The Methylosome, a 20S Complex Containing JBP1 and pICln, Produces Dimethylarginine-Modified Sm Proteins (original) (raw)
Abstract
snRNPs, integral components of the pre-mRNA splicing machinery, consist of seven Sm proteins which assemble in the cytoplasm as a ring structure on the snRNAs U1, U2, U4, and U5. The survival motor neuron (SMN) protein, the spinal muscular atrophy disease gene product, is crucial for snRNP core particle assembly in vivo. SMN binds preferentially and directly to the symmetrical dimethylarginine (sDMA)-modified arginine- and glycine-rich (RG-rich) domains of SmD1 and SmD3. We found that the unmodified, but not the sDMA-modified, RG domains of SmD1 and SmD3 associate with a 20S methyltransferase complex, termed the methylosome, that contains the methyltransferase JBP1 and a JBP1-interacting protein, pICln. JBP1 binds SmD1 and SmD3 via their RG domains, while pICln binds the Sm domains. JBP1 produces sDMAs in the RG domain-containing Sm proteins. We further demonstrate the existence of a 6S complex that contains pICln, SmD1, and SmD3 but not JBP1. SmD3 from the methylosome, but not that from the 6S complex, can be transferred to the SMN complex in vitro. Together with previous results, these data indicate that methylation of Sm proteins by the methylosome directs Sm proteins to the SMN complex for assembly into snRNP core particles and suggest that the methylosome can regulate snRNP assembly.
The neuromuscular disease spinal muscular atrophy (SMA) is characterized by degeneration of motor neurons of the spinal cord, leading to muscular weakness and atrophy (40). The survival-of-motor-neurons gene (SMN) is present as an inverted repeat on chromosome 5 at 5q13, and over 98% of SMA patients have mutations or deletions of the telomeric copy of the gene (SMN1), resulting in reduced levels of the survival motor neuron (SMN) protein (31; reviewed in reference 8).
snRNP core particles assemble in the cytoplasm from newly exported snRNAs and the core Sm proteins (SmB, SmD1, SmD2, SmD3, SmE, SmF, and SmG). Cap hypermethylation of the U snRNAs requires that the core Sm proteins assemble on the Sm sites of the U1, U2, U4, and U5 snRNAs. snRNP Sm core particles are believed to be constructed of a seven-member ring containing each of the Sm proteins with a single U snRNA bound in the center of the ring (28). The presence of the properly assembled Sm core as well as the 2,2,7-trimethylguasnosine (m3G) cap is required for snRNP import to the nucleus (13, 16, 17, 27, 38, 39, 41). Regions conserved in all of the Sm proteins (Sm motifs 1 and 2) (51) are most likely required for proper folding of these proteins and their reciprocal interactions (28). In the cytoplasm, SMN is associated with the Sm proteins (9, 36). In Xenopus oocytes, microinjection of anti-SMN complex antibodies inhibits or stimulates snRNP core particle formation, and transient expression of a dominant-negative mutant of SMN in mammalian cells sequesters Sm proteins and snRNA in large cytoplasmic aggregates (7, 15, 45). These results indicated that the SMN complex has a crucial role in snRNP core particle assembly.
Another protein, pICln, has been suggested to be a negative regulator of snRNP assembly. This conclusion was based on inhibition of snRNP assembly upon injection of large quantities of recombinant pICln into Xenopus oocytes. PICln was also shown to bind to the Sm proteins B′, D1, D2, D3, and E (47) as well as to a protein described as IBP72 (30). Recently, IBP72 was shown to interact with Janus kinases (JAK1 and JAK2) and was renamed JBP1 (for JAK-binding protein 1) (46). JBP1 has been shown to be a protein arginine methyltransferase (46, 49). The yeast homologue of JBP1 (skb1) appears to be involved in the osmotic response and in regulation of mitosis (4, 26). However, the function of mammalian JBP1 is not known.
SMN oligomerizes and is found in a large complex with Gemin2 (formerly SIP1) (36), Gemin4 (10), and the DEAD box RNA helicase Gemin3 (9). The conserved YG domain (amino acids 276 to 279 in human SMN) is responsible for SMN oligomerization (37, 44), which greatly increases SMN′s affinity for SmD1, SmD3, and SmB (44). The SmD1 and SmD3 arginine- and glycine-rich (RG) carboxyl-terminal domains are necessary and sufficient for SMN binding (20). In contrast, SmB has a much longer carboxyl-terminal RG domain (approximately 151 amino acids long) which has stretches of prolines and dispersed RG repeats. This domain is required, but not sufficient, for SMN binding (20). All of the thus-far-tested SMN mutants found in SMA patients who do not have the SMA gene deleted are defective in Sm protein binding, providing evidence that a defect in these interactions may play a role in the pathology of SMA (7, 20, 44).
Specific arginines in the carboxyl-terminal RG domains of SmD1 and SmD3 are posttranslationally modified to symmetrical dimethylarginines (sDMAs) (5). SMN preferentially binds to the sDMA-modified forms of SmD1 and SmD3, suggesting that protein methylation may be a general mechanism for regulating protein-protein interaction (21). Proteins were shown to contain dimethylarginines over 30 years ago (42, 43), yet knowledge of the molecular functions of protein arginine methylation remains limited. Type I protein arginine methyltransferase (PRMT) activity produces asymmetrical dimethylarginine (aDMA), while type II PRMT activity produces sDMA (reference 24 and references therein). A number of type I PRMTs have been characterized, and it appears that the majority of cellular PRMT activity is from the type I enzymes (19, 25). Asymmetrical dimethylation of hnRNP proteins has been suggested to play a role in their nuclear export (52), and the type I methyltransferases CARM1 and PRMT1 were shown to be involved in transcriptional activation by a nuclear hormone receptor (11, 29). No cloned PRMTs have been conclusively shown to be type II enzymes, and besides SmD1 and SmD3, myelin basic protein (3) is the only other protein known to contain sDMAs.
Here, we demonstrate that JBP1 is a type II PRMT which symmetrically dimethylates the RG domains of Sm proteins and associates, along with the pICln protein, with RG domain-containing Sm proteins in a 20S methyltransferase complex. We refer to this large 20S complex as the methylosome and show that SmD3 from the methylosome, but not that from a 6S pICln complex, can be transferred to the SMN complex. These findings suggest that the methylosome functions to symmetrically dimethylate Sm proteins prior to their association with SMN and snRNP assembly and therefore that the methylosome could regulate snRNP assembly.
MATERIALS AND METHODS
DNA constructs and recombinant proteins.
Flag-pcDNA3 was constructed by ligating a linker encoding the Flag epitope into _Hin_dIII-_Bam_HI-cleaved pcDNA3 (Invitrogen). The JBP1 cDNA was a kind gift from Stevan Marcus (26) and was cloned in frame with the Flag epitope in Flag-pcDNA3 to make Flag-JBP1pcDNA3. The arginine 368-to-alanine mutation in JBP1 was constructed by PCR site-directed mutagenesis by overlapping PCR using primer pairs encoding the desired amino acid change and restriction sites for cloning back into Flag-pcDNA3. Constructs for bacterial expression and recombinant protein purification of glutathione _S_-transferase (GST)–D1, GST-D1c29, GST-D3, and GST-D3c32 and constructs for mammalian cell expression of myc-D3, myc-D1, myc-D1Δc29, and myc-D3Δc32 were described previously (20). Maltose-binding protein (MBP) fused to the alpha subunit of β-galactosidase (MBP-βGalα) and MBP fused to the RGG domain of hnRNP A2 (MBP-A2-RGG) were produced from pMAL-c2 (New England Biolabs) and purified according to the manufacturer's recommendation. MBP-A2-RGG was constructed by subcloning a PCR fragment encoding hnRNP A2 amino acids 266 to 341 in frame with MBP in pMAL-c2.
Cell culture and transfection.
293 cells were cultured in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum. 293 cells growing on 100-mm-diameter culture dishes (about 40% confluent) were transfected with 5 to 10 μg of DNA using the CalPhos Mammalian Transfection Kit (Clontech Laboratories) according to the manufacturer's recommendation.
Affinity chromatography and cell extract preparation.
Extract preparation and cell fractionation were done as described previously (54). GST fusion proteins (3 to 8 μg) immobilized on 30 μl of glutathione-Sepharose beads (Amersham) or peptides (1 nmol) immobilized on High Performance streptavidin-Sepharose (Amersham) were incubated with 3 to 4 mg of extracted cellular protein (50 to 150 μl) in 1 ml of binding buffer (50 mM Tris [pH 7.5], 200 mM NaCl, 0.2 mM EDTA, 0.05% NP-40, 2 mM dithiothreitol, and one tablet of complete EDTA-free protease inhibitor cocktail per 50 ml). Beads were washed seven times with 1 ml of binding buffer, boiled for 5 min in sodium dodecyl sulfate (SDS) sample buffer, and separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE). The peptides used were described previously (21).
Sucrose gradient centrifugation.
Cellular extracts were separated on 5 to 20% sucrose gradients at 33,000 rpm in an SW41 rotor at 5°C for 15 h 20 min. Fractions (0.5 ml) were collected, and 6% of each fraction was separated by SDS-PAGE and analyzed by Western blotting as indicated. The remainder of the fractions were pooled as indicated and used directly for immunoprecipitation or interaction analysis. Five 100-mm-diameter plates of 293 cells transfected with the indicated expression constructs were used for each sucrose gradient.
Purification of the SMN complex.
For purification of the SMN complex, Flag-Gemin2, myc-SMN, myc-Gemin3, and myc-Gemin4 were transiently expressed in 293 cells, and cytoplasmic extract was prepared from these cells and incubated with anti-Flag Sepharose (Sigma) for 2 h at 4°C. After extensive washing with RSB-200 (10 mM Tris-HCl [pH 7.5], 200 mM NaCl, 2.5 mM MgCl2) with 0.01% NP-40, the complex thus purified was used in the experiment presented in Fig. 9.
FIG. 9.
SmD3 is transferred from the methylosome to the SMN complex. Cytoplasmic extract prepared from myc-D3-expressing 293 cells was separated on a sucrose gradient, and fractions 2 to 5 (6S) and 12 to 15 (20S) were pooled and incubated with SMN complex (FLAG-SMN) which had been purified on anti-Flag beads from cytoplasmic extract transiently expressing myc-SMN, Flag-Gemin2, myc-Gemin3, and myc-Gemin4 or with anti-Flag beads incubated with cytoplasmic extract prepared from vector-transfected cells (FLAG-vector) as indicated. After washing, retained proteins were analyzed by Western blotting to detect myc-D3 and JBP1 (arrows). The total lanes show 10% of the pooled fractions used in each binding.
Immunoprecipitation, Western blotting, and antibodies.
Immunoprecipitations, SDS-PAGE, and Western blot analysis were done as previously described (36). Antibodies used in these experiments were as follows: anti-SMN (2B1) (35), anti-Gemin2 (2E17) (36), anti-Gemin3 (11G9) (9), anti-Gemin4 (22C10) (10), anti-myc (9E10), anti-pICln (Transduction Laboratories), anti-Sm protein (Y12) (33), anti-JBP1 (a kind gift from Michael Nunn and Gary Zieve), nonimmune antibody SP2/0 (12), and anti-Flag (Sigma). Immunoprecipitation on anti-Flag beads and elution with Flag peptide (Sigma) were done according to the manufacturer's recommendation.
Mass spectrometric identification of JBP1.
The 70-kDa band of GST-D3c32 was excised, reduced, alkylated, and digested using trypsin as described previously (53). The acidified supernatant was placed on a thin layer of 4-hydroxy-α-cinnamic acid as a matrix (56). Spectra were acquired on a Reflex III instrument (Bruker Daltonics, Bremen, Germany). The data were interpreted and searched against a nonredundant database containing over 600,000 protein entries using the Protein and Peptide Software Suite from MDS Proteomics (Odense, Denmark).
Methyltransferase activity assay.
Flag-JBP1pcDNA3, Flag-JBP1R368ApcDNA3, and Flag-pcDNA3 DNAs (7.5 μg) were each transfected into one 100-mm-diameter plate of 293 cells. Total cellular extracts were made in RSB-100 (10 mM Tris [pH 7.5], 100 mM NaCl, and 2.5 mM MgCl2) with 1% Empigen, incubated with 30 μl of anti-Flag M2 Agarose Affinity Gel (Sigma), and washed five times with 1 ml of RSB-200 with 1% Empigen. Flag-JBP1 and Flag-JBP1R368A were eluted from the beads with Flag peptide (Sigma) into RSB-100 according to the manufacturer's recommendation, and about 200 ng of eluted protein was taken for Western blot analysis. For methylation, approximately 100 ng of Flag-JBP1 or Flag-JBP1R368A (dialyzed into buffer D [14]) was incubated with 2 μCi of adenosyl-l-[_methyl_-3H]methionine (3H-SAM) and 600 ng of each recombinant protein in 30 μl at 30°C for 30 min. Reactions were stopped by the addition of SDS sample buffer, and products were separated by SDS-PAGE. Following Coomassie blue staining, radioactive signals were amplified by treatment with Amplify (Amersham) and exposed to film for 3 h. HeLa cytoplasmic extract was separated by sucrose centrifugation, and fractions 12 to 25 were pooled and immunoprecipitated with 30 μl of anti-pICln antibody or 30 μl of SP2/0 immobilized on 60 μl of GammaBind G Sepharose (Amersham). One-sixth of the immunoprecipitation product was used for Western blot analysis, and the remainder was split evenly and used for methylation. Immunoprecipitation product from one gradient was used for 12 methyltransferase reactions. Methyltransferase activity was analyzed as the anti-Flag immunoprecipitates were analyzed except that films were exposed for 16 to 18 h. Purification of the methylosome and analysis of its methyltransferase activity were done as described above except that 20S fractions from cytoplasmic extract separated on a sucrose gradient were immunoprecipitated with anti-Flag antibody with 0.01% NP-40 instead of Empigen. For analysis of methylated arginine products, 1 μg of GST-TEV-D3c32 was incubated with Flag-JBP1 or His-PRMT1 (500 ng) with 20 μl of 3H-SAM (20 μCi, 0.4 nmol) and 1 nmol of cold SAM for 1.5 h at 30°C. Binding buffer (500 μl) was added, and GST-TEV-D3c32 was captured on 20 μl of glutathione-Sepharose. After washing five times with 1 ml of binding buffer and two times with 1 ml of TEV cleavage buffer (Gibco BRL), the peptide was cleaved in a 50-μl reaction mixture with 5 U of TEV protease (Gibco BRL) for 1 h at 30°C. TEV cleavage buffer was removed (the peptide remained associated with the Sepharose in TEV cleavage buffer). The peptide was eluted with 50 μl of 1 M triethylammonium bicarbonate (pH 8.5) and dried in a Speed-Vac. After resuspension in 50 μl of 6 M constant boiling (Pierce) HCl, the peptide was hydrolyzed under vacuum at 110°C for 20 h, and acid was removed by drying. The hydrolyzed peptide was resuspended in 10 μl of water, and 5 μl was mixed with 30 nmol each of sDMA and monomethylarginine (MMA) (JBP1 methylated) or aDMA and MMA (His-PRMT1 methylated). The standard amino acids were purchased from Calbiochem. This mixture was loaded on LK6DF silica gel 60 thin-layer chromatography plates (Whatman) and separated with ammonium hydroxide-chloroform-methanol-water (2:0.5:4.5:1). Standard amino acids were visualized with ninhydrin, and radiolabeled methylated arginine products were visualized by phosphorimager analysis.
RESULTS
The RG domains of SmD1 and SmD3 bind the methyltransferase JBP1.
We have recently shown that the RG domains of SmD1 and SmD3 are necessary and sufficient for SMN binding (20). Furthermore, SMN binds preferentially to the sDMA-modified forms of these domains (21). To further investigate the interactions of the RG domains of SmD1 and SmD3 and to identify the potential methyltransferase that modifies them, HeLa cell extract was incubated with immobilized GST fused to the RG domain of SmD3 (GST-D3c32) or to GST alone as a control. After extensive washing, bound proteins were eluted with SDS sample buffer, resolved by SDS-PAGE, and visualized by Coomassie blue staining (Fig. 1). The immobilized proteins themselves (without incubation with cellular extract) were also resolved and stained to distinguish recombinant proteins from bound cellular proteins. A specific profile of cellular proteins was bound by immobilized GST-D3c32. One of the prominent bands, a ca. 70-kDa protein which binds to GST-D3c32 but not GST alone, was excised from the gel and identified unambiguously by matrix-assisted laser desorption ionization–time-of-flight (MALDI-TOF) mass spectrometry as JBP1 (Fig. 1). The sequence coverage was 32% at a mass accuracy below 30 ppm (the 22 matching peptides covered 205 of the 637 amino acids of JBP1). The JBP1-binding protein pICln was also bound to GST-D3c32 but not to GST alone (see below).
FIG. 1.
Affinity chromatography with the RG domain of SmD3 isolates the methyltransferase JPB1. The indicated recombinant proteins (8 μg) were incubated with (+) or without (−) total HeLa extract. Following extensive washing, proteins were separated by SDS-PAGE and visualized by Coomassie blue staining. The methyltransferase JBP1 (arrow) was identified by MALDI-TOF mass spectrometry, and pICln was identified by Western blot analysis (see Fig. 2). Numbers on the left are molecular masses in kilodaltons.
JBP1 is associated with SmD1 and SmD3.
To confirm that JBP1 is in fact the methyltransferase that binds SmD1 and SmD3, HeLa cell extract was incubated with immobilized GST-D3c32 and GST fused to SmD1 (GST-D1), SmD3 (GST-D3), and the RG domain of SmD1 (GST-D1c29), and bound proteins were resolved by SDS-PAGE and Western blotted with anti-SMN, -JBP1, and -pICln specific antibodies. JBP1 and the JBP1-binding protein pICln bound to the immobilized Sm proteins and their RG domains but not to GST alone. As was previously shown, SMN was not bound to the unmodified proteins (21) (Fig. 2A). To examine the role of sDMA modification in these interactions, previously described peptides (21) corresponding to the RG domains of SmD1 and SmD3 without (D1c29 and D3c32) or with (D1c29-sDMA and D3c32-sDMA) the specific sDMA modifications formed were immobilized and incubated with HeLa cytoplasmic extract. After washing, bound proteins were eluted, resolved by SDS-PAGE, and probed by Western blotting to detect SMN, pICln, and JBP1 (Fig. 2B). Strikingly, JBP1 and pICln bound only to the unmodified peptides, while SMN bound only to the sDMA-modified peptides. This demonstrates that JBP1 and pICln associate with the unmethylated RG domains of SmD1 and SmD3, whereas SMN preferentially binds the dimethylarginine-modified RG domains of SmD1 and SmD3.
FIG. 2.
JBP1 and pICln associate with unmodified Sm proteins. (A) The indicated immobilized GST fusion proteins were incubated with HeLa cell extract, and bound proteins were analyzed by Western blotting to detect the indicated proteins (arrows). The total lane shows 10% of the extract used in each binding. (B) Streptavidin-immobilized biotin-linked peptides with (D1c29-sDMA and D3c32-sDMA) or without (D1c29 and D3c32) the specific sDMA modifications formed in vivo and biotin alone were incubated with HeLa cytoplasmic extract, and bound proteins were analyzed by Western blotting to detect the indicated proteins (arrows). The total lane shows 10% of the extract used in each binding. (C) Anti-SMN (2B1), anti-Sm protein (Y12), anti-pICln (α-pICln), and nonimmune (SP2/0) antibodies were used for immunoprecipitation of HeLa cytoplasmic extract, and bound cellular proteins were analyzed by Western blotting to detect the proteins indicated by arrows. The total lane shows 10% of the extract used in each immunoprecipitation.
Because pICln has been previously shown to bind to JBP1 (then termed IBP72) (30), the results presented in Fig. 2 suggest that a JBP1-pICln complex and the SMN complex associate with different forms of the same Sm proteins. This suggested that these two complexes are separate and that the SMN complex binds the Sm proteins after they have been modified by JBP1. To examine these complexes directly and to determine if JBP1 is indeed associated with Sm proteins, immunoprecipitations were performed from HeLa cell cytoplasmic lysate using monoclonal antibodies against the Sm proteins SMN and pICln. As a negative control, a nonimmune antibody (SP2/0) was also used (Fig. 2C). The anti-JBP1 antibody does not efficiently immunoprecipitate JBP1 (data not shown) and was therefore not used for these experiments. As expected, the anti-SMN antibodies coimmunoprecipitated SMN, Gemin2, Gemin3, Gemin4, and SmB (9, 10, 36) but not pICln or JBP1. Conversely, anti-pICln antibodies coimmunoprecipitated pICln, JBP1, and SmB but not SMN, Gemin2, Gemin3, or Gemin4. Anti-Sm protein antibodies coimmunoprecipitated pICln, JBP1, SMN, Gemin2, Gemin3, Gemin4, and SmB. SP2/0 nonimmune antibodies did not coimmunoprecipitate any of these proteins. These results demonstrate that JBP1 is associated with Sm proteins and that the SMN complex and a JBP1-pICln complex exist as two separate complexes, both of which contain Sm proteins.
A 20S complex containing pICln, JBP1, and the RG domain Sm proteins.
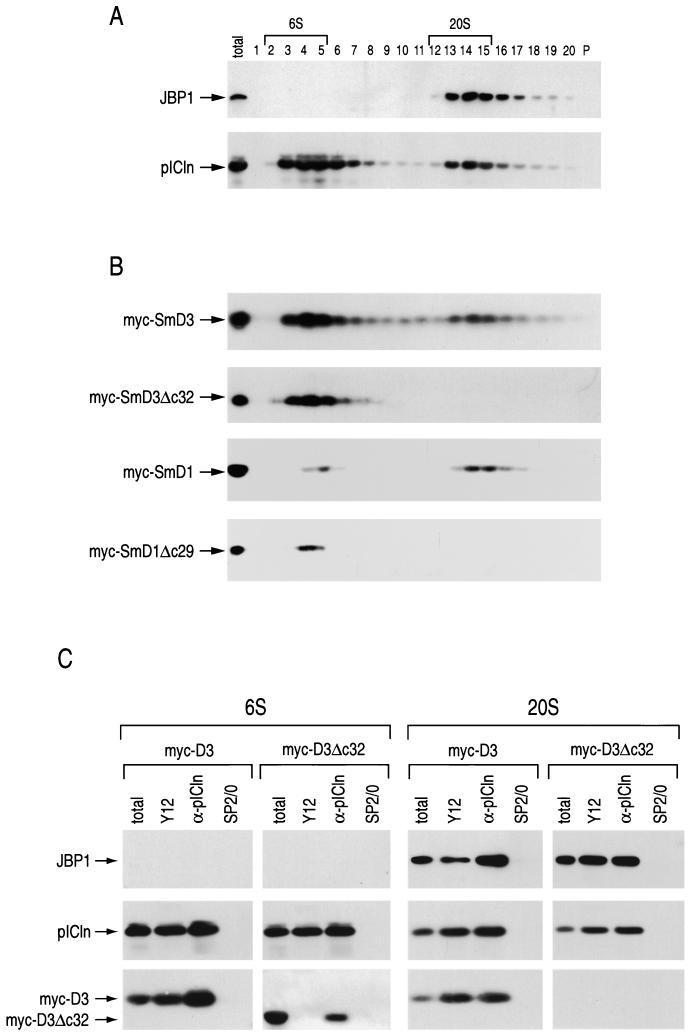
The sizes of cytoplasmic intermediates in snRNP assembly containing Sm proteins have been studied by sucrose gradient centrifugation (22, 23, 50). To examine the fractionation of cytoplasmic pICln and JBP1 on sucrose gradients in relation to that of the Sm proteins, cytoplasmic extracts were prepared and separated on sucrose gradients. After centrifugation, fractions were collected, resolved by SDS-PAGE, and analyzed by Western blotting with antibodies to pICln and JBP1 (Fig. 3A). pICln was detected in two peaks of approximately 6S and 20S. Interestingly, JPB1 was found only in the 20S pICln-containing peak. Thus, a distinct 20S complex containing JBP1 and pICln, which we term the methylosome (see below), is present in the cytoplasm of mammalian cells.
FIG. 3.
The carboxyl-terminal RG domains of SmD1 and SmD3 are required for association with a 20S pICln-JBP1 complex. (A) 293 cell cytoplasmic extract was separated on a 5 to 20% sucrose gradient. Fractions (indicated by numbers) were collected, separated by SDS-PAGE, and Western blotted to detect the proteins indicated by arrows. (B) 293 cells transiently expressing myc-D3, myc-D3Δc32, myc-D1, or myc-D1Δc29 (as indicated) were separated on sucrose gradients, and fractions were collected, separated by SDS-PAGE, and immunoblotted to detect the myc-tagged proteins. The presence of the myc-tagged proteins did not affect the sucrose gradient migration pattern of pICln or JBP1 (data not shown). In both panels A and B, lanes P contain 5% of the pellet from the gradients. The total lanes contain 5% of the extract loaded on each gradient. (C) Fractions 2 to 5 and 12 to 15 (6S and 20S as indicated, respectively, in panel A) from the sucrose gradient separation of myc-D3- and myc-D3Δc32-expressing cytoplasmic extracts were pooled and immunoprecipitated with anti-Sm protein (Y12), anti-pICln (α-pICln), and nonimmune (SP2/0) antibodies as indicated. Immunoprecipitated proteins were separated by SDS-PAGE and immunoblotted to detect the indicated proteins (arrows). The total lanes show 10% of the pooled fractions used in each immunoprecipitation.
To directly measure the sucrose gradient migration patterns of SmD1 and SmD3 in relation to those of JBP1 and pICln, myc-tagged SmD1 (myc-D1) and SmD3 (myc-D3) were transiently expressed in 293 cells and sedimented in sucrose gradients, and fractions were analyzed by Western blotting with anti-myc antibodies (Fig. 3B). myc-D3 and myc-D1 migrated in two peaks that coincide almost exactly with the peak of pICln at 6S and with the 20S methylosome peak. Because JBP1 and pICln bound to the RG domains of SmD1 and SmD3, we examined the sucrose gradient migration patterns of RG domain deletions of SmD1 and SmD3. For this, cytoplasmic extracts from cells transiently transfected with myc-tagged RG domain deletions of SmD1 (myc-D1Δc29) and SmD3 (myc-D3Δc32) were separated on sucrose gradients, and fractions were analyzed by Western blotting with anti-myc antibodies. Strikingly, myc-D3Δc32 and myc-D1Δc29 were detected only in the 6S region of the gradient (Fig. 3B). The myc-tagged proteins did not affect the sucrose gradient migration pattern of pICln or JBP1 (data not shown). To determine if these myc-tagged Sm proteins and their RG domain deletions are associated with the 6S peak of pICln and the 20S methylosome, fractions 2 to 5 and 12 to 15, respectively, were pooled and immunoprecipitated with anti-pICln, anti-Sm protein, or SP2/0 nonimmune antibodies (Fig. 3C). Western blotting with anti-myc, anti-pICln, or anti-JBP1 antibodies revealed that myc-D3 and myc-D3Δc32 were both associated with pICln in the 6S peak. The Y12 antibody binds the RG domains of SmD3 and SmD1 (5). Thus, myc-D3Δc32 is not immunoprecipitated by Y12 antibody, while pICln can be immunoprecipitated by Y12 via SmD1-pICln complexes that do not contain D3Δc32. In contrast, myc-D3, but not myc-D3Δc32, was associated with the methylosome in the 20S peak. This was also the case for myc-D1 and myc-D1Δc29 (data not shown). These results demonstrate that SmD1 and SmD3 are associated with the methylosome at 20S and with pICln at 6S. Furthermore, the RG domains of SmD1 and SmD3 are required for their association with the 20S methylosome but not for association with 6S pICln.
The methylosome binds the RG-containing Sm proteins.
To determine which native Sm proteins are associated with pICln at 6S and the methylosome at 20S, cells were metabolically labeled with [35S]methionine and [35S]cysteine, and cytoplasmic extract was prepared and separated on sucrose gradients. As before, fractions 2 to 5 (6S) and fractions 12 to 15 (20S) were pooled and used for immunoprecipitation with anti-Sm protein, anti-pICln, and SP2/0 nonimmune antibodies. The immunoprecipitates were subjected to SDS-PAGE, and the immunoprecipitated proteins were visualized by fluorography (Fig. 4). Immunoprecipitated Sm proteins have a well-defined pattern on SDS-PAGE (32, 58), and their positions are indicated in Fig. 4. All of the Sm proteins were detected in both pooled fractions. However, consistent with previous reports (2, 22, 50) SmD1, SmD2, SmE, SmF, and SmG were prominent in the 6S fractions, and SmD3 and SmB were prominent in the 20S fractions. Similar to the results presented in Fig. 3, anti-pICln antibodies immunoprecipitated pICln and JBP1 from the 20S fractions and pICln from the 6S fractions. SmD3 and to a lesser extent SmD1/D2 (in our hands SmD1 and SmD2 could not be fully separated using this gel system) were detected in the anti-pICln immunoprecipitate from the 6S and 20S fractions. SmB was detected only in the anti-pICln immunoprecipitate from the 20S fractions. In contrast, the non-RG-containing Sm proteins, SmE, SmF, and SmG, were not associated with the 6S or 20S pICln-containing complexes. Anti-pICln also coimmunoprecipitated several unknown proteins from the 6S and 20S regions of the gradient (Fig. 4). Two prominent proteins from the 20S fractions (p50 and p37) are likely to be components of the methylosome (Fig. 4). These results indicate that the 20S methylosome binds the RG-containing Sm proteins SmB, SmD1/D2, and SmD3 but not the non-RG-containing Sm proteins SmE, SmF, and SmG.
FIG. 4.
Cytoplasmic SmD1/D2, SmD3, and SmB are associated with the 20S methylosome. Cytoplasmic extract was prepared from HeLa cells metabolically labeled with [35S]methionine and [35S]cysteine and fractionated on a 5 to 20% sucrose gradient as for Fig. 3. Fractions 2 to 5 (6S) and 12 to 15 (20S) were pooled, immunoprecipitated with the anti-Sm (Y12), anti-pICln (α-pICln), and nonimmune (SP2/0) antibodies as indicated, and separated by SDS-PAGE. Radioactive signals were enhanced with Amplify (Amersham), and the gel was exposed to film. The Sm proteins SmB/B′, SmD3, SmD1/D2, SmE, SmF, and SmG, identified based on their molecular masses and presence in Y12 immunoprecipitates, are indicated. JBP1 was identified based its molecular mass and presence in the 20S anti-pICln immunoprecipitate. Similarly, pICln was identified based its size and presence in the 6S and 20S pICln antibody immunoprecipitates. Relatively minor unidentified proteins coimmunoprecipitated by anti-pICln from both the 6S and 20S regions of the gradient are indicated with black dots. Prominent proteins (p50 and p37) immunoprecipitated by anti-pICln in the 20S fraction are also indicated. The positions of molecular mass markers (in kilodaltons) are shown on the left.
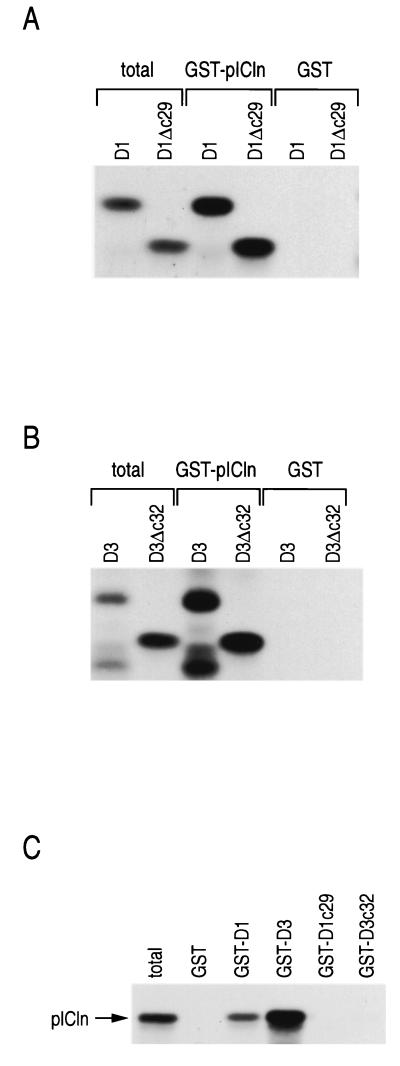
pICln binds the Sm domains of SmD1 and SmD3.
Previously, it was shown that pICln binds the RG domain-containing Sm proteins SmB, SmD1, and SmD3 (47). To determine if the RG domains mediate these interactions, radiolabeled myc-D1, myc-D3, myc-D1Δc29, and myc-D3Δc32 were produced by in vitro translation and incubated with GST fused to pICln (GST-pICln) or GST alone. After washing, bound proteins were separated by SDS-PAGE and visualized by fluorography (Fig. 5A and B). Deletion of the RG domains of SmD1 and SmD3 did not affect their binding to GST-pICln. In the reverse experiment, in vitro-translated pICln bound to immobilized SmD1 and SmD3 but did not bind to immobilized RG domains of SmD1 and SmD3 (Fig. 5C). Thus, pICln interacts with the Sm domains of SmD1 and SmD3. Furthermore, because the RG domains of SmD1 and SmD3 are required for these proteins to associate with the 20S methylosome (Fig. 3), these results suggest that the binding of JBP1 to RG domains is important for the interaction of SmD1 and SmD3 with the methylosome. This also suggests that pICln is pulled down by GST-D3c32 from cellular extract (Fig. 1) through JBP1 which is bound to GST-D3c32.
FIG. 5.
pICln binds the Sm domains of SmD1 and SmD3. (A and B) The indicated in vitro-translated and [35S]methionine-labeled proteins were incubated with GST or GST-pICln, and after washing, bound proteins were separated by SDS-PAGE and visualized by fluorography. (C) Binding of in vitro-translated pICln to the indicated immobilized proteins was performed as for panels A and B.
The methylosome methylates Sm proteins.
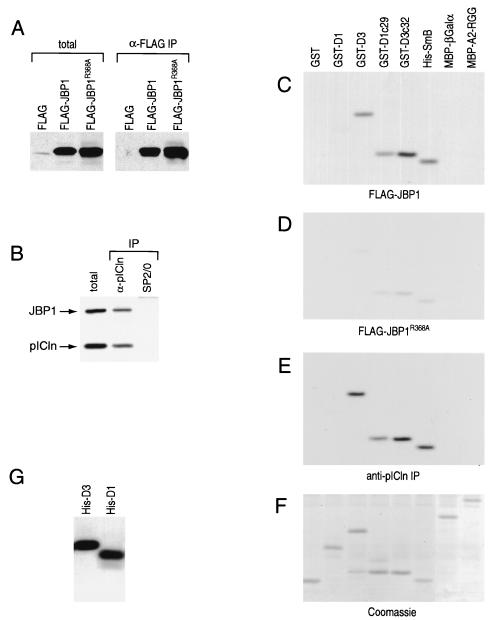
To test specifically if the methylosome can methylate Sm proteins, we purified the methylosome and tested for methyltransferase activity towards Sm proteins. For this, cytoplasmic extract was prepared from 293 cells transiently expressing Flag-JBP1 and separated on a sucrose gradient. Fractions were immunoblotted to detect Flag-JBP1, native JBP1, and pICln (Fig. 6A). While a considerable amount of Flag-JBP1 was found in the pellet of the gradient, some Flag-JBP1 was found in the methylosome. The 20S peak (fractions 12 to 15) was immunoprecipitated with anti-Flag beads and eluted with Flag peptide. Western blotting of the eluate revealed that both Flag-JBP1 and native JBP1 as well as pICln were immunoprecipitated (Fig. 6B). This is consistent with previous results showing that JBP1 oligomerizes (49). The purified methylosome could efficiently methylate GST-D3, GST-D3c32, GST-D1c29, and His-SmB but not GST, GST-D1, MBP-βGalα, and MBP-A2-RGG (Fig. 6C). This indicates that the methylosome methylates Sm proteins.
FIG. 6.
The methylosome. (A) Cytoplasmic extracts were prepared from 293 cells transiently expressing Flag-JBP1 and separated on a sucrose gradient. Fractions were collected, separated by SDS-PAGE, and Western blotted to detect Flag-JBP1, native JBP1, and pICln (arrows). (B) Fractions 12 to 15 (20S) were immunoprecipitated with anti-Flag beads, and the Flag peptide-eluted methylosome was separated by SDS-PAGE and Western blotted to detect Flag-JBP1, native JBP1, and pICln (arrows). (C) The purified methylosome was incubated with 3H-SAM and each protein substrate in 50 μl of binding buffer at 30°C for 30 min. The samples were then separated by SDS-PAGE, stained with Coomassie blue to check protein substrates, and exposed to film after treatment with Amplify to enhance radioactive signals.
JBP1 produces sDMA-modified Sm proteins.
We could not detect methyltransferase activity from bacterially produced recombinant JBP1 (rJBP1). Others have shown that rJBP1 has methyltransferase activity which is at least 200-fold lower than that of JBP1 produced in cultured cells (49). Thus, two independent methods were used to immunopurify JBP1 from cellular extracts. First, we employed a technique previously used to demonstrate that JBP1 is indeed a methyltransferase (46). To do so, we transiently expressed Flag epitope-tagged JBP1 (Flag-JBP1) and a JBP1 mutant carrying a point mutation (arginine 368 to alanine) in the highly conserved putative SAM binding domain previously shown to reduce JBP1 methyltransferase activity (46) (Flag-JBP1R368A). Flag-tagged proteins were purified from cytoplasmic extract on anti-Flag beads and eluted with Flag peptide. As shown by Western blotting with anti-JBP1 specific antibodies, anti-Flag immunoprecipitation of extracts prepared from Flag-JBP1- and Flag-JBP1R368A-expressing cells contained significant amounts of Flag-JBP1 and Flag-JBP1R368A, respectively (Fig. 7A). In contrast, anti-Flag immunoprecipitation products from Flag-expressing cells (vector transfected) did not contain any JBP1. Anti-Flag antibody immunoprecipitation was carried out in the presence of the detergent Empigen, which strips off coimmunoprecipitating proteins. This was done because we wished to determine if JBP1 itself, rather than a contaminating protein, can methylate Sm proteins. These preparations of Flag-JBP1 are over 90% pure as determined by silver staining after SDS-PAGE (data not shown). We also immunopurified native JBP1 from HeLa cytoplasmic extract. For this, HeLa cytoplasmic extract was separated on sucrose gradients, and the 20S fractions containing JBP1 and pICln were incubated with immobilized anti-pICln or SP2/0 as a negative control. As detected by Western blotting, JBP1 and pICln were present in the anti-pICln immunoprecipitate and not present in the SP2/0 precipitate (Fig. 7B).
FIG. 7.
JBP1 has methyltransferase specificity for SmD1, SmD3, and SmB. (A) Cytoplasmic extracts prepared from 293 cells transiently expressing Flag-JBP1, Flag-JBP1R368A, or Flag alone (as indicated) were used for immunoprecipitation with immobilized anti-Flag antibody (α-flag IP). After elution with Flag peptide, a fraction of each immunoprecipitate was Western blotted with anti-JBP1 antibody. The total lanes contain 10% of the total extract used in each immunoprecipitation lane. (B) HeLa cytoplasmic extract was separated on a 5 to 20% sucrose gradient. Fractions 12 to 15 (20S) (see Fig. 3A) were pooled and immunoprecipitated with anti-pICln (α-pICln) and nonimmune (SP2/0) antibodies. A fraction of each immunoprecipitate was Western blotted with specific antibodies to detect JBP1 and pICln (arrows). The total lane contains 10% of the total extract blotted in each immunoprecipitation lane. (C to F) Flag-JBP1 (C), Flag-JBP1R368A (D), or anti-pICln antibody immunoprecipitate from sucrose gradient fractions 12 to 15 (E) was incubated with 3H-SAM and each protein substrate in 50 μl of binding buffer at 30°C for 30 min. The samples were then separated by SDS-PAGE, stained with Coomassie blue, and treated with Amplify to enhance radioactive signals. (F) A representative Coomassie blue-stained gel showing the positions of the recombinant proteins. (G) His-tagged SmD1 and SmD3 were methylated with immunopurified Flag-JBP1, separated by SDS-PAGE, and visualized by fluorography as in panels C to F.
To test for methyltransferase activity, the eluates Western blotted in Fig. 7A and B were incubated with 3H-SAM and each recombinant protein substrate. Each sample was then separated by SDS-PAGE, and the recombinant proteins were visualized by Coomassie blue staining followed by fluorography. A representative Coomassie blue-stained gel showing the migration patterns of the recombinant protein substrates used is shown in Fig. 7F. Both the anti-Flag immunoprecipitate from Flag-JBP1-expressing cells (Fig. 7C) and the anti-pICln immunoprecipitate from 20S sucrose gradient fractions (Fig. 7E) had methyltransferase specificity toward Sm protein substrates. Similar to the purified methylosome, both of these immunoprecipitates efficiently methylated GST-D3, GST-D3c32, GST-D1c29, and His-SmB. However, these immunoprecipitates could not efficiently methylate GST, GST-D1, MBP-βGalα, and MBP-A2-RGG. Neither SP2/0 immunoprecipitate from sucrose gradient fractions nor anti-Flag immunoprecipitate from vector-transfected cells showed significant methyltransferase activity toward any of the substrates used (data not shown). Anti-Flag immunoprecipitate from Flag-JBP1R368A-expressing cells had significantly reduced methyltransferase activity compared to immunopurified Flag-JBP1 (Fig. 7D). The experiments with Flag-JBP1 indicate that JBP1 can methylate Sm proteins. We note that native JBP1 is in a 20S complex which is associated with Sm proteins (Fig. 3 and 4). Immunoprecipitation of this complex with anti-Flag antibody (in the case of Flag-JBP1 expression) (Fig. 6) and anti-pICln antibody (Fig. 7) isolates an activity which specifically methylates Sm proteins, strongly suggesting that JBP1 functions in the context of this methylosome.
Surprisingly, immunopurified JBP1 methylated the RG domain from SmD1 fused to GST but did not methylate full-length SmD1 fused to GST. It is possible that when SmD1 is fused to GST it is a poor substrate for JBP1. To address this, we produced recombinant His-tagged SmD1 (His-D1) and SmD3 (His-D3) and found that, under the same conditions used above, Flag-JBP1 could indeed methylate His-D1 with efficiency similar to that for His-D3 (Fig. 7G). These results show that JBP1 can specifically methylate SmD1, SmD3, and SmB.
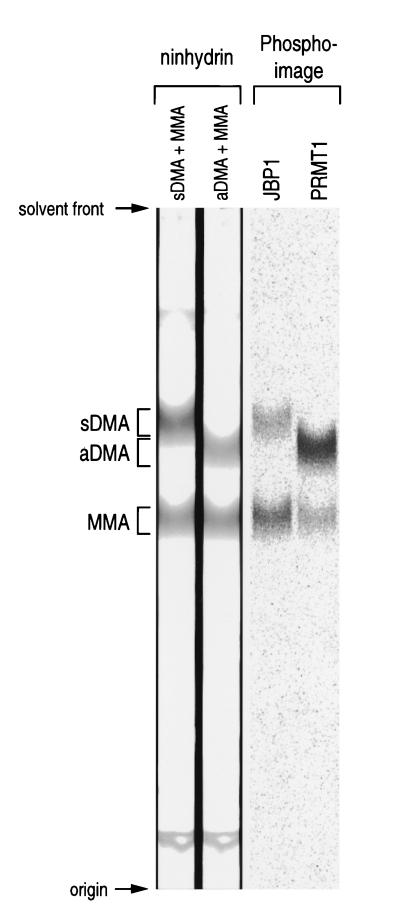
If JBP1 is the Sm protein arginine methyltransferase, it should form sDMAs rather than aDMAs on the RG domains of the Sm proteins. To test this, Flag-JBP1 was incubated with 3H-SAM and GST fused to a TEV protease site fused to the carboxyl terminus of SmD3 (GST-TEV-D3c32). As a control, highly active (55) recombinant His-tagged PRMT1 (His-PRMT1) was also used to methylate GST-TEV-D3c32. Following methylation, the fusion protein was captured on glutathione-Sepharose, washed, and cleaved with TEV protease. Acid hydrolysates of the released peptides were mixed with standards (sDMA, aDMA, and MMA) and resolved by thin-layer chromatography on silica gel 60 plates (Fig. 8). Standard amino acids were visualized by ninhydrin staining, and the radiolabeled arginine products were visualized on a phosphorimager. As expected, the known type I methyltransferase PRMT1 (34) produced aDMA and MMA. In contrast, JBP1 produced sDMA and MMA. These results indicate that JBP1 is a type II methyltransferase and that it modifies the RG domains of SmD1 and SmD3 to form sDMA.
FIG. 8.
JBP1 is a type II protein arginine methyltransferase. GST-TEV-D3c32 was labeled with [3H]methyl groups by JBP1 or PRMT1 (as indicated). Following purification on glutathione-Sepharose beads and cleavage with TEV protease, the D3c32 peptides were hydrolyzed in acid; mixed with aDMA, sDMA, and MMA standard amino acids (as indicated); and separated on silica gel 60 thin-layer chromatography plates. The methylated arginine standards were visualized with ninhydrin, and 3H-methylated arginine residues were visualized on a phosphorimager.
The methylosome-associated SmD3 can be transferred to the SMN complex in vitro.
The experiments up to this point indicated that the two complexes, SMN and the methylosome, bind different forms of the Sm proteins. The methylosome binds unmodified Sm proteins and produces sDMA-modified Sm proteins, and SMN binds preferentially the sDMA-modified Sm proteins. Thus, Sm proteins may be transferred to the SMN complex after sDMA modification by the methylosome. To determine if there is a difference in the ability of Sm proteins associated with 6S pICln versus those associated with the methylosome to bind the SMN complex, we determined if myc-D3 from these two regions of a gradient could differentially associate with SMN. To do this, cytoplasmic extract was prepared from cells transiently expressing myc-D3 and separated on a sucrose gradient. Fractions containing the 6S and 20S peaks of myc-D3 (Fig. 3B) were pooled and incubated with SMN complex which had been immobilized on anti-Flag beads. After washing, proteins bound by the immobilized SMN complex were analyzed by Western blotting (Fig. 9). Strikingly, myc-D3 from the methylosome, but not from the 6S fractions, bound to the SMN complex. In contrast, JBP1 from the methylosome did not bind to the SMN complex. This suggests that the methylosome can transfer Sm proteins to the SMN complex.
DISCUSSION
Here we describe a methylosome that serves to symmetrically dimethylate Sm proteins. We demonstrate that the SMN complex and the methylosome are separate complexes and bind different forms of the same Sm proteins. We show that SmD3 can be transferred from the methylosome to the SMN complex but that SmD3 from pICln at 6S cannot (Fig. 9). Based on the results presented here and on the fact that sDMA modification of the RG domains of SmD1 and SmD3 is important for their association with SMN (21), we propose a model in which methylation of SmD1 and SmD3 (and possibly SmB) by the methylosome targets these proteins to the SMN complex for assembly into snRNP core particles (Fig. 10). The binding of JBP1 to unmodified RG domains and the binding of pICln to the Sm domains of the RG-containing Sm proteins (Fig. 3 and 5) appear to recruit these proteins to JBP1 for methylation. Based on inhibition of snRNP assembly after injection of recombinant pICln in Xenopus oocytes, it was proposed that pICln functions as an inhibitor of snRNP assembly (47). Our results argue that pICln functions, at least in part, as a component of a JBP1 methyltransferase complex which produces methylated RG-containing Sm proteins for the SMN complex and thereby for assembly into snRNPs. Thus, excess pICln would be expected to perturb this pathway prior to SMN-Sm protein binding, making it appear that pICln is a general inhibitor of snRNP assembly when it actually functions in the methylation of Sm proteins.
FIG. 10.
Methylosome methylation of RG-containing Sm proteins targets them to the SMN complex for assembly into snRNP core particles. A schematic depicting Sm RG domain posttranslational methylation by the methylosome is shown. After sDMA modification, the Sm proteins associate with the SMN complex and, along with the other Sm proteins (which also bind SMN), are assembled on snRNA to form an snRNP core particle.
Sm proteins are believed to be stored in the cytoplasm (50), suggesting that core assembly may be regulated, and our findings indicate that the methylosome could serve to regulate interaction of RG-containing Sm proteins with SMN, thus regulating snRNP assembly. Interestingly, JBP1 has been shown to interact with JAK kinases (46), raising the possibility that phosphorylation of JBP1 may modulate its activity. JAK kinases have been shown to be involved in signal transduction of many cytokines, hormones, and growth factors (reviewed in reference 57), and they may provide cells the capacity to modulate the rate of snRNP biogenesis. It will be of interest to determine if JAK kinases can phosphorylate JBP1 and if this can change the activity of JBP1.
Immunopurified JBP1 produces sDMA-modified residues in the carboxyl RG terminus of SmD3, indicating that JBP1 is a type II methyltransferase. It is not likely that the methyltransferase activity in the JBP1 preparations is a result of a contaminating methyltransferase, because a single point mutation (R368A) in the highly conserved putative SAM binding domain greatly reduced this methyltransferase activity (Fig. 7). Others have shown that bacterially produced rJBP1 has arginine methyltransferase activity which is at least 200-fold lower than the activity of mammalian cell-produced JBP1 (49). We were unable to detect methyltransferase activity in our preparation of rJBP1. It is possible that disulfide-linked homo-oligomerization or specific phosphorylation is important for JBP1 activity (46, 49). Furthermore, because JBP1 functions as part of a large complex, it is likely that its activity requires other components of the methylosome such as the Sm domain-binding protein pICln. Type I methyltransferases produce aDMA and constitute the majority of cellular arginine methyltransferase activity (25, 55). Only three proteins have been shown to contain sDMA: myelin basic protein (25) and SmD1 and SmD3 (5). Two of these, SmD1 and SmD3, associate with JBP1 in the methylosome. While this paper was in preparation, Branscombe et al. reported that JBP1 is a type II methyltransferase (6). However, those authors have not demonstrated that JBP1 methylates Sm proteins, nor have they shown that it functions in the context of the methylosome, as we show here.
We show that pICln is present in a 6S JBP1-free complex and a 20S JBP1-containing complex in the cytoplasm. SmD3 and SmD1 and/or SmD2 are found in both 20S and 6S complexes (Fig. 4). Based on pulse-chase experiments, it was proposed that the Sm proteins accumulate in 4S-6S complexes prior to assembly into snRNP core particles of approximately 11S (18). Subsequent work also identified SmB and SmD3 in an approximately 20S complex (1, 22, 50). Anti-Sm protein antibody immunoprecipitation experiments and Sm protein purification identified pre-snRNP Sm protein complexes of B/D3, D1/D2, and E/F/G (48, 50). Our results show that in a 6S complex SmD3, but not SmB, is associated with pICln, and this is consistent with previous work showing that B and D3 are associated with each other in a 20S complex but not in a 6S complex (22, 50, 58). Our data show that SmD1 and SmD3 can interact with the 6S, JBP1-free pICln complex, and these associations do not require the carboxyl-terminal RG domains. This is consistent with our data showing that pICln binds to the Sm domains of SmD1 and SmD3 (Fig. 5). In contrast, the RG domains of SmD1 and SmD3 are required for association of these proteins with the methylosome, and this association may be mediated, at least partially, by the direct binding of JBP1 to RG domains.
The methylosome is important for modifying Sm proteins so that they have higher affinity for the SMN complex. This raises the possibility that reduced activity of the methylosome may reduce the level of Sm protein-SMN interaction, which may have consequences similar to those from having reduced levels of or mutations in SMN, as is the case in SMA. Thus, it is conceivable that defects in or suboptimal function of the methylosome may also result in degeneration of motor neurons or further aggravate the severity of SMA.
In summary, we present evidence that modification of the RG domains of SmD1 and SmD3 to form sDMAs is carried out by a novel 20S complex, the methylosome. The activity of the methylosome depends on the methyltransferase JBP1. The function of the methylosome is to produce sDMA-modified SmD1 and SmD3, which drastically increase their affinity for the SMN complex. These findings suggest a pathway of snRNP assembly in which methylation of Sm proteins by the methylosome regulates snRNP core particle assembly.
ACKNOWLEDGMENTS
We thank Gary Zieve and members of our laboratory, especially Livio Pellizzoni, Zissimos Mourelatos, and Amelie Gubitz, for helpful discussions and comments on the manuscript.
This work was supported by a grant from the National Institutes of Health to G.D. J.R. is a Marie Curie Fellow. Work in M.M.'s laboratory is supported by a fund of the Danish National Research Foundation to the Center of Experimental Bioinformatics. G.D. and G.V. are Investigators of the Howard Hughes Medical Institute.
REFERENCES
- 1.Andersen J, Feeney R J, Zieve G W. Identification and characterization of the small nuclear ribonucleoprotein particle D′ core protein. Mol Cell Biol. 1990;10:4480–4485. doi: 10.1128/mcb.10.9.4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen J, Zieve G W. Assembly and intracellular transport of snRNP particles. Bioessays. 1991;13:57–64. doi: 10.1002/bies.950130203. [DOI] [PubMed] [Google Scholar]
- 3.Baldwin G S, Carnegie P R. Specific enzymic methylation of an arginine in the experimental allergic encephalomyelitis protein from human myelin. Science. 1971;171:579–581. doi: 10.1126/science.171.3971.579. [DOI] [PubMed] [Google Scholar]
- 4.Bao S, Qyang Y, Yang P, Kim H, Du H, Bartholomeusz G, Henkel J, Pimental R, Verde F, Marcus S. The highly conserved protein methyltransferase, Skb1, is a mediator of hyperosmotic stress response in the fission yeast Schizosaccharomyces pombe. J Biol Chem. 2001;276:14549–14552. doi: 10.1074/jbc.C100096200. [DOI] [PubMed] [Google Scholar]
- 5.Brahms H, Raymackers J, Union A, de Keyser F, Meheus L, Luhrmann R. The C-terminal RG dipeptide repeats of the spliceosomal Sm proteins D1 and D3 contain symmetrical dimethylarginines, which form a major B-cell epitope for anti-Sm autoantibodies. J Biol Chem. 2000;275:17122–17129. doi: 10.1074/jbc.M000300200. [DOI] [PubMed] [Google Scholar]
- 6.Branscombe T L, Frankel A, Lee J H, Cook J R, Yang Z, Pestka S, Clarke S. PRMT5 (the Janus kinase-binding protein 1) catalyzes the formation of symmetric dimethylarginine residues in proteins. J Biol Chem. 2001;18:18. doi: 10.1074/jbc.M105412200. [DOI] [PubMed] [Google Scholar]
- 7.Buhler D, Raker V, Luhrmann R, Fischer U. Essential role for the tudor domain of SMN in spliceosomal U snRNP assembly: implications for spinal muscular atrophy. Hum Mol Genet. 1999;8:2351–2357. doi: 10.1093/hmg/8.13.2351. [DOI] [PubMed] [Google Scholar]
- 8.Burghes A H. When is a deletion not a deletion? When it is converted. Am J Hum Genet. 1997;61:9–15. doi: 10.1086/513913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Charroux B, Pellizzoni L, Perkinson R A, Shevchenko A, Mann M, Dreyfuss G. Gemin3: a novel DEAD box protein that interacts with SMN, the spinal muscular atrophy gene product, and is a component of gems. J Cell Biol. 1999;147:1181–1194. doi: 10.1083/jcb.147.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charroux B, Pellizzoni L, Perkinson R A, Yong J, Shevchenko A, Mann M, Dreyfuss G. Gemin4: a novel component of the SMN complex that is found in both Gems and nucleoli. J Cell Biol. 2000;148:1177–1186. doi: 10.1083/jcb.148.6.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen D, Ma H, Hong H, Koh S S, Huang S M, Schurter B T, Aswad D W, Stallcup M R. Regulation of transcription by a protein methyltransferase. Science. 1999;284:2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- 12.Choi Y D, Dreyfuss G. Monoclonal antibody characterization of the C proteins of heterogeneous nuclear ribonucleoprotein complexes in vertebrate cells. J Cell Biol. 1984;99:1197–1204. doi: 10.1083/jcb.99.6.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeRobertis E M. Nucleoplasmic segregation of proteins and RNAs. Cell. 1983;32:1021–1025. doi: 10.1016/0092-8674(83)90285-4. [DOI] [PubMed] [Google Scholar]
- 14.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischer U, Liu Q, Dreyfuss G. The SMN-SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell. 1997;90:1023–1029. doi: 10.1016/s0092-8674(00)80368-2. [DOI] [PubMed] [Google Scholar]
- 16.Fischer U, Luhrmann R. An essential signaling role for the m3G cap in the transport of U1 snRNP to the nucleus. Science. 1990;249:786–790. doi: 10.1126/science.2143847. [DOI] [PubMed] [Google Scholar]
- 17.Fischer U, Sumpter V, Sekine M, Satoh T, Luhrmann R. Nucleo-cytoplasmic transport of U snRNPs: definition of a nuclear location signal in the Sm core domain that binds a transport receptor independently of the m3G cap. EMBO J. 1993;12:573–583. doi: 10.1002/j.1460-2075.1993.tb05689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fisher D E, Conner G E, Reeves W H, Wisniewolski R, Blobel G. Small nuclear ribonucleoprotein particle assembly in vivo: demonstration of a 6S RNA-free core precursor and posttranslational modification. Cell. 1985;42:751–758. doi: 10.1016/0092-8674(85)90271-5. [DOI] [PubMed] [Google Scholar]
- 19.Frankel A, Clarke S. PRMT3 is a distinct member of the protein arginine N-methyltransferase family. Conferral of substrate specificity by a zinc-finger domain. J Biol Chem. 2000;275:32974–32982. doi: 10.1074/jbc.M006445200. [DOI] [PubMed] [Google Scholar]
- 20.Friesen W J, Dreyfuss G. Specific sequences of the Sm and Sm-like (Lsm) proteins mediate their interaction with the spinal muscular atrophy disease gene product (SMN) J Biol Chem. 2000;275:26370–26375. doi: 10.1074/jbc.M003299200. [DOI] [PubMed] [Google Scholar]
- 21.Friesen W J, Massenet S, Paushkin S, Wyce A, Dreyfuss G. SMN, the product of the spinal muscular atrophy gene, binds preferentially to dimethylarginine-containing protein targets. Mol Cell. 2001;7:1111–1117. doi: 10.1016/s1097-2765(01)00244-1. [DOI] [PubMed] [Google Scholar]
- 22.Fury M, Andersen J, Ponda P, Aimes R, Zieve G W. Thirteen anti-Sm monoclonal antibodies immunoprecipitate the three cytoplasmic snRNP core protein precursors in six distinct subsets. J Autoimmun. 1999;12:91–100. doi: 10.1006/jaut.1998.0266. [DOI] [PubMed] [Google Scholar]
- 23.Fury M G, Zhang W, Christodoulopoulos I, Zieve G W. Multiple protein: protein interactions between the snRNP common core proteins. Exp Cell Res. 1997;237:63–69. doi: 10.1006/excr.1997.3750. [DOI] [PubMed] [Google Scholar]
- 24.Gary J D, Clarke S. RNA and protein interactions modulated by protein arginine methylation. Prog Nucleic Acid Res Mol Biol. 1998;61:65–131. doi: 10.1016/s0079-6603(08)60825-9. [DOI] [PubMed] [Google Scholar]
- 25.Gary J D, Lin W J, Yang M C, Herschman H R, Clarke S. The predominant protein-arginine methyltransferase from Saccharomyces cerevisiae. J Biol Chem. 1996;271:12585–12594. doi: 10.1074/jbc.271.21.12585. [DOI] [PubMed] [Google Scholar]
- 26.Gilbreth M, Yang P, Bartholomeusz G, Pimental R A, Kansra S, Gadiraju R, Marcus S. Negative regulation of mitosis in fission yeast by the shk1 interacting protein skb1 and its human homolog, Skb1Hs. Proc Natl Acad Sci USA. 1998;95:14781–14786. doi: 10.1073/pnas.95.25.14781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamm J, Darzynkiewicz E, Tahara S M, Mattaj I W. The trimethylguanosine cap structure of U1 snRNA is a component of a bipartite nuclear targeting signal. Cell. 1990;62:569–577. doi: 10.1016/0092-8674(90)90021-6. [DOI] [PubMed] [Google Scholar]
- 28.Kambach C, Walke S, Young R, Avis J M, de la Fortelle E, Raker V A, Luhrmann R, Li J, Nagai K. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell. 1999;96:375–387. doi: 10.1016/s0092-8674(00)80550-4. [DOI] [PubMed] [Google Scholar]
- 29.Koh S S, Chen D, Lee Y H, Stallcup M R. Synergistic enhancement of nuclear receptor function by p160 coactivators and two coactivators with protein methyltransferase activities. J Biol Chem. 2000;276:1089–1098. doi: 10.1074/jbc.M004228200. [DOI] [PubMed] [Google Scholar]
- 30.Krapivinsky G, Pu W, Wickman K, Krapivinsky L, Clapham D E. pICln binds to a mammalian homolog of a yeast protein involved in regulation of cell morphology. J Biol Chem. 1998;273:10811–10814. doi: 10.1074/jbc.273.18.10811. [DOI] [PubMed] [Google Scholar]
- 31.Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 32.Lehmeier T, Foulaki K, Luhrmann R. Evidence for three distinct D proteins, which react differentially with anti-Sm autoantibodies, in the cores of the major snRNPs U1, U2, U4/U6 and U5. Nucleic Acids Res. 1990;18:6475–6484. doi: 10.1093/nar/18.22.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lerner M R, Steitz J A. Antibodies to small nuclear RNAs complexed with proteins are produced by patients with systemic lupus erythematosus. Proc Natl Acad Sci USA. 1979;76:5495–5499. doi: 10.1073/pnas.76.11.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin W J, Gary J D, Yang M C, Clarke S, Herschman H R. The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J Biol Chem. 1996;271:15034–15044. doi: 10.1074/jbc.271.25.15034. [DOI] [PubMed] [Google Scholar]
- 35.Liu Q, Dreyfuss G. A novel nuclear structure containing the survival of motor neurons protein. EMBO J. 1996;15:3555–3565. [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Q, Fischer U, Wang F, Dreyfuss G. The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell. 1997;90:1013–1021. doi: 10.1016/s0092-8674(00)80367-0. [DOI] [PubMed] [Google Scholar]
- 37.Lorson C L, Strasswimmer J, Yao J M, Baleja J D, Hahnen E, Wirth B, Le T, Burghes A H, Androphy E J. SMN oligomerization defect correlates with spinal muscular atrophy severity. Nat Genet. 1998;19:63–66. doi: 10.1038/ng0598-63. [DOI] [PubMed] [Google Scholar]
- 38.Mattaj I W. Cap trimethylation of U snRNA is cytoplasmic and dependent on U snRNP protein binding. Cell. 1986;46:905–911. doi: 10.1016/0092-8674(86)90072-3. [DOI] [PubMed] [Google Scholar]
- 39.Mattaj I W, De Robertis E M. Nuclear segregation of U2 snRNA requires binding of specific snRNP proteins. Cell. 1985;40:111–118. doi: 10.1016/0092-8674(85)90314-9. [DOI] [PubMed] [Google Scholar]
- 40.Melki J. Spinal muscular atrophy. Curr Opin Neurol. 1997;10:381–385. doi: 10.1097/00019052-199710000-00005. [DOI] [PubMed] [Google Scholar]
- 41.Neuman H E de Vegvar, Dahlberg J E. Nucleocytoplasmic transport and processing of small nuclear RNA precursors. Mol Cell Biol. 1990;10:3365–3375. doi: 10.1128/mcb.10.7.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paik W K, Kim S. Enzymatic methylation of protein fractions from calf thymus nuclei. Biochem Biophys Res Commun. 1967;29:14–20. doi: 10.1016/0006-291x(67)90533-5. [DOI] [PubMed] [Google Scholar]
- 43.Paik W K, Kim S. Protein methylase. I. Purification and properties of the enzyme. J Biol Chem. 1968;243:2108–2114. [PubMed] [Google Scholar]
- 44.Pellizzoni L, Charroux B, Dreyfuss G. SMN mutants of spinal muscular atrophy patients are defective in binding to snRNP proteins. Proc Natl Acad Sci USA. 1999;96:11167–11172. doi: 10.1073/pnas.96.20.11167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pellizzoni L, Kataoka N, Charroux B, Dreyfuss G. A novel function for SMN, the spinal muscular atrophy disease gene product, in pre-mRNA splicing. Cell. 1998;95:615–624. doi: 10.1016/s0092-8674(00)81632-3. [DOI] [PubMed] [Google Scholar]
- 46.Pollack B P, Kotenko S V, He W, Izotova L S, Barnoski B L, Pestka S. The human homologue of the yeast proteins Skb1 and Hsl7p interacts with Jak kinases and contains protein methyltransferase activity. J Biol Chem. 1999;274:31531–31542. doi: 10.1074/jbc.274.44.31531. [DOI] [PubMed] [Google Scholar]
- 47.Pu W T, Krapivinsky G B, Krapivinsky L, Clapham D E. pICln inhibits snRNP biogenesis by binding core spliceosomal proteins. Mol Cell Biol. 1999;19:4113–4120. doi: 10.1128/mcb.19.6.4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raker V A, Plessel G, Luhrmann R. The snRNP core assembly pathway: identification of stable core protein heteromeric complexes and an snRNP subcore particle in vitro. EMBO J. 1996;15:2256–2269. [PMC free article] [PubMed] [Google Scholar]
- 49.Rho J, Choi S, Seong Y R, Cho W K, Kim S H, Im D S. Prmt5, which forms distinct homo-oligomers, is a member of the protein-arginine methyltransferase family. J Biol Chem. 2001;276:11393–11401. doi: 10.1074/jbc.M008660200. [DOI] [PubMed] [Google Scholar]
- 50.Sauterer R A, Goyal A, Zieve G W. Cytoplasmic assembly of small nuclear ribonucleoprotein particles from 6 S and 20 S RNA-free intermediates in L929 mouse fibroblasts. J Biol Chem. 1990;265:1048–1058. [PubMed] [Google Scholar]
- 51.Seraphin B. Sm and Sm-like proteins belong to a large family: identification of proteins of the U6 as well as the U1, U2, U4 and U5 snRNPs. EMBO J. 1995;14:2089–2098. doi: 10.1002/j.1460-2075.1995.tb07200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shen E C, Henry M F, Weiss V H, Valentini S R, Silver P A, Lee M S. Arginine methylation facilitates the nuclear export of hnRNP proteins. Genes Dev. 1998;12:679–691. doi: 10.1101/gad.12.5.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins with silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 54.Siomi M C, Eder P S, Kataoka N, Wan L, Liu Q, Dreyfuss G. Transportin-mediated nuclear import of heterogeneous nuclear RNP proteins. J Cell Biol. 1997;138:1181–1192. doi: 10.1083/jcb.138.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tang J, Frankel A, Cook R J, Kim S, Paik W K, Williams K R, Clarke S, Herschman H R. PRMT1 is the predominant type I protein arginine methyltransferase in mammalian cells. J Biol Chem. 2000;275:7723–7730. doi: 10.1074/jbc.275.11.7723. [DOI] [PubMed] [Google Scholar]
- 56.Vorm O, Roepstorff P, Mann M. Improved resolution and very high sensitivity in MALDI TOF of matrix surfaces made by fast evaporation. Anal Chem. 1994;66:3281–3287. [Google Scholar]
- 57.Wilks A F, Oates A C. The JAK/STAT pathway. Cancer Surv. 1996;27:139–163. [PubMed] [Google Scholar]
- 58.Zieve G W, Sauterer R A. Cell biology of the snRNP particles. Crit Rev Biochem Mol Biol. 1990;25:1–46. doi: 10.3109/10409239009090604. [DOI] [PubMed] [Google Scholar]