Orphan Nuclear Receptor LRH-1 Is Required To Maintain Oct4 Expression at the Epiblast Stage of Embryonic Development (original) (raw)
Abstract
Oct4 plays an essential role in maintaining the inner cell mass and pluripotence of embryonic stem (ES) cells. The expression of Oct4 is regulated by the proximal enhancer and promoter in the epiblast and by the distal enhancer and promoter at all other stages in the pluripotent cell lineage. Here we report that the orphan nuclear receptor LRH-1, which is expressed in undifferentiated ES cells, can bind to SF-1 response elements in the proximal promoter and proximal enhancer of the Oct4 gene and activate Oct4 reporter gene expression. LRH-1 is colocalized with Oct4 in the inner cell mass and the epiblast of embryos at early developmental stages. Disruption of the LRH-1 gene results in loss of Oct4 expression at the epiblast stage and early embryonic death. Using _LRH-1_−/− ES cells, we also show that LRH-1 is required to maintain Oct4 expression at early differentiation time points. In vitro and in vivo results show that LRH-1 plays an essential role in the maintenance of Oct4 expression in ES cells at the epiblast stage of embryonic development, thereby maintaining pluripotence at this crucial developmental stage prior to segregation of the primordial germ cell lineage at gastrulation.
Pluripotence in embryonic stem (ES) cells and the early mouse embryo is controlled by several key transcription factors such as Oct4, Sox2, Foxd3, Nanog, and signaling molecules such as STAT3, FGF4, FGF4 receptor, and LIF (4, 11, 18, 22, 32, 35, 51). The regulation of expression of these key factors is thus crucial for the maintenance of pluripotence and embryonic development. However, little is known about the upstream factors that regulate the expression of these genes. Oct4, a member of the POU homeodomain family of transcription factors, is the best studied of these factors. Recent studies have shown that Oct4 plays a critical role in embryonic development and cellular differentiation (38, 43). Oct4 is expressed in totipotent and pluripotent stem cells of the pregastrulation embryo, primordial germ cells, and oocytes (37, 44, 47). Oct4 is also highly expressed in ES and embryonic carcinoma cell lines, such as P19 and F9 cells, and is rapidly down-regulated by differentiation induced with retinoic acid (RA) (6, 31). In ES cells, a less than twofold increase in the level of Oct4 mRNA causes the cells to differentiate into primitive endoderm and mesoderm, whereas reduction to less than 50% of normal levels triggers differentiation into trophectoderm (36). Moreover, targeted disruption of the Oct4 gene in mice results in embryonic death at the blastocyst stage and compacted morula cells that do not differentiate along the pluripotent inner cell mass lineage but instead differentiate into trophectodem (32). Thus, the level of Oct4 expression is crucial not only to the maintenance of pluripotence but also to early cell differentiation decisions (36).
Previous studies have shown that several members of the nuclear receptor family, including SF-1, GCNF, RAR/RXR, and COUP TF I/II, regulate Oct4 expression by binding to its proximal promoter region (5, 6, 19, 42, 49). In addition to the proximal promoter (PP), the proximal enhancer (PE) of the Oct4 regulatory region is essential for expression of the Oct4 gene (56). The PE is specifically activated at the epiblast stage of embryonic development. In contrast, the distal enhancer (DE) is active in the blastocyst and the germ line (31, 56). However, it is not known which transcription factors are involved in the regulation of Oct4 expression through these different enhancers.
Of the nuclear receptors implicated in the regulation of Oct4, germ cell nuclear receptor (GCNF; NR6A1) is the best validated. The repression function of GCNF on the Oct4 promoter is driven by binding of GCNF to a DR0 element located in the Oct4 PP (19). In _GCNF_−/− embryos, Oct4 expression is not repressed efficiently in somatic cells and thus is no longer restricted to primordial germ cells after gastrulation, and the embryos die around embryonic day 10.5 (E10.5) (12, 19).
The DR0 motif within the Oct4 promoter is also a binding site for the orphan nuclear receptor steroidogenic factor 1 (SF-1; NR5A1). SF-1 is expressed in P19 cells and activates Oct4 expression (5). On treatment of P19 cells with RA, SF-1 expression is decreased to undetectable levels. SF-1 knockout mice lack adrenal glands and gonads and die due to adrenal insufficiency within the first week of birth (30, 46). Comparison of SF-1 and Oct4 knockout mouse models suggests that although SF-1 can regulate Oct4 expression in P19 cells, it is essential only during late organogenesis and thus there must be another factor that regulates Oct4 expression during early embryogenesis.
The orphan nuclear receptor liver receptor homologue 1 (LRH-1; NR5A2), also termed fetoprotein transcription factor (FTF), is closely related to SF-1, particularly in the DNA-binding domain, and has the same DNA response element as SF-1 (34). SF-1 and LRH-1 are differentially expressed in the ovary (15, 23) and activate the transcription of genes encoding steroidogenic enzymes (50). LRH-1 is expressed in endoderm-derived tissues such as the liver, pancreas, and intestine in the adult and developing embryos (2, 34, 45) and is involved in regulating bile acid metabolism (13, 16, 48). Recently, LRH-1 and β-catenin have been shown to synergistically regulate intestinal cell proliferation through cyclin G1 (9). Inactivation of the LRH-1 gene results in early embryonic death in part due to misregulation of endodermal genes (9, 13, 39). All of these studies focused on the function of LRH-1 in endoderm development and gene regulation. However, LRH-1 is also expressed in the morula and inner cell mass and the _LRH-1_−/− embryos died at early stages (E6.5 to 7.5) (39). The similar DNA-binding properties and complementary expression patterns between SF-1 and LRH-1 suggest that LRH-1 may regulate Oct4 expression during early embryonic development and in ES cells similar to SF-1-dependent regulation of Oct4 expression in P19 cells (5, 19).
In this study, we demonstrate that LRH-1 binds directly to response elements located in the PP and PE of the Oct4 gene and activates Oct4 reporter gene expression. We also show that the expression pattern of LRH-1 in ES cells and early embryonic development stages overlaps with Oct4 expression. Moreover, targeted inactivation of the LRH-1 gene results in embryonic death around E6.5, loss of Oct4 gene expression at the epiblast stage of development, and more rapid down-regulation of the Oct4 gene in _LRH-1_−/− ES cells on differentiation. These results demonstrate that LRH-1 plays an important role in the regulation of Oct4 gene expression and pluripotence in ES cells at the epiblast stage of embryonic development.
MATERIALS AND METHODS
Plasmids and antibodies.
The mouse SF-1 expression plasmids T7-Myc-SF-1 and pCEP4SF-1 were provided by David Moore. The full-length cDNA of mouse LRH-1 was obtained by reverse transcription-PCR from undifferentiated ES cell total RNA. The sequence of PCR primers used are 5′CAAGAATTTCCGCTAAGAATGTCTGAG 3′ (the forward primer) and 5′AGTAACTTCCAGGGGTGC 3′ (the reverse primer). For mammalian expression and in vitro translation, plasmids pHA-mLRH-1 and pGBK-mLRH-1 were generated by insertion of mLRH-1 cDNA into pCMV-HA and pGBK-T7 (Clontech, Sparks, Md.), respectively. The Oct4 luciferase reporter vectors have been described previously (19). The Oct4 reporter PP-Luc contains a 0.4-kb XbaI-BanI fragment from the proximal-promoter region, and PP* is identical to PP except for the point mutations inserted in the DR0 element (19). The reporter PE-PP-Luc contains a 1.2-kb BamHI-BanI fragment from the Oct4 promoter region, and PE-PP* is identical to PE-PP except for the insertion of point mutations in the DR0 element (19). PE-SV40-Luc was constructed by the insertion of a 0.8-kb BamHI-XbaI PE fragment of the Oct4 enhancer into pGL3-SV40-Luc (Promega, Madison, Wis.). Anti-mSF-1 antibody was purchased from Upstate (Charlottesville, Va.; catalog no. 13702). Anti-Oct4 (no. 8628) and anti-actin (no. sc-1616) antibodies and all of the horseradish peroxidase-conjugated secondary antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, Calif.). The anti-mouse LRH-1 antibody used in Western blot analysis and supershift assays, which was produced in our laboratory, was raised specifically against the N terminus of the recombinant LRH-1 mouse protein expressed in bacteria. The portion of the LRH-1 cDNA encoding the N-terminal 100 amino acids was inserted into a His-tagged bacterial expression vector, pRSET-A (Invitrogen, Carlsbad, Calif.).
Cell culture and transient transfection.
P19 cells and COS-1 cells were maintained in Dulbecco's medium supplemented with 10% fetal calf serum. Wild-type and _LRH-1_−/− ES cells were maintained on 0.1% gelatin-coated plates in Dulbecco's medium supplemented with 15% fetal calf serum, 110 μM 2-mercapoethanol, 1,000 U of recombinant murine leukemia inhibitory factor (LIF; Chemicon, Temecula, Calif.), 100 U per ml, of penicillin per ml, and 100 μg of streptomycin per ml. For reverse transcription-PCR using differentiated P19 and ES cells, cells were differentiated with 1 μM RA in the absence of LIF and harvested at the indicated times. For Northern blot analysis using differentiated ES cells, wild-type or _LRH-1_−/− ES cells were cultured on 1% gelatin-coated bacterial plates in the absence of LIF. Cells were harvested on the indicated days, and RNA was isolated. Day 0 cells were harvested 24 h after seeding. All the transient transfections were performed with Fugene 6 as specified by the manufacturer (Roche, Indianapolis, Ind.). The expression vector and reporter DNA were cotransfected into P19 cells plated in six-well plates (2 × 105 cells per well) for 24 h prior to transfection. The Renilla RL-Luc reporter (Promega) DNA was also cotransfected as an internal control to correct for differences in transfection efficiency. Total plasmid DNAs were balanced with empty vector pCMV-HA. After 48 h of incubation, the cells were harvested and the luciferase activity was analyzed using the dual-luciferase assay kit (Promega) as specified by the manufacturer.
Electrophoretic mobility shift assays.
In vitro- translated mLRH-1 and mSF-1 were produced using a T7/TNT in vitro translation kit as specified by the manufacturer (Promega). COS-1-overexpressed proteins and P19 and ES cell nuclear extracts were extracted with 2× binding buffer (25 mM HEPES [pH 7.9], 150 mM KCl, 0.4 mM EDTA, 2 mM dithiothreitol, 20% glycerol, 1 mM phenylmethylsulfonyl fluoride, 1× proteinase inhibitor cocktail [Roche]). The sequence of the DR0 probe and electrophoretic mobility shift assay were performed by to methods described previously (19). The sequence of PE1 was as follows: sense, 5′GCATCCTGGCCATTCAAGGGTTGAGTACTTGTT 3′; antisense, 5′GCTAAACAAGTACTCAACCCTTGAATGGGCCAGG 3′ (−928 to −919). The sequence of PE2 was as follows: sense, 5′CTAGGATTGTCCAAGCCAAGGCCATTGTCCTGCCC 3′; antisense, 5′CTGAGGGCAGGACAATGGCCTTGGCTTGGACAATC 3′ (−867 to −858).
Chromatin immunoprecipitation (ChIP) assay.
Chromatin immunoprecipitation ChIP assays were performed by the on-line protocol provided by the company Upstate. Undifferentiated and differentiated P19 and ES cells were treated with 1% formaldehyde. Cross-linked DNA/protein was extracted and disrupted by sonication. The recovered DNA was amplified with gene-specific primers. The sequences of the primers surrounding the DR0 site in the Oct4 PP (from −127 to +95) are CCTCCGTCTGGAAGACACAGGCAGATAGCG for forward and CGAAGTCTGAAGCCAGGTGTCCAGCCATGG for reverse; the sequence of the primers surrounding the PE1 and PE2 sites of the PE of the Oct4 gene (from −1000 to −824) are GCTGGGGAAGTCTTGTGTGA (forward) and R:GCTTCCAGCCTAGTTCCTGG (reverse).
Northern blot analysis.
Total RNAs from different time points of differentiated P19 and ES cells were isolated using Trizole reagent (Invitrogen). Northern blot analysis was performed as specified in protocol PT1190-1 (Clontech). Blots were hybridized with 32P-labeled cDNAs corresponding to LRH-1 (bases 712 to 1296 [GenBank NM_030676]), Sox2 (bases 1239 to 1903, [GenBank U31967]), Oct4 (bases 908 to 1260 [GenBank NM_013633]), FGF4 (bases 1 to 609 [GenBank NM_010202]), UTF1 (bases 324 to 918 [GenBank NM_009482]), and REX1 (bases 461 to 2073 [GenBank NM_009556]). To ensure equal loading of RNA samples, the blots were probed with radiolabeled cDNA corresponding to the rat 18S rRNA (18S rRNA, bases 293 to 970 [GenBank X01117]).
Immunofluorescent and immunohistochemical staining.
Collection of blastocysts and immunofluorescent staining were performed by methods described previously (24, 28). Female mice were superovulated by treatment with pregnant mare serum gonadotropin and human chorionic gonadotropin, and the blastocysts were flushed out of the uterus at E3.5 to 4.0. The zona pellucida of the embryos was removed by brief exposure to Tyrode's solution (pH 2.1 to 2.5) (Irvine Scientific, Santa Ana, Calif.). The embryos were fixed with 4% paraformaldehyde and permeabilized by incubation with 0.2% Triton X-100 in phosphate-buffered saline (PBS). After being blocked in 10% donkey serum plus 2% bovine serum albumin, the embryos were incubated with normal rabbit and goat immunoglobulin G (IgG) or rabbit anti-LRH-1 antibody and goat anti-Oct4 antibody in blocking buffer at 4°C overnight. They were then washed with PBS and incubated with fluorescein isothiocyanate FITC-conjugated donkey anti-rabbit IgG and Texas Red-conjugated donkey anti-goat IgG (Jackson ImmunoResearch Laboratories, Inc., West Grove, Pa.) and washed with PBS. They were stained with 0.5 μg of Hoechst 33258 (Sigma, St. Louis, Mo.) per ml and mounted on glass slides with Vectashield mounting medium for fluorescence (Vector Laboratories, Burlingame, Calif.) or kept in PBS buffer and examined with an Axioskop 2 immunofluorescence microscope (Carl Zeiss).
The embryos at E6.5 were fixed in 10% formaldehyde in PBS and embedded in parafilm. Antigens were unmasked by boiling the sections (7.5 μm) and treating them with 3% H2O2 and were blocked in 10% normal goat serum. An immunostaining kit from Santa Cruz Biotechnology was used to detect the LRH-1 or Oct4 protein. After being stained, the slides were counterstained in 0.05% methyl green to visualize histological structures.
Whole-mount and section in situ hybridization.
Whole-mount in situ hybridization was carried out as described previously (19). The LRH-1 cDNA fragment corresponding to nucleotides 1155 to 1540 and Oct4 cDNA (19) were labeled with digoxigenin (Dig) (In vitro translation kit; Promega) as cRNA probes. E6.5 embryos were fixed in 4% paraformadehyde and embedded in paraffin. Sections were probed with 35S-labeled sense or antisense cRNA probes against the LRH-1 cDNA fragment.
Genotyping and identification of _LRH-1_−/− embryos.
The LRH-1+/− mice were purchased from Lexicon Genetics Inc. The accession number corresponding to the OmniBank mice that were obtained is NM_030676. Genotypes of weaned mice or embryos were determined by PCR analysis of tail-cut DNA samples or yolk sacs. The primer set for the wild-type LRH-1 allele (forward F1, 5′ TCTGCTAGTTTGGATACTGG 3′; forward F2, 5′ AAAGGACTGCCAATAATTTCGCT 3′; reverse R1, 5′ TTACAGAGTGAAGTTCCAGG 3′; reverse R2, 5′ AAGTGGATCTCTGAGTCTGAG 3′) was derived from the sequence of the first exon and intron; the reverse primer for the mutant LRH-1 allele was the same as that for the wild-type, and the forward primer (R4, 5′ GCAGCGCATCGCCTTCTATC 3′) was derived from the neo gene within the targeting vector. Genomic DNA was extracted from tails of pups or from the yolk sacs of embryos after E7.0, from the whole blastocysts after immunofluorescent staining, or from scraped embryonic tissues from stained sections. Primers F1 plus R1 and R4 were used to genotype the pups, and the nested PCR strategy was used for genotyping the blastocysts and scraped embryonic tissues.
Generation of _LRH-1_−/− ES cells.
_LRH-1_−/− ES cells were generated by deletion of exon 6 on both chromosomes, using a two-step targeting strategy (see Fig. 7A). A mouse ES cell line was first generated in which loxP sites were placed in the introns surrounding exon 6. ES cells that underwent homologous recombination were selected with G418 and then transfected with a plasmid encoding Cre recombinase; clones in which exon 6 and the neomycin resistance gene were excised to allow selection for a second targeting event were identified by Southern blot analysis. These cells were then subjected to a second round of mutagenesis, using a targeting vector in which exon 6 was replaced by the neomycin resistance gene, and were reselected with G418. Southern blot analysis identified clones no longer containing a wild-type LRH-1 gene. The absence of LRH-1 transcript was verified by Northern blot analysis (see Fig. 7C).
FIG. 7.
Loss of Oct4 gene expression in _LRH-1_−/− ES cells. (A) Schematic representation of the two-step targeting strategy to inactivate the LRH-1 gene in ES cells. The schematic shows exons 4 to 7 of the LRH-1 gene, the two targeting vectors, and the two disrupted LRH-1 alleles. Solid boxes indicate exons, and solid arrows indicate the PGK-tk and PGK-neo cassettes. Selected restriction enzyme sites are indicated. After disruption of the first LRH-1 allele, the neo cassette was excised with Cre recombinase prior to targeted disruption of the second LRH-1 allele. The second targeting vector did not contain lox P sites. (B) Southern blot analysis of HindIII-digested DNA with the indicated probe. The wild-type allele yields a 17.7-kb fragment, the first targeted LRH-1 allele yields a 14.3-kb fragment, and the second targeted LRH-1 allele yields 4.6 kb on HindIII digestion. (C) Loss of expression of the Oct4 gene and other pluripotency factors in differentiated _LRH-1_−/− ES cells. Northern blot analyses of the indicated genes were performed with RNA prepared from LRH-1+/+ or _LRH-1_−/− ES cells differentiated for 0, 4, 8, or 12 days by withdrawal of LIF.
Detection and display of LRH-1/SF-1 response elements in the mouse genome.
A set of 19 LRH-1/SF-1 responses elements (LRE) was collected from the published data (see Table 1). These sites were used to train a hidden Markov model (HMM) using HMMER v.2.32 by Eddy (14). All mouse chromosomes were scanned using the HMM with an expected value cutoff set to 0.01, and the chromosomal coordinates of the resulting hits were saved. The chromosomal locations of genes of interest were taken from the KnownGenes tables of the UCSC Genome Browser database (26). The LREs were visualized by uploading the list of hits to the UCSC Genome Browser (27), using the “custom tracks” feature (see Fig. 2A).
TABLE 1.
List of published LRH-1 response elements
| Gene | Binding elements | Species | Reference |
|---|---|---|---|
| α-Fetoprotein (AFP) | TCAAGGACA | Rabbit | 21 |
| Fetoprotein transcription factor (FTF) | TCAAGTCCA | Mouse | 40 |
| Hepatic nuclear factor 1 (HNF1) | CCAAGGTTC | Mouse | 8 |
| Hepatic nuclear factor 3 (HNF3) | TCAAGGTTA | Mouse | 40 |
| 3-β-Hydroxysteroid dehydrogenase | TCAAGGTTC | Human | 41 |
| (3β-HSD) | ACAAGGACA | Human | |
| Cytochrome P450 19 (Cyp19) | CCAAGGTCA | Human | 23 |
| TCAAGGGCA | Human | ||
| TCAAGGGCG | Mouse | ||
| Carboxyl ester lipase (CEL) | CCAAGGTCA | Human | 17 |
| Cytochrome P450 7A1 (Cyp7A1) | TCAAGGCCA | Human | 34 |
| TCAAGGCCG | Rabbit | ||
| TCAAGGCTG | Hamster | ||
| Cytochrome P450 8B1 (Cyp8B1) | GCAAGGTCC | Rabbit | 55 |
| CCAAGGGCA | Rabbit | ||
| Scavenger receptor class B1 (SR-B1) | CCAAGGCTG | Human | 48 |
| Octamer-binding transcription factor 3/4 (Oct4) | TCAAGGCTA | Mouse | 49 |
| Phospholipase A2 (PLA-2) | TCAAGGCTG | Mouse | 3 |
FIG. 2.
Direct binding of SF-1/LRH-1 to the Oct4 proximal enhancer and promoter. (A) Localization of the LRH-1- binding sites, PE1, PE2, and DR0 elements in the Oct4 proximal enhancer and promoter. The SF-1/LRH-1-binding sites were identified by HMMER and labeled as black bars in the mouse Oct4 gene. The relative conservation between the mouse and human Oct4 gene derived from the UCSC genome browser was plotted as the gray curve. Arrows indicate the three regulatory regions DE, PE, and PP. The highly conserved regions CR1, 2, 3 and 4 are depicted as open boxes. The LRH-1- binding sites PE1, PE2, and DR0 that were tested are shown as black bars in CR2 and CR1. (B) COS1-overexpressed SF-1 and LRH-1 binding to the PE1, PE2, and DR0 probes in the absence or presence of antibodies. (C) Total proteins extracted from undifferentiated or differentiated ES cells were incubated with DR0 probe in the absence or presence of LRH-1 or SF-1 antibodies. The arrowheads mark the SF-1- and LRH-1-binding activity. Five-point stars mark the position of antibody-supershifted binding signal, and four-point stars mark the GCNF TRIF complex binding to the DR0. (D) ChIP analysis of direct binding of endogenous SF-1 and LRH-1 to the Oct4 PE and PP in vivo. Cross-linked genomic DNA from RA-differentiated P19 and ES cells was immunoprecipitated with anti-LRH-1 or anti-SF-1 antibody or preimmune serum IgG and amplified with Oct4 PP and PE specific primers.
RESULTS
Expression of LRH-1 in ES cells.
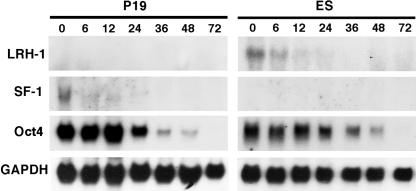
To investigate whether LRH-1 regulates Oct4 expression during early embryonic development, we compared LRH-1 and SF-1 expression patterns in ES cells and P19 cells by Northern analysis (Fig. 1). As previously shown, SF-1 was coexpressed with Oct4 in undifferentiated P19 cells and expression of both genes decreased during differentiation (19). LRH-1 was not expressed in P19 cells. In contrast, LRH-1 and Oct4 were coexpressed in undifferentiated ES cells whereas SF-1 was not (Fig. 1). Expression of LRH-1 decreased to undetectable levels in ES cells after 12 h of RA treatment. The expression of Oct4 also decreased with RA treatment and was almost undetectable after 36 h of RA treatment. Thus, Northern blot analysis showed that SF-1 is selectively expressed in P19 cells and LRH-1 is expressed in ES cells (Fig. 1). The concomitant expression pattern of LRH-1 and Oct4 in ES cells suggested that LRH-1 might regulate Oct4 expression.
FIG. 1.
Northern blot analysis of SF1, LRH-1, and Oct4 expression in P19 and ES cells at different time points of RA-induced differentiation. The numbers above the figures indicate the RA (1 μM) treatment time (hours). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is an internal control for loading and RNA integrity.
Direct binding of LRH-1 to the Oct4 PE and PP.
It was reported that there are two SF-1 response elements in the Oct4 promoter: DR0 (AGGTCAAGGCTA [from −42 to −31]) and the SF1β site in the Oct4 PP region (5, 19). Expression of Oct4 at the epiblast stage is regulated by the proximal enhancer in addition to the proximal promoter (31, 56). Therefore, we searched the Oct4 promoter for additional SF1/LRH-1 response elements. To do this, we generated a matrix of known LRH-1-binding sites (Table 1) from which a Hidden Markov Model was generated using HMMER and used to search the entire mouse genome. This search identified six LRH-1 binding sites. Site 1 is in a nonconserved region outside the proximal enhancer. Two sites, 2 and 3, are located in the evolutionally conserved region (CR2) that corresponds to the Oct4 PE. Sites 4 and 5 (site 5 corresponds to SF1β site in reference 5) are in a moderately conserved region, and site 6 is the DR0 element in a highly conserved region (CR1) of the (PP) (Fig. 2A).
To address the function of LRH-1 in the regulation of Oct4 expression, we tested the ability of LRH-1 protein to bind to these predicted response elements. We focused on the LRH-1-binding sites in the evolutionarily conserved region, sites 2, 3 and 6, which were termed PE1, PE2, and DR0, respectively. The binding of SF-1 and LRH-1 to the PE1, PE2, and DR0 sites was confirmed by supershift analysis (Fig. 2B). Both SF-1 and LRH-1 bound with high affinity to the DR0 and PE2 probes and weakly to the PE1 probe. The anti-SF-1 antibody disrupted the binding of COS1-overexpressed SF-1 proteins with the probes (lanes 2, 6, and 10). The anti-LRH-1 antibody supershifted the binding complex generated by LRH-1 protein (lanes 4, 8, and 12). To detect endogenous LRH-1 DNA-binding activity, we extracted nuclear proteins from undifferentiated ES cells or cells that had been treated with RA for 36 h, and electrophoretic mobility shift assay was performed. A fast-migrating complex appeared when nuclear extracts from undifferentiated ES cells were used; this complex was lost when RA-differentiated cell extracts were used (Fig. 2C). LRH-1 antibodies supershifted the complex in ES cells (lane 5), but SF-1 antibodies did not (lane 4). The antibody supershift results agreed with the LRH-1 expression results for ES cells using Northern analysis (Fig. 1). As previously reported (19, 20), a transient retinoid acid induced factor complex formed with the Oct4 DR0 probe in extracts from differentiated P19 cells; a complex with similar mobility was detected using extracts prepared from differentiated ES cells (Fig. 2C, lane 2).
To demonstrate binding of endogenous LRH-1 to the Oct4 promoter in ES cells, ChIP was employed. The results of ChIP analysis demonstrated direct binding of LRH-1 to the Oct4 PE and PP in vivo (Fig. 2D). Using Oct4 PP- and PE-specific ChIP primers, strong PCR-amplified signals were generated from undifferentiated ES cell DNA coimmunoprecipitated with anti- LRH-1 antibodies, demonstrating that LRH-1 binds to these two regions of the Oct4 promoter in vivo. Similar binding activities were observed in P19 cells with anti-SF-1 antibodies but not anti-LRH-1 antibodies. Binding of LRH-1 and SF-1 decreased on RA-induced differentiation in ES cells and P19 cells, respectively. This DNA-binding pattern is consistent with the expression profiles of SF-1 and LRH-1 in P19 and ES cells (Fig. 1). These results clearly demonstrate that endogenous LRH-1 in undifferentiated ES cells is directly bound to the Oct4 PP and PE in vivo and support a role for LRH-1 in the regulation of Oct4 expression in ES cells.
LRH-1 activation of Oct4 through the PE and PP.
To determine whether LRH-1 can regulate expression of the Oct4 gene, a set of luciferase reporters was constructed as illustrated in Fig. 3A and described in Materials and Methods. The various Oct4 luciferase reporter constructs were transfected into undifferentiated and differentiated P19 cells. Insertion of the Oct4 PE upstream of the PP enhanced reporter activity about fivefold in undifferentiated P19 cells (Fig. 3B). When the LRH-1-binding site in the PP was mutated (PP*), the PE enhancer activity was lost in P19 cells (Fig. 3B). Regulation by the Oct4 PE could be bestowed on a heterologous promoter since a similar effect was seen when the PE was placed before the simian virus 40 (SV40) basic promoter (Fig. 3B). In RA-differentiated P19 cells, the PE had no enhancer activity in either the wild-type context of the Oct4 promoter or linked to the SV40 promoter (Fig. 3B). These results show that transcription factors present in undifferentiated P19 cells but not in differentiated P19 cells (i.e., SF-1) act on the Oct4 PE.
FIG. 3.
Activation of the Oct4 PE and PP by SF-1 and LRH-1. (A) Illustration of the Oct4 PE and PP luciferase reporter constructs. (B) Activation of the Oct4 PE-luciferase reporters in the P19 cells and loss of the Oct4 PE-luciferase reporter activation in RA-differentiated P19 cells. (C) Activation of the Oct4 PE-luciferase reporters in differentiated P19 cells by transiently transfected SF-1 and LRH-1 expression vectors (200 ng/well). Relative fold activation of the PE was calculated by dividing by the activity of corresponding luciferase reporter activity in the absence of the PE, which was set at 1. Relative activation by transfected SF-1 or LRH-1 was calculated by dividing by the activity of corresponding luciferase reporter activity in the absence of SF-1 and LRH-1, which was set at 1. Results represent the mean and standard deviation of data from triplicate experiments.
To further address the function of SF-1 and LRH-1 on the PE enhancer, we cotransfected SF-1 and LRH-1 expression vectors with the different Oct4 luciferase reporters in RA-treated P19 cells (Fig. 3C). The transfection results showed that SF-1 and LRH-1 increased the PP and PE-PP reporter activities fourfold whereas SF-1 and LRH-1 had no effect on the mutated PP* and PE-PP* reporter activities. Regulation of a heterologous reporter by SF-1 and LRH-1 could be bestowed by introduction of the PE into the SV40 promoter. Transfected SF-1 and LRH-1 had no activity on the control SV40 promoter but produced twofold activation of the PE-SV40 reporter. These in vitro results demonstrated that endogenous SF-1 and transfected SF-1 or LRH-1 in P19 cells can activate reporter activity through the proximal enhancer of the Oct4 gene by binding to elements in the PE; such activation is dependent on the proximal promoter.
LRH-1 expression during early murine embryonic development.
To investigate the physiological relevance of LRH-1 to the regulation of Oct4 expression in vivo, we examined the expression pattern of LRH-1 in mouse embryos at early developmental stages when the Oct4 gene is expressed. Using LRH-1-specific antibodies generated against the N-terminal 100 amino acids of mouse LRH-1, the inner cell mass (ICM) and primitive endoderm of the blastocyst (E3.5 to E4.0) were specifically immunostained; the trophectoderm layer and blastocoelic cavity were only weakly stained (Fig. 4A, panel g). No staining was seen in experiments performed with control preimmune IgG (panel c). At this stage, Oct4 is also strongly expressed in the ICM and primitive endoderm (32) (panel h). The expression pattern of LRH-1 protein in the blastocyst was identical to that of β-galactosidase staining in a previously reported LRH-1:lacZ knockout mouse model (39). The expression of LRH-1 in embryos from E6.5 to E7.5 was detected by whole-mount in situ hybridization with Dig-labeled LRH-1 cRNA probe. At the advanced egg cylinder stage (E6.5 to E7.0), LRH-1 mRNA was detected throughout the embryonic and extraembryonic regions, including the visceral endoderm and embryonic ectoderm (Fig. 3B, panel a). During the early primitive streak stage embryo (E7.5), LRH-1 mRNA was still expressed throughout the embryo (Fig. 4B, panel b). There are conflicting reports of LRH-1 expression in the embryonic ectoderm (39, 45). Therefore, to confirm that LRH-1 was expressed in the epiblast and to demonstrate coexpression of LRH-1 and Oct4 proteins in the embryonic ectoderm, embryos at E6.5 to E7.0 were dissected and hybridized with cRNA probe against LRH-1 cDNA and immunostained with anti-LRH-1 and anti-Oct4 antibodies. Results from section in situ showed that LRH-1 was expressed throughout the entire embryo at E6.5 including the embryonic and extraembryonic cell layers (Fig. 4C). Consistent with the in situ results, LRH-1 protein was detected throughout the embryo, including the visceral endoderm, embryonic ectoderm, and the extraembryonic region (Fig. 4D, panel b). Oct4 was specifically located in the ectoderm of the epiblast (panel c). Thus, the LRH-1 gene is coexpressed with the Oct4 gene in the ICM and epiblast during early developmental stages.
FIG. 4.
Expression of LRH-1 in early mouse embryonic development. (A) Immunofluorescent detection of LRH-1 and Oct4 proteins in blastocysts. DNA in blastocysts was stained with Hoechst 33258 and appears blue (panels b and f), whereas LRH-1 protein stained green with a fluorescein isothiocyanate conjugate (g) and Oct4 appeared red after staining with Texas Red conjugates (h). At least 30 blastocysts from more than three wild-type mice were stained. (B) Whole-mount in situ hybridization of embryos at E6.5 to E7.0 and E7.5 with Dig-labeled antisense LRH-1 cRNA probe. (C) In situ hybridization with sagittal sections of mouse E6.5 embryos and surrounding deciduas performed with sense and antisense 35S-labeled LRH-1 RNA probe. (D) Immunohistochemical detection of LRH-1 and Oct4 proteins in sagittal sections of mouse embryos on day E6.5 with preimmune IgG (a), anti-LRH-1 antibody (b), and anti-Oct4 antibody (c). bc, blastocoel; eec, embryonic ectoderm; em, embryonic mesoderm; ex, extraembryo; exec, extraembryonic ectoderm; exed, extraembryonic endoderm; exem, extraembryonic mesoderm; pe, primitive endoderm; tp, trophectoderm; ved, visceral endoderm.
Disruption of LRH-1 gene expression leads to embryonic death.
The in vitro and in vivo results suggested that LRH-1 probably plays a key role in regulating Oct4 expression during early embryonic development. To further understand the regulation of Oct4 expression by LRH-1 in vivo, the LRH-1 gene was disrupted by gene targeting. Disruption of the LRH-1 gene was undertaken by insertion of the β-geo cassette into the first exon of LRH-1 gene via homologous recombination in ES cells (Fig. 5A) (54). Southern blot and PCR analysis demonstrated the insertion of the β-geo gene (Fig. 5B and C). A cDNA probe that spanned exons 6 to 9, encoding the ligand-binding domain, was used as probe to detect LRH-1 mRNA in mutant embryos. LRH-1 mRNA was present in wild-type E6.5 to E7.0 embryos but not in _LRH-1_−/− embryos, demonstrating that the LRH-1 gene was successfully disrupted (Fig. 5D) through the replacement of the first exon with the β-geo gene. The genotype of the embryos was further confirmed by genomic PCR analysis.
FIG. 5.
Loss of LRH-1 expression in _LRH-1_−/− embryos. (A) LRH-1 gene-targeting strategy. Approximately 20 kb of the LRH-1 locus is shown. The KpnI restriction sites are labeled (K). The first exon, which was disrupted, contains the LRH-1 ATG start codon (depicted as a solid box). The targeting vector pKOS51 with homologous arms and the lacZ and neo genes are shown relative to the genomic structure. The first exon of the recombinant allele was disrupted by the insertion of the β-geo gene. (B) Southern blot analysis was used to genotype the ES cell clones by using a 5′probe upstream of the targeting vector. Digestion with KpnI yielded a 15-kb band for the wild-type allele and a 10.5-kb band for the targeted allele. The clone 2B5 was injected into recipient blastocysts. (C) Genotyping of the offspring by genomic PCR with primers F1 plus R1 and R4 produced a 350-bp wild-type fragment and 490-bp neo DNA fragment. (D) Analysis of LRH-1 expression in _LRH-1_−/− embryos at E6.5-7.0 by whole-mount in situ hybridization with an LRH-1 cRNA probe showed complete loss of LRH-1 expression in _LRH-1_−/− embryos.
Breeding results showed that matings of heterozygous offspring failed to produce pups homozygous for the mutant LRH-1 allele. In total, 248 pups were genotyped (Table 2); 119 of the mice were wild type and 129 were heterozygous, but no homozygous pups were detected. These data confirm previous data that disruption of the LRH-1 gene causes embryonic lethality (13, 15, 39). The lower ratio (1.1:1) of LRH-1+/− to wild-type pups (expected 2:1 based on Mendelian genetics) indicates haploinsufficiency (13). To determine precisely when homozygous LRH-1 mutants die, timed-mated heterozygous females were sacrificed at different gestational stages and the blastocysts or embryos were genotyped by PCR analysis (Table 2). _LRH-1_−/− blastocysts were found at E3.5 to E4.0 at a wild-type/heterozygote/homozygote ratio of 23:42:21, which is close to the expected Mendelian ratio of 1:2:1. The _LRH-1_−/− blastocysts appeared morphologically normal (Fig. 6A, panel i). Mutant embryos were also found at E6.5 to E7.0, but the ratio (12%) was much lower than expected. No _LRH-1_−/− embryos were detected at E7.5 to E9.5. These data demonstrate that _LRH-1_−/− embryos die around E6.5.
TABLE 2.
Targeted disruption of the LRH-1 gene leads to embryonic death
| Age | No. (%) of deaths in: | ||
|---|---|---|---|
| +/+ mice | +/− mice | −/− mice | |
| Pups | 119 (48%) | 129 (52%) | 0 |
| E3.5-E4.0 | 23 (27%) | 42 (49%) | 21 (24%) |
| E6.5 | 31 (33%) | 51 (55%) | 11 (12%) |
| E7.5 | 4 | 10 | 0 |
| E8.5 | 3 | 6 | 0 |
| E9.5 | 1 | 5 | 0 |
FIG. 6.
Analysis of Oct4 expression in embryos on days E3.5 to E7.0 from LRH-1+/− intercrosses. (A) Immunofluorescent detection of LRH-1 and Oct4 proteins in blastocysts (E3.5 to E4.0) with anti-LRH-1 and anti-Oct4 antibodies. About 90 blastocysts flushed from six heterozygous females were stained and genotyped. (B) Whole-mount in situ analysis of Oct4 mRNA in the embryos at E6.5 to E7.0 with a Dig-labeled Oct4 RNAprobe. Panels a to d shows four individual embryos. (C) Histology and Oct4 protein analysis in E6.5 to E7.0 embryos. LRH-1+/− (a to c) and _LRH-1_−/− embryos (d to f) were dissected, and the sections were stained with hemotoxylin and eosin (a, b, d, and e) or immunostained with anti-Oct4 antibody and counterstained with methyl green (c and f). After staining, blastocysts were recovered or embryonic tissue was scraped off the sections for genotyping. ec, exocoelomic cavity; eec, embryonic ectoderm; em, embryonic mesoderm exec, extraembryonic ectoderm; exed, extraembryonic endoderm; exem, extraembryonic mesoderm; pac, proamniotic cavity; ved, visceral endoderm.
Loss of Oct4 expression in the LRH-1−/− embryos.
The activation of Oct4 reporters by LRH-1 and the early embryonic death of the _LRH-1_−/− embryos led us to examine Oct4 expression in the _LRH-1_−/− embryos. Analysis of Oct4 expression showed a strong signal in the ICM of blastocysts at E3.5 to E4.0 and epiblast of wild-type embryos at E6.5 (Fig. 4 and 6). Oct4 was expressed in the ICM (Fig. 6A, panels h and l) in the LRH-1+/− or _LRH-1_−/− blastocysts even though the LRH-1 staining signal was weak (panel g) or absent (panel k), respectively. However, at E6.5 to E7.0, Oct4 mRNA was undetectable in epiblasts of _LRH-1_−/− embryos (Fig. 6B, panels c and d). The mutant embryos were much smaller than the wild-type controls. Further analysis revealed that histologic abnormalities in _LRH-1_−/− embryos were apparent as early as E6.5 to E7.0 (Fig. 6C). At this stage, the extraembryonic and embryonic tissues in wild-type embryos were clearly distinguished from each other (Fig. 6C, panels a to c); in contrast, they were difficult to distinguish in the _LRH-1_−/− embryos (panels d to f). Moreover, in _LRH-1_−/− embryos, the visceral endoderm was expanded and the cells were loosely arranged; the embryonic ectoderm showed a marked disorganization, and the proamniotic cavity was malformed. The loss of Oct4 expression in the epiblast of _LRH-1_−/− embryos was further confirmed by immunohistochemical staining (panel f). These results indicated that LRH-1 is not required to maintain the expression of the Oct4 gene in the ICM but is required to maintain its expression in the epiblast of developing embryos.
Rapid down-regulation of Oct4 expression in differentiating _LRH-1_−/− ES cells.
To determine if the effects of loss of LRH-1 on Oct4 expression were cell autonomous, an _LRH-1_−/− ES cell line was established. Due to the inability to generate _LRH-1_−/− ES cell lines from blastocyst outgrowths from our LRH-1 knockout mice, a sequential knockout strategy using two different targeting vectors was used in which exon 6, which encodes the T-box region of the DNA-binding domain and part of the hinge region, was disrupted in both LRH-1 alleles (Fig. 7A). Southern blot analysis confirmed deletions in both LRH-1 alleles (Fig. 7B). As expected, Northern blot analysis of the _LRH-1_−/− ES cells failed to detect LRH-1 mRNA in these cells (Fig. 7C). Monolayer cultures of LRH-1+/+ and _LRH-1_−/− ES cells were induced to differentiate for 4, 8, and 12 days by withdrawal of LIF. No RA was added to the ES cell cultures, so that the kinetics of Oct4 repression was slower (comparing Fig. 1A with 6C). No differences were seen in the number or morphology of wild-type and knockout cells during differentiation (data not shown).
Expression of Oct4 was compared in wild-type and _LRH-1_−/− ES cells induced to differentiate for 4, 8, and 12 days. Oct4 expression decreased much more rapidly in _LRH-1_−/− ES cells than in wild-type cells (Fig. 6C). LRH-1 expression was lost and the expression of Oct4 decreased more rapidly in the mutant ES cells than in differentiating wild-type ES cells. Expression of Sox2, FGF4, UTF1, and REX1, which are dependent on Oct4 (1, 7, 33, 52), were maintained in wild-type ES cells but dropped in _LRH-1_−/− ES cells after removal of LIF (Fig. 6C). Thus, in the absence of LRH-1, there is a failure to maintain Oct4 expression with a concomitant loss of expression of genes involved in maintaining pluripotence in ES cells.
DISCUSSION
In contrast to lower organisms, segregation of the mammalian germ cell lineage occurs relatively late during embryonic development as the embryo gastrulates. To facilitate this novel evolutionary mechanism, pluripotence must be maintained in all cells prior to this stage to allow segregation of the pluripotent cells destined to become primordial germ cells. To achieve this goal, two extraordinary events have to occur. One is that pluripotence has to be maintained as the embryo proper expands from an ICM to the epiblast. Oct4 is an essential transcription factor that regulates pluripotence, and thus its expression has to be maintained in the epiblast in the face of burgeoning signals to differentiate and pattern the embryo. Second, Oct4 expression has to be susceptible to repression in cells destined to become somatic cells and maintained in the small population that are destined to become PGCs. It is known that two enhancers in the Oct4 promoter regulate cell-specific expression of the Oct4 gene (31, 56). However, little is known about the transcription factors that specifically regulate Oct4 gene expression in different cell types or at different developmental stages. In this study, we demonstrate that LRH-1 can regulate Oct4 expression in ES cells and is required to maintain its expression at the epiblast stage of mouse embryonic development.
Complementary expression and function of SF-1 and LRH-1 in P19 and ES cells.
Based on work with P19 cells, it was previously hypothesized that the orphan receptor SF-1 regulates Oct4 expression. However, the survival of SF-1 knockout mice till birth demonstrates that SF-1 is not a key factor in the maintenance of Oct4 expression during early embryonic development (30). Expression of SF-1 in P19 cells appears to be a quirk of their derivation. ES and P19 cells are derived from different embryonic developmental stages: the former is a nontumor cell line derived from blastocyst ICM (about E4.5), whereas the latter is a teratocarcinoma cell line derived from transplanted epiblast stage cells at E7.5 (10, 53). P19 cells are considered a representative cell system to study events that occur during gastrulation stages of embryonic development. Unlike P19 cells, ES cells do not express SF-1 but do express LRH-1 (Fig. 1 and 2C and D), which more faithfully reflects the in vivo situation in mouse embryos (10). Thus, LRH-1 and not SF-1 regulates Oct4 expression during early embryonic development.
Regulation of Oct4 expression.
Despite its crucial role in development and maintenance of the germ cell lineage, relatively little is known about how the Oct4 gene is regulated. The cis regulation of Oct4 expression throughout the pluripotent life cycle has been defined using a LacZ transgenic reporter system (56). Three important regulatory regions were defined in the Oct4 promoter by this study: the PP element and the PE and DE. The transgenic analysis established that the DE is essential for the expression of Oct4 in all stages of the pluripotent life cycle except the epiblast stage, when Oct4 expression is regulated by the proximal enhancer. The proximal promoter is essential for Oct4 expression at all stages of the pluripotent life cycle. There are likely to be several important transcription factors binding to each of these elements at different stages. To date the Sp factors Sp1 and Sp3 and several nuclear receptors, including retinoid receptors and the orphan receptors COUP-TF and SF-1, have been implicated in the regulation of Oct4 expression (5, 6, 31, 42, 51). However, KO mouse models have not substantiated a functional link between any of these factors and regulation of Oct4 expression. In contrast, the orphan receptor GCNF binds to the DR0 element in the PP and regulates Oct4 expression. The functional consequence of inactivation of the GCNF gene is loss of proper repression of the Oct4 gene in somatic cells at gastrulation (19).
In this study we established that the orphan nuclear receptor LRH-1 also regulates Oct4 expression by binding to elements in the PP and PE. The consequence of inactivation of the LRH-1 gene is loss of Oct4 expression at the epiblast stage of development. The requirement of LRH-1 to maintain Oct4 expression in ES cells of the epiblast correlates directly with the requirement for the elements to which it binds for epiblast expression (56). Phylogenetic comparison of the sequence of the Oct4 promoter shows that the elements regulating Oct4 expression are conserved across species (38). Importantly, the conservation of the DR0, PE1, and PE2 sequences between mice and humans suggests that LRH-1 probably will play an essential role in regulating human Oct4 expression in differentiating ES cells and embryonic development.
Developmental roles of LRH-1.
The broad expression of LRH-1 during embryonic development suggests that it probably likely impact multiple developmental processes (17). Belanger's group generated an LRH-1:lacZ knock in reporter allele and observed β-galactosidase staining in heterozygous morulae and blastocysts (39). We have confirmed the expression of LRH-1 protein in the ICM of blastocysts. It was reported that LRH-1 mRNA was detectable only in the visceral endoderm and not in the embryonic ectoderm from E5.5 to E6.5 (39). In our experiments, LRH-1 mRNA and protein are clearly detectable throughout the entire embryo at E6.5 (Fig. 4). This discrepancy may be due to the use of different detection systems. Unfortunately, we were unable to detect the expression of the lacZ reporter gene in our β-geo knock-in allele. This is due to deletion of 70 bp upstream of the LRH-1 ATG initiation codon caused by the insertion of the β-geo gene, which disrupts LRH-1 and LacZ reporter expression.
Inactivation of the LRH-1 gene results in embryonic lethality at a later stage than in Oct4 mutant embryos, which die on day E5.5 lacking an ICM as the morula differentiated along the trophectodermal lineage (32). This suggests that LRH-1 is not required to maintain Oct4 expression in the ICM of blastocysts but, rather, is required to maintain its expression in the epiblast. Consistent with the results obtained in the embryo, Oct4 expression is maintained in undifferentiated _LRH-1_−/− ES cells. LRH-1 may not play a significant role in early preimplantation development. Its expression in the zygote and morula may simply represent expression of the factor before the time when it is required to maintain Oct4 expression.
The LRH-1 expression pattern at later embryonic stages supports a role for LRH-1 in regulating endodermal development and expression of endodermal genes, such as α-fetoprotein, _HNF1_α, _HNF3_β, and _HNF4_α during early hepatic and pancreatic development (2, 29, 39, 40, 49). Likewise, LRH-1 may regulate these genes in the visceral endoderm. It was previously reported that inactivation of the LRH-1 gene leads to defective visceral endoderm development (39). Thus, there are likely to be multiple defects in the _LRH-1_−/− embryos that contribute to embryonic lethality.
Role of LRH-1 in ES cells.
Several pluripotency factors in addition to Oct4, including Sox2, FGF4, UTF1, and REX1, are more rapidly down-regulated upon withdrawal of LIF in the _LRH-1_−/− ES cell line than in wild-type ES cells. Previous studies showed that Oct4 functions alone or as a complex with other factors to regulate the expression of these genes (1, 7, 33, 52). Northern analysis of the wild-type ES cells showed that although Oct4 expression was decreased along with LRH-1 levels after withdrawal of LIF, expression of Sox2, FGF4, UTF1, and REX1 was maintained. These data suggest that there is adequate Oct4 to maintain their expression. However, in _LRH-1_−/− ES cells, a more rapid loss of Oct4 expression coincided with decreased expression of Sox2, FGF4, UTF1, and REX1. The decreased expression of these genes is unlikely to be a direct consequence of the absence of LRH-1, since no LRH-1-binding sites have been identified in their promoters. Decreased expression of Sox2, FGF4, UTF1, and REX1 is not a generalized, nonspecific effect due to unhealthy ES cells, since most genes are expressed normally as measured by microarray analysis (data not shown). The early stages of ES cell differentiation probably approximate what is occurring in the epiblast in that pluripotence must be maintained even in the presence of nascent differentiation signals.
Maintenance of Oct4 expression is unlikely to be the only role of LRH-1 in regulating ES cell function. In addition to pluripotence, another property of ES cells is a high proliferative capacity. LRH-1 plays an important role in regulating proliferation through regulation of cyclin G1 expression (9). Interestingly it was found that LRH-1 and β-catenin synergistically regulate intestinal cell proliferation through cyclin G1 (9). Inactivation of the β-catenin gene is lethal to the embryo at the same stage as LRH-1 and presents a similar phenotype (25). Thus, LRH-1 and β-catenin may cooperate to regulate ES cell proliferation and expansion from an ICM to an epiblast.
Novel regulatory switch for Oct4 expression.
Based on our findings in this study and our previous studies, we propose a novel regulatory switch for Oct4 gene expression during ES cell differentiation and early embryonic development (Fig. 8). LRH-1 activates or maintains Oct4 gene expression in the epiblast of mouse embryos and undifferentiated ES cells. Maintenance of Oct4 expression by LRH-1 is mediated through binding to LRH-1 elements in the proximal enhancer and promoter. Upon differentiation of ES cells or gastrulation of embryos, GCNF expression is induced and LRH-1 is down-regulated, leading to the replacement of LRH-1 by GCNF at the Oct4 promoter. This switch in transcription factors blocks the DR0 in the proximal promoter and represses Oct4 gene expression in somatic cells. In the ICM and primordial germ cells, other transcription factors (X) bind to the distal enhancer and proximal promoter to maintain Oct4 expression. The reciprocal regulation of Oct4 by GCNF and LRH-1 through binding to common sequence elements has profound implications for the maintenance of pluripotence and differentiation of ES cells. Both LRH-1 and GCNF are orphan nuclear receptors that may be regulated by ligands, which provides another potential mechanism for regulating pluripotency in ES cells. The demonstration that LRH-1 is required to maintain Oct4 expression in the epiblast of mouse embryos opens a new window to view ES cell maintenance, pluripotence, and differentiation and a new target with which to manipulate these processes.
FIG. 8.
Reciprocal regulatory model for Oct4 expression by the orphan nuclear receptors LRH-1 and GCNF during early embryonic development. Oct4 is expressed in the ICM and epiblast (EP) and restricted to the primordial germ cells (PGC) after gastrulation. LRH-1 maintains Oct4 gene expression in the epiblast and early differentiated ES cells by direct binding to the PE and PP. After gastrulation, GCNF replaces LRH-1, binds to the PP, and represses Oct4 gene expression in somatic cells. Unknown factors (X) regulate the expression of Oct4 in the blastocyst and PGCs through the DE and PP. TP: trophectoderm; PE: primitive ectoderm; PC: proamnion cavity; EX: extraembryo; EP epiblast; PGC: primordial germ cell; VED: visceral endoderm.
Acknowledgments
We thank Kathy J. Jackson for ES cell culture. We thank David Moore for kindly providing the mouse SF-1 expression plasmids. LRH-1 knockout mice were generated by Lexicon Genetics Inc.
This work was supported by grants NIH DK57743 and U54HD07495-31 to A.J.C. S.A.K. and L.P. were supported by grants from the NIH (DK62434), the Robert A. Welch Foundation (I-1558), the Jensen Charitable Lead Annuity Trust, and Mr. and Mrs. Irwin Grossman.
REFERENCES
- 1.Ambrosetti, D. C., C. Basilico, and L. Dailey. 1997. Synergistic activation of the fibroblast growth factor 4 enhancer by Sox2 and Oct-3 depends on protein-protein interactions facilitated by a specific spatial arrangement of factor binding sites. Mol. Cell. Biol. 17**:**6321-6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Annicotte, J. S., E. Fayard, G. H. Swift, L. Selander, H. Edlund, T. Tanaka, T. Kodama, K. Schoonjans, and J. Auwerx. 2003. Pancreatic-duodenal homeobox 1 regulates expression of liver receptor homolog 1 during pancreas development. Mol. Cell. Biol. 23**:**6713-6724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antonio, V., B. Janvier, A. Brouillet, M. Andreani, and M. Raymondjean. 2003. Oxysterol and 9-_cis_-retinoic acid stimulate the group IIA secretory phospholipase A2 gene in rat smooth-muscle cells. Biochem. J. 376**:**351-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avilion, A. A., S. K. Nicolis, L. H. Pevny, L. Perez, N. Vivian, and R. Lovell- Badge. 2003. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 17**:**126-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnea, E., and Y. Bergman. 2000. Synergy of SF1 and RAR in activation of Oct-3/4 promoter. J. Biol. Chem. 275**:**6608-6619. [DOI] [PubMed] [Google Scholar]
- 6.Ben-Shushan, E., H. Sharir, E. Pikarsky, and Y. Bergman. 1995. A dynamic balance between ARP-1/COUP-TFII, EAR-3/COUP-TFI, and retinoic acid receptor:retinoid X receptor heterodimers regulates Oct-3/4 expression in embryonal carcinoma cells. Mol. Cell. Biol. 15**:**1034-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ben-Shushan, E., J. R. Thompson, L. J. Gudas, and Y. Bergman. 1998. Rex- 1, a gene encoding a transcription factor expressed in the early embryo, is regulated via Oct-3/4 and Oct-6 binding to an Octamer site and a novel protein, ROX-1, binding to an adjacent site. Mol. Cell. Biol. 18**:**1866-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernier, D., H. Thomassin, D. Allard, M. Guertin, D. Hamel, M. Blaquiere, M. Beauchemin, H. LaRue, M. Estable-Puig, and L. Belanger. 1993. Functional analysis of developmentally regulated chromatin-hypersensitive domains carrying the alpha 1-fetoprotein gene promoter and the albumin/alpha 1-fetoprotein intergenic enhancer. Mol. Cell. Biol. 13**:**1619-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Botrugno, O. A., E. Fayard, J. S. Annicotte, C. Haby, T. Brennan, O. Wendling, T. Tanaka, T. Kodama, W. Thomas, J. Auwerx, and K. Schoonjans. 2004. Synergy between LRH-1 and beta-catenin induces G1 cyclin-mediated cell proliferation. Mol. Cell 15**:**499-509. [DOI] [PubMed] [Google Scholar]
- 10.Bradley, A., R. Ramirez-Solis, H. Zheng, P. Hasty, and A. Davis. 1992. Genetic manipulation of the mouse via gene targeting in embryonic stem cells. Ciba Found. Symp. 165**:**256-269; discussion, 269-276. [DOI] [PubMed] [Google Scholar]
- 11.Chambers, I., D. Colby, M. Robertson, J. Nichols, S. Lee, S. Tweedie, and A. Smith. 2003. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113**:**643-655. [DOI] [PubMed] [Google Scholar]
- 12.Chung, A. C., D. Katz, F. A. Pereira, K. J. Jackson, F. J. DeMayo, A. J. Cooney, and B. W. O'Malley. 2001. Loss of orphan receptor germ cell nuclear factor function results in ectopic development of the tail bud and a novel posterior truncation. Mol. Cell. Biol. 21**:**663-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Del Castillo-Olivares, A., J. A. Campos, W. M. Pandak, and G. Gil. 2004. The Role of {alpha} 1-fetoprotein transcription factor/LRH-1 in bile acid biosynthesis. J. Biol. Chem. 279**:**16813-16821. [DOI] [PubMed] [Google Scholar]
- 14.Eddy, S. 1998. HMMER: profile HMMs for protein sequence analysis. Bioinformatics 14**:**755-763.9918945 [Google Scholar]
- 15.Falender, A. E., R. Lanz, D. Malenfant, L. Belanger, and J. S. Richards. 2003. Differential expression of steroidogenic factor-1 and FTF/LRH-1 in the rodent ovary. Endocrinology 144**:**3598-3610. [DOI] [PubMed] [Google Scholar]
- 16.Fayard, E., J. Auwerx, and K. Schoonjans. 2004. LRH-1: an orphan nuclear receptor involved in development, metabolism and steroidogenesis. Trends Cell Biol. 14**:**250-260. [DOI] [PubMed] [Google Scholar]
- 17.Fayard, E., K. Schoonjans, J. S. Annicotte, and J. Auwerx. 2003. Liver receptor homolog 1 controls the expression of carboxyl ester lipase. J. Biol. Chem. 278**:**35725-35731. [DOI] [PubMed] [Google Scholar]
- 18.Feldman, B., W. Poueymirou, V. E. Papaioannou, T. M. DeChiara, and M. Goldfarb. 1995. Requirement of FGF-4 for postimplantation mouse development. Science 267**:**246-249. [DOI] [PubMed] [Google Scholar]
- 19.Fuhrmann, G., A. C. Chung, K. J. Jackson, G. Hummelke, A. Baniahmad, J. Sutter, I. Sylvester, H. R. Scholer, and A. J. Cooney. 2001. Mouse germline restriction of Oct4 expression by germ cell nuclear factor. Dev. Cell 1**:**377-387. [DOI] [PubMed] [Google Scholar]
- 20.Fuhrmann, G., I. Sylvester, and H. R. Scholer. 1999. Repression of Oct-4 during embryonic cell differentiation correlates with the appearance of TRIF, a transiently induced DNA-binding factor. Cell. Mol. Biol. 45**:**717-724. [PubMed] [Google Scholar]
- 21.Galarneau, L., J. F. Pare, D. Allard, D. Hamel, L. Levesque, J. D. Tugwood, S. Green, and L. Belanger. 1996. The alpha1-fetoprotein locus is activated by a nuclear receptor of the Drosophila FTZ-F1 family. Mol. Cell. Biol. 16**:**3853-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanna, L. A., R. K. Foreman, I. A. Tarasenko, D. S. Kessler, and P. A. Labosky. 2002. Requirement for Foxd3 in maintaining pluripotent cells of the early mouse embryo. Genes Dev. 16**:**2650-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hinshelwood, M. M., J. J. Repa, J. M. Shelton, J. A. Richardson, D. J. Mangelsdorf, and C. R. Mendelson. 2003. Expression of LRH-1 and SF-1 in the mouse ovary: localization in different cell types correlates with differing function. Mol. Cell. Endocrinol. 207**:**39-45. [DOI] [PubMed] [Google Scholar]
- 24.Hogan B, B. R., Costantini F, Lacy E. 1994. Manipulating the mouse embryo: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Huelsken, J., R. Vogel, V. Brinkmann, B. Erdmann, C. Birchmeier, and W. Birchmeier. 2000. Requirement for beta-catenin in anterior-posterior axis formation in mice. J Cell Biol 148**:**567-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karolchik, D., et al. 2003. The UCSC Genome Browser Database. Nucleic Acids Res. 31**:**51-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kent, W. J., C. W. Sugnet, T. S. Furey, K. M. Roskin, T. H. Pringle, A. M. Zahler, and D. Haussler. 2002. The human genome browser at UCSC. Genome Res. 12:996-1006. [DOI] [PMC free article] [PubMed]
- 28.Lan, Z. J., P. Gu, X. Xu, and A. J. Cooney. 2003. Expression of the orphan nuclear receptor, germ cell nuclear factor, in mouse gonads and preimplantation embryos. Biol. Reprod. 68**:**282-289. [DOI] [PubMed] [Google Scholar]
- 29.Lu, T. T., M. Makishima, J. J. Repa, K. Schoonjans, T. A. Kerr, J. Auwerx, and D. J. Mangelsdorf. 2000. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol. Cell 6**:**507-515. [DOI] [PubMed] [Google Scholar]
- 30.Luo, X., Y. Ikeda, and K. L. Parker. 1994. A cell-specific nuclear receptor is essential for adrenal and gonadal development and sexual differentiation. Cell 77**:**481-490. [DOI] [PubMed] [Google Scholar]
- 31.Minucci, S., V. Botquin, Y. I. Yeom, A. Dey, I. Sylvester, D. J. Zand, K. Ohbo, K. Ozato, and H. R. Scholer. 1996. Retinoic acid-mediated down-regulation of Oct3/4 coincides with the loss of promoter occupancy in vivo. EMBO J. 15**:**888-899. [PMC free article] [PubMed] [Google Scholar]
- 32.Nichols, J., B. Zevnik, K. Anastassiadis, H. Niwa, D. Klewe-Nebenius, I. Chambers, H. Scholer, and A. Smith. 1998. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95**:**379-391. [DOI] [PubMed] [Google Scholar]
- 33.Nishimoto, M., A. Fukushima, A. Okuda, and M. Muramatsu. 1999. The gene for the embryonic stem cell coactivator UTF1 carries a regulatory element which selectively interacts with a complex composed of Oct-3/4 and Sox-2. Mol. Cell. Biol. 19**:**5453-5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nitta, M., S. Ku, C. Brown, A. Y. Okamoto, and B. Shan. 1999. CPF: an orphan nuclear receptor that regulates liver-specific expression of the human cholesterol 7alpha-hydroxylase gene. Proc. Natl. Acad. Sci. USA 96**:**6660-6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niwa, H., T. Burdon, I. Chambers, and A. Smith. 1998. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 12**:**2048-2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niwa, H., J. Miyazaki, and A. G. Smith. 2000. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 24**:**372-376. [DOI] [PubMed] [Google Scholar]
- 37.Palmieri, S. L., W. Peter, H. Hess, and H. R. Scholer. 1994. Oct-4 transcription factor is differentially expressed in the mouse embryo during establishment of the first two extraembryonic cell lineages involved in implantation. Dev. Biol. 166**:**259-267. [DOI] [PubMed] [Google Scholar]
- 38.Pan, G. J., Z. Y. Chang, H. R. Scholer, and D. Pei. 2002. Stem cell pluripotency and transcription factor Oct4. Cell Res. 12**:**321-329. [DOI] [PubMed] [Google Scholar]
- 39.Pare, J. F., D. Malenfant, C. Courtemanche, M. Jacob-Wagner, S. Roy, D. Allard, and L. Belanger. 2004. The fetoprotein transcription factor (FTF) gene is essential to embryogenesis and cholesterol homeostasis and is regulated by a DR4 element. J. Biol. Chem. 279**:**21206-21216. [DOI] [PubMed] [Google Scholar]
- 40.Pare, J. F., S. Roy, L. Galarneau, and L. Belanger. 2001. The mouse fetoprotein transcription factor (FTF) gene promoter is regulated by three GATA elements with tandem E box and Nkx motifs, and FTF in turn activates the Hnf3beta, Hnf4alpha, and Hnf1alpha gene promoters. J. Biol. Chem. 276**:**13136-13144. [DOI] [PubMed] [Google Scholar]
- 41.Peng, N., J. W. Kim, W. E. Rainey, B. R. Carr, and G. R. Attia. 2003. The role of the orphan nuclear receptor, liver receptor homologue-1, in the regulation of human corpus luteum 3beta-hydroxysteroid dehydrogenase type II. J. Clin. Endocrinol. Metab. 88**:**6020-6028. [DOI] [PubMed] [Google Scholar]
- 42.Pesce, M., M. Marin Gomez, S. Philipsen, and H. R. Scholer. 1999. Binding of Sp1 and Sp3 transcription factors to the Oct-4 gene promoter. Cell Mol. Biol. 45**:**709-716. [PubMed] [Google Scholar]
- 43.Pesce, M., and H. R. Scholer. 2001. Oct-4: gatekeeper in the beginnings of mammalian development. Stem Cells 19**:**271-278. [DOI] [PubMed] [Google Scholar]
- 44.Pesce, M., X. Wang, D. J. Wolgemuth, and H. Scholer. 1998. Differential expression of the Oct-4 transcription factor during mouse germ cell differentiation. Mech. Dev. 71**:**89-98. [DOI] [PubMed] [Google Scholar]
- 45.Rausa, F. M., L. Galarneau, L. Belanger, and R. H. Costa. 1999. The nuclear receptor fetoprotein transcription factor is coexpressed with its target gene HNF-3beta in the developing murine liver intestine and pancreas. Mech. Dev. 89**:**185-188. [DOI] [PubMed] [Google Scholar]
- 46.Sadovsky, Y., P. A. Crawford, K. G. Woodson, J. A. Polish, M. A. Clements, L. M. Tourtellotte, K. Simburger, and J. Milbrandt. 1995. Mice deficient in the orphan receptor steroidogenic factor 1 lack adrenal glands and gonads but express P450 side-chain-cleavage enzyme in the placenta and have normal embryonic serum levels of corticosteroids. Proc. Natl. Acad. Sci. USA 92**:**10939-10943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scholer, H. R., R. Balling, A. K. Hatzopoulos, N. Suzuki, and P. Gruss. 1989. Octamer binding proteins confer transcriptional activity in early mouse embryogenesis. EMBO J. 8**:**2551-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schoonjans, K., J. S. Annicotte, T. Huby, O. A. Botrugno, E. Fayard, Y. Ueda, J. Chapman, and J. Auwerx. 2002. Liver receptor homolog 1 controls the expression of the scavenger receptor class B type I. EMBO Rep. 3**:**1181-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schoorlemmer, J., A. van Puijenbroek, M. van Den Eijnden, L. Jonk, C. Pals, and W. Kruijer. 1994. Characterization of a negative retinoic acid response element in the murine Oct4 promoter. Mol. Cell. Biol. 14**:**1122-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sirianni, R., J. B. Seely, G. Attia, D. M. Stocco, B. R. Carr, V. Pezzi, and W. E. Rainey. 2002. Liver receptor homologue-1 is expressed in human steroidogenic tissues and activates transcription of genes encoding steroidogenic enzymes. J. Endocrinol. 174**:**R13-R17. [DOI] [PubMed] [Google Scholar]
- 51.Stewart, C. L. 1994. Leukaemia inhibitory factor and the regulation of pre- implantation development of the mammalian embryo. Mol. Reprod. Dev. 39**:**233-238. [DOI] [PubMed] [Google Scholar]
- 52.Tomioka, M., M. Nishimoto, S. Miyagi, T. Katayanagi, N. Fukui, H. Niwa, M. Muramatsu, and A. Okuda. 2002. Identification of Sox-2 regulatory region which is under the control of Oct-3/4-Sox-2 complex. Nucleic Acids Res. 30**:**3202-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van der Heyden, M. A., and L. H. Defize. 2003. Twenty one years of P19 cells: what an embryonal carcinoma cell line taught us about cardiomyocyte differentiation. Cardiovasc. Res. 58**:**292-302. [DOI] [PubMed] [Google Scholar]
- 54.Wattler, S., M. Kelly, and M. Nehls. 1999. Construction of gene targeting vectors from lambda KOS genomic libraries. BioTechniques 26**:**1150-1156, 1158, 1160. [DOI] [PubMed] [Google Scholar]
- 55.Yang, Y., M. Zhang, G. Eggertsen, and J. Y. Chiang. 2002. On the mechanism of bile acid inhibition of rat sterol 12alpha-hydroxylase gene (CYP8B1) transcription: roles of alpha-fetoprotein transcription factor and hepatocyte nuclear factor 4alpha. Biochim. Biophys. Acta 1583**:**63-73. [DOI] [PubMed] [Google Scholar]
- 56.Yeom, Y. I., G. Fuhrmann, C. E. Ovitt, A. Brehm, K. Ohbo, M. Gross, K. Hubner, and H. R. Scholer. 1996. Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development 122**:**881-894. [DOI] [PubMed] [Google Scholar]