Poly(A) Polymerase Modification and Reverse Transcriptase PCR Amplification of Environmental RNA (original) (raw)
Abstract
We describe a combination of two established techniques for a novel application for constructing full-length cDNA clone libraries from environmental RNA. The cDNA was cloned without the use of prescribed primers that target specific genes, and the procedure did not involve random priming. Purified RNA was first modified by addition of a poly(A) tail and then was amplified by using a commercially available reverse transcriptase PCR (RT-PCR) cDNA synthesis kit. To demonstrate the feasibility of this approach, a cDNA clone library was constructed from size-fractionated RNA (targeting 16S rRNA) purified from a geothermally heated soil in Yellowstone National Park in Wyoming. The resulting cDNA library contained clones representing Bacteria and Eukarya taxa and several mRNAs. There was no exact clone match between this library and a separate cDNA library generated from an RT-PCR performed with unmodified rRNA and _Bacteria_-specific forward and universal reverse primers that were designed from cultivated organisms; however, both libraries contained representatives of the Firmicutes and the α-Proteobacteria. Unexpectedly, there were no Archaea clones in the library generated from poly(A)-modified RNA. Additional RT-PCRs performed with universal and _Archaea_-biased primers and unmodified RNA demonstrated the presence of novel Archaea in the soil. Experiments with pure cultures of Sulfolobus solfataricus and Halobacterium halobium revealed that some Archaea rRNA may not be a suitable substrate for the poly(A) tail modification step. The protocol described here demonstrates the feasibility of directly accessing prokaryote RNA (rRNA and/or mRNA) in environmental samples, but the results also illustrate potentially important problems.
The PCR is a popular tool used for studying microbial communities and is most often used for amplifying 16S rRNA genes and functional genes from DNA templates to facilitate the study of structure-function relationships. However, discovery of novel genes for either of these applications is potentially constrained by primer design. For example, 16S rRNA gene primers originate from signature sequence studies of cultivated organisms (7, 25; for reviews see references 18 and 20). If, as is popularly believed, cultivated microorganisms actually represent a small fraction of the earth's microbial biota (24), then the use of such primers could potentially lead to profound underestimation of microbial diversity and perhaps limit the discovery of novel organisms. An example of a novel microorganism that escaped PCR detection is the apparently parasitic archaeon recently discovered by Huber et al. (10). This organism is sufficiently phylogenetically divergent from other known prokaryotes that PCR primers thought to be universal or considered general for the Archaea failed to amplify its 16S rRNA gene. Furthermore, fluorescence in situ hybridization probes targeting Crenarchaeota and Euryarchaeota failed to hybridize to cellular RNA of this organism (10). The discovery of this novel archaeon prompted Boucher and Doolittle (3) to suggest that “even weirder” organisms await discovery by techniques that depart from current PCR protocols.
The same constraint applies to the PCR cloning of prokaryotic protein-encoding genes from the environment, and in contrast to eukaryotic mRNA, which has poly(A) tails and thus can be accessed by using commercially available cDNA cloning kits, environmental prokaryote mRNA is not directly accessible by general cloning. In the present study we demonstrate the feasibility of a technique that, in theory, should allow the discovery of additional phylogenetically divergent microorganisms, as well as provide direct access to prokaryote community mRNA. Here we describe a protocol that couples poly(A) polymerase with a multifunctional reverse transcriptase (RT) for targeted cloning of environmental prokaryotic cDNA. In essence, oligonucleotides are synthesized on each end of purified environmental RNA molecules and become priming sites for a subsequent PCR step. Modification of native RNA molecules with oligonucleotides whose sequences are known allows cDNA amplification to proceed without primers designed from previously known and characterized genes. Advantages and apparent molecular biases of this alternative PCR approach are described and discussed.
MATERIALS AND METHODS
General description of the technique.
All of the steps used to modify and amplify the environmental RNA are summarized at http://www.tbi.montana.edu/faculty/mcdermottpolyARNA.html. In the first step, poly(A) polymerase is used to synthesize poly(A) tails at the 3′ termini of purified soil RNA molecules. The RNA molecules with the poly(A) tails are then the starting templates for a single-step, multiple-stage reaction performed with a commercially available cDNA cloning kit (SMART cDNA construction kit; Clontech, Palo Alto, Calif.). First-strand cDNA synthesis is accomplished by using the Moloney murine leukemia virus (MMLV) RT with a poly(T) oligonucleotide primer [primer 1; 5′-AAGCAGTGGTATCAACGCAGAGTACT30N*N-3′, where N is A, C, G, or T and N* is A, G, or C; the nucleotide triphosphate modifications help direct primer annealing to the junction of the RNA 3′ terminus with the poly(A) tail]. This MMLV RT has terminal transferase and template-switching activities (13, 27). The terminal transferase activity adds primarily CTPs to the 3′ end of the first-strand cDNA (equivalent to the 5′ end of the rRNA and mRNA), which then serves as the primary annealing site for a second oligonucleotide (primer 2; 5′-AAGCAGTGGTATCAACGCAGAGTACGCGGG-3′) that base pairs with the C-rich region, allowing the MMLV RT to then switch templates and continue replicating to the end of this oligonucleotide template. The result of the terminal transferase and template-switching activities is the addition of an oligonucleotide to the 3′ end of the cDNA (corresponding to the 5′ terminus of the RNA molecule). A final PCR, performed with approximately 20 ng of cDNA, primes from these synthesized oligonucleotide regions (primer 3; 5′-AAGCAGTGGTATCAACGCAGAGT-3′), yielding a cDNA library for cloning, transformation, and sequencing.
RNA extraction.
RNA used for the development of this technique was extracted from a geothermally heated acidic soil (65 to 92°C, pH 3.9) located in Yellowstone National Park in Wyoming. Soil samples for RNA analysis were collected with autoclaved spatulas and mixed approximately 1:1 with sterile distilled water in a sterile disposable 50-ml tube. Aliquots (0.5 ml) of the soil slurry were dispensed into sterile 2.0-ml screw-cap microcentrifuge tubes, flash frozen in liquid nitrogen, transported to the laboratory on dry ice, and stored at −75°C. For RNA extraction, acid-washed glass beads (0.5 g; diameter, 106 μm; Sigma, St. Louis, Mo.), 33.3 μl of 20% sodium dodecyl sulfate (SDS), 167 μl of 3% diatomaceous earth (Sigma), and 583 μl of Tris-HCl-buffered phenol (pH 8.0) were added to each frozen soil slurry sample. After rapid thawing in warm tap water, the sample was shaken for 45 s on a bead mill and centrifuged in the cold for 15 min at 14,000 × g. The aqueous layer was transferred to a fresh tube to precipitate the nucleic acids at −20°C with 3 M sodium acetate (pH 5.2) and 95% ethanol. After centrifugation (14,000 × g), the nucleic acid pellet was washed with 70% ethanol and suspended in 100 μl of nuclease-free water. RNA was further purified and treated with DNase by using the SV total RNA isolation system (Promega, Madison, Wis.). Purified RNA was eluted with 100 μl of nuclease-free water, and the RNA integrity was verified in 1% denaturing agarose gels containing formaldehyde (0.66%, wt/vol) as the denaturant. As a final purification step, RNA that was the size of 16S rRNA was excised from a 1.2% low-melting-point agarose gel and eluted from the agarose by using the AgarACE agarose-digesting enzyme from Promega.
Poly(A) tailing reaction and oligonucleotide synthesis.
Approximately 2 μg of gel-purified RNA was added as a substrate to a 100-μl reaction mixture containing 0.5 mM ATP, 0.5 mM MnCl2, 1× poly(A) polymerase buffer, 50 μg of bovine serum albumin, and 4 U of poly(A) polymerase (Ambion Inc., Austin, Tex.). Poly(A) polymerase is specific for single-stranded RNA and initiates synthesis of poly(A) tails only from the 3′ OH position of the 3′ terminal ribose. The reaction mixture was incubated at 37°C for 1 h, and then successful poly(A) tailing of the RNA was verified by using denaturing formaldehyde-agarose gels (as described above); i.e., reduced migration of the RNA corresponded to increased RNA molecular size. For RT-PCR amplification of the RNAs with poly(A) tails, the instructions provided by the manufacturer of the SMART cDNA construction kit (Clontech) were followed. cDNA products were then cloned into the vector pCR2.1-TOPO and transformed into Escherichia coli TOP10 (Invitrogen, Carlsbad, Calif.).
Dot blot analysis.
A preliminary sequence analysis of 25 cDNA clones suggested that the cDNA library contained a significant number of 23S rRNA sequences of an _Acetobacter_-like organism. Therefore, an oligonucleotide designed to detect Acetobacter 23S rRNA genes (5′-TGGGTCAGCGAGTTTCTGTT-3′) was end labeled with T4 polynucleotide kinase (Promega) and [32P]ATP (3,000 Ci/mmol; NEN Life Science Products, Boston, Mass.) and then used to probe the entire library. cDNA clones were blotted onto nylon membranes (GeneScreen Plus; NEN Life Science Products), which were prehybridized at 65°C for 1 h in 1% SDS-5.8% NaCl-10% dextran sulfate (sodium salt). A probe was added (106 cpm per probe), and the membranes were incubated overnight at 65°C and washed with 2× SSC (1× SSC is 15 mM NaCl plus 1.5 mM sodium citrate · 2H2O) containing 1% SDS at 60°C to remove nonspecifically bound probe. Hybridizing clones were identified by autoradiography.
RT-PCRs with unmodified RNA.
For one aspect of the study, RT-PCRs with purified but unmodified soil RNA were carried out with either the _Bacteria_-specific forward primer 8F (positions 8 to 27) (2) or the _Archaea_-biased forward primer 2F (2) paired with the universal primer 1510R (positions 1492 to 1510) (14). Each 50-μl reaction mixture contained 5 U of avian myeloblastosis virus (AMV) RT and 5 U of Tfl DNA polymerase (both enzymes were obtained from Promega), 2 μl of total RNA, 10 pmol of each primer, 1× AMV/Tfl reaction buffer, 2.5 mM MgSO4, 50 mM dATP, 50 mM dCTP, 50 mM dGTP, and 50 mM dTTP. Bovine serum albumin (fraction V; Fischer Scientific, Fair Lawn, N.J.) was included (5 μg) as a nonspecific inhibitor-binding protein in the reaction mixtures. The absence of DNA contamination was verified by performing the same reactions without AMV RT. All RT-PCRs and PCRs were performed with a Gene Amp PCR system 9700 thermal cycler (Applied Biosystems, Foster City, Calif.). RT-PCR fidelity was examined with agarose gels by verifying that the RT-PCR product contained a single amplicon and by comparing amplicon size with the sizes of known molecular weight standards. RT-PCR products were cloned directly into pCR2.1-TOPO and transformed into E. coli TOP10 as described by the manufacturer (Invitrogen). Restriction fragment length polymorphism (RFLP) analysis (digestion with RsaI and with HaeIII plus MspI) of the clones was used to define operational taxonomic units in order to help focus sequencing efforts.
Sequencing and phylogenetic analysis.
Sequencing of the RT-PCR clones was accomplished with an ABI310 DNA sequencer (Applied Biosystems, Norwalk, Conn.) by using synthetic primers complementary to the vector plasmid sequences flanking the multiple cloning site. When appropriate, sequences were first screened for chimeras by using the CHECK_CHIMERA program of the Ribosome Database Project (5). For phylogenetic analysis, an initial BLAST (Basic Local Alignment Search Tool) (1) search of public databases revealed the approximate phylogenetic affiliations of clones and identified closely related sequences, which were downloaded and aligned by using ClustalW (22). Phylogenetic trees were constructed by using the parsimony and maximum-likelihood programs in the PAUP software package (http://paup.csit.fsu.edu).
RESULTS
Protocol development.
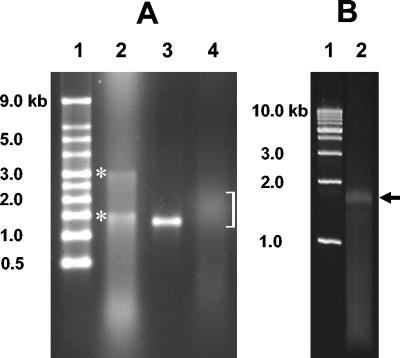
To demonstrate the feasibility of the technique described here, we targeted 16S rRNA. Gel purification of the RNA (Fig. 1A, lanes 2 and 3), whose size corresponded to the size of 16S rRNA (as predicted by using molecular weight standards and purified RNA), was found to be essential because of the extreme sensitivity of the poly(A) polymerase to soil contaminants and/or inhibitors (results not shown). In preliminary assays we determined that 2 μg of gel-purified RNA was sufficient for the poly(A) tailing reaction, which consistently yielded RNAs that had poly(A) tails ranging primarily from 200 to 600 A residues long and that in aggregate appeared as a short smear in the denaturing agarose gel (Fig. 1A, lane 4). The RNA with the poly(A) tail was the starting substrate for a single reaction mixture containing primers 1 and 2 (described in Materials and Methods) along with the MMLV RT. The multifunctional feature of MMLV RT is essential for the efficiency and simplicity of this protocol.
FIG. 1.
Demonstration of RNA purification, poly(A) tail modification, and RT-PCR amplification of environmental RNA. (A) Denaturing agarose (1%) gel containing electrophoresed samples resulting from RNA extraction, purification, and modification with poly(A)polymerase. Lane 1, molecular weight markers; lane 2, RNA extracted from soil and purified; lane 3, DNase-treated and gel-purified RNA; lane 4, soil RNA after poly(A) tailing reaction. The bracket indicates the position of the various tailed RNA molecules. The locations of 23S and 16S RNAs are indicated by asterisks and were determined by examining numerous reproducible gels in which these bands were aligned with the corresponding bands of RNA purified from E. coli (data not shown) and by molecular weight, as shown. (B) Final PCR product electrophoresed in a 1% agarose gel. Lane 1, molecular weight markers; lane 2, cDNA product from PCR amplification. The arrow indicates the position of the PCR product. In both panels, the sizes of all molecular weight markers are indicated on the left.
The oligonucleotide-modified single-strand cDNA was the template for the final PCR. Determining the optimum PCR cycle number was an empirical exercise; however, 15 to 18 cycles was typically optimal (results not shown). An excessive cycle number resulted in significant amplification of templates whose sizes differed substantially from the sizes of the targeted 16S RNA size templates and was indicated by smearing throughout the entire lane in an agarose gel (results not shown). When an optimum number of PCR cycles was used, the final PCR product was typically represented by a band that indicated a relatively narrow size range and was consistent with the starting template size range (Fig. 1B, lane 2). In the experiment in which PCR clones were characterized in the present study, 346 clones were obtained from transformation of the entire PCR product.
Clone analysis with poly(A)-modified RNA.
Preliminary sequence analysis of a clone sample suggested that the cDNA library was comprised of a significant number of 23S rRNA clones from an _Acetobacter_-like organism. Because our efforts were focused on 16S rRNA cDNA clones, the entire library was dot blot screened to identify 23S rRNA clones. An oligonucleotide designed to detect an internal portion of the 23S rRNA gene sequence (corresponding to nucleotides 556 to 575 of Acetobacter xylinus; accession number X89812) of this _Acetobacter_-like organism was used as the probe. Of the 346 clones, 95 (∼27%) hybridized with the 23S rRNA probe. The probe-positive clones were set aside, and in the subsequent sequence analysis we focused on the remaining clones.
Sequence analysis (∼600 to 650 bp) of the remaining 251 clones showed that the library was primarily (211 clones) comprised of prokaryotic 16S rRNA gene sequences (Table 1; also see data at http://www.tbi.montana.edu/faculty/mcdermottpolyARNA.html for a complete summary of the cDNA clones identified in this study). Seventy of these clones were found to lack various portions of the 5′ end of the 16S cDNA and so were also set aside because an important goal was to determine whether these cDNA clones contained _Bacteria_- or _Archaea_-specific or universal sequences normally used as priming sites for current PCR protocols. Approximately 94% of the remaining 16S rRNA clones represented the α-Proteobacteria, the β-Proteobacteria, and the Firmicutes (Table 1). Of the 58 α-Proteobacteria clones, 32 were most closely related (99 to 100% sequence identity) to an unclassified gram-negative, heterotrophic acidothermophile recently isolated from various acidic geothermal environments in Yellowstone National Park (11), and 10 clones were most closely related (98 to 100% sequence identity) to an Acidocella sp. All 24 β-Proteobacteria clones were 97% identical to a _Thiomonas_-like organism, and more than one-half of the Firmicutes clones were 97 to 99% identical to an _Alicyclobacillus acidiphilus_-like organism (Table 1).
TABLE 1.
Summary of major cDNA clone groups obtained from poly(A)-modified and RT-PCR-amplified environmental RNAa
| RNA group | Sample clone (accession no.) | No. of clones | % Identity | Genus or organism represented by closest phylogenetic neighbor (accession no.) |
|---|---|---|---|---|
| Prokaryote 16S rRNA | ||||
| α-Proteobacteria | 58 | |||
| NBFF62 (AY389855) | 32 | 99-100 | Gram-negative acidothermophilic Yellowstone National Park isolate Y008 (AY140238) | |
| NBFF56 (AY389853) | 10 | 98-100 | Acidocella sp. (D86510) | |
| NBFF21 (AB006711) | 6 | 99 | Acidophilum multivorum (AY389838) | |
| NBFF217 (AY389839) | 2 | 97 | Geothermal spring PCR clone (AF232921) | |
| NBFF48 (AY389852) | 1 | 99 | Yellowstone thermal soil PCR clone (AF391980) | |
| β-Proteobacteria | NBFF120 (AY389832) | 24 | 97 | Thiomonas sp. (AJ549220) |
| γ-Proteobacteria | NBFF110 (AY389831) | 6 | 97 | Frateuria sp. (AF376025) |
| Firmicutes | 50 | |||
| NBFF81 (AY389857) | 28 | 97-99 | Alicyclobacillus acidiphilus (AB076660) | |
| NBFF85 (AY389858) | 4 | 97-98 | Alicyclobacillus acidoterrestris (AB042058) | |
| Aquificae | NBFF44 (AY389849) | 1 | 99 | Hydrogenobaculum sp. (AJ320225) |
| Acidobacteria | NBFF308 (AY389843) | 1 | 97 | PCR clone from methanol soil enrichment (AF200696) |
| Actinobacteria | NBFF223 (AY389840) | 1 | 95 | Bacterium Ellin301 (AF498683) |
| Prokaryote 23S rRNA | ||||
| α-Proteobacteria | NBFF15 (AY389866) | 106b | 88-93 | Acetobacter xylinus (X89812) |
| NBFF16 (AY389868) | Acetobacter europaeus (X89771) | |||
| γ-Proteobacteria | NBFF164 (AY389881) | 1 | 90 | Xanthomonas campestris (AEO12540) |
| Firmicutes | NBFF50 (AY389882) | 7 | 90-93 | Bacillus sp. (X60981) |
| Aquificae | NBFF332 (AY389873) | 3 | 75-80 | Aquifex pyrophilus (APY15785) |
| Eukaryote 18S rRNA | ||||
| Fungi | 15 | |||
| NBFF72 (AY389879) | 1 | 99 | Trichoderma (AY180319) | |
| NBFF144 (AY389867) | 1 | 96 | Eupenicillium (U21298) | |
| NBFF211 (AY389869) | 1 | 99 | Aphanoascus (ABO15788) | |
| Eukaryote 28S rRNA | ||||
| Fungi | NBFF108 (AY389862) | 1 | 99 | Hypocrea lutea (ABO27384) |
| Eukaryote 25S rRNA | ||||
| Bryophyta | NBFF46 (AY389876) | 1 | 90 | Diphyscium foliosum (AJ271116) |
| Eukaryote 28S rRNA | ||||
| Metazoa | NBFF113 (AY389863) | 1 | 87 | Austrobilharzia variglandis (AY157250) |
| Eukaryote chloroplast large-subunit rRNA | NBFF106 (AY389861) | 2 | 97 | Chlorococcum (L44123) |
| NBFF121 (AY389864) | 89 | Pylaiella (X61179) | ||
| Eukaryote chloroplast small-subunit rRNA | NBFF337 (AY389874) | 1 | 89 | Odontella (Z67753) |
In addition to the 23S rRNA clones identified by using dot blots with a 23S RNA-specific probe, 32 other 23S rRNA cDNA clones were identified by sequence analysis. Some of these clones represented random regions of the 23S rRNA gene distant from the portion targeted by the probe, suggesting that RNA shearing occurred during soil extraction, generating random fragments whose sizes were similar to the size of the 16S rRNA targeted in the gel purification step. In addition, some of these clones represented 23S rRNAs of other microorganisms (although the levels of identity for clones exhibiting the closest resemblance to Aquifex pyrophilus were very low), and the data resulted in a total of 127 cDNA clones containing cDNA corresponding to some portion of a 23S rRNA gene, a significant majority of which (91%) apparently represented an organism similar (88 to 93% identity) to A. xylinus, an α-proteobacterium (Table 1).
Finally, several 18S and 28S rRNA gene sequences representing various fungi (97 to 100% similarity), a moss, a metazoan, and chloroplast rRNA from algae (Table 1) were also identified in the cDNA clone library, but they accounted for a minor proportion (∼7%) of the total cDNA clones (Table 1).
The cDNA library also contained clones apparently representing mRNAs (Table 2). The inferred amino acid sequences for clones NBFF33, NBFF306, NBFF413, and NBFF429 exhibited relatively high identity scores with the amino acid sequences encoded by various protein-encoding genes from documented thermophilic archaea. Clone NBFF58 contained a cDNA that exhibited homology with an _S_-adenosylmethionine-dependent methyltransferase gene from Bacillus halodurans, and two other clones exhibited significant inferred amino acid identity with hypothetical proteins in the fungi Neurospora crassa and Aspergillus nidulans (Table 2).
TABLE 2.
Descriptive information for cDNA clones with sequences matching sequences of protein-encoding genes: clone reference numbers, descriptions of closest database matches (and accession numbers), and identity and similarity scores for the inferred amino acid sequences as well as number of inferred amino acids used in the alignment
| Clone (accession no.) | Database match (accession no.) | % Identity/% similarity (no. of inferred amino acids) |
|---|---|---|
| NBFF33 (AY389884) | Aeropyrum pernix thermosome subunit (NP_147591) | 75/90 (214) |
| NBFF58 (AY389888)a | Bacillus halodurans S-adenosylmethionine-dependent methyltransferase (NP_623400) | 40/58 (164) |
| NBFF306 (AY389883) | Methanothermobacter thermautotrophicus glutamate-1-semialdehyde aminotransferase (NP_275371) | 42/57 (164) |
| NBFF413 (AY389886) | Pyrobaculum aerophilum glutamine synthetase (NP_560736) | 52/71 (221) |
| NBFF429 (AY389887) | Pyrobaculum aerophilum conserved protein (toprim domain) (NP_559773) | 66/86 (201) |
| NBFF349 (AY389885) | Neurospora crassa probable DFG5 protein (XP_323602) | 67/77 (225) |
| NBFF28 (AY389882) | Aspergillus nidullans hypothetical protein (AN12742) | 51/67 (198) |
Clone analysis with unmodified RNA.
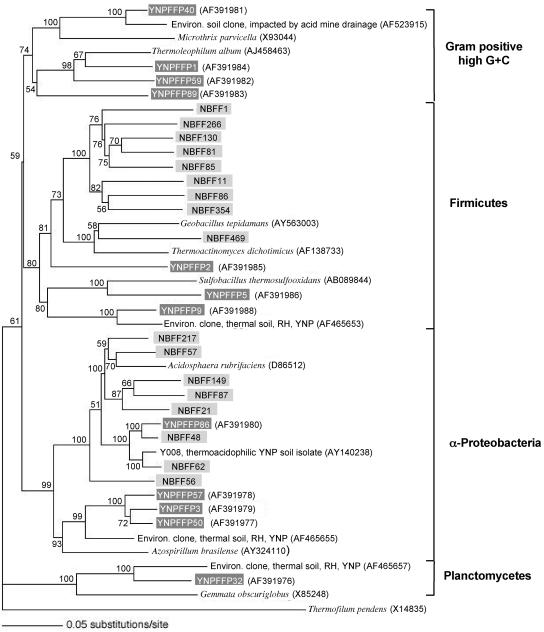
Additional RT-PCRs were conducted to compare the clones described above with clones amplified from unmodified RNA templates by using previously described primers designed from cultivated organisms. RFLP screening of approximately 200 clones generated from RT-PCRs with the _Bacteria_-specific and universal reverse primers yielded 12 clone groups. Samples of each RFLP group were sequenced and then examined to determine the phylogenetic position and relationship in comparison with phylum-related clones obtained with poly(A)-modified RNA (see above). There was phylogenetic overlap between the libraries in that both contained clones that belong to the α-Proteobacteria and the Firmicutes, but there were no exact sequence matches in the two clone libraries (Fig. 2). However, both libraries contained sequences very similar to the sequence of the isolated acidothermophilic heterotroph Y008 (11) mentioned above.
FIG. 2.
Inferred evolutionary relationships of Bacteria sequences RT-PCR amplified from RNA purified from the same Yellowstone National Park geothermal soil. All clones obtained from unmodified rRNA (amplified by using the 8F [_Bacteria_-specific] and 1510R [universal] primers) are indicated by dark gray boxes and are compared to taxonomically relevant clones derived from rRNAs with poly(A) tails, which are indicated by light gray boxes. Phyla are indicated by brackets on the right. The tree was constructed by maximum-likelihood analysis, and the cloned sequences (approximately 1,400 nucleotides) were aligned with and compared to the most similar (closest BLAST matches) 16S rRNA gene sequences of cultured and characterized bacteria and, where informative, to the most similar environmental (Environ.) clones (RH, Ragged Hills; YNP, Yellowstone National Park). Bootstrap values resulting from analysis of 100 pseudoreplicates are indicated at the nodes, and a nucleotide substitution scale bar is below the tree. The archaeon Thermofilum pendens was used as an outgroup.
Another contrast between these libraries was that the majority (94%) of the clones derived from RNAs with poly(A) tails exhibited at least 97% identity with cultivated and characterized microorganisms, which provided a high probability of predictable form and function (see data at http://www.tbi.montana.edu/faculty/mcdermottpolyARNA.html). In contrast,only about one-half of the clones amplified from unmodified RNA exhibited similarly high levels of sequence identity with characterized organisms. Clones YNPFFP2, YNPFFP5, and YNPFFP9 were phylogenetically related to the Firmicutes. The BLAST match values for these three clones were poor to moderate, ranging from 88.2 to 95.6% with other environmental clones; the best match was between YNPFFP9 and another Yellowstone National Park thermal soil clone that we previously encountered at Ragged Hills in the Norris Geyser Basin (Fig. 2) (16). The closest cultivated relative of YNPFFP9 was found to be the acidothermophile Sulfobacillus thermosulfooxidans (8). Sequences of the other three Bacteria clone groups obtained by using unmodified rRNA were associated with three other phyla (Fig. 2). Clone group YNPFFP32 was most similar (94%) to other thermal soil clones that we previously amplified from geothermally heated soils at Ragged Hills in Norris Geyser Basin (16) and phylogenetically placed in the phylum Planctomyces (Fig. 2). Clone group YNPFFP50 was most closely related to clone groups YNPFFP3 and YNPFFP57 (97.9 and 97.8% identity, respectively), and all of these organisms were placed in the α-Proteobacteria subdivision. Together, the latter three clone groups likely represent different species of the same uncharacterized genus. The next most closely related organism (94.8% sequence identity) is represented by an environmental clone (accession number AF465655) also obtained from a geothermally heated soil at Ragged Hills in Norris Geyser Basin (16). Organisms represented by clone group YNPFFP86 are also α-proteobacteria and are highly related (98.4% sequence identity) to clone group NBFF48 derived from poly(A)-modified rRNA, and both groups are closely related to the acidothermophilic heterotroph isolate Y008 mentioned above (Table 1). Finally, four clone groups were placed in the gram-positive, high-G+C-content division. YNPFFP40 exhibited the highest level of identity (96%) with an environmental clone recovered from a forest soil perturbed by acid mine drainage from a coal reject materials pile (4), whereas YNPFFP1 and YNPFFP59 were more closely related to the thermophile Thermoleophilum album (26), as was the more distantly related clone YNPFFP89 (Fig. 2).
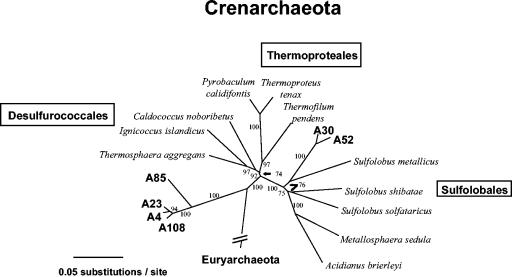
We were also interested in further examining this soil for Archaea, whose presence not only was expected but was suggested by a few of the mRNA cDNA clones (Table 2). RT-PCRs performed with purified soil total RNA as the template and with the 2F (biased for Archaea) and 1510R (universal) primers yielded an amplicon that was a uniform length and the appropriate size (results not shown). Restriction analysis of a portion of the resulting clone library (66 clones) resulted in identification of six RFLP groups that, after sequencing and phylogenetic analysis, were found to represent organisms in the Crenarchaeota division (Fig. 3). BLAST searches with sequences of clones A4, A23, A85, and A108 showed that all of these clones were more closely related to each other than to other sequences in the database. As a group, the closest matches were also environmental clones; however, the level of sequence similarity was still low (84 to 86% identity) for the best three-gap alignments. The phylogenetic relationship of these clones was inferred by distance and parsimony analyses (Fig. 3). The clones formed a small clade emerging as a branch near the base of the Crenarchaeota division (Fig. 3); A85 was separate from the other three clones but shared the same main branch (Fig. 3). Clones A30 and A52 were likewise most similar to each other (∼99% identity) and shared a node with Sulfolobus metallicus in the order Sulfolobales (Fig. 3). While isolation of these novel archaea awaits further study, RT-PCR amplification of their 16S rRNAs from purified but unmodified soil RNA clearly suggested that these organisms were present in this extreme thermal soil environment but that their RNAs were not detected by the RNA modification technique described in this study.
FIG. 3.
Inferred evolutionary relationships of Archaea sequences RT-PCR amplified from Yellowstone National Park geothermal soil total RNA (unmodified) by using the 2F (_Archaea_-biased) and 1510R (universal) primers. The tree was constructed by using distance analysis (parsimony analysis yielded a very similar tree), and the cloned archaeal sequences (at approximately 1,400 positions) were compared to 16S rRNA gene sequences of cultured and characterized archaea representing separate lines of decent within the Crenarchaeota division. The taxonomic orders within the Crenarchaeota are enclosed in boxes. Cloned sequences are indicated by boldface type. The bootstrap value resulting from 500 pseudoreplicates is shown for each branch, and a nucleotide substitution scale bar is below the tree. Clones A4, A23, A30, A52, A85, and A108 correspond to GenBank accession numbers AF391990, AF391991, AF391995, AF391996, AF391992, and AF391993, respectively.
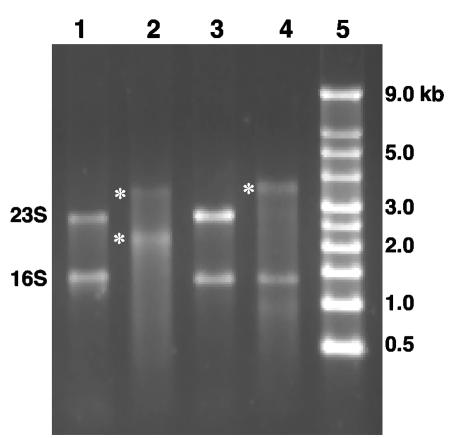
Additional experiments were then undertaken to determine why the archaeal 16S rRNAs were not cloned and identified by the RNA modification approach. Total RNAs from pure cultures of Sulfolobus solfataricus strain P2 and Halobacterium halobium strain ATCC 43214 were used as substrates for the same poly(A) polymerase reaction used with purified soil RNA. S. solfataricus 16S rRNA and 23S rRNA appeared to be suitable substrates for the poly(A) polymerase, as judged by molecular weight shifts for both rRNA moieties in denaturing agarose gels (Fig. 4, compare lanes 1 and 2). However, with H. halobium RNA preparations, only the 23S rRNA appeared to be a suitable substrate for poly(A) polymerase, based on the same criterion (Fig. 4, lanes 3 and 4). Poly(A) tailing of the 23S rRNA in the H. halobium RNA reaction mixture clearly suggested that the lack of 16S rRNA tailing was not due to some inhibitory substance in the reaction mixture.
FIG. 4.
Variation in poly(A) tailing of purified RNA from S. solfataricus and H. halobium. Lane 1, _S. solfataricu_s untreated RNA; lane 2, S. solfataricus RNA treated with poly(A) polymerase; lane 3, H. halobium untreated RNA; lane 4, H. halobium RNA treated with poly(A) polymerase; lane 5, nucleic acid fragment length standards. The positions of size standards are indicated on the right. The positions of 16S and 23S rRNAs are indicated on the left, and asterisks indicated the positions of poly(A)-modified RNA.
DISCUSSION
The experiments summarized here provided an alternative cloning approach that allows RT-PCR amplification of environmental RNA without a requirement for primers designed from known and characterized genes. The technique was used to examine the microbial diversity in an extreme thermal soil environment, and gel fractionation techniques were used to target the small-subunit rRNA molecule. RNA-based studies are particularly important for examining geothermal soil environments. Nonthermophilic microorganisms introduced into high-temperature environments (e.g., by wind, water erosion, etc.) are quickly heat killed; however, DNA is relatively stable even in dead cells (12), and so soil community diversity estimates could be affected by nonresident microorganisms. In contrast, this would likely not be a problem in most aquatic geothermal environments, where heat-killed organisms would be removed by spring flow. RNA-based RT-PCR amplification might also be expected to provide a more direct link to the metabolically active fraction (15, 16, 17), which is thought to be engaged in ribosome synthesis proportional to growth (23).
The cDNA library amplified from poly(A)-modified RNA contained 16S rRNA cDNA clones very similar to previously isolated and characterized thermophilic bacteria (e.g., clone groups NBFF62 and NBFF120) (Table 1). Also, several of these clones were very similar to 16S rRNA gene PCR clones previously amplified from Yellowstone's thermal environments (e.g., clone groups NBFF48 and NBFF217) (Table 1). Furthermore, a majority (64%) of the Firmicutes clones were closely related to _Alicyclobacillus_-like organisms (clone groups NBFF81 and NBFF85), and Alicyclobacillus is a genus that we have encountered frequently in our efforts to cultivate organisms from samples from acidic environments throughout Yellowstone (unpublished data). The presence of acidophiles in this low-pH soil was also implied by clones having high sequence identity with characterized acidophiles or with environmental clones derived from other acidic environments (e.g., clone groups NBFF62, NBFF56, and NBFF21).
In addition to being represented by 16S rRNA cDNA clones, the α-Proteobacteria, the γ-Proteobacteria, and the Firmicutes were also represented by 23S rRNA cDNA clones (Table 1). Interestingly, the 23S rRNA cDNA clones were dominated by sequences from _Acetobacter_-like organisms (although the levels of sequence identity were very low). This may have resulted from greater stability in vivo relative to the rRNA turnover in other organisms present, or it is also possible that unanticipated bias was again encountered at the poly(A) tailing step. It was somewhat surprising that the apparent identity of the organisms represented by the 23S rRNA sequences did not directly overlap the apparent identity of the organisms represented by the 16S rRNA sequences (particularly organisms such as the uncharacterized Yellowstone National Park isolate Y008 [see reference 11]). However, this may have been due in part to the relative paucity of 23S rRNA sequences available in public databases, which could have limited our ability to match 16S rRNA and 23S rRNA sequences with specific cultivated organisms.
For comparative purposes, a separate cDNA library was constructed from an RT-PCR that was performed with unmodified rRNA as the template and was primed by using oligonucleotides designed from cultivated organisms belonging to the domain Bacteria. The clones contained in this library exhibited limited overlap with the clones derived from poly(A)-modified RNAs. The latter library represented 18S, 23S, 25S, and 28S rRNAs (Table 1), as well as a few mRNAs (Table 2). Furthermore, the phylogenetic relatedness between the libraries was limited to two phyla, the Firmicutes and the α-Proteobacteria (Fig. 2), and there were no exact clone matches. The closest agreement between these libraries involved clones that were highly related to an acidothermophilic heterotroph isolated from acid thermal environments throughout Yellowstone National Park (11) (Fig. 2). The general lack of better agreement between libraries was not unexpected, as primer bias (6, 9) can account for such variations even between traditionally constructed libraries. However, we noted that the generally much greater division-level diversity encountered with the novel technique described in this paper implied that there was a potentially greater level of utility within at least the domain Bacteria.
The presence of viable, metabolically active, eukaryotic microorganisms in this extreme geothermal environment was implied by a small number of cDNA clones representing 18S and 28S rRNAs from fungi, clones exhibiting relatively low levels of sequence identity with 28S rRNA from an apparent metazoan, and 25S rRNA from a bryophyte (Table 1). The occurrence of these clones, along with cDNA representing chloroplast rRNA, presents interesting questions about thermal tolerance in eukaryotes (see below). The fungal RNA could be associated with heat-resistant spore structures; however, presumably this would not be possible for the other eukaryotes suggested to be present by the RNA clone analysis.
Even though the RNA purification step was intentionally biased towards obtaining 16S rRNA, the occurrence of mRNA clones in the final library was not surprising (Table 2). Theoretically, mRNA should be accessible with this technique, and when the gel purification techniques described above were used, mRNA in the size range from 1,400 to 1,600 bp was presumably coextracted with the targeted 16S rRNA. The putative identities of six of the mRNA cDNA clones suggested that there was in situ expression of protein-encoding genes whose sequences were similar to the sequences previously characterized for known thermophilic prokaryotes, primarily archaea (Table 2). The presence of apparent fungal gene expression (N. crassa and A. nidulans coding sequences) (Table 2) also suggested that the fungal 18S rRNA cloned in this study (see above) (Table 1) may not necessarily have been associated with heat-resistant spore structures but rather may have been from metabolically active, extremely thermophilic eukaryotes. The presence of highly labile RNAs from such organisms in this high-temperature environment is intriguing and at least suggests that there is potential for discovering eukaryotes capable of growth at temperatures exceeding current known limits. Thermophilic fungi have been described (21), although so far fungi that grow at this extreme temperature have not been described. It is also possible that these putative metabolically active fungi, as well as putative metabolically active phototrophs (as implied from chloroplast rRNA cDNA clones) (Table 1), may have inhabited a tolerable temperature niche at one extreme of a steep temperature gradient that potentially occurs at the soil-air interface in thermal soil environments.
Unexpectedly, RT-PCRs performed with oligonucleotide-modified RNA failed to generate archaeal 16S rRNA cDNAs. Independent RT-PCRs performed with an archaeon-biased primer and purified but unmodified soil RNA readily and reproducibly amplified archaeal sequences. While clearly not the focus of this study, these archaea are extremely interesting due to their apparent phylogenetic novelty (Fig. 3). Our initial cultivation efforts yielded enrichments (at 75°C with ferrous iron as the electron donor and oxygen as the electron acceptor) containing an archaeon corresponding to clone A23 (unpublished data). Thus, the novel archaeal sequences discovered in the control experiments appear to represent bona fide extremely thermophilic archaea. Other evidence of archaea in this environment comes from the identification of mRNA cDNA clones having inferred amino acid sequences that exhibit high levels of identity and similarity with the amino acid sequences encoded by protein-encoding genes from cultivated archaea (Table 2).
The primary goals of the experiments described in this paper were to demonstrate the feasibility of cDNA cloning of prokaryotic RNA from the environment and to attempt to avoid the bias inherent in selecting a primer sequence based on preexisting sequence information obtained from cultivated microorganisms. Unexpectedly, a different type of apparent molecular bias was encountered, and it appeared to involve organisms belonging to the domain Archaea. One potential source of the hypothesized molecular bias could have been the fact that some rRNA molecules were poor substrates for poly(A) polymerase. Experiments aimed at investigating this possibility demonstrated that poly(A) tailing was achieved with purified S. solfataricus 23S and 16S rRNAs (Fig. 4); however, with H. halobium RNA preparations, the 16S rRNA moiety was a poor substrate even though the 23S rRNA was poly(A) tailed in the same reactions (Fig. 4). Archaea RNA is extensively modified relative to the RNA observed thus far in bacteria (19); any modification of the 3′-terminal ribose moiety at the 3′ OH position would prohibit poly(A) tailing. Also, RNA modifications resulting in rRNA folding could sterically hinder poly(A) polymerase accessibility to the RNA molecule 3′ terminus and thus would also be expected to reduce poly(A) tailing efficiency.
This limitation notwithstanding, we suggest that the methodology described in this paper still has significant potential for discovery of novel bacterial 16S rRNA gene sequences that cannot be amplified by PCR primers currently in use. A recent example is the discovery of a nanoarchaeon having a significantly divergent 16S rRNA gene sequence (10). In the high-temperature soil sample used in the present study, we did not encounter novelty of this type, and indeed each of the Bacteria 16S rRNA cDNA clones obtained via the poly(A) tailing approach contained sequences that should have allowed their amplification with the 8F and 522F Bacteria primers. Furthermore, random sampling (∼20%) of the 16S rRNA cDNA clones suggested that all of the clones could be amplified by PCR with the 1510R universal primer (results not shown).
The methodology described here may also facilitate transcriptome-based studies, in which prokaryote community gene expression can be probed without a requirement for inconvenient arrays and without random priming and can yield full-length transcript information. We believe that the small number of mRNA cDNA clones found in this study represents a very minor fraction of the total microbial community gene expression profile. Presumably, this low proportional representation could be significantly increased by screening and/or fractionation steps that target the removal of rRNA prior to the poly(A) tailing step. Such techniques are currently being investigated.
Acknowledgments
We express our gratitude for the enthusiastic support of Christie Hendrix and John Varley, Yellowstone Center for Resources, Yellowstone National Park, Wyo.
This work was supported by the National Science Foundation (grants DEB-9809360 and MCB-0132022), by the National Aeronautics and Space Administration (grant NAG5-8807), and by the Montana Agricultural Experiment Station (MAES 911310).
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. L. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25**:**3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59**:**143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boucher, Y., and W. F. Doolittle. 2002. Something new under the sea. Nature 417**:**27-28. [DOI] [PubMed] [Google Scholar]
- 4.Brofft, J. E., J. V. McArthur, and L. J. Shimkets. 2002. Recovery of novel bacterial diversity from a forested wetland impacted by reject coal. Environ. Microbiol. 4**:**764-769. [DOI] [PubMed] [Google Scholar]
- 5.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31**:**442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El Fantroussi, S., L. Verschuere, W. Verstraete, and E. M. Top. 1999. Effect of phenylurea herbicides on soil microbial communities estimated by analysis of 16S rRNA gene fingerprints and community-level physiological profiles. Appl. Environ. Microbiol. 65**:**982-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fox, G. E., K. R. Pechman, and C. R. Woese. 1977. Comparative cataloging of 16S ribosomal ribonucleic acid: molecular approach to procaryotic systematics. Int. J. Syst. Bacteriol. 27:44-57. [Google Scholar]
- 8.Goto, K., K. Mochida, M. Asahara, M. Suzuki, H. Kasai, and A. Yokota. 2003. Alicyclobacillus pomorum sp. nov., a novel thermo-acidophilic, endospore-forming bacterium that does not possess omega-alicyclic fatty acids, and emended description of the genus Alicyclobacillus. Int. J. Syst. Evol. Microbiol. 53**:**1537-1544. [DOI] [PubMed] [Google Scholar]
- 9.Hansen, M. C., T. T. Nielsen, M. Givskov, and S. Molin. 1998. Biased 16S rDNA PCR amplifications caused by interference from DNA flanking the template region. FEMS Microbiol. Ecol. 26**:**141-149. [Google Scholar]
- 10.Huber, H., M. J. Hohn, R. Rachel, T. Fuchs, V. C. Wimmer, and K. O. Stetter. 2002. A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. Nature 417**:**63-67. [DOI] [PubMed] [Google Scholar]
- 11.Johnson, D. B., N. Okibe, and F. F. Roberto. 2003. Novel thermo-acidophilic bacteria isolated from geothermal sites in Yellowstone National Park: physiological and phylogenetic characteristics. Arch. Microbiol. 180**:**60-68. [DOI] [PubMed] [Google Scholar]
- 12.Lindahl, T. 1993. Instability and decay of the primary structure of DNA. Nature 362**:**709-715. [DOI] [PubMed] [Google Scholar]
- 13.Matz, M., D. Shagin, E. Bogdanova, O. Britanova, S. Lukyanov, L. Diatchenko, and A. Chenchik. 1999. Amplification of cDNA ends based on template-switching effect and step-out PCR. Nucleic Acids Res. 27**:**1558-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muyzer, G., A. Teske, C. O. Wirsen, and H. W. Jannasch. 1995. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch. Microbiol. 164**:**165-172. [DOI] [PubMed] [Google Scholar]
- 15.Nogales, B., E. R. Moore, W. R. Abraham, and K. N. Timmis. 1999. Identification of the metabolically active members of a bacterial community in a polychlorinated biphenyl-polluted moorland soil. Environ. Microbiol. 1**:**199-212. [DOI] [PubMed] [Google Scholar]
- 16.Norris, T. B., J. Wraith, R. C. Castenholz, and T. R. McDermott. 2002. Soil microbial community structure across a thermal gradient following a recent geothermal heating event. Appl. Environ. Microbiol. 68**:**6300-6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogram, A., W. Sun, F. J. Brockman, and J. K. Fredrickson. 1995. Isolation and characterization of RNA from low-biomass deep-subsurface sediments. Appl. Environ. Microbiol. 61**:**763-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olsen, G. J., D. J. Lane, S. J. Giovannoni, and N. R. Pace. 1986. Microbial ecology and evolution: a ribosomal RNA approach. Annu. Rev. Microbiol. 40**:**337-365. [DOI] [PubMed] [Google Scholar]
- 19.Omer, A. D., S. Ziesche, W. A. Decatur, M. J. Fournier, and P. P. Dennis. 2003. RNA-modifying machines in archaea. Mol. Microbiol. 48**:**617-629. [DOI] [PubMed] [Google Scholar]
- 20.Pace, N. R., D. A. Stahl, D. J. Lane, and G. J. Olsen. 1986. The analysis of natural microbial populations by ribosomal RNA sequences. Adv. Microb. Ecol. 9**:**1-55. [Google Scholar]
- 21.Tansey, M. R., and T. D. Brock. 1973. Dactylaria gallopava, a cause of avian encephalitis, in hot spring effluents, thermal soils, and self-heated coal waste piles. Nature 242**:**202-203. [DOI] [PubMed] [Google Scholar]
- 22.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22**:**4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wagner, R. 1994. The regulation of ribosomal RNA synthesis and bacterial cell growth. Arch. Microbiol. 161**:**100-109. [DOI] [PubMed] [Google Scholar]
- 24.Wayne, L. G., D. J. Brenner, R. R. Colwell, P. A. D. Grimont, O. Kandler, M. I. Krichevsky, L. H. Moore, W. E. C. Moore, R. G. E. Murray, E. Stackebrandt, M. P. Starr, and H. G. Truper. 1987. Report of the Ad Hoc Committee on Reconciliation of Approaches to Bacterial Systematics. Int. J. Syst. Bacteriol. 37**:**463-464. [Google Scholar]
- 25.Woese, C. R., E. Stackebrandt, T. J. Macke, and G. E. Fox. 1985. A phylogenetic definition of the major eubacterial taxa. Syst. Appl. Microbiol. 6**:**143-151. [DOI] [PubMed] [Google Scholar]
- 26.Yakimov, M. M., H. Lunsdorf, and P. N. Golyshin. 2003. Thermoleophilum album and Thermoleophilum minutum are culturable representatives of group 2 of the Rubrobacteridae (Actinobacteria). Int. J. Syst. Evol. Microbiol. 53:377-380. [DOI] [PubMed] [Google Scholar]
- 27.Zhu, Y. Y., E. M. Machleder, A. Chenchik, R. Li, and P. D. Siebert. 2001. Reverse transcriptase template switching: a SMART approach for full-length cDNA library construction. BioTechniques 30**:**892-897. [DOI] [PubMed] [Google Scholar]