Lamin-dependent Localization of UNC-84, A Protein Required for Nuclear Migration in Caenorhabditis elegans (original) (raw)
Abstract
Mutations in the Caenorhabditis elegans unc-84 gene cause defects in nuclear migration and anchoring. We show that endogenous UNC-84 protein colocalizes with Ce-lamin at the nuclear envelope and that the envelope localization of UNC-84 requires Ce-lamin. We also show that during mitosis, UNC-84 remains at the nuclear periphery until late anaphase, similar to known inner nuclear membrane proteins. UNC-84 protein is first detected at the 26-cell stage and thereafter is present in most cells during development and in adults. UNC-84 is properly expressed in unc-83 and anc-1 lines, which have phenotypes similar to unc-84, suggesting that neither the expression nor nuclear envelope localization of UNC-84 depends on UNC-83 or ANC-1 proteins. The envelope localization of Ce-lamin, Ce-emerin, Ce-MAN1, and nucleoporins are unaffected by the loss of UNC-84. UNC-84 is not required for centrosome attachment to the nucleus because centrosomes are localized normally in unc-84 hyp7 cells despite a nuclear migration defect. Models for UNC-84 localization are discussed.
INTRODUCTION
The wholesale movement or “migration” of nuclei is required for the growth and development of all eukaryotes. Genes involved in nuclear migration have been identified and characterized in Saccharomyces cerevisiae, filamentous fungi, Caenorhabditis elegans, Drosophila, and vertebrates (reviewed in Morris, 2000). Most of these genes encode molecular motors (dynein, or dynein-associated proteins) or proteins that mediate microtubule assembly or disassembly.
In _C. elegan_s, the development of several specific cell types depends on nuclear migration events, and these migrations in turn depend on the unc-83 and unc-84 genes. For example, during the formation of the embryonic hypodermal syncytium, 17 of the 23 hyp7 nuclei undergo migration from one side of the cell to the other (Malone et al., 1999). Likewise, in the ventrolateral layer of P cells, nuclei migrate toward the ventral cord, allowing the P cells to form a single row along the ventral cord. In both unc-83 and unc-84 lines, these nuclear migration events fail to occur (Horvitz and Sulston, 1980; Malone et al., 1999). The UNC-84 protein is predicted to be split approximately in half by a single transmembrane domain, and its C-terminal region was found to be homologous to the Schizosaccharomyces pombe Sad1 gene in a region termed the SUN (_S_ad/_UN_c) domain, the function of which is unknown. Sad1 is an integral membrane protein associated during meiosis and mitosis with the spindle pole body in S. pombe, and its overexpression results in its accumulation at the nuclear periphery (Hagan and Yanagida, 1995). Expression of an UNC-84:GFP transgene suggested that UNC-84 is present in many cells and is localized to the nuclear periphery. Based on this nuclear peripheral localization and the presence of the SUN domain, it was proposed that UNC-84 is localized to the outer nuclear membrane, where it is associated with centrosomes (Malone et al., 1999; Raff, 1999; Morris, 2000).
The nuclear envelope is the boundary between the nucleus and the cytoplasm. The outer nuclear membrane is continuous with the endoplasmic reticulum. The outer nuclear membrane is separated from the inner nuclear membrane by the perinuclear space. The nuclear lamina is a filamentous meshwork that lies between the inner nuclear membrane and the peripheral chromatin. The lamina also extends throughout the nuclear interior (Broers et al., 1999; Liu et al., 2000; Moir et al., 2000). An increasing number of integral and peripheral membrane proteins have been identified that associate with lamins. Thus, we define the term “nuclear lamina” broadly to include lamins and lamin-binding proteins (Cohen et al., 2001).
Because the nuclear outer membrane is continuous with ER membranes, all known proteins present in the outer membrane are also distributed throughout the ER. Membrane proteins that localize exclusively to the nuclear envelope are believed to do so by a retention mechanism, in which they diffuse along the “pore membrane domain” to the inner membrane, where they are retained by binding to intranuclear components such as lamins (Soullam and Worman, 1995). There is currently no precedent for a protein localized exclusively at the outer membrane of the nucleus. We therefore decided to further examine the localization of the UNC-84 protein. Our results suggest that UNC-84 is a nuclear lamina protein whose nuclear envelope localization requires lamin.
MATERIALS AND METHODS
Strains and Antibodies
All unc-84, unc-83, and anc-1 strains were described previously (Malone et al., 1999). Antiserum against Ce-lamin was described previously (Liu et al., 2000). Mouse polyclonal antipeptide antibodies against Ce-emerin (serum 3272) and Ce-MAN1 (serum 3266) are described (Lee et al., 2000). To obtain polyclonal antibodies against UNC-84, one mouse (serum 3398) and one rat (serum 3595) were immunized at 3-week intervals with synthetic peptides conjugated to keyhole limpet hemocyanin (KLH; peptide synthesis and conjugation were done by Boston BioMolecules, Woburn, MA). Immunizations and serum production were done by Covance Research Products (Denver, PA). The peptide antigen was CAVWKWIGNQSQKRW-COOH, which corresponds to the first 14 residues of UNC-84 plus an N-terminal Cys residue. Both antisera against UNC-84 worked well for protein blots and indirect immunofluorescence staining. Antiserum against the mtf-1 gene product (matefin) was obtained by immunizing rats with a synthetic peptide corresponding to the matefin N-terminus.
MAb414, which recognizes several different nucleoporins, was purchased from BAbCO (Richmond, CA). An mAb against tubulin was purchased from Sigma Chemical Co. (catalogue number T-9026; St. Louis, MO). Cy3-conjugated goat anti-mouse and goat anti-rat antibodies and FITC-conjugated goat anti-rabbit antibodies were purchased from Jackson Laboratories (West Grove, PA). G. Hermann and J. Priess (Fred Hutchinson Cancer Research Center, Seattle, WA) kindly provided mAb IFA, which was used to detect embryonic centrosomes (Leung et al., 1999).
Immunostaining
Immunostaining was performed as described (Lee et al., 2000; Liu et al., 2000). Nematodes were fixed for 20 min at −20°C in methanol and 10 min at −20°C in acetone. Antibodies were diluted in PBS (1:200 for UNC-84, Ce-emerin, or Ce-MAN1, 1:400 for lamin and 1:1000 for mAb414). Nematodes were viewed with a Zeiss (Thornwood, NY) Axioskop microscope equipped with epifluorescence illumination with the use of a 63×/numerical aperture 1.4 Apochromat objective lens. Confocal samples were acquired with the Noran Oz confocal laser scanning microscope system with the use of Intervision Software (version 6.3) on a Silicon Graphics Indy R5000 platform (Silicon Graphics Inc., Mountain View, CA). A krypton–argon laser (Omnichrome series 43, Noran Instruments Inc., Middleton, WI) that excites at wavelengths of 488 and 568 nm was used to obtain optical sections. Narrow-band emission filters (525 and 605 nm) were used to eliminate channel cross-talk, and 0.5-mm _z_-plane sections (as determined by full-width half-maximum intensity values) were collected using a 10-mm fixed slit. Slides were imaged with the use of a 100× oil-immersion planar apochromatic objective lens (numerical aperture = 1.35) through an Olympus (Tokyo, Japan) IX-50 inverted microscope.
Centrosomes were immunostained using mAb IFA as described (Leung et al., 1999). Images were collected using an Axioplan2 microscope (Carl Zeiss Inc.) and a Hamamatsu C4742–95 CCD camera (Hamamatsu Photonics KK, Bridgewater, NJ). A stack of images taken in the z plane at 0.5-mm intervals was deconvolved and analyzed using Openlab 2.0.7 (Improvision, Lexington, MA) software.
Protein Extracts
To prepare protein samples for SDS-PAGE, mixed-stage nematodes were boiled for 5 min in 2× SLB solution (25 mM Tris-HCl, pH 6.8, 20% glycerol, 0.2 M β-mercaptoethanol, 4% SDS, 0.001% bromphenol blue), and the extract was then passed through a 25-gauge, 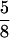 -inch syringe. Protein extracts were subjected to SDS-PAGE, transferred to nitrocellulose membranes, and immunoblotted with specific antibodies.
-inch syringe. Protein extracts were subjected to SDS-PAGE, transferred to nitrocellulose membranes, and immunoblotted with specific antibodies.
Cell Extracts
C. elegans nuclei were prepared essentially as described (Lee et al., 2000). For chemical extraction, 1 volume of nuclei was either used directly or thawed on ice, washed once in PBS-Inh (PBS containing 1 mM PMSF,1 mg/ml leupeptin, and 1 mg/ml aprotinin), centrifuged at 4000 × g for 1 min at 4°C, and then extracted for 30 min at 4°C in 10 volumes of PBS-Inh plus the extraction reagent (e.g.,1 M NaCl, 1% Triton X-100, or 8 M urea). After extraction, the residual nuclear pellet was separated from the supernatant by centrifugation at 9000 × g for 1 min at 4°C. The nuclear pellet was washed in PBS. The supernatant was further purified by centrifugation at 14,000 × g for 5 min at 4°C.
To prepare protein samples for SDS-PAGE, we boiled each sample for 5 min in 2× SLB solution (25 mM Tris-HCl, pH 6.8, 20% glycerol, 0.2 M β-mercaptoethanol, 4% SDS, 0.001% bromphenol blue) and then passed the extract through a 25-gauge, 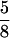 -inch syringe. Protein extracts were subjected to SDS-PAGE, transferred to polyvinylidene difluoride membrane, and immunoblotted with specific antibodies.
-inch syringe. Protein extracts were subjected to SDS-PAGE, transferred to polyvinylidene difluoride membrane, and immunoblotted with specific antibodies.
RESULTS
UNC-84 Protein Colocalizes with the Nuclear Lamina
Polyclonal antibodies were raised in both rat and mouse against an N-terminal peptide of UNC-84 (see MATERIALS AND METHODS) and used to localize endogenous UNC-84 in C. elegans embryos by indirect immunofluorescence. Both the mouse antibodies (Figure 1, left) and the rat antibodies (Figure 2) localized UNC-84 to the nuclear envelope. This localization was specific for UNC-84, because envelope staining was not detected with preimmune sera or in unc-84(n369) null embryos (see below).
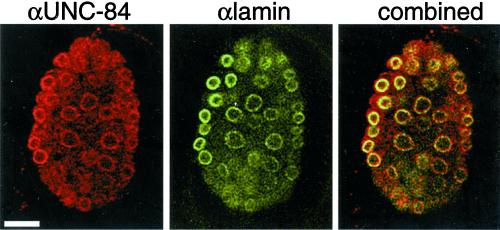
Figure 1.
Colocalization of endogenous UNC-84 and Ce-lamin at the nuclear envelope in wild-type embryos. C. elegans embryos were double-stained by indirect immunofluorescence using antibodies against endogenous UNC-84 (red) and endogenous Ce-lamin (green) and viewed by confocal microscopy. Overlap between UNC-84 and Ce-lamin appears yellow in the combined panel. Bar, 10 μm (all panels).
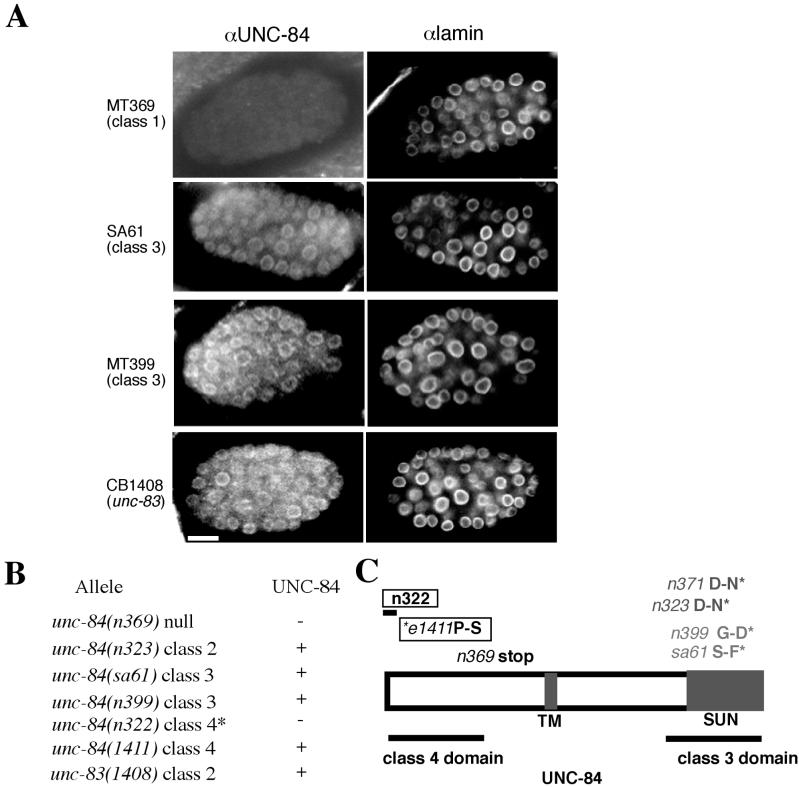
Figure 2.
Immunolocalization of endogenous UNC-84 and Ce-lamin in unc-84 and unc-83 mutant lines. (A) Embryos were double-stained by indirect immunofluorescence using antibodies against UNC-84 (left panels) and Ce-lamin (right panels) and viewed by confocal microscopy. Ce-lamin staining was detected in all nuclei and was not affected by the unc-84 mutation. Bar, 10 μm (all panels). (B) Summary of UNC-84 localization in all tested unc-84 alleles. Because unc-84(n322) is a small deletion that removes the antigen that the UNC-84 antibody was raised against, we expect a negative result. (C) The molecular lesions in tested alleles of unc-84. Class 1 mutations are written in black, class 2 mutations in dark gray, class 3 mutations in light gray, and class 4 mutations are boxed.
The localization of UNC-84 was similar to that of nuclear lamins (Liu et al., 2000). We therefore used confocal microscopy to analyze embryos double-stained for both UNC-84 and Ce-lamin and found that UNC-84 and Ce-lamin colocalized at the nuclear envelope (Figure 1, right). To determine if UNC-84 behaves as an integral membrane protein, we tested its resistance to extraction by detergents, salt, and chaotrophic agents (Singer, 1974). Isolated C. elegans nuclei were extracted with PBS containing 1% Triton X-100, 1 M NaCl, or 8 M urea reagent and analyzed as described (Lee et al., 2000). UNC-84 and Ce-lamin both pelleted after treatment with 1% Triton X-100. As predicted for an integral membrane protein, UNC-84, but not Ce-lamin, pelleted after extraction with 1 M NaCl or 8 M urea, similar to the extraction properties of Ce-MAN1 and Ce-emerin (Lee et al., 2000), suggesting that UNC-84 is an integral membrane protein.
Staining Endogenous UNC-84 in Wild-type, unc-84, unc-83, and anc-1 Lines
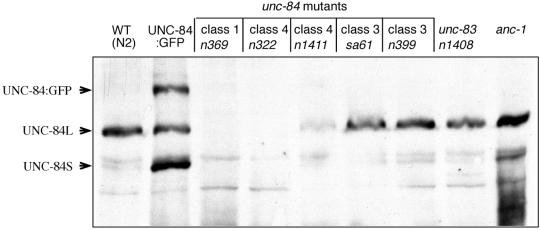
We used Western blotting to detect endogenous UNC-84 protein on blots containing total protein extracts from mixed-stage wild-type (N2) C. elegans. Antibodies against UNC-84 recognized one major band and several minor bands (Figure 3). The major band and one minor band were absent in mixed-stage extracts from unc-84(n369) (arrows in Figure 3), which is a predicted null with a nonsense mutation early in the coding region of UNC-84 (Malone et al., 1999). We therefore concluded that the large and small bands were both UNC-84 proteins. The two UNC-84–specific bands migrated on SDS-PAGE close to the predicted masses (125 and 99 kDa) of the two alternatively spliced products of unc-84. The large (125 kDa) isoform is sufficient for UNC-84 function (Malone et al., 1999).
Figure 3.
Immunodetection of UNC-84 protein in unc-84, unc-83, and anc-1 mutant lines. Western blot analysis of mixed stages: wild-type (WT), UNC-84: GFP, unc-84(n369) (class 1), unc-84(n322) (class 4), unc-84(n1411) (class 4), unc-84(sa61) (class 3), unc-84(n399) (class 3), unc-83(n1408) and anc-1 lines. The positions of UNC-84 long (L, 125 kDa) and short (S, 98 kDa) protein isoforms in wild-type nematodes are indicated. A band corresponding in size to the UNC-84: GFP fusion protein is marked by an arrow.
The UNC-84 antibodies were also used to determine if genetically identified point mutations in the N-terminal or SUN-domains of UNC-84, which disrupt UNC-84 activity (Malone et al., 1999), also interfered with the expression or localization of each mutant protein. Both isoforms of UNC-84 were present in nematodes carrying mutations in the SUN-domain (class 3; unc-84(sa61) and unc-84(n399); Malone et al., 1999), as shown by immunoblotting (Figure 3), and the mutant proteins were properly localized at the nuclear envelope (Figure 2). UNC-84 protein was also present in nematodes carrying mutations in the N-terminal domain (class 4; unc-84(n1411); Malone et al., 1999; Figure 3). The class 2 alleles unc-84(n371) and unc-84(n323) (Malone et al., 1999) also did not disrupt UNC-84 localization to the nuclear envelope. As expected, UNC-84 was not detected in unc-84(n322) embryos_,_ which lack the peptide epitope, nor was UNC-84 protein detected in immunoblots (Figure 3) or by antibody staining (Figure 2) in the unc-84(n369) null line. These results confirmed that unc-84(n369) is devoid of UNC-84 protein and demonstrated that mutant UNC-84 proteins were expressed and properly localized in lines carrying unc-84 mutant alleles from classes 2, 3, and 4.
Unc-84 and unc-83 have significantly overlapping nuclear migration defects and they were proposed to be involved in the same pathway (Horvitz and Sulston, 1980). In addition, a recent study shows that UNC-83 requires the SUN domain of UNC-84 for its nuclear envelope localization (Starr et al., 2001). It was therefore interesting to analyze if UNC-84 localization depends on UNC-83 activity. We found that UNC-84 does not depend on UNC-83 for its expression or envelope localization, because UNC-84 was expressed (Figure 3) and localized (Figure 2) normally in lines carrying unc-83(e1408), a null allele (Malone et al., 1999). Anc-1, mutations in which cause anchoring defects similar to unc-84, was also not required for UNC-84 expression, because normal levels of UNC-84 protein were detected in extracts from the anc-1 null line (Figure 3). Thus, UNC-84 is likely to act upstream of both UNC-83 and ANC-1 or in parallel pathways.
The Pattern of UNC-84 Expression during C. elegans Development
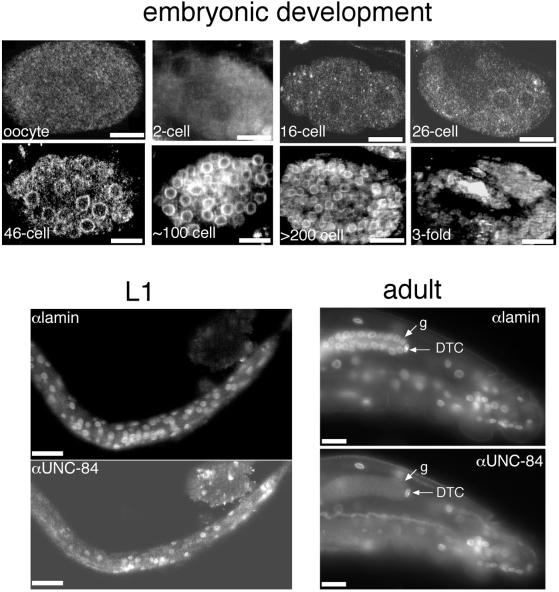
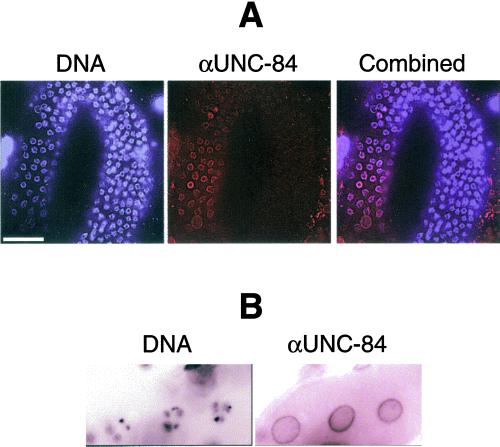
We used rat and mouse anti–UNC-84 antibodies to independently determine the pattern of UNC-84 expression during C. elegans development; both sera gave similar results. UNC-84 was not detected in the nuclear envelope from fertilized oocytes until just before the 26-cell stage (Figure 4). Weak staining was first detected in all nuclear envelopes at the 26-cell stage (Figure 4). This signal became stronger as embryonic development proceeded, with UNC-84 localized in every nuclear envelope. We also double-stained larval stages L1–L4 for both endogenous UNC-84 and Ce-lamin (Figure 4). We stained for Ce-lamin because it is present in the nuclear envelope of every cell except spermatocytes (Liu et al., 2000), and therefore served as a positive control for antibody penetration. Almost all nuclear envelopes in L1–L4 stage larvae stained positively with UNC-84 antibodies. Likewise, all somatic adult cells, including the distal tip cell (DTC) in the gonad, stained positive for UNC-84 (see Figure 4 for anterior view of C. elegans adult). Germ cells in the mitotic and transition zones of the gonad stained negative for UNC-84 (Figures 4 and 5), whereas Ce-lamin staining was normal (Figure 4; see also Liu et al., 2000). UNC-84 was detectable in nuclear envelopes after the pachytene stage and continued to be positive in oocytes before fertilization (Figure 5). These results showed that endogenous UNC-84 is expressed in the nuclear envelopes of essentially all adult and embryonic cells, except between fertilization and the 26-cell stage. This result confirms the localization of UNC-84::GFP (Malone et al., 1999), but was surprising, given that loss of UNC-84 affects only a small number of migrating nuclei in the developing embryo, when the protein is expressed in most cell types.
Figure 4.
Immunolocalization of UNC-84 during development. Animals at different stages of development were double-stained with antibodies against UNC-84 and Ce-lamin and viewed by confocal microscopy. UNC-84 was first detected at low levels at the 26-cell stage and was present in all cells (panels marked “embryonic development”). The UNC-84 signal became stronger as development progressed, and was present in all embryonic cells. UNC-84 was detected in most (but not all) cells at the L1/L2 larvae stages, whereas all cells stained positive for lamins, as expected (Liu et al., 2000). The UNC-84 and Ce-lamin signals colocalized at the nuclear envelopes of essentially all somatic adult cells, including the distal tip cell (DTC). The only adult cells that lacked UNC-84 signal were germ cells at the distal part of the gonad (g). Bar, 10 μm (each panel).
Figure 5.
UNC-84 is first expressed in pachytene cells (A). UNC-84 continues to be expressed in oocytes (B). DNA staining, blue; UNC-84 staining, red. Bar, 10 μm (all panels).
UNC-84 in C. elegans Remains in the Nuclear Envelope until Late Anaphase
We recently showed that during mitosis in C. elegans embryos, the nuclear membranes and lamina are completely disassembled only during late anaphase (Lee et al., 2000). To determine if UNC-84 fits this same pattern, we used antibodies to follow the fate of endogenous UNC-84 protein during the different stages of mitosis in 64 to 200-cell embryos. UNC-84 maintained a nuclear rim-staining pattern during interphase, prophase, prometaphase, metaphase, and early anaphase. UNC-84 was completely released from chromatin only during late anaphase, and began to reaccumulate around chromatin in early telophase (Figure 6). This unusual pattern was strikingly similar to the mitotic disassembly and reassembly dynamics of the inner nuclear membrane proteins, Ce-emerin and Ce-MAN1, which are integral proteins of the inner nuclear membrane (Lee et al., 2000).
Figure 6.
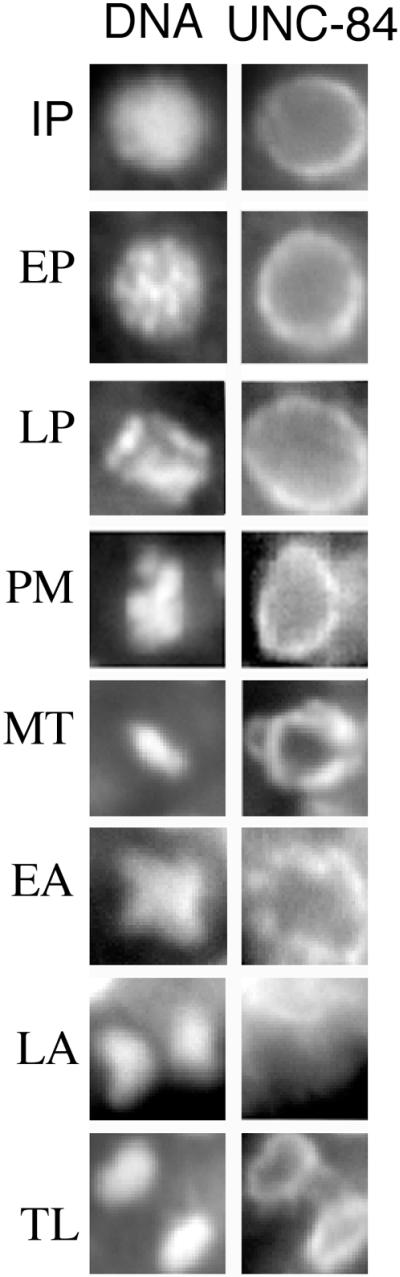
UNC-84 localization during mitosis. Wild-type embryos (100–300 cells) were stained with rat anti–UNC-84 antibodies (right panels) and Hoechst 33258 to stain DNA (left panels), viewed by fluorescence microscopy and photographed at different stages of the cell cycle. A representative nucleus from each stage is shown: IP, interphase; EP, early prophase; LP, late prophase; PM, prometaphase; MT, metaphase; EA, early anaphase; LA, late anaphase and TL, telophase. Nuclear staining was detected in all stages of mitosis except late anaphase, consistent with other nuclear membrane markers (Lee et al., 2000).
Ce-lamin, Ce-emerin, Ce-MAN1, and Nucleoporins Do Not Depend on UNC-84 for Their Nuclear Envelope Localization
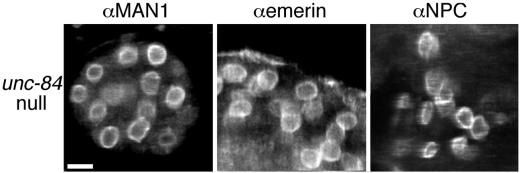
The unc-84(n369) line, which has no detectable UNC-84 protein (Figures 2 and 3), was used to determine if UNC-84 is required for the nuclear envelope localization of other known lamina proteins. unc-84(n369) embryos containing between 50 and a few hundred nuclei were stained pair-wise for UNC-84 and either nucleoporins (using mAb414) or each of three other nuclear lamina proteins: Ce-lamin (Liu et al., 2000), Ce-emerin, and Ce-MAN1 (Lee et al., 2000). The nucleoporins and all three lamina proteins remained properly localized to the nuclear envelope in the absence of UNC-84 (Figure 7).
Figure 7.
MAN1, emerin, and nucleoporins localize normally in the unc-84(n369) line (unc-84 null). Embryos (left and middle) or adults (right) from the unc-84(n369) line were double-stained with antibodies against Ce-MAN1, Ce-emerin, a family of FG-repeat nucleoporins (αNPC), and viewed by confocal microscopy. Bar, 10 μm (all panels).
UNC-84 Requires Ce-lamin for Its Nuclear Envelope Localization
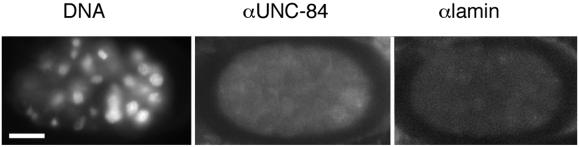
In mammals, lamins are essential for the efficient localization of at least one tested nuclear membrane protein, emerin (Sullivan et al., 1999; Olins et al., 2001). We used indirect immunofluorescence to localize endogenous UNC-84 in lamin-deficient lmn-1(RNAi) embryos, created by injecting double-stranded lamin RNA into the syncytial gonad of adult hermaphrodites (Liu et al., 2000). Embryos with more than 50 cells were triple-stained for DNA, endogenous Ce-lamin, and endogenous UNC-84 (Figure 8). UNC-84 staining at the nuclear envelope was not detectable in lamin-deficient embryos, demonstrating that UNC-84 requires Ce-lamin for stable retention at the nuclear envelope (Figure 8). In parallel, lmn-1(RNAi) embryos were stained for nucleoporins (mAb414), Ce-emerin, and matefin, a novel nuclear membrane protein. Nucleoporins were clustered to one side of the nucleus as expected in lamin-deficient cells (Liu et al., 2000). Ce-emerin was displaced from the nuclear envelope, similar to UNC-84. In contrast, matefin remained localized to the nuclear envelope in the lmn-1(RNAi) embryos (K. Lee, J. Liu, K. Wilson, and Y. Gruenbaum, unpublished observations). These results demonstrate the dependence of UNC-84 on Ce-lamin for its nuclear envelope localization.
Figure 8.
UNC-84 depends on Ce-lamin for its nuclear envelope localization. Lamin-deficient lmn-1 (RNAi) embryos were triple-stained with DAPI (left), and antibodies against endogenous UNC-84 (middle) and Ce-lamin (right). Bar, 10 μm (all panels).
Centrosome Attachment to the Nuclear Envelope Is Not Disrupted in unc-84 Mutants
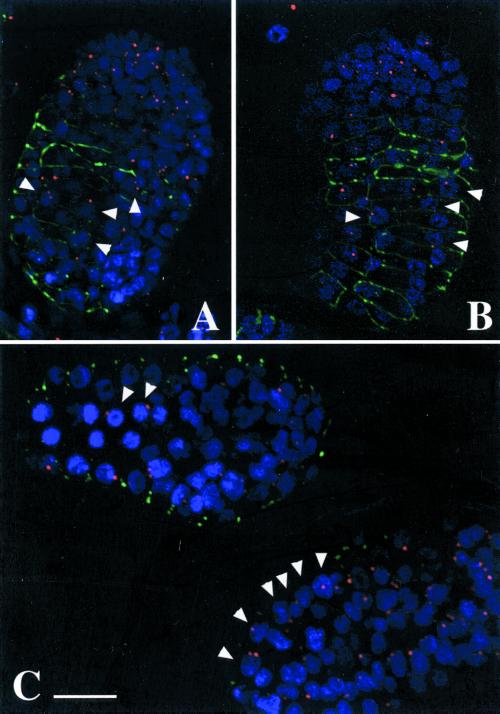
UNC-84 was previously proposed to reside in the outer nuclear membrane and mediate attachment to centrosomes or microtubules during nuclear migration (Malone et al., 1999; Raff, 1999). This model predicted that in an unc-84 null background, centrosomes would migrate normally across the hyp-7 cell, but the nucleus would fail to follow because of lack of UNC-84. Detachment of centrosomes was found in one-cell stage C. elegans embryos mutated in the heavy chains of the molecular motor, dynein, resulting in migration defects (Gonczy et al., 1999). Likewise, cytoplasmic dynein is required for the nuclear attachment and migration of centrosomes during mitosis in Drosophila embryos (Robinson et al., 1999). To test the hypothesis that loss of UNC-84 might similarly uncouple the nucleus from the centrosome, we localized the centrosomes and nuclei in migrating hyp7 cells in wild-type and unc-84(n369) null embryos. In wild-type hyp7 cells, centrosomes associated closely with migrating nuclei (Figure 9, A and B). These centrosomes did not associate with the leading edge of nuclei, but instead were associated with random sides, suggesting that the force that pulls (or pushes) migrating nuclei may not be exerted through the centrosome. Also surprisingly, centrosome localization in unc-84 null embryonic hyp7 cells was indistinguishable from wild-type (Figure 9C). This result strongly suggested that UNC-84 is not required for centrosome attachment to the nuclear envelope.
Figure 9.
Centrosome localization in wild-type (N2) and unc-84(n369) hyp7 cells. Single, deconvoluted sections through postgastrulation, preelongation embryos are shown. (A and B) Nearly dorsal views of wild-type embryos. (C) A dorsal view (left) and a lateral view (right) of unc-84(n369) embryos. Centrosomes, identified by mAb IFA, are pseudocolored red, DAPI stained nuclei are blue, and JAM-1: GFP is shown in green to identify the hypodermal cell–cell boundaries. Arrowheads point to hyp7 nuclei and their associated centrosomes. Bar, 10 μm (all panels).
DISCUSSION
As the structural scaffold for the nucleus, the nuclear lamina, which includes lamins and lamin-associated proteins, is involved directly or indirectly in many biological functions. The lamina provides structural support for chromosomes, maintains nuclear shape, and spaces nuclear pore complexes. The lamina is also required for DNA replication and is proposed to mediate transcriptional regulation (Cohen et al., 2001). Our findings for UNC-84 add a novel role to the nuclear lamina in regulating nuclear migration and nuclear, the possible mechanisms of which are discussed below.
UNC-84 Is Probably a Nuclear Lamina Protein
The number of integral and peripheral membrane proteins that localize to the nuclear inner membrane is growing steadily (Cohen et al., 2001). Most inner membrane proteins bind directly to nuclear lamins and are therefore defined as part of the lamina (Gruenbaum et al., 2000). Each lamin-binding protein is likely to have a unique function, none of which are currently understood; in some cases lamin-binding activity may only be needed to localize the protein. A previous study suggested that UNC-84 is part of the nuclear envelope (Malone et al., 1999). Our current results suggest in several independent ways that UNC-84 directly or indirectly interacts with the nuclear lamina: UNC-84 colocalizes with the single C. elegans lamin protein, Ce-lamin, during interphase and exhibits the same distinct dynamics as inner membrane proteins Ce-MAN1 and Ce-emerin during mitosis (Lee et al., 2000). UNC-84 also depends on lamins for its nuclear envelope localization in vivo. In addition, a recent study reveals that the mouse homolog of UNC-84 is localized in the nuclear envelope, most probably to the inner nuclear membrane (Dreger et al., 2001). Finally, ectopic expression of C. elegans UNC-84 in mouse NIH-3T3 cells resulted in its association with the nuclear envelope. Thus, UNC-84 probably joins the growing number of lamin-associated proteins and adds new functions to this structure. Because the loss of UNC-84 had no effect on the localization of any other tested envelope proteins (Ce-emerin, Ce-MAN1, nucleoporins), we propose that UNC-84 associates with lamins and organizes factors important for nuclear migration and anchorage, independent of any complexes formed by other known nuclear membrane proteins. One of these proteins is UNC-83, which depends on UNC-84 for its nuclear envelope localization (Starr et al., 2001).
UNC-84 Expression during C. elegans Development
UNC-84 is expressed in most cells during C. elegans development. This result is consistent with a previous report showing UNC-84::GFP expression in most larval and adult cells (Malone et al., 1999). In contrast, loss of UNC-84 affects only a small number of migrating nuclei. We therefore hypothesize that UNC-84 functions may overlap with other protein(s), which is not present in migrating nuclei. Alternatively, UNC-84 might have a binding partner that is expressed uniquely in migrating nuclei, and depends on UNC-84 for its function.
UNC-84 staining was not detected in germ cells before the pachytene stage, or between fertilization and the 26-cell stage. This is a unique expression pattern for a nuclear envelope protein. The lack of staining between fertilization and the 26-cell stage could be due to 1) degradation of UNC-84 after fertilization, 2) release and diffusion in the ER resulting in cytoplasmic staining which is too weak to be detected, or 3) loss of antibody recognition due to posttranslational modification at the UNC-84 N-terminus.
UNC-84 Expression in Different Mutant Lines
Existing unc-84 mutations have been divided into four distinct complementation groups, termed classes 1–4 (Malone et al., 1999). Alleles in class 3 and class 4 completely complement each other. It was therefore suggested that class 3 and class 4 mutations, which cluster in the C- and N-terminal halves of UNC-84, respectively, do not grossly affect protein levels or protein structure. We verified this hypothesis by showing that UNC-84 expression levels in class 3 and 4 mutants were roughly similar to wild-type animals. We also showed that the envelope localization of UNC-84 was normal in these mutants. Because both class 3 and class 4 alleles affect nuclear migration, we suggest that mutations in either domain specifically disrupt interactions between UNC-84 and unknown factors required for nuclear migration. The best such candidate is UNC-83, because the nuclear migration phenotypes in unc-83 and unc-84 lines overlap and UNC-83 localization depends on UNC-84 (Malone et al., 1999; Starr et al., 2001). UNC-84 expression and envelope localization were also normal in class 2 (unc-84 (n371)) mutants. Class 3 mutations lie in the C-terminal SUN domain and disrupt nuclear anchoring. We therefore suggest that the SUN-domain of UNC-84 interacts with factors involved in nuclear anchoring. One candidate is ANC-1, because mutations in the anc-1 gene cause nuclear anchoring phenotypes similar to class 3 alleles. Because UNC-84 expression is normal in unc-83 and anc-1 mutant lines, we suggest that both UNC-83 and ANC-1 act downstream or in parallel to UNC-84.
Centrosome Localization Does Not Depend on UNC-84
Nuclear migration is required for development in both animals and plants (reviewed by Morris, 2000). Although relatively little is known about nuclear migration, it usually requires microtubules, microtubule-dependent motors, and, in many cases, the centrosomes (Morris, 2000). In a previous article (Malone et al., 1999) and reviews (Raff, 1999; Morris, 2000), it was suggested that UNC-84 might directly anchor cytoplasmic dynein and centrosomes to nuclei. However, we found that centrosomes maintained their wild-type localization and attachment to the nuclear envelope in unc-84 null hyp7 cells. Furthermore, because our data suggest that UNC-84 could be in the inner nuclear membrane and centrosomes are located in the cytoplasm, it is unlikely that UNC-84 interacts directly with centrosomes because the outer membrane and lumenal space separate them. The topology of UNC-84 in the membrane has not been determined, so it is not known which domain (N-terminus or C-terminus) is facing the perinuclear space.
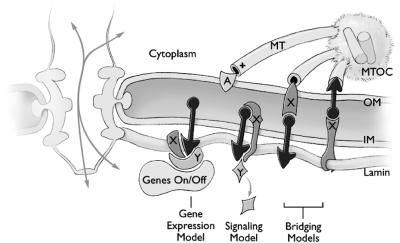
Models for UNC-84 in Gene Expression, Signaling, or Nuclear Envelope “Bridging”
Several possible models for UNC-84 function are consistent with its lamin-dependent localization and nuclear migration function, as diagrammed in Figure 10. Because UNC-84 is expressed in nearly all cells but its loss causes a phenotype in only a few cells, we speculate that UNC-84 may have multiple binding partners, some of which are cell specific in their expression. Putative binding partners are depicted as proteins X and Y in Figure 10. One such partner is UNC-83, which is a nuclear envelope protein expressed only in cells with migrating nuclei (Starr et al., 2001). In the gene expression model, we propose that UNC-84 and its binding partners might play an active role in regulating the transcription of proteins required for nuclear migration and anchoring, similar to the manner in which mammalian transcriptional regulators Rb and Oct-1 are proposed to repress transcription when associated with the nuclear lamina (Cohen et al., 2001). In the signaling model, we propose that binding to UNC-84 might serve a signaling role by retaining, sequestering, or activating downstream proteins that mediate nuclear migration and anchorage. Finally, we propose a bridging model in which UNC-84 interacts with the lumenal domain of an outer membrane protein, to form a structural “bridge” through the nuclear envelope; this bridge would connect UNC-84 to outer membrane proteins that interact with microtubule-based motors or the centrosome. Alternatively, it is also possible that UNC-84 is actually located in the outer membrane and its lamin-dependent localization depends on linking to an inner membrane protein that binds lamins (Figure 10). In this case, UNC-84 would represent a completely novel outer-nuclear-membrane–specific protein, which remains connected to the nuclear lamina until late anaphase during mitosis. To our knowledge, there is no precedent for such a translumenal structural bridge through the nuclear envelope, but this model is attractive because it provides a mechanism to structurally link the pushing or pulling forces of nuclear migration to the nuclear lamina, via UNC-84. In theory, the attachment of cytoskeletal elements to nuclear pore complexes, which are anchored in the lamina, would achieve the same goal, but there is currently no evidence for functional links between pore complexes and the cytoskeleton. Determining the orientation of UNC-84 in the inner membrane and identifying and localizing its binding partners will be essential for distinguishing between these models.
Figure 10.
Three potential models for UNC-84 function during nuclear migration. A cross section of the nuclear envelope is depicted with one nuclear pore complex. The two domains of UNC-84 protein are depicted as a black circle and black triangle, one on each side of the inner nuclear membrane (INM). The plus ends of three microtubules (MT) are shown extending from the centrosome. In the gene expression model, UNC-84 forms a complex with one or more partners (X, Y), to either activate or repress genes that directly mediate nuclear migration. In the signaling model, UNC-84 could act alone or with partner X (depicted as an integral membrane protein) to regulate protein Y, which either activates or inhibits migration. In the bridging model, UNC-84 is located on either the inner or outer nuclear membrane, and interacts with the lumenal domain of protein X, in the opposing membrane. In either case, the protein on the inner membrane binds directly to lamins.
ACKNOWLEDGMENTS
The authors thank Dieter Riemer and Klaus Weber for antibodies to Ce-lamin and G. Hermann and J. Priess for the mAb IFA. This work was supported by grants from the USA-Israel Binational Science Foundation, the Israel Science Foundation, and the German-Israel Foundation GIF 1–573-036.13 (to Y.G), by grants from the W.W. Smith Charitable Trust and National Institutes of Health (NIH) grant RO1GM48646 (to K.L.W). This work was also supported by the Howard Hughes Medical Institute where M.H. is an assistant investigator and by NIH postdoctoral fellowship F32GM20127 (to D.S.) and NIH postdoctoral fellowship F32HD08331 (to J.L.).
Footnotes
REFERENCES
- Broers JL, Machiels BM, van Eys GJ, Kuijpers HJ, Manders EM, van Driel R, Ramaekers FC. Dynamics of the nuclear lamina as monitored by GFP-tagged A-type lamins. J Cell Sci. 1999;112:3463–3475. doi: 10.1242/jcs.112.20.3463. [DOI] [PubMed] [Google Scholar]
- Cohen M, Lee KK, Wilson KL, Gruenbaum Y. Transcriptional repression, apoptosis, human disease, and the functional evolution of the nuclear lamina. Trends Biochem Sci. 2001;26:41–47. doi: 10.1016/s0968-0004(00)01727-8. [DOI] [PubMed] [Google Scholar]
- Dreger M, Bengtsson L, Schoneberg T, Otto H, Hucho F. Nuclear envelope proteomics. Novel integral membrane proteins of the inner nuclear membrane. Proc Natl Acad Sci USA. 2001;98:11943–11948. doi: 10.1073/pnas.211201898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonczy P, Pichler S, Kirkham M, Hyman AA. Cytoplasmic dynein is required for distinct aspects of MTOC positioning, including centrosome separation, in the one cell stage Caenorhabditis elegans embryo. J Cell Biol. 1999;147:135–150. doi: 10.1083/jcb.147.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum Y, Wilson KL, Harel A, Goldberg M, Cohen M. Nuclear lamins-structural proteins with fundamental functions. J Struct Biol. 2000;129:313–323. doi: 10.1006/jsbi.2000.4216. [DOI] [PubMed] [Google Scholar]
- Hagan I, Yanagida M. The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J Cell Biol. 1995;129:1033–1047. doi: 10.1083/jcb.129.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvitz HR, Sulston JE. Isolation and genetic characterization of cell-lineage mutants of the nematode Caenorhabditis elegans. Genetics. 1980;96:435–454. doi: 10.1093/genetics/96.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KK, Gruenbaum Y, Spann P, Liu J, Wilson KL. C. elegans nuclear envelope proteins emerin, MAN1, lamin, and nucleoporins reveal unique timing of nuclear envelope breakdown during mitosis. Mol Biol Cell. 2000;11:3089–3099. doi: 10.1091/mbc.11.9.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung B, Hermann G, Priess J. Organogenesis of the Caenorhabditis elegans intestine. Dev Biol. 1999;216:114–134. doi: 10.1006/dbio.1999.9471. [DOI] [PubMed] [Google Scholar]
- Liu J, Rolef-Ben Shahar T, Riemer D, Spann P, Treinin M, Weber K, Fire A, Gruenbaum Y. Essential roles for Caenorhabditis elegans lamin gene in nuclear organization, cell cycle progression, and spatial organization of nuclear pore complexes. Mol Biol Cell. 2000;11:3937–3947. doi: 10.1091/mbc.11.11.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CJ, Fixsen WD, Horvitz HR, Han M. UNC-84 localizes to the nuclear envelope and is required for nuclear migration and anchoring during C. elegans development. Development. 1999;126:3171–3181. doi: 10.1242/dev.126.14.3171. [DOI] [PubMed] [Google Scholar]
- Moir RD, Spann TP, Lopez-Soler RI, Yoon M, Goldman AE, Khuon S, Goldman RD. The dynamics of the nuclear lamins during the cell cycle- relationship between structure, and function. J Struct Biol. 2000;129:324–334. doi: 10.1006/jsbi.2000.4251. [DOI] [PubMed] [Google Scholar]
- Morris NR. Nuclear migration. from fungi to mammalian brain. J Cell Biol. 2000;148:1097–1101. doi: 10.1083/jcb.148.6.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olins AL, Herrmann H, Lichter P, Kratzmeier M, Doenecke D, Olins DE. Nuclear envelope, and chromatin compositional differences comparing undifferentiated, and retinoic acid-, and phorbol ester-treated HL-60 cells. Exp Cell Res. 2001;268:130–142. doi: 10.1006/excr.2001.5269. [DOI] [PubMed] [Google Scholar]
- Raff JW. Nuclear migration: the missing (L)UNC. Curr Biol. 1999;9:R708–R710. doi: 10.1016/s0960-9822(99)80446-1. [DOI] [PubMed] [Google Scholar]
- Robinson JT, Wojcik EJ, Sanders MA, McGrail M, Hays TS. Cytoplasmic dynein is required for the nuclear attachment and migration of centrosomes during mitosis in Drosophila. J Cell Biol. 1999;146:597–608. doi: 10.1083/jcb.146.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer SJ. The molecular organization of membranes. Annu Rev Biochem. 1974;43:805–833. doi: 10.1146/annurev.bi.43.070174.004105. [DOI] [PubMed] [Google Scholar]
- Soullam B, Worman HJ. Signals and structural features involved in integral membrane protein targeting to the inner nuclear membrane. J Cell Biol. 1995;130:15–27. doi: 10.1083/jcb.130.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr D, Hermann GJ, Malone CJ, Fixsen W, Priess JR, Horvitz HR, Han M. unc-83 encodes a novel component of the nuclear envelope, and is essential for proper nuclear migration. Development. 2001;128:5039–5050. doi: 10.1242/dev.128.24.5039. [DOI] [PubMed] [Google Scholar]
- Sullivan T, Escalente-Alcalde D, Bhatt H, Anver M, Naryan B, Nagashima K, Stewart CL, Burke B. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol. 1999;147:913–920. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]