Ecological fitness, genomic islands and bacterial pathogenicity: A Darwinian view of the evolution of microbes (original) (raw)
Abstract
The compositions of bacterial genomes can be changed rapidly and dramatically through a variety of processes including horizontal gene transfer. This form of change is key to bacterial evolution, as it leads to ‘evolution in quantum leaps’. Horizontal gene transfer entails the incorporation of genetic elements transferred from another organism—perhaps in an earlier generation—directly into the genome, where they form ‘genomic islands’, i.e. blocks of DNA with signatures of mobile genetic elements. Genomic islands whose functions increase bacterial fitness, either directly or indirectly, have most likely been positively selected and can be termed ‘fitness islands’. Fitness islands can be divided into several subtypes: ‘ecological islands’ in environmental bacteria and ‘saprophytic islands’, ‘symbiosis islands’ or ‘pathogenicity islands’ (PAIs) in microorganisms that interact with living hosts. Here we discuss ways in which PAIs contribute to the pathogenic potency of bacteria, and the idea that genetic entities similar to genomic islands may also be present in the genomes of eukaryotes.
Introduction
Bacteria, which have existed for more than 3 billion years, represent the most ancient forms of life on the earth. The enormous evolutionary potential of these organisms is illustrated by the fact that the innumerable species currently living differ in many properties including metabolic capacities, cell surface compositions, life styles, ecological niches and host specificities (Doolittle, 1999). From a Darwinian point of view, every living organism is a result of the driving forces of evolution, which include the plasticity of the genome and the rate of phenotype generation, as well as the selective pressures exerted by the environment (Arber, 2000). The capacity for change, as determined by these factors, forms the basis of evolutionary progress.
In eukaryotes, genetic variability is primarily the result of sexual reproduction, which involves chromosomal recombination during meiosis. In prokaryotes, where this form of shuffling is not available, other factors determine the rate of evolution. These include the frequent occurrence of point mutants, high levels of recombination and gene silencing, and the transfer of genetic material between different bacterial species—even genera. In particular the latter process, referred to as horizontal gene transfer, represents a cornerstone of bacterial evolution, and it has led to dramatic changes in the composition of microbial genomes over relatively short time periods (Ochman et al., 2000).
Prokaryotic genomes: core and flexible gene pools
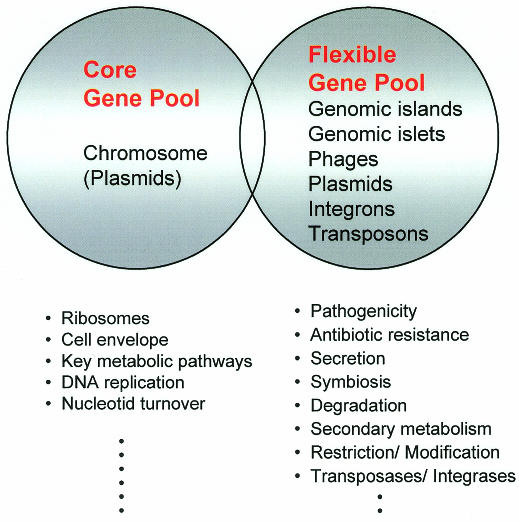
Bacterial genes may be transmitted between different organisms via conjugation, transduction and natural transformation. The former two processes require specific gene ferries, such as plasmids or bacterial viruses, which transport bacterial DNA along with their own sequences from donor to recipient cells. The majority of the horizontally transferred DNA is part of the flexible bacterial gene pool. In addition to phages and plasmids, the flexible gene pool comprises conjugative transposons, ‘simple’ transposons, integrons, ‘genomic islets’ (<10 kb), and ‘genomic islands’ (>10 kb) (Figure 1). In contrast, the core gene pool is restricted to genes that are part of the bacterial chromosome, except in a few species (e.g. in Streptomyces) for which plasmids also may be included (del Solar et al., 1995). The majority of the genes of the core gene pool encode proteins that play roles in basic cellular functions (e.g. translation, metabolism, architecture) and exhibit rather homogeneous G+C contents and codon usage. DNA elements from the flexible gene pool, on the other hand, often have features characteristic of transferred elements (different G+C content and codon usage, presence of mobility genes) and encode additional functions that are not essential for bacterial growth but provide advantages under particular conditions (changes in the environment, entry into a new host, etc.). Although the majority of the genes of the flexible gene pool seem to confer selective advantages to their bacterial recipients, a few (IS elements, prophages, restriction/modification systems) represent ‘selfish’ DNA-molecules, whose only mission is to promote their own spread (Lilley et al., 2000). As indicated in Figure 1, some genes may belong to both the core and flexible pools, but the majority belong to either one or the other. Perhaps not surprisingly, the number of genes within a cell that belong to the flexible pool may vary from 18% (Escherichia coli K-12) to <1% (Mycoplasma) of the total genome (Lawrence and Ochman, 1998; Ochman et al., 2000).
Fig. 1. Model of the DNA pools in the genomes of prokaryotes. The DNA elements comprising the core as well as the flexible gene pools are presented in the circles. Functions encoded by the pools are given in the lower part of the diagram.
Genomic islands: elements of the flexible gene pool
In recent years, ‘pathogenicity islands’ (PAIs) have attracted a great deal of attention (Kaper and Hacker, 1999). First described in the genomes of pathogenic E. coli, they were subsequently also found in other pathogens, where they form specific entities associated with bacterial pathogenicity (Blum et al., 1994). Sequencing of several entire genomes revealed that PAIs are much more widespread than previously thought, and represent a paradigm of more general genetic entities that are present in the genomes of many bacterial species and are termed genomic islands (Strauss and Falkow, 1997; Hacker and Kaper, 2000).
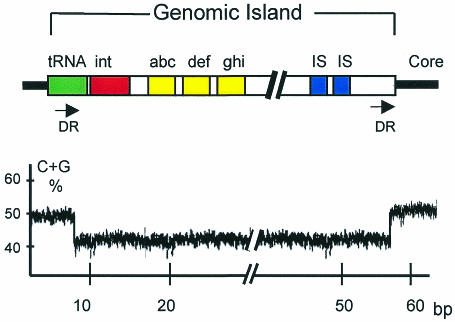
Genomic islands are part of the flexible bacterial gene pool and are somewhere between 10 and 100 kilobases (kb) in length (see Figure 2). They frequently harbor phage- and/or plasmid-derived sequences, including transfer genes or integrases and IS elements. These particular blocks of DNA are most often inserted into tRNA genes and may be unstable. This instability appears to be mediated by flanking direct repeats which are often homologous to phage attachment sites and promote integration into, and excision out of, the bacterial genome (Hacker et al., 1997; Buchrieser et al., 1998). In addition to mobility loci, genomic islands carry gene clusters with specific functions. As for other elements of the flexible gene pool, the majority of these islands differ from the core genome with respect to their G+C content and codon usage. The evolutionary advantage of genomic islands over smaller inserts (‘islets’) is that a large number of genes (e.g. operons, gene clusters encoding related functions) may be transferred and incorporated en bloc into the recipient genome. This transfer may lead to dramatic changes in the behavior of the organism, resulting ultimately in ‘evolution in quantum leaps’ (Groisman and Ochman, 1996; Finlay and Falkow, 1997).
Fig. 2. Schematic model of a genomic island of bacteria (upper part). The formerly transferred DNA block is linked to a tRNA gene and flanked by direct repeats (DR). The guanine plus cytosine (G+C) content of the genomic island is different from that of the core genome (lower part). Other abbreviations: int, integrase gene; abc, def and ghi, genes encoding specific functions; IS, insertion sequence element; bp, base pair.
A wide range of functions
Sequence analysis revealed that genomic islands carry selfish genes, especially of the type that encode proteins with transfer, recombination and restriction/modification properties. However, the majority of the clusters located on these genetic elements encode functions that can be useful for the survival and transmission of the microbes (Table I). Thus, they may provide a selective advantage to the island-carrying organisms within a population. For instance, DNA elements encoding sucrose-uptake in Salmonella senftenberg are necessary for metabolic adaptation of these bacteria to their hosts (Hochhut et al., 1997). Other genomic islands encode iron-uptake systems which enhance the capacity of bacteria to grow and disseminate in the soil or in a host. This holds true for many enterobacteria and for bacteria of the Pseudomonas group, which are part of the plant rhizosphere. Other Pseudomonas strains carry genomic islands that encode enzymes involved in degradation of phenolic compounds (Ravatn et al., 1998). Genomic islands may also carry genes encoding factors that confer resistance to antimicrobial substances. For example, the _mec_A-region of staphylococci enhances survival of its carriers, both in soil compartments in which antibiotic-producing microbes exist, and in hospitals with strong antibiotic pressure (Ito et al., 1999). In addition, the symbiosis islands of rhizobia carry nitrogen fixation genes whose products are necessary for the interactions of the bacteria with plant cells (Sullivan and Ronson, 1998). Other genomic islands encode toxins or adherence factors involved in pathogenicity.
Table I. Functions encoded by fitness islands.
| Subtypes of fitness islands | Function | Organism | Increased pathogenicity |
|---|---|---|---|
| PAI | iron uptake | Yersinia spp. | + |
| SAI | iron uptake | fecal E. coli | – |
| ECI | iron uptake | Klebsiella spp. | – |
| ECI | sucrose uptake | Salmonella senftenberg | – |
| ECI | degradation of phenols | Pseudomonas putida | – |
| PAI | toxin production | Vibrio cholerae | + |
| SAI | adhesins | fecal E. coli | – |
| PAI | adhesins | urinary E. coli | |
| ECI | methicillin resistance | Staphylococcus aureus | – |
| ECI | multi-resistance | Shigella flexneri | – |
| SYI | nitrogen fixation | Mesorhizobium loti | – |
| PAI | type III-system | Salmonella enterica | + |
| type III-system | Shigella flexneri | ||
| type III-system | Yersinia spp. | + | |
| SYI | type III-system | Sinorhizobium fredii | – |
| PAI | type IV-system | Helicobacter pylori | + |
| type IV-system | Legionella pneumophila | + | |
| EAI | type IV-system | E. coli F-plasmid | – |
Increasing bacterial fitness and driving evolution
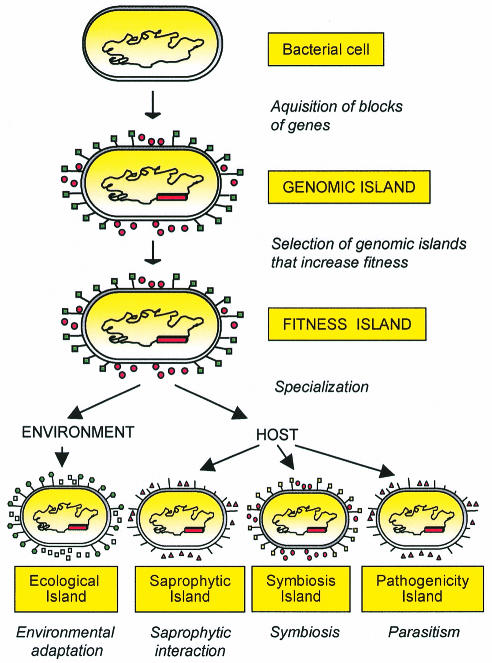
The progress of evolution is determined by an increase in the fitness of the organism. Fitness, in this context, is considered to be a set of properties that enhance the survival, spread, and/or transmission of an organism within a specific ecological niche (Preston et al., 1998). The Darwinian laws (‘survival of the fittest’) are valid for the development of eukaryotes as well as for prokaryotes (Arber, 2000). Therefore, carrying a genomic island may provide a selective advantage under specific environmental conditions (stress, in vivo conditions, exposure to antibacterial substances) because it enhances microbial transmission, survival or colonization within a niche. From a functional point of view, then, genomic islands that increase the fitness of the recipient microbes should be termed ‘fitness islands’, as already suggested by Preston et al. (1998) (Figure 3). Under these circumstances, genomic, fitness islands confer new properties which enhance the adaptational capacity of their bacterial host.
Fig. 3. Model for the development of genomic islands. Following acquisition of transferred DNA and/or deletion of genetic material, a genomic island is selected. If the gene products of the island enhance the fitness of the recipient, the island-harboring bacteria will be positively selected. The gene products of these foreign blocks of DNA can contribute to survival in the environment, saprophytic life, symbiosis or pathogenicity.
Fitness islands can be subdivided into different subsets, depending on the life-style of the microbe (its niche) (Figure 3), rather than on the intrinsic composition of the islands. Fitness islands that help microorganisms to live in the environment or to persist as saprophytes in a host may be considered ‘ecological islands’ and ‘saprophytic islands’, respectively. Other bacteria reside temporarily or permanently in a host (another microorganism, a plant or an animal), where they either provide some benefits to the host-organism (symbiont) or cause damage to it (pathogen). Accordingly, a ‘symbiosis island’ is a specific type of fitness island that helps bacteria to positively interact with their hosts, while a fitness island that participates directly or indirectly in the induction of lesions is a true pathogenicity island.
Pathogenicity islands may influence microbial evolution
PAIs represent a subset of genomic islands and share the same general composition and organization (Hacker and Kaper, 2000). Like the saprophytic islands and the symbiosis islands, they exert their action in a host. However, in this case, their gene products contribute, directly or indirectly, to the pathogenic potency of bacteria and generate lesions in the infected host (Groisman and Ochman, 1996; Hacker et al., 1997). PAIs are components of the genomes of many Gram positive, as well as Gram negative, bacteria. As the Darwinian laws are also valid for the generation of PAIs, the presence of PAIs should contribute to the in vivo fitness of the PAI-positive bacteria and increase their survival and/or transmission to new hosts.
In certain cases, ‘fitness properties’ are directly related to the clinical symptoms caused by the pathogenic bacteria. An example of a direct contribution of PAI-encoded functions to the fitness of bacteria is found in enteropathogenic organisms. Both Vibrio cholerae and enterotoxigenic E. coli stimulate efflux of water from the gut of infected individuals and the resulting bacterial spread via feces directly contributes to microbial transmission.
More generally, enhanced microbial transmission is often a direct consequence of the action of pathogenicity factors such as adhesins and toxins, which are encoded by PAIs, phages, or plasmids (Waldor and Mekalanos, 1996; Karaolis et al., 1998). The actions of these pathogenicity factors seems to result from direct evolutionary pressures. This is true not only for enteric, but also respiratory pathogens, where the action of pathogenicity factors supports their transmission and therefore positively influences microbial evolution.
Contributions to ecological adaptation and to pathogenesis
As already mentioned, the division of fitness islands into different subtypes is not based on their intrinsic genetic composition, but on their effects in a specific niche and within a particular organism. In other words, the same fitness island may act as an ecological island when the bacterial recipient resides outside of a host, but become a pathogenicity island when the bacterium enters a host. For example, the genes encoding an iron-uptake system termed yersiniabactin are part of a genomic island that was first identified in highly pathogenic strains of the genus Yersinia (Carniel et al., 1996). This ‘high pathogenicity island’ (HPI), however, is not only present in pathogenic yersiniae, but also in harmless E. coli of the intestinal flora and in Klebsiella from the soil (Schubert et al., 1998; Bach et al., 2000). The iron-uptake system seems to have evolved to adapt certain enterobacteria specifically to iron-limiting conditions. In bacteria that reside in the environment, this island can be considered as an ecological island with a role in cellular metabolism. If, on the other hand, the island is present in a bacterium with a host, and it carries additional virulence features, it is a pathogenicity island. If it is integrated in the chromosome of a non-virulent bacterium, it may constitute a saprophytic island.
Like the iron-uptake system, adhesins may exhibit ‘dual’ functions in bacteria (Finlay and Falkow, 1997). For example, certain adherence factors in E. coli (e.g. P-, S-, and F1C-fimbriae) are encoded by genomic islands and are produced by commensal strains that are part of the normal human gut flora (Hacker, 2000). If the adhesins are involved in colonization of the gut, the genetic entity is a saprophytic island. Under special circumstances, however, P-, S- or F1C-positive E. coli may reach the urinary tract, where they cause infections of the bladder or the kidney (Khan et al., 2000), becoming true PAIs. In other words, the PAIs of uropathogenic E. coli were originally selected as ‘pure’ fitness islands in the gut, but then helped a particular bacterial pathotype to emerge as the microbes colonized a new niche, the kidney or the bladder.
Other PAIs carry genes whose products form secretion systems of type III or IV. Again, if these secretion systems transport proteins involved in the infectious process, they can be considered PAIs. This is true for strains of the Salmonella- (Galán and Collmer, 1999), Shigella- (Parsot and Sansonetti, 1999), and _Yersinia_-groups (Cornelis et al., 1998) for the type III system, and for Legionella pneumophila (Vogel et al., 1998) and Helicobacter pylori (Cesini et al., 1996) for the type IV system (Table I). If, however, the secretion systems transport proteins or even DNA molecules of non-pathogenic organisms, as in the case of the type III system of rhizobia, or the type IV system of F plasmids, they do not form PAIs but rather symbiotic islands or ecological islands which enhance the fitness of bacteria in their natural niche (Preston et al., 1998). Therefore, the subtypes of fitness islands depend on several criteria including not only the genetic composition of the island itself, but also the genetic background of its bacterial host, and the ecological habitat of the microorganism.
Driving bacterial evolution
PAIs have been selected during evolution because their presence conferred selective advantages to their bacterial host. However, in some instances, their acquisition might subsequently have oriented the evolution of their host bacteria. This has probably been the case for Y. pestis, the agent of plague. Y. pestis is a highly clonal species that emerged recently (1500 to 20 000 years ago) from Y. pseudotuberculosis (Achtmann et al., 1999). In contrast to its progenitor which uses the oral route to contaminate human and animal hosts, Y. pestis is transmitted by flea bites and the septicemia that systematically occurs in the host at the pre-mortem stage of plague is a prerequisite for Y. pestis transmission by fleas. By promoting the systemic dissemination and thereby the efficient transmission of the bacteria in vivo, the HPI presumably served as one of the key factors in the emergence of this highly dangerous microorganism. In other words, Y. pestis would probably not have evolved from Y. pseudotuberculosis if the genome of the latter had not already harbored the HPI.
Genomic islands in eukaryotes?
Horizontal gene transfer represents an important mechanism in the evolution of eubacteria, and genomic islands belong to the group of genetic elements that are involved in evolutionary progress. Recently, it has become evident that laterally transferred DNA is also present in the genomes of archeabacteria (Doolittle, 1999), and questions regarding a role for horizontal gene transfer in the evolution of eukaryotes (de la Cruz and Davis, 2000; Kurland, 2000) have arisen. From our point of view, there are good indications that this is the case, and genetic elements with features of genomic islands have been found in eukaryotic genomes. First, mobile genetic elements such as retrotransposons are present even in the genomes of mammals, where they have the capacity to jump into 3′ ends of tRNA genes, a process that was first identified in bacterial genomes. Secondly, the well-characterized Ti plasmid is able to transfer genes from a prokaryotic organism, Agrobacterium tumefaciens, to the genomes of plants, and the transferred DNA (T-region) was recently assimilated to a PAI (Winans et al., 1999). Thirdly, mitochondria and plastids exhibit DNA-signatures that are also found in the genomes of prokaryotes such as rickettsia or cyanobacteria. There is speculation that these organelles were derived from genomes of former bacterial endosymbionts (Hentschel et al., 2000), and are thus special types of genomic islands. Last, but not least, large ‘pathogenicity loci’, which share common features with bacterial PAIs have been identified in the pathogenic fungus, Ustilago hordei (Lee et al., 1999). Further analysis of eukaryotic genomes will certainly define more clearly the roles of these elements in eukaryotic evolution by quantum leaps.
Acknowledgments
Acknowledgements
We thank Cesare Montecucco (Padua) and Ute Hentschel (Würzburg) for discussions, Claudia Borde and Hilde Merkert for editorial assistance. Part of the article was composed during a sabbatical of J.H. in the laboratory of E.C. at the Institut Pasteur, Paris. Our own work, related to the subject of the article is supported by the DFG (SFB 479) and the Fonds der Chemischen Industrie.
References
- Achtmann M., Zurth, K., Morelli, G., Torrea, G., Guiyoule, A. and Carniel, E. (1999) Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl Acad. Sci. USA, 96, 14043–14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arber W. (2000) Genetic variation: molecular mechanisms and impact on microbial evolution. FEMS Microbiol. Rev., 24, 1–7. [DOI] [PubMed] [Google Scholar]
- Bach S., de Almeida, A. and Carniel, E. (2000) The Yersinia high-pathogenicity island is present in different members of the family Enterobacteriaceae. FEMS Microbiol. Lett., 183, 289–294. [DOI] [PubMed] [Google Scholar]
- Blum G., Ott, M., Lischewski, A., Ritter, A., Imrich, H., Tschäpe, H. and Hacker, J. (1994) Excision of large DNA regions termed pathogenicity islands from tRNA-specific loci in the chromosome of an Escherichia coli wild-type pathogen. Infect. Immun., 62, 606–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchrieser C., Brosch, R., Bach, S., Guiyoule, A. and Carniel, E., (1998) The high pathogenicity island of Yersinia pseudotuberculosis can be inserted into any of the three chromosomal asn tRNA genes. Mol. Microbiol., 30, 965–978. [DOI] [PubMed] [Google Scholar]
- Carniel E., Guilvout, I. and Prentice, M., (1996) Characterization of a large chromosomal ‘high-pathogenicity island’ in biotype 1B Yersinia enterocolitica. J. Bacteriol., 178, 6743–6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesini S. et al. (1996) cag, a pathogenicity island of Helicobacter pylori, encodes type-I specific and disease-associated virulence factors. Proc. Natl Acad. Sci. USA, 93, 14648–14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis G.R. et al. (1998) The virulence plasmid of Yersinia, an antihost genome. Microbiol. Mol. Biol. Rev., 62, 1315–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz F. and Davies, J. (2000) Horizontal gene transfer and the origin of species: lessons from bacteria. Trends Microbiol., 8, 128–133. [DOI] [PubMed] [Google Scholar]
- del Solar G., Alonso, J.C., Espinosa, M. and Diaz-Orejas, R. (1995) Broad-host-range plasmid replication: an open question. Mol. Microbiol., 21, 661–666. [DOI] [PubMed] [Google Scholar]
- Doolittle W.F. (1999) Phylogenetic classification and the universal tree. Science, 284, 2124–2129. [DOI] [PubMed] [Google Scholar]
- Finlay B.B. and Falkow, S. (1997) Common themes in microbial pathogenicity revised. Microbiol. Mol. Biol. Rev., 61, 136–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galán J.E. and Collmer, A. (1999) Type III secretion machines: bacterial devices for protein delivery into host cells. Science, 284, 1322–1328. [DOI] [PubMed] [Google Scholar]
- Groisman E.A. and Ochman, H. (1996) Pathogenicity islands: bacterial evolution in quantum leaps. Cell, 87, 791–794. [DOI] [PubMed] [Google Scholar]
- Hacker J. (2000) Urinary tract infection: From basic science to clinical application. In Emödy, L., Blum, G., Hacker, J. and Pal, T. (eds), Genes and Proteins Underlying Microbial Urinary Tract Virulence: Basic Aspects and Applications. Advances in Experimental Medicine and Biology, Plenum Press, New York, NY, pp. 1–8. [DOI] [PubMed]
- Hacker J. and Kaper, J.B. (2000) Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol., 54, 641–679. [DOI] [PubMed] [Google Scholar]
- Hacker J., Blum-Oehler, G., Mühldorfer, I. and Tschäpe, H. (1997) Pathogenicty island of virulent bacteria: Structure, function and impact on microbial evolution. Mol. Microbiol., 23, 1089–1097. [DOI] [PubMed] [Google Scholar]
- Hentschel U., Steinert, M. and Hacker, J. (2000) Common molecular mechanisms of symbiosis and pathogenesis. Trends Microbiol., 8, 226–231. [DOI] [PubMed] [Google Scholar]
- Hochhut B., Jahreis, K., Lengeler, J.W. and Schmid, K. (1997) CT_nscr94_, a conjugative transposon found in enterobacteria. J. Bacteriol., 179, 2097–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Katayama, Y. and Hiramatsu, K. (1999) Cloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant Staphylococcus aureus N315. Antimicrob. Agents Chemother., 43, 1449–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaper J. and Hacker, J. (1999) Pathogenicity islands and other mobile virulence elements. ASM-Press, Washington DC, pp. 1–352.
- Karaolis D.K.R., Johnson, J.A., Bailey, C.C., Boedeker, E.C., Kaper, J.B. and Reeves P.R. (1998) A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc. Natl Acad. Sci. USA, 95, 3132–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A.S., Kniep, B., Ölschläger, A.T., Van Die, I., Korhonen, T. and Hacker, J. (2000) The receptor structure for F1C fimbriae of uropathogenic Escherichia coli. Infect. Immun., 68, 3541–3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland C.G. (2000) Something for everyone. EMBO Rep., 1, 92–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence J.G. and Ochman, H. (1998) Molecular archaeology of the Escherichia coli genome. Proc. Natl Acad. Sci. USA, 95, 9413–9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N., Bakkeren, G., Wong, K., Sherwood, J.E. and Kronstad, J.W. (1999) The mating-type and pathogenicity locus of the fungus Ustilago hordei spans a 500-kb region. Proc. Natl Acad. Sci. USA, 96, 15026–15031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley A., Young, P. and Bailey, M. (2000) Bacterial population genetics: Do plasmids maintain bacterial diversity and adaptation? In Thomas, C.M. (ed.), The Horizontal Gene Pool. Harvard Academic Publisher, pp. 287–300.
- Ochman H., Lawrence, J.G. and Groisman, E.A. (2000) Lateral gene transfer and the nature of bacterial innovation. Nature, 405, 299–304. [DOI] [PubMed] [Google Scholar]
- Parsot C. and Sansonetti, P.J. (1999) The virulence plasmid of Shigellae: an archipelago of pathogenicity islands? In Kaper, J. and Hacker, J. (eds), Pathogenicity Islands and Other Mobile Virulence Elements. ASM Press, Washington, DC, pp. 151–65.
- Preston G.M., Hauboldt, B. and Rainey, P.B. (1998) Bacterial genomics and adaptation to life on plants: implications for the evolution of pathogenicity and symbiosis. Curr. Op. Microbiol., 589–597. [DOI] [PubMed] [Google Scholar]
- Ravatn R., Studer, S., Springael, D., Zehnder, A.J.B. and van der Meer, J.R. (1998) Chromosomal integration, tandem amplification, and deamplification in Pseudomonas putida F1 of a 105-kilobase genetic element containing the chlorocatechol degrative genes from Pseudomonas sp. strain B13. J. Bacteriol., 180, 4360–4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert S., Rakin, A., Karch, H., Carniel, E. and Heesemann, J. (1998) Prevalence of the ‘high pathogenicity island’ of Yersinia species among Escherichia coli strains that are pathogenic to humans. Infect. Immun., 66, 480–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss E.J. and Falkow, S. (1997) Microbial pathogenesis: genomics and beyond. Science, 276, 707–712. [DOI] [PubMed] [Google Scholar]
- Sullivan J.T. and Ronson, C.W. (1998) Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a phe-tRNA gene. Proc. Natl Acad. Sci. USA, 95, 5145–5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J.P., Andrews, H.L., Wong, S.K. and Isberg, R.R. (1998) Conjugative transfer by the virulence system of Legionella pneumophila. Science, 279, 873–876. [DOI] [PubMed] [Google Scholar]
- Waldor M.K. and Mekalanos, J.J. (1996) Lysogenic conversion by a filamentous phage encoding cholera toxin. Science, 272, 1910–1914. [DOI] [PubMed] [Google Scholar]
- Winans S.C., Kalogeraki, V., Jafri, S., Akakura, R. and Xia, Q. (1999) Diverse roles of Agrobacterium Ti plasmid-borne genes in the formation and colonization of plant tumors. In Kaper, J. and Hacker, J. (eds), Pathogenicity Islands and Other Mobile Virulence Elements. ASM Press, Washington, DC, pp. 289–308.