Mudd M H, Govan J R, Holloway B W, Deretic V. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc Natl Acad Sci USA. 1993;90:8377–8381. doi: 10.1073/pnas.90.18.8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McElvaney N G, Nakamura H, Birrer P, Hebert C A, Wong W L, Alphonso M, Baker J B, Catalano M A, Crystal R G. Modulation of airway inflammation in cystic fibrosis. In vivo suppression of interleukin-8 levels on the respiratory epithelial surface by aerosolization of recombinant secretory leukoprotease inhibitor. J Clin Invest. 1992;90:1296–1301. doi: 10.1172/JCI115994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGuffie E M, Frank D W, Vincent T S, Olson J C. Modification of Ras in eukaryotic cells by Pseudomonas aeruginosa exoenzyme S. Infect Immun. 1998;66:2607–2613. doi: 10.1128/iai.66.6.2607-2613.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakamura H, Yoshimura K, McElvaney N G, Crystal R G. Neutrophil elastase in respiratory epithelial lining fluid of individuals with cystic fibrosis induces interleukin-8 gene expression in a human bronchial epithelial cell line. J Clin Invest. 1992;89:1478–1484. doi: 10.1172/JCI115738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olson J C, Fraylick J E, McGuffie E M, Dolan K M, Yahr T L, Frank D W, Vincent T S. Interruption of multiple cellular processes in HT-29 epithelial cells by Pseudomonas aeruginosa exoenzyme S. Infect Immun. 1999;67:2847–2854. doi: 10.1128/iai.67.6.2847-2854.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olson J C, McGuffie E M, Frank D W. Effects of differential expression of the 49-kilodalton exoenzyme S by Pseudomonas aeruginosa on cultured eukaryotic cells. Infect Immun. 1997;65:248–256. doi: 10.1128/iai.65.1.248-256.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pederson K J, Vallis A J, Aktories K, Frank D W, Barbieri J T. The amino-terminal domain of Pseudomonas aeruginosa ExoS disrupts actin filaments via small-molecular-weight GTP-binding proteins. Mol Microbiol. 1999;32:393–401. doi: 10.1046/j.1365-2958.1999.01359.x. [DOI] [PubMed] [Google Scholar]
- 23.Sawa T, Yahr T L, Ohara M, Kurahashi K, Gropper M A, Wiener-Kronish J P, Frank D W. Active and passive immunization with the Pseudomonas V antigen protects against type III intoxication and lung injury. Nat Med. 1999;5:392–398. doi: 10.1038/7391. [DOI] [PubMed] [Google Scholar]
- 24.Toussaint B, Delic-Attree I, Vignais P M. Pseudomonas aeruginosa contains an IHF-like protein that binds to the algD promoter. Biochem Biophys Res Commun. 1993;196:416–421. doi: 10.1006/bbrc.1993.2265. [DOI] [PubMed] [Google Scholar]
- 25.West S E, Schweizer H P, Dall C, Sample A K, Runyen-Janecky L J. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene. 1994;148:81–86. doi: 10.1016/0378-1119(94)90237-2. [DOI] [PubMed] [Google Scholar]
- 26.Yahr T L, Frank D W. Transcriptional organization of the trans-regulatory locus which controls exoenzyme S synthesis in Pseudomonas aeruginosa. J Bacteriol. 1994;176:3832–3838. doi: 10.1128/jb.176.13.3832-3838.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yahr T L, Goranson J, Frank D W. Exoenzyme S of Pseudomonas aeruginosa is secreted by a type III pathway. Mol Microbiol. 1996;22:991–1003. doi: 10.1046/j.1365-2958.1996.01554.x. [DOI] [PubMed] [Google Scholar]
- 28.Yahr T L, Hovey A K, Kulich S M, Frank D W. Transcriptional analysis of the Pseudomonas aeruginosa exoenzyme S structural gene. J Bacteriol. 1995;177:1169–1178. doi: 10.1128/jb.177.5.1169-1178.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yahr T L, Vallis A J, Hancock M K, Barbieri J T, Frank D W. ExoY, an adenylate cyclase secreted by the Pseudomonas aeruginosa type III system. Proc Natl Acad Sci USA. 1998;95:13899–13904. doi: 10.1073/pnas.95.23.13899. [DOI] [PMC free article] [PubMed] [Google Scholar]