Modulation of Transcriptional Regulation by LEF-1 in Response to Wnt-1 Signaling and Association with β-Catenin (original) (raw)
Abstract
Wnt signaling is thought to be mediated via interactions between β-catenin and members of the LEF-1/TCF family of transcription factors. Here we study the mechanism of transcriptional regulation by LEF-1 in response to a Wnt-1 signal under conditions of endogenous β-catenin in NIH 3T3 cells, and we examine whether association with β-catenin is obligatory for the function of LEF-1. We find that Wnt-1 signaling confers transcriptional activation potential upon LEF-1 by association with β-catenin in the nucleus. By mutagenesis, we identified specific residues in LEF-1 important for interaction with β-catenin, and we delineated two transcriptional activation domains in β-catenin whose function is augmented in specific association with LEF-1. Finally, we show that a Wnt-1 signal and β-catenin association are not required for the architectural function of LEF-1 in the regulation of the T-cell receptor α enhancer, which involves association of LEF-1 with a different cofactor, ALY. Thus, LEF-1 can assume diverse regulatory functions by association with different proteins.
Since the initial identification of Wnt-1 as a proto-oncogene (45), Wnt-1 and Wnt-related genes have been shown to encode secreted signaling molecules that regulate many developmental processes in both vertebrates and invertebrates (reviewed in references 19, 39, 44, 45, 50, and 64). The molecular mechanism of the Wnt signaling pathway has been elucidated by genetic analysis in Drosophila melanogaster and Caenorhabditis elegans and by biochemical studies in Xenopus laevis and mammalian tissue culture cells. In response to a Wnt signal in vertebrates or to a wingless signal in Drosophila, a receptor encoded by the frizzled gene family (4, 73) is activated and conveys the signal to cytoplasmic downstream components (reviewed in reference 25). These components included dishevelled (dsh), zeste-white 3 kinase (zw3), and armadillo (arm), which are homologous to the vertebrate proteins Dsh (63), GSK3 (20, 63), and β-catenin (33), respectively. In response to a Wnt signal, Dishevelled inhibits, through an as-yet-unknown mechanism, the function of the serine/threonine kinase GSK3 (53), resulting in the cytoplasmic accumulation of hypophosphorylated β-catenin and the transmission of the Wnt signal to more downstream targets (52, 53, 74).
In addition to its function in the Wnt signaling pathway, β-catenin is also an essential component of adhesion complexes (18, 24). β-Catenin is directly associated with cadherins and couples these calcium-dependent, membrane-bound adhesion molecules to components of the cytoskeleton (1, 23). Distinct functions of β-catenin in Wnt signaling and cell adhesion have been implicated by studies with Drosophila and Xenopus (11, 48, 60). β-Catenin is also associated with the adenomatous polyposis coli (APC) protein, which regulates the half-life of β-catenin that is not complexed with cadherins (42, 49). Wnt signaling has been shown to increase the pool of β-catenin–APC complexes and the pool of free, or uncomplexed, β-catenin (49, 58). The free β-catenin has been proposed to transmit the Wnt signal by interactions with downstream effectors (36).
A potential mechanism for the function of free β-catenin in Wnt signaling has been suggested by the observation that β-catenin can associate with the transcription factor LEF-1 (2, 22) and the closely related XTCF-3 protein (38). A Drosophila ortholog of LEF-1, termed pangolin or dTCF, has been shown genetically to act downstream of armadillo (7, 69). Murine LEF-1 can bind the wingless-responsive enhancer of the Drosophila ultrabithorax gene and regulate the enhancer in an armadillo-dependent manner in vivo (56). Likewise, binding sites for LEF-1/TCF proteins coincide with Wnt-responsive sequence elements in the promoters of the Xenopus siamois and the nodal related 3 genes (5, 34). However, these previous studies have not established whether and how a Wnt signal results in transcriptional activation by LEF-1 in cells expressing normal levels of β-catenin.
Lymphoid enhancer-binding factor (LEF-1) is a member of the family of high-mobility group (HMG) proteins that recognizes the nucleotide sequence 5′CCTTTGAACT (68, 72) and is expressed in lymphoid cells and at multiple sites of organogenesis in the developing mouse (47, 68, 71). LEF-1 is closely related to T-cell factor 1 (TCF-1), which has a similar expression pattern in the mouse (47, 70), and to TCF-3 and -4 (26, 38). Functional and biochemical characterization of LEF-1 has indicated that this protein has no transcriptional activation potential by itself and is unable to stimulate transcription from a synthetic enhancer containing multimerized LEF-1 binding sites (68, 72). However, LEF-1 stimulates the function of the T-cell receptor α (TCRα) enhancer in collaboration with other DNA-binding proteins (6, 15, 32, 68, 72). LEF-1 acts as an architectural transcription factor that introduces a sharp bend in the DNA (13, 31) and contains a context-dependent activation domain which functions only in a specific context of other transcription factors (8, 14).
Three distinct regions of LEF-1 have been shown to interact with other proteins. The amino-terminal 76 residues of LEF-1 can associate directly with β-catenin (2, 22). A similar β-catenin interaction domain has been identified in the LEF-1-related proteins TCF-1, TCF-3, and TCF-4, which in combination with overexpressed β-catenin can stimulate transcription from multimerized LEF/TCF binding sites (26, 38). In addition, the protein ALY was shown to interact with the context-dependent activation domain of LEF-1 and with AML-1, another TCRα-binding protein, to stimulate the function of the TCRα enhancer (6). Finally, Pendulin, a protein distantly related to β-catenin, can interact with a carboxyl-terminal nuclear localization signal of LEF-1 and may regulate the nuclear localization of LEF-1 (55). The interaction of LEF-1 with multiple proteins raises the questions of whether they confer distinct regulatory properties upon LEF-1, or whether they collaborate with, or antagonize, each other in mediating a particular transcriptional response.
In this study, we examine the regulation of transcription by LEF-1 in response to Wnt-1 signaling and association of LEF-1 with endogenous, rather than overexpressed, β-catenin. We identify specific residues in LEF-1 that are required for interaction with β-catenin, and we study the mechanism by which β-catenin augments transcriptional activation by LEF-1. Finally, we address the question of whether the association with β-catenin is obligatory for the function of LEF-1.
MATERIALS AND METHODS
Plasmids and oligonucleotides.
The pLEF-CAT reporter gene construct was generated by inserting a _Xho_I-_Xba_I fragment, containing seven multimerized wild-type (wt) LEF-1 binding sites, into a plasmid, p301fosCAT, containing the minimal fos promoter linked to the chloramphenicol acetyltransferase (CAT) gene (3). pmLEF-CAT was created by inserting six multimerized oligonucleotides containing mutated LEF-1 binding sites into p301fosCAT. The nucleotide sequences of the wt and mutated LEF-1 binding sites are as follows: wt, 5′GCACCCTTTGAAGCTC; mutated, 5′GCACCaaTTcAAGCTC.
Point mutations in LEF-1 and β-catenin were generated by site-directed mutagenesis with primers that contain mismatches to alter specific amino acids and introduce restriction sites. The mutated codons in the primers are underlined and the mutated nucleotides are in lowercase letters, as follows: m1LEF-1, 5′CTCTGCGCCACCGcgGcGATGATCCCCTTC; m2LEF-1, 5′GATGATCCCCTTCgcGGcCGcAGGCGATCCCCAG; m3LEF-1, 5′GGCGATCCCCAGgcGGccgcGATCTTCGCCGAG; m4LEF-1, 5′GAGGAGGGCGACgcAGtCGcgATCgcGTCATCTTTGGTTAACG; and DPβ-catenin, 5′GGATTGCCTTTACCAgctAGcGcAGGAGCTGTGGTAGcGGCACCAGcATGGATTCCAGcGT CCAGGTAAGACTG. The expression plasmids for LEF-1 or β-catenin were generated by linking the appropriate cDNAs as _Xba_I/_Kpn_I fragments to the cytomegalovirus enhancer-promoter in the pCG vector (14). Subclones containing the amino-terminal deletion mutant LEF-1Δ56 and the LEF-1HMG domain were constructed by using naturally occurring restriction enzyme sites, _Hpa_I and _Nde_I, respectively. For the construction of pCMV-DPβ-catenin, we first introduced point mutations in Bluescript–β-catenin by site-directed mutagenesis. Subsequently, we used a PCR-generated mutant DNA fragment (encoding amino acids 1 to 90 of β-catenin) to replace the corresponding DNA fragment in pCMV-β-catenin. All β-catenin constructs used in this study contain a bacteriophage T7 epitope tag at the amino terminus and were derived from a β-actin promoter–β-catenin construct (2). The construction of amino- and carboxyl-terminal deletion mutants of β-catenin was performed in two steps. First, we prepared DNA fragments encoding amino acids 132 to 781 (ΔN) and 1 to 695 (ΔC) by PCR and inserted the fragments in frame into pEVRFT7 vectors (6). To avoid the introduction of mutations by PCR, we subsequently subcloned an _Sna_BI fragment (ΔN) and a _Bst_BI-_Xba_I fragment (ΔC) and replaced the corresponding regions in pCMV-β-catenin. The chimeric β-catenin–LEF-1 and β-catenin–GAL4 plasmids were constructed with PCR-generated fragments. In brief, DNA fragments encoding amino acids 1 to 131 or amino acids 696 to 781 of β-catenin were obtained by PCR. The purified PCR products were subcloned either into the _Xba_I site at codon 3 of LEF-1 in plasmid pCMV-LEF-1 or into the _Bam_HI site in the polylinker of pGAL4DBD (6). For the construction of the plasmid encoding the chimeric CD8–β-catenin, we prepared a DNA fragment encoding amino acids 1 to 211 of CD8 by PCR from the plasmid pSp65.F1.1T8 (30) and subcloned it into the _Xba_I and _Sma_I sites of the pCG vector. Subsequently, a full-length β-catenin fragment was isolated from the β-actin promoter–β-catenin construct by digestion with _Bam_HI and inserted into the pCG-CD8 plasmid. Nucleotide sequences of the coding regions for all expression plasmids were verified by sequencing.
Cell culture, transient transfections, and reporter gene assays.
NIH 3T3, HeLa, and Cos7 cells were cultured at 37°C in Dulbecco’s modified Eagle medium, supplemented with 5% fetal bovine serum. Neuro2A cells were grown in Eagle’s minimal essential medium with Earle’s balanced salt solution medium containing 5% fetal bovine serum. Transient transfections were generally performed by the DEAE-dextran–chloroquine procedure (16). A Rous sarcoma virus–β-galactosidase control plasmid was included in each transfection experiment to control for the efficiency of transfection. The total DNA concentration in each transfection experiment was kept constant by adding vector plasmid DNA if necessary. CAT and β-galactosidase assays were performed as described in the work of Starr et al. (66). After background subtraction, the CAT activity was normalized to β-galactosidase activity as an internal transfection control. Transfections of NIH 3T3 cells for immunoprecipitation and immunoblot analysis were performed by electroporation.
Cell lysis, immunoprecipitation, and immunoblot analysis.
For immunoprecipitations, cytoplasmic cell extracts were prepared from transfected cells by adding lysis buffer (100 mM NaCl, 50 mM Tris-HCl [pH 7.5], 0.5% Nonidet P-40, 0.5 mg of leupeptin per ml, 1 mg of pepstatin per ml, and 0.2 mM phenylmethylsulfonyl fluoride) and preclearing the lysates by centrifugation. The cell extracts were preabsorbed with protein G-Sepharose and normal mouse immunoglobulin G (IgG) (Pharmacia) for 1 h at 4°C prior to the addition of β-catenin monoclonal antibody (Signal Transduction). After incubation for 1 h at 4°C, the immunoprecipitates were washed three times with lysis buffer, solublized in sodium dodecyl sulfate (SDS)-sample buffer, and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE).
Immunoblots were incubated in Tris-buffered saline containing 0.1% Tween 20, including 5% nonfat dried milk during the blocking and antibody incubation steps. The bound antibodies were detected with an alkaline phosphatase-conjugated goat anti-mouse antibody (Promega) and subsequently developed with nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate (Promega). For the experiment shown in Fig. 4, the ECL system (Amersham Corp.), in conjunction with horseradish peroxidase-conjugated goat anti-mouse antibodies (Organon Teknika Corp.), was used to visualize proteins recognized by the antibodies.
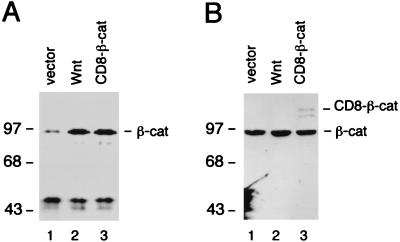
FIG. 4.
Wnt and CD8–β-catenin increase the pool of cytosolic β-catenin but do not affect the total amount of β-catenin. (A) NIH 3T3 cells were transfected with an expression plasmid encoding Wnt-1 or CD8–β-catenin fusion protein. Cytosolic cell extract was prepared, and β-catenin was immunoprecipitated with a monoclonal anti-β-catenin antibody and visualized by immunoblot analysis. (B) Whole-cell lysates from transfected NIH 3T3 cells were prepared, and β-catenin expression was analyzed as described above. Numbers to the left of each panel show molecular mass in kilodaltons.
For the preparation of whole-cell extracts, transfected cells were lysed in 2× sample buffer (120 mM Tris-HCl [pH 6.8], 4% SDS, 20% glycerol, 200 mM dithiothreitol, 0.01% bromophenol blue), boiled for 5 min and clarified by centrifugation. Protein concentrations were determined by bicinchoninic acid assays (Pierce).
Immunocytochemistry.
For immunocytochemistry to detect LEF-1 and β-catenin, NIH 3T3 cells were allowed to settle on poly-D-lysine-coated coverslips the day before transfection by the Lipofectamine procedure (GIBCO-BRL). Cells on the coverslips were fixed in freshly made 4% paraformaldehyde in phosphate-buffered saline (PBS), 36 to 48 h after transfection. After fixation, cells were blocked in Block Buffer (1× PBS, 0.1% Tween 20, 20% normal goat serum [Sigma], 3% bovine serum albumin [Sigma]) for 30 min at room temperature. For immunodetection, rabbit polyclonal anti-LEF-1 antibody or anti-β-catenin monoclonal antibody (Transduction Laboratories) was used at a 1:750 or 1:100 dilution, respectively. Washes were performed in 1× PBS containing 0.1% Tween 20, and fluorescein isothiocyanate-conjugated donkey anti-rabbit IgG (heavy plus light) and Texas Red-conjugated donkey anti-mouse IgG (heavy plus light) (Jackson Immunochemicals) were used at a 1:250 dilution as secondary antibodies. After washing, the coverslips were mounted in Fluoromount G and the cells were photographed.
In vitro translation, protein purification, nuclear extracts, and DNA binding analysis.
In vitro-translated proteins were synthesized with a coupled transcription and translation (TnT) kit (Promega). wt and mutant LEF-1 proteins were expressed from Bluescript KS+ constructs, and the efficiencies of in vitro translation reactions were assessed by SDS-PAGE of parallel translation reactions performed in the presence of [35S]methionine (Amersham). To study the interaction between LEF-1 and β-catenin, in vitro-translated LEF-1 was incubated with recombinant six-His-tagged β-catenin in the presence of a 32P-labeled duplex oligonucleotide probe, containing a LEF-1 binding site. The binding reaction contained 20 mM HEPES buffer (pH 7.9), 75 mM NaCl, 1 mM dithiothreitol, 2 mM MgCl2, 10% glycerol, 0.1 mg of bovine serum albumin per ml, 10 μg of salmon sperm DNA per ml, and ∼40,000 cpm of labeled DNA probe. The protein-DNA complexes were analyzed by electrophoretic mobility shift assays as described in the work of Travis et al. (68). The six-His-tagged protein β-catenin was expressed in bacteria and purified with Ni-nitrilotriacetic acid agarose beads (Qiagen) according to the manufacturer’s instructions. Nuclear extracts from Cos7 cells were prepared according to the method of Schreiber et al. (62).
RESULTS
Wnt-1 augments transcriptional activation by LEF-1.
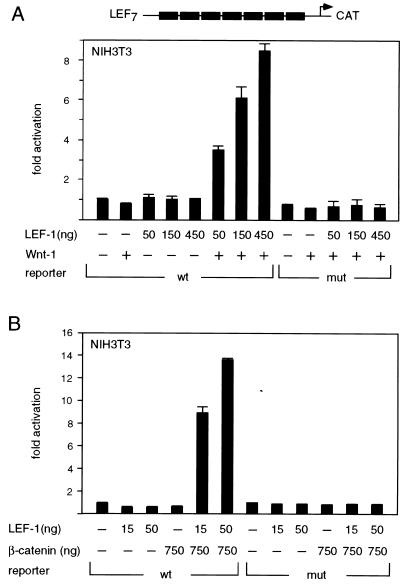
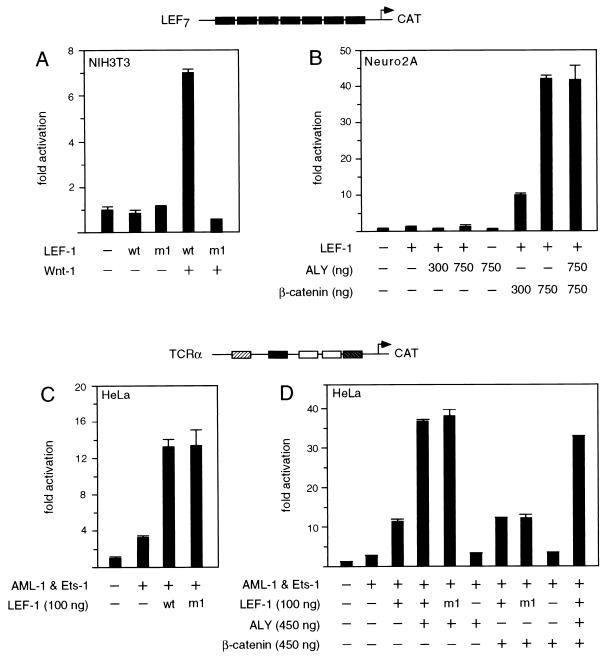
To examine the effects of Wnt signaling on transcriptional activation by LEF-1 in a mammalian tissue culture system, we used murine NIH 3T3 cells, which contain endogenous β-catenin but lack LEF-1 (68). Cells were transfected transiently with a CAT reporter gene, containing multimerized LEF-1 binding sites linked to the minimal fos promoter (LEF-CAT), together with increasing amounts of a LEF-1 expression plasmid (Fig. 1A). As anticipated from previous experiments (68), expression of LEF-1 alone did not augment the activity of the reporter gene. However, coexpression of LEF-1 and Wnt-1 stimulated reporter gene activity in a dose-dependent manner, with a maximal level of eightfold over that with LEF-1 alone. This transcriptional activation was not observed with a reporter gene containing multimerized mutant LEF-1 binding sites, indicating that the transcriptional response to Wnt-1 signaling is dependent on the presence of functional LEF-1 binding sites.
FIG. 1.
Transcriptional activation by LEF-1 in response to Wnt-1 signaling. (A) LEF-1 stimulates reporter gene activity in the presence of Wnt-1. NIH 3T3 cells were transiently transfected with 0.8 μg of a LEF-1–CAT reporter gene construct, together with expression plasmids encoding Wnt-1 (1.5 μg) or LEF-1 (amount as indicated). The LEF-1–CAT reporter gene contained either multimerized wt or mutant LEF-1 binding sites. For these and subsequent transfection assays, the levels of CAT activity were normalized for the expression of a cotransfected Rous sarcoma virus–β-galactosidase expression plasmid. Fold activation was quantitated relative to the level of CAT activity from cells transfected with the reporter gene alone. All the transfection experiments were performed at least twice, and the results of a representative experiment are shown here. Error bars represent standard errors of the mean. (B) β-Catenin activates transcription in collaboration with LEF-1. NIH 3T3 cells were transiently transfected with a LEF-CAT reporter gene, together with LEF-1 and β-catenin cDNA expression plasmids as indicated. mut, mutant.
β-Catenin is a component of the Wnt signaling pathway and has been shown to interact directly with LEF-1 (2, 22). Coexpression of LEF-1 and β-catenin in NIH 3T3 cells augmented reporter gene activity by a factor of 14 relative to the level observed with expression of LEF-1 alone (Fig. 1B). This transcriptional activation is independent of a Wnt signal, consistent with a role for β-catenin as a downstream component of the Wnt signaling pathway.
Identification of amino acids in LEF-1 that are important for association with β-catenin.
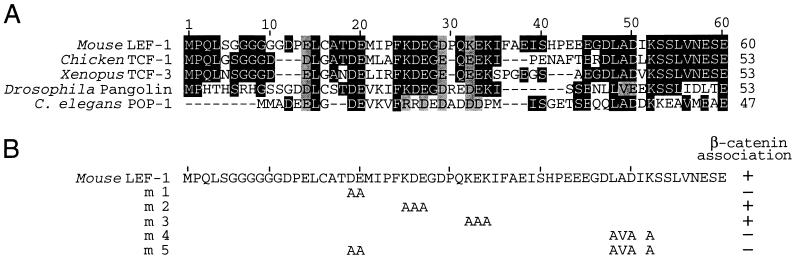
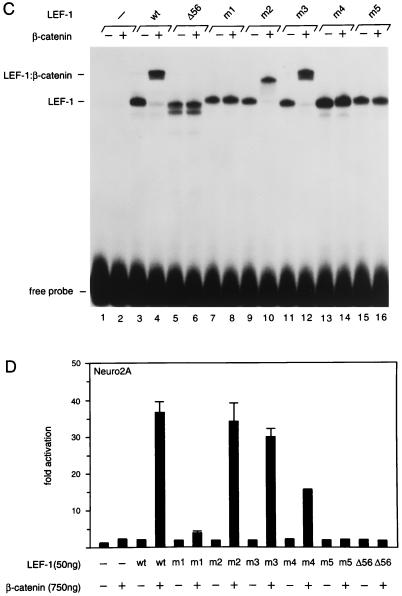
Association assays in a yeast two-hybrid system and in glutathione _S_-transferase pull-down experiments have indicated that the amino-terminal 56 residues of LEF-1 are necessary and sufficient for mediating the interaction with β-catenin (2, 22). The amino terminus of LEF-1 is highly conserved among members of the LEF-1/TCF family in various species (Fig. 2A). To further define the β-catenin interaction domain of LEF-1, we used site-directed mutagenesis to introduce point mutations into the amino terminus of the protein (Fig. 2B). We examined the interaction of purified recombinant β-catenin with in vitro-translated wt and mutant LEF-1 polypeptides in an electrophoretic mobility shift assay, with a radiolabeled LEF-1 binding site as a DNA probe. Incubation of β-catenin with wt LEF-1 yielded a complex that migrated at a lower mobility than that of the complex generated with LEF-1 alone, consistent with the formation of a ternary complex (Fig. 2C, lanes 3 and 4). In contrast, the formation of a ternary complex was not observed with the LEF-1 deletion mutant Δ56, in which the β-catenin interaction domain had been removed, although this deletion did not alter DNA binding by LEF-1 (lanes 5 and 6). The mutant LEF-1 proteins m1, m4, and m5 also failed to interact with β-catenin at detectable levels (lanes 7, 8, and 13 to 16), whereas the mutations in m2 and m3 had no significant effect on the association with β-catenin (lanes 9 to 12). Together, these point mutations define two distinct regions in the amino terminus of LEF-1 that are important for association with β-catenin.
FIG. 2.
Delineation of the β-catenin interaction domain in LEF-1. (A) Sequence comparison of the amino termini of mouse LEF-1 (64), chicken TCF-1 (9), Xenopus TCF-3 (38), Drosophila pangolin/dTCF (7, 69), and C. elegans POP-1 (29). Identical residues are indicated by white letters, and conserved residues are shaded. (B) Alanine substitution mutagenesis of the β-catenin interaction domain in LEF-1. The mutations in the amino terminus of LEF-1 are indicated in the diagram, and their effects on the interaction with β-catenin in vitro are summarized. (C) Electrophoretic mobility shift assay of DNA binding by wt LEF-1 and various mutant forms of LEF-1, alone or in association with purified recombinant His6–β-catenin. In vitro-translated LEF-1 polypeptides were incubated with a 32P-labeled DNA probe containing a LEF-1 binding site in the absence or presence of β-catenin as indicated. The protein-DNA complexes were separated by electrophoresis through a native 6% polyacrylamide gel and visualized by autoradiography. Translation of the Δ56 mutant of LEF-1 also yielded a slightly smaller protein product, presumably by initiation at a downstream ATG. (D) Mutations in the β-catenin interaction domain of LEF-1 reduce the transcriptional activation by LEF-1 in association with β-catenin. Neuro2A cells were transiently cotransfected with a LEF-CAT reporter gene and wt or mutant LEF-1 expression plasmids, alone or together with a β-catenin expression plasmid.
We also examined the effects of the point mutations in LEF-1 on its transcriptional activation potential. The LEF-CAT reporter gene was cotransfected, together with LEF-1 and β-catenin expression plasmids, into Neuro2A cells, which contain low levels of endogenous β-catenin. The LEF-1 mutations in m1 and m4 reduced transcriptional activation by a factor of 10 and 3, respectively. Transcriptional activation was not detected with the double mutant m5 and the deletion mutant Δ56. Thus, the potential of LEF-1 to interact with β-catenin correlates with transcriptional activation, although the effects of the m1 and m4 mutations in the transfection assays are less pronounced than those observed in the association assays in vitro. The molecular basis for this difference is unclear, but these observations raise the possibility that modifications or additional factors may contribute to the transcriptional activation by LEF-1 and β-catenin in vivo.
Nuclear translocation of β-catenin in response to Wnt-1 signaling.
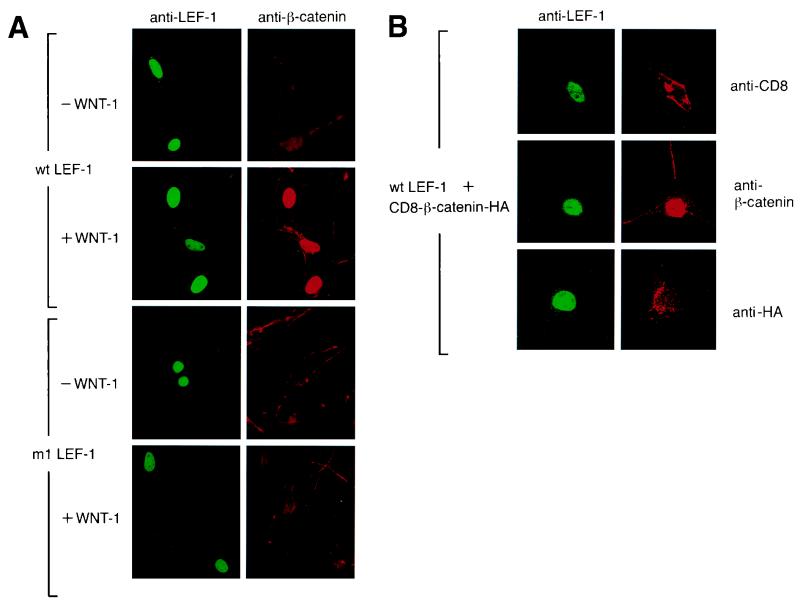
Overexpression of β-catenin and LEF-1 results in nuclear translocation of β-catenin (2, 22). To examine whether the expression of LEF-1 is sufficient for the nuclear translocation of endogenous β-catenin, we performed indirect immunocytochemistry on NIH 3T3 cells that had been transfected with LEF-1 (Fig. 3A, first panel). Antibodies directed against LEF-1 detected strong nuclear expression of LEF-1 in transfected cells, whereas anti-β-catenin antibodies detected β-catenin only in the cytoplasm, even in cells that contained nuclear LEF-1. In contrast, expression of Wnt-1 resulted in nuclear localization of β-catenin in cells that expressed LEF-1 (Fig. 3A, second panel). Nuclear β-catenin was not detected in control cells that were transfected with Wnt-1 alone (data not shown). To confirm the specificity of nuclear translocation of endogenous β-catenin in response to Wnt signaling, we expressed the mutant m1 LEF-1 protein, alone or in combination with Wnt-1. In cells expressing the mutant LEF-1 protein, a markedly reduced nuclear accumulation of β-catenin was observed in the presence of a Wnt-1 signal (Fig. 3A, third and fourth panels). Thus, expression of LEF-1 and a Wnt-1 signal are both required for the nuclear localization of endogenous β-catenin.
FIG. 3.
Nuclear localization of endogenous β-catenin in NIH 3T3 cells is dependent on expression of LEF-1 and on Wnt signaling. (A) NIH 3T3 cells were transiently transfected with an expression plasmid encoding wt or mutant (m1) LEF-1. The LEF-1 expression plasmids were transfected alone (−Wnt-1) or together with a Wnt-1 expression plasmid (+Wnt-1). LEF-1 and endogenous β-catenin were detected by indirect immunofluorescence. (B) Transient expression of LEF-1 and a CD8–β-catenin fusion protein in NIH 3T3 cells. The fusion protein was detected by indirect immunofluorescence with anti-CD8 antibody (top panel), anti-β-catenin antibody (middle panel), or anti-HA antibody (bottom panel). Endogenous β-catenin was also visualized with anti-β-catenin antibody (middle panel).
Recent experiments indicated that the expression of a membrane-tethered form of plakoglobin, a member of the family of catenin proteins, can mediate Wnt responsiveness in Xenopus embryos (35). This observation raised the possibility that nuclear localization of catenin-like proteins might not be necessary to mediate a nuclear response to Wnt signaling. To examine whether a membrane-tethered form of β-catenin can collaborate with LEF-1 to activate transcription in the absence of nuclear β-catenin, we generated a fusion protein in which the transmembrane protein CD8 was linked to the amino terminus of β-catenin (see below and Fig. 5A and B). This fusion protein also contains a hemagglutinin (HA) epitope tag at the carboxyl terminus. Expression of the CD8–β-catenin fusion protein together with LEF-1 stimulated the expression of a LEF-CAT reporter gene in Neuro2A cells to a level three- to fivefold higher than that observed with wt β-catenin (see below and Fig. 5C).
FIG. 5.
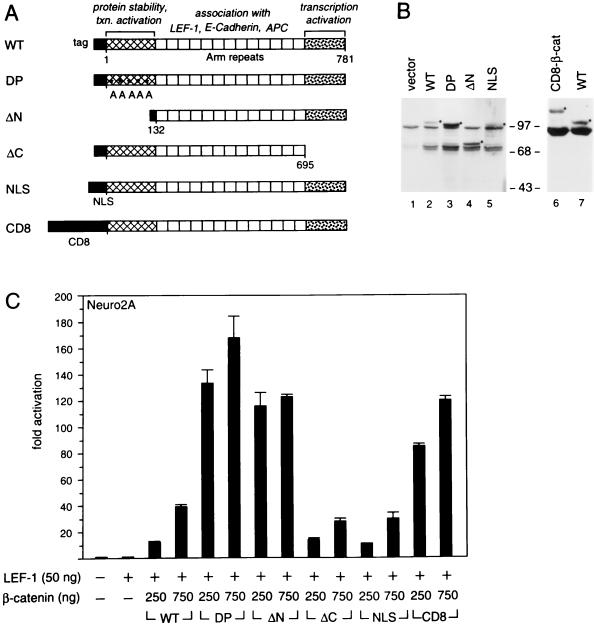
Analysis of transcriptional activation domains in β-catenin. (A) Schematic diagram of the structure of wt and mutant forms of β-catenin with individual domains highlighted. The numbers indicate the amino acid positions in β-catenin. The proteins contain one or two epitope tags at their amino termini. The DP mutant form of β-catenin contains substitutions of Ser/Thr residues in the amino terminus that are phosphorylated and regulate protein stability (74). The amino acid changes, indicated by asterisks, are S33A, S37A, T40A, S45A, and S47A. The mutant nuclear localization signal (NLS) β-catenin protein contains a nuclear localization sequence at the amino terminus. (B) Steady-state protein expression of wt and mutant β-catenins in transfected Cos7 cells. β-Catenin expression plasmids were transiently transfected into Cos7 cells, and cytotosolic (lanes 1 to 5) or whole-cell (lanes 6 and 7) extracts were prepared and analyzed by SDS–7.5% PAGE. A monoclonal antibody against β-catenin was used to detect the level of protein expression. Closed circles indicate the positions of the exogenous β-catenins. The altered electrophoretic mobility of the exogenous β-catenins is likely due to the amino-terminal epitope tags. The molecular size markers are shown in kilodaltons. (C) Transcriptional properties of various forms of β-catenin. Neuro2A cells were transiently cotransfected with a LEF-CAT reporter gene together with expression plasmids encoding LEF-1 and one of the various forms of β-catenin as indicated.
For immunocytochemical analysis of the subcellular localization of CD8–β-catenin and endogenous β-catenin in transfected NIH 3T3 cells expressing LEF-1, we used an anti-CD8, anti-β-catenin, or anti-HA antibody (Fig. 3B). With the anti-CD8 antibody, we detected abundant expression of the fusion protein at the plasma membrane (Fig. 3B, first panel), whereas with the anti-β-catenin antibody, β-catenin was also found in the nucleus of cells expressing LEF-1 (Fig. 3B, second panel). The β-catenin detected in the nucleus of these cells could represent either a proteolytic degradation product of the CD8–β-catenin fusion protein or alternatively endogenous β-catenin. However, nuclear staining was not detected with the anti-HA antibody (third panel). This observation suggests that expression of a membrane-tethered form of β-catenin can result in nuclear localization of endogenous β-catenin in cells expressing LEF-1.
Signaling by Wnt-1 has been shown to increase the free cytosolic pool of β-catenin (49). To examine the accumulation of endogenous β-catenin in response to either Wnt-1 signaling or the expression of the CD8–β-catenin fusion protein, we performed an immunoblot analysis with cytosolic lysates from transfected NIH 3T3 cells. In cells transfected with a Wnt-1 expression plasmid, the level of endogenous cytosolic β-catenin was significantly increased relative to the level observed in cells transfected with vector DNA (Fig. 4A). Notably, a similar increase in the level of cytosolic β-catenin was detected in cells expressing the CD8–β-catenin fusion protein. The molecular mass of β-catenin that accumulated at high levels in cells expressing the fusion protein was identical to that of endogenous β-catenin, suggesting an increase in the pool of free endogenous β-catenin rather than accumulation of a degradation product of the fusion protein. We also determined the amount of total cellular β-catenin and found that it was similar for all lysates examined (Fig. 4B). Thus, the increase in the pool of cytosolic β-catenin correlates with the immunological detection of β-catenin in the nucleus and transcriptional activation by LEF-1. This can occur either naturally in response to Wnt signaling or artificially as a consequence of the presence of a membrane-tethered β-catenin.
β-Catenin contains two transcriptional activation domains.
Mutations in the amino terminus of β-catenin (74) and mutations in the β-catenin-interacting protein APC (41, 59) increase the pool of free β-catenin in the absence of Wnt signaling. To examine whether a constitutive increase in the pool of free β-catenin alters transcriptional activation by LEF-1, we deleted the amino terminus of β-catenin (ΔN) or generated a DP form of β-catenin by mutating serine residues that have been previously shown to affect protein stability (74) (Fig. 5A). Immunoblot analysis of cytosolic extracts from COS cells transfected with wt or mutant β-catenin expression plasmids indicated a higher abundance of the mutant forms of β-catenin than of the wt protein (Fig. 5B). In transfection experiments with Neuro2A cells, the mutant DP and ΔN β-catenins augmented transcription by LEF-1 to a significantly larger extent (up to 10-fold) than did wt β-catenin (Fig. 5C).
Genetic and biochemical data suggested that β-catenin contains sequences that mediate transcriptional activation. In particular, the carboxyl terminus of armadillo, the Drosophila ortholog of β-catenin, has been shown genetically to be involved in signaling by wingless (51). Moreover, the carboxyl-terminal region of β-catenin functions as a transcriptional activation domain in fusion with the GAL4 DNA-binding domain (69). To examine whether the carboxyl terminus of β-catenin is the sole determinant for transcriptional activation in association with LEF-1, we generated the deletion mutant ΔC (Fig. 5A). Expression of this mutant β-catenin with LEF-1 in transfected Neuro2A cells stimulated the activity of the LEF-CAT reporter gene to a level similar to that with the wt β-catenin (Fig. 5C). Since this deletion removed the epitope recognized by the anti-β-catenin antibody, we could not compare the accumulation of the mutant protein to that of the wt protein. Nevertheless, the data suggest that β-catenin may contain additional sequences that contribute to transcriptional activation. Alternatively, overexpression of exogenous forms of β-catenin may result in an increase in the pool of free cytosolic endogenous β-catenin which would obscure the mapping of transcriptional activation domains.
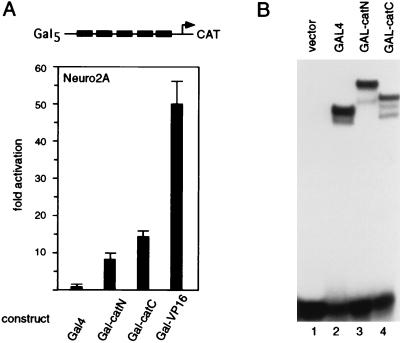
To identify transcriptional activation domains in β-catenin, we generated fusion proteins in which various portions of β-catenin were linked to the GAL4 DNA-binding domain. Expression of GAL4 fusion proteins containing the amino- or carboxyl-terminal region of β-catenin stimulated transcription of a GAL-CAT reporter gene in transfected Neuro2A cells by a factor of 8 or 14, respectively (Fig. 6A). In comparison, a 50-fold activation of the reporter gene was observed with a GAL4-VP16 expression plasmid in which the transcriptional activation domain of the viral protein VP16 is linked to GAL4 as a positive control. We confirmed a similar accumulation of both GAL4–β-catenin fusion proteins by electrophoretic mobility shift assays with nuclear extracts from COS cells transfected with the corresponding expression plasmids (Fig. 6B). Thus, β-catenin contains two distinct transcription activation domains that can function in a heterologous context. However, the levels of transcriptional activation by the GAL4–β-catenin fusion proteins are significantly lower than those observed in cotransfections of Neuro2A cells with LEF-1 and β-catenin, raising the possibility that the transcriptional activation domains of β-catenin collaborate with one another or with LEF-1.
FIG. 6.
Both amino- and carboxyl-terminal regions of β-catenin contain transcriptional activation domains. (A) Amino- and carboxyl-terminal regions of β-catenin were fused in frame to the DNA-binding domain of the yeast GAL4 activator. Neuro2A cells were transiently transfected with a GAL-CAT reporter gene construct together with a plasmid expressing the GAL4 fusion protein as indicated. (B) Analysis of DNA binding by GAL4–β-catenin fusion proteins. Shown are the results of an electrophoretic mobility shift assay of 4 μg of nuclear extracts from Cos7 cells, transiently expressing GAL4–β-catenin fusion proteins, and a probe containing a GAL4 DNA binding site in the presence of 500 ng of salmon sperm DNA and 1 μg of poly(dI-dC).
Functional collaboration between LEF-1 and the activation domains of β-catenin.
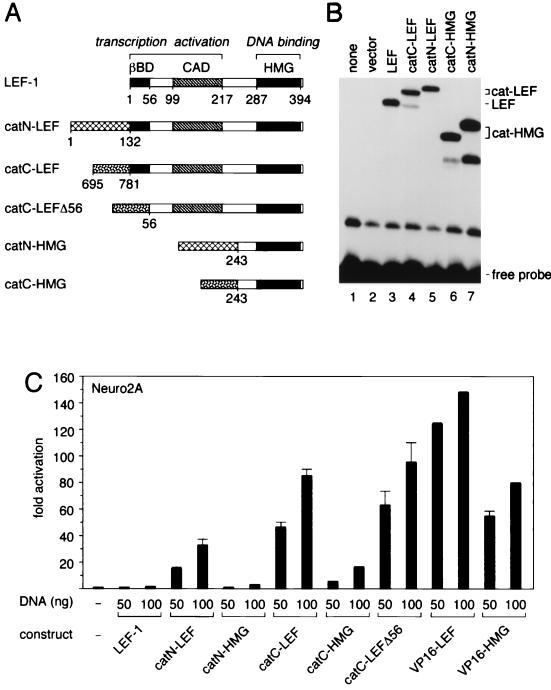
The association of β-catenin with LEF-1 could simply serve to tether the transcriptional activation domains of β-catenin to specific sites in DNA. Alternatively, the transcriptional activation domains of β-catenin could collaborate with other regions in LEF-1 to regulate transcription. To distinguish between possibilities, we linked the amino- or carboxyl-terminal domains of β-catenin to either full-length LEF-1 (catN-LEF and catC-LEF) or the HMG domain of LEF-1, which mediates DNA binding (catN-HMG and catC-HMG [Fig. 7A]). We examined the steady-state accumulation and DNA binding abilities of these fusion proteins by transient transfection of COS cells and electrophoretic mobility assays of nuclear extracts (Fig. 7B). Protein-DNA complexes that migrated with different mobilities corresponding to the relative molecular masses of the fusion proteins were detected. The levels of the complexes containing LEF-1 or the β-catenin–LEF fusion proteins were similar but were slightly lower than those of the complexes containing the β-catenin–HMG fusion proteins. The specificity of DNA binding by these complexes was confirmed by competition experiments with excess unlabeled LEF-1 binding site oligonucleotide (data not shown).
FIG. 7.
Transcriptional activation by β-catenin–LEF-1 fusion proteins. (A) Schematic diagrams of β-catenin–LEF-1 fusion proteins in which amino- or carboxyl-terminal domains of β-catenin were linked to either full-length LEF-1 (catN-LEF and catC-LEF) or the HMG domain of LEF-1 (catN-HMG and catC-HMG). The β-catenin-binding domain (βBD), context-dependent activation domain (CAD), and high-mobility-group domain (HMG) are indicated. The carboxyl-terminal domain of β-catenin was also linked to LEF-1 lacking the amino-terminal 56 residues (catC-LEFΔ56). (B) DNA binding by LEF-1 and β-catenin–LEF-1 fusion proteins. Shown are results of an electrophoretic mobility shift assay with 4 μg of nuclear extracts of Cos7 cells transfected with control vector (lane 2) or expression plasmids encoding LEF-1 (lane 3) or β-catenin–LEF-1 fusion proteins (lanes 4 to 7), catN-LEF (lane 5), catC-HMG (lane 6), and catN-HMG (lane 7). (C) Transcriptional activation by β-catenin–LEF-1 fusion proteins. Neuro2A cells were transiently transfected with a LEF-CAT reporter gene together with expression plasmids encoding the LEF-1 fusion proteins as indicated.
We transfected Neuro2A cells with expression plasmids encoding LEF-1 or the various fusion proteins at two concentrations to ensure a linear range of the transcriptional activation of the LEF-CAT reporter gene. Efficient transcriptional activation by both β-catenin–LEF-1 fusion proteins was detected, although the level of activation by the catC-LEF protein was approximately twofold higher than that by the catN-LEF protein (Fig. 7C). Notably, transcriptional activation by the catN-HMG and catC-HMG proteins was reduced by factors of 9 and 12 relative to the corresponding β-catenin–LEF fusion proteins. In comparison, a fusion protein in which the VP16 activation domain was linked to the HMG domain stimulated the reporter gene at levels only twofold lower than those observed with a VP16–LEF-1 fusion protein. A fusion protein of glutathione _S_-transferase and full-length LEF-1 was used as a negative control and did not result in transcriptional activation (data not shown). To rule out that endogenous β-catenin can interact with the β-catenin–LEF fusion proteins, we also examined transcriptional activation by a chimeric protein in which the carboxyl-terminal domain of β-catenin was linked to the Δ56 LEF-1 protein (catC-LEFΔ56). The level of transcriptional activation was comparable to that observed with the catC-LEF protein. Taken together, these data suggest that the transcriptional activation domains of β-catenin can functionally collaborate with protein domains of DNA-bound LEF-1 to stimulate transcription.
Distinct regulatory properties of LEF-1 in association with β-catenin and ALY.
In response to Wnt signaling and association with β-catenin, LEF-1 can activate transcription from a synthetic enhancer containing multimerized binding sites. In contrast, LEF-1 has been shown to activate the natural TCRα enhancer in a specific context of other transcription factors (8, 14, 68, 72). In the TCRα enhancer, LEF-1 collaborates with the lymphoid-specific proteins AML-1 and Ets-1 and with the ubiquitous protein CREB (15, 32). The potential of LEF-1 to activate the TCRα enhancer is further augmented by the protein ALY, which associates with the context-dependent activation domains of LEF-1 and AML-1 (6). Thus, the question arises as to whether the LEF-1-interacting proteins ALY and β-catenin play overlapping or distinct roles in transcriptional activation and Wnt signaling.
To confirm that the transcriptional activation by LEF-1 in response to Wnt signaling is dependent on interaction with endogenous β-catenin, we expressed the wt or the m1 mutant LEF-1 protein, alone or in combination with Wnt-1 (Fig. 8A). In transfected NIH 3T3 cells, the m1 mutation, which impairs association of LEF-1 with β-catenin in vitro (Fig. 2), abrogated transcriptional activation by LEF-1 in response to Wnt signaling. We also examined whether ALY, by itself or in combination with β-catenin, could modulate the function of LEF-1. Using a LEF-CAT reporter gene in transfected Neuro2A cells, we found that ALY neither stimulated transcription by LEF-1 alone nor altered its activation potential in combination with β-catenin (Fig. 8B).
FIG. 8.
LEF-1-interacting proteins confer distinct regulatory properties upon LEF-1. (A) Requirement of the β-catenin interaction domain in LEF-1 for transcriptional activation in response to Wnt signaling. NIH 3T3 cells were transiently cotransfected with a LEF-CAT reporter gene (0.8 μg) and a wt or mutant (m1) LEF-1 expression plasmid (450 ng), alone or together with a cytomegalovirus–Wnt-1 expression plasmid (1.5 μg). (B) LEF-1 in collaboration with ALY fails to activate the LEF-CAT reporter gene. Neuro2A cells were transiently transfected with the LEF-CAT reporter gene and 50 ng of LEF-1 expression plasmid, alone or together with ALY or β-catenin expression vectors as indicated. (C) The β-catenin interaction domain of LEF-1 is dispensible for the activation of the TCRα enhancer. HeLa cells were transiently transfected with a TCRα-CAT reporter gene (250 ng), expression plasmids for AML-1 and Ets-1 (200 ng each), and a wt or mutant (m1) LEF-1 expression plasmid (100 ng). (D) β-Catenin has no effect on TCRα enhancer function. A TCRα-CAT reporter gene was cotransfected with expression plasmids for AML-1, Ets-1, and LEF-1, as described for panel C, in the absence or presence of expression plasmids encoding ALY or β-catenin.
We also examined the effects of the m1 mutation in LEF-1 on the regulation of the TCRα enhancer. The function of the TCRα enhancer in HeLa cells was reconstituted by transfecting expression plasmids encoding AML-1, Ets-1, and LEF-1. Expression of wt LEF-1 protein augmented the activity of the enhancer to a similar extent as did expression of mutant LEF-1 protein (Fig. 8C). We further compared the regulation of the TCRα enhancer by LEF-1 in association with ALY with that by LEF-1 in association with β-catenin. As anticipated from previous experiments (6), LEF-1, in combination with ALY, augmented the activity of the enhancer by a factor of 14 (Fig. 8D). This transcriptional activation was not significantly changed by using the mutant m1 LEF-1 protein or by coexpression of β-catenin. In contrast, a fourfold stimulation of the TCRα enhancer was observed with LEF-1 alone or in combination with β-catenin. Notably, ALY and β-catenin appear to act independently to modify the function of LEF-1, and both proteins interacted with LEF-1 independently of each other in an in vitro association assay (data not shown). Taken together, these data suggest that LEF-1 can exert at least two distinct regulatory roles. In association with β-catenin, LEF-1 can mediate transcriptional activation in a context-independent manner, whereas in association with ALY, LEF-1 regulates transcription only in a particular context of other proteins.
DISCUSSION
In this study, we examine the transcriptional regulation by LEF-1 in response to a Wnt-1 signal under conditions of normal endogenous β-catenin expression. We find that that Wnt signaling augments transcriptional activation by LEF-1 in association with β-catenin and show that a Wnt signal is necessary but not sufficient for the nuclear localization of endogenous β-catenin. We have identified amino- and carboxyl-terminal activation domains in β-catenin whose function is specifically augmented by juxtaposition with LEF-1 but not with the DNA-binding domain of LEF-1, suggesting a reciprocal interaction between these proteins. Finally, we show that association of LEF-1 with different proteins, β-catenin and ALY, confers distinct regulatory properties upon LEF-1.
In the NIH 3T3 tissue culture model, we find that endogenous β-catenin is localized in the nucleus only in cells that both express LEF-1 and receive a Wnt-1 signal, or in cells in which an artificial membrane-tethered form of β-catenin is present. However, overexpressed β-catenin is localized to the nucleus of Neuro 2A or NIH 3T3 cells that express LEF-1, independent of a Wnt signal (2, 22; also data not shown). Thus, expression of LEF-1 may result only in nuclear localization of free β-catenin which is generated either by activation of the Wnt signaling pathway or by overexpression of β-catenin. Likewise, overexpression of Wnt-1 in a mammary epithelial cell line leads to the formation of a nuclear complex between β-catenin and TCF-4 protein (27). None of these data, however, bear on the mechanism by which β-catenin is translocated into the nucleus. β-Catenin could either associate with newly synthesized LEF-1 protein in the cytoplasm, resulting in nuclear translocation via LEF-1, or alternatively be translocated by other means into the nucleus and accumulate through association with LEF-1. Recently, β-catenin has been shown to be imported into the nucleus by binding directly to the nuclear pore complex, similar to importin β-like factors, suggesting that β-catenin localizes to the nucleus independently of interaction with LEF-1/TCF proteins (12).
The stimulation of transcription by LEF-1 in collaboration with a membrane-tethered form of β-catenin raises the question of the mechanism of stimulation of transcription by LEF-1. In cells expressing both the CD8–β-catenin fusion protein and LEF-1, we detected a significant amount of nuclear β-catenin that has the molecular mass of endogenous β-catenin. Thus, overexpression of a membrane-tethered β-catenin may competitively liberate endogenous β-catenin from either E-cadherin or APC. Consistent with this interpretation, expression of a membrane-tethered form of β-catenin in Xenopus embryos increases the pool of free β-catenin that is associated with APC rather than with E-cadherin (37). This interpretation may also account for the observation that a membrane-tethered form of plakoglobin induces neural axis duplication in Xenopus embryos (35).
Nuclear β-catenin has also been detected under conditions of overexpression in vivo. For example, expression of a dominant negative form of GSK3 in Xenopus embryos results in nuclear localization of β-catenin on the dorsal side of the embryo (74). Likewise, TCF–β-catenin or LEF-1–β-catenin complexes have been identified in nuclear extracts from mammalian cell lines that either are defective in APC or carry mutations in the amino terminus of β-catenin, conditions which both result in accumulation of endogenous free β-catenin (41, 54, 59). In addition, a small group of cells in the dorsalizing center of normal Xenopus embryos contains nuclear β-catenin (61). However, to date, immunocytochemical analysis has failed to detect nuclear β-catenin in mouse embryos, even in cells known to express LEF-1 or in tissues that show a developmental defect in LEF-1-deficient mice, for example, tooth germs and hair follicles (12a, 28, 71). Possibly, the techniques used are not sensitive enough to detect small amounts of nuclear β-catenin that may suffice for a functional response in normal cells. Alternatively, only a small subset of yet-unidentified LEF-1-expressing cells may contain nuclear β-catenin.
Our functional analysis of point mutations in the β-catenin interaction domain of LEF-1 indicates that the transcriptional response to Wnt signaling is dependent upon association of LEF-1 with β-catenin. Moreover, the ability of LEF-1 to interact with recombinant β-catenin in vitro correlates with transcriptional activation by LEF-1 in response to a Wnt-1 signal in transfected NIH 3T3 cells. The mechanism of transcriptional activation by LEF-1–β-catenin complexes is still obscure. Consistent with previous reports (47, 54, 65), we identified a domain in the carboxyl-terminal region of β-catenin that mediates transcriptional activation when fused to the GAL4 or LEF-1 DNA-binding domain. In addition, we show that the amino-terminal 131 residues of β-catenin, which regulate protein stability (41, 43, 59, 74), can confer transcriptional activation upon heterologous DNA-binding domains. Unlike the prototypic transcriptional activation domain of VP16, the function of both activation domains of β-catenin is significantly augmented by fusion with full-length LEF-1 compared to fusions with the DNA-binding domain of LEF-1 or GAL4. This observation suggests that the transcriptional activation domains of β-catenin functionally collaborate with specific domains in LEF-1. A transcriptional activation domain which interacts with ALY and activates transcription only in a specific context of other DNA-binding proteins has been identified in a region between amino acids 99 and 256 of LEF-1 (6, 8, 14). However, the β-catenin–LEF fusion protein does not show a dependence on a specific context of other transcription factors, and therefore, sequences in LEF-1 that are distinct from this context-dependent activation domain may collaborate with β-catenin.
In principle, the activation domains of β-catenin, when tethered to LEF-1 by protein-protein interaction or by covalent protein fusion, could augment transcription through direct interactions with components of the basal transcription machinery. According to this view, the association of β-catenin with LEF-1 would generate a composite transcriptional activation surface. Alternatively, the activation domains in β-catenin could stimulate transcription indirectly by bringing LEF-1 to a particular compartment of the nucleus or by recruiting enzymes that posttranslationally modify the complex with LEF-1. Finally, β-catenin could mediate interaction with other transcription factors, although we consider this scheme unlikely because LEF-1–β-catenin complexes can act at synthetic enhancers containing only multimerized LEF-1 binding sites.
Are all effects of Wnt signaling mediated by LEF-1 and TCF proteins? In Drosophila, mutations in the pangolin/dTCF gene suppress the phenotype of constitutively activated armadillo (7, 69). This observation has been interpreted to suggest that all signaling by wingless is mediated by the action of this transcription factor. However, signaling by Wnt proteins involves diverse physiological responses that include inductive and mitogenic events as well as changes in cell adhesion and mitotic spindle orientation (10, 17, 21, 45, 57, 67). Wnt-1 signaling in mammalian cells can induce cell growth and transcriptional activation by LEF-1 and TCF proteins (27, 54, 75; also this study). Likewise, mutations in APC or β-catenin in colon carcinomas and melanomas correlate with constitutively active TCF-4–β-catenin complexes (26, 41, 59). However some responses to Wnt signaling may be independent of LEF/TCF–β-catenin complexes. For example, overexpression of β-catenin in Xenopus embryos can increase cell-cell adhesion in the absence of zygotic transcription (17). Moreover, a mutant form of β-catenin (S37A), which can mediate transcriptional activation of a LEF/TCF reporter gene in Rat-1 cells, fails to elicit the proliferative response that is normally observed in cells expressing wt β-catenin or Wnt-1 (75). Finally, Wnt signaling in C. elegans can induce changes in mitotic spindle orientation and cytoskeletal polarity that are independent of the β-catenin-like protein WRM-1 (57, 67).
Different Wnt proteins may also elicit distinct physiological responses. Cellular transformation assays in epithelial cells and axis duplication assays in Xenopus suggest that the 16 currently known Wnt genes can be classified into at least two groups (reviewed in reference 39). Xenopus Wnt-1, XWnt-8, and other Wnt proteins can induce a secondary axis, whereas XWnt-5a and related Wnt proteins decrease cell-cell adhesion but fail to induce a secondary axis (10, 40). Although our data indicate that signaling by Wnt-1 results in transcriptional activation by LEF-1 in association with β-catenin, we cannot extrapolate to a general function of LEF-1/TCF protein in signaling by other Wnt proteins.
The association of β-catenin with LEF-1 also raises the question of whether this protein-protein interaction is obligatory for transcriptional activation by LEF-1. Our study indicates that Wnt signaling may not be required for all functions of LEF-1. The TCRα gene is a target for regulation by LEF-1 and TCF-1 in vivo because experimentally induced mutations in both transcription factor genes in the mouse result in a severe defect in endogenous TCRα gene expression in purified immature CD8 single positive T cells (46). However, we found that regulation of the TCRα enhancer by LEF-1, which is augmented by ALY, is independent of association with β-catenin because mutations in the β-catenin interaction domain of LEF-1 have no detectable effect on transactivation. In contrast, LEF-1 can stimulate the activity of a synthetic enhancer containing multimerized LEF-1 binding sites in association with β-catenin, but not with ALY. Thus, LEF-1 can assume different regulatory functions in association with β-catenin and ALY. In association with ALY and in the context of the TCRα enhancer, LEF-1 appears to play an architectural role in the assembly and function of a higher-order nucleoprotein complex in which DNA bending and a context-dependent activation domain facilitate interactions between multiple DNA-binding proteins (6, 8, 13–15, 32). In association with β-catenin, LEF-1 can activate transcription independent of a specific context of other DNA-binding proteins.
Multiple target genes in which LEF-1/TCF proteins function in collaboration with β-catenin or armadillo have been recently identified. The Ubx enhancer in Drosophila contains a functionally important binding site for LEF/TCF proteins, and expression of LEF-1 in transgenic embryos regulates this enhancer in an armadillo-dependent manner (56). Moreover, the regulation of the Ubx gene is impaired in pangolin/dTCF-deficient embryos (69). Recently, Wnt-responsive sequence elements in the siamois and Xnr3 promoters of Xenopus have been found to be regulated by LEF-1/XTCF-3 proteins in embryo injection assays (5, 34). Notably, the regulation of the wingless/Wnt response in these systems is also dependent upon a particular context of other transcription factor binding sites. The regulation of the Ubx enhancer in response to wingless signaling requires a juxtaposition of the LEF/TCF binding site with a response element for decapentaplegic signaling (56). Likewise, the Wnt responsiveness of the Xnr3 promoter is dependent upon both a LEF/TCF binding site and a binding site for a putative homeodomain protein (34). Thus, the regulation of Wnt-responsive target genes by LEF/TCF proteins in vivo may require a specific context of transcription factor binding sites. Wnt signaling has the interesting property of collaborating with other signaling pathways such as sonic hedgehog, fibroblast growth factor, activins, and others (39). The ability of LEF-1 to function as an architectural protein that stimulates transcription only in collaboration with other proteins may contribute to the integration of signaling pathways and allow for diversity in the transcriptional response.
ACKNOWLEDGMENTS
We thank W. Birchmeier for providing human β-catenin cDNA and Neuro2A cells, and we are grateful to Jackie Papkoff for providing cytomegalovirus–Wnt-1. We also thank Jan Kitajewski for communicating results prior to publication and Danesh Moazed, Tannishtha Reya, and Mary O’Riordan for their helpful comments on the manuscript. We thank members of the Grosschedl laboratory for valuable discussions.
This work was supported by the Howard Hughes Medical Institute. Shu-Chi Hsu was supported by a postdoctoral fellowship from the Leukemia Society of America.
REFERENCES
- 1.Aberle H, Schwartz H, Kemler R. Cadherin-catenin complex: protein interactions and their implications for cadherin function. J Cell Biochem. 1996;61:514–523. doi: 10.1002/(SICI)1097-4644(19960616)61:4%3C514::AID-JCB4%3E3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 2.Behrens J, von Kries J, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Functional interaction of β-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 3.Berkowitz L A, Riabowol K T, Gilman M Z. Multiple sequence elements of a single functional class are required for cyclic AMP responsiveness of the mouse c-fos promoter. Mol Cell Biol. 1989;9:4272–4281. doi: 10.1128/mcb.9.10.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhanot P, Brink M, Samos C H, Hsieh J C, Wang Y, Macke J P, Andrew D, Nathans J, Nusse R. A new member of the frizzled family from Drosophila functions as a Wingless receptor. Nature. 1996;382:225–230. doi: 10.1038/382225a0. [DOI] [PubMed] [Google Scholar]
- 5.Brannon M, Gomperts M, Sumoy L, Moon R T, Kimelman D. A beta-catenin/XTcf-3 complex binds to the siamois promoter to regulate dorsal axis specification in Xenopus. Genes Dev. 1997;11:2359–2370. doi: 10.1101/gad.11.18.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruhn L, Munnerlyn A, Grosschedl R. ALY, a context-dependent coactivator of LEF-1 and AML-1, is required for TCRalpha enhancer function. Genes Dev. 1997;11:640–653. doi: 10.1101/gad.11.5.640. [DOI] [PubMed] [Google Scholar]
- 7.Brunner E, Peter O, Schweizer L, Basler K. pangolin encodes a Lef-1 homologue that acts downstream of Armadillo to transduce the Wingless signal in Drosophila. Nature. 1997;385:829–833. doi: 10.1038/385829a0. [DOI] [PubMed] [Google Scholar]
- 8.Carlsson P, Waterman M L, Jones K A. The hLEF/TCF-1 alpha HMG protein contains a context-dependent transcriptional activation domain that induces the TCR alpha enhancer in T cells. Genes Dev. 1993;7:2418–2430. doi: 10.1101/gad.7.12a.2418. [DOI] [PubMed] [Google Scholar]
- 9.Castrop J, Hoevenagel R, Young J R, Clevers H C. A common ancestor of the mammalian transcription factors TCF-1 and TCF-1 alpha/LEF-1 expressed in chicken T cells. Eur J Immunol. 1992;22:1327–1330. doi: 10.1002/eji.1830220531. [DOI] [PubMed] [Google Scholar]
- 10.Du S J, Purcell S M, Christian J L, McGrew L L, Moon R T. Identification of distinct classes and functional domains of Wnts through expression of wild-type and chimeric proteins in Xenopus embryos. Mol Cell Biol. 1995;15:2625–2634. doi: 10.1128/mcb.15.5.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fagotto F, Funayama N, Gluck U, Gumbiner B M. Binding to cadherins antagonizes the signaling activity of beta-catenin during axis formation in Xenopus. J Cell Biol. 1996;132:1105–1114. doi: 10.1083/jcb.132.6.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fagotto F, Gluck U, Gumbiner B M. Nuclear localization signal-independent and importin/karyopherin-independent nuclear import of β-catenin. Curr Biol. 1998;8:181–190. doi: 10.1016/s0960-9822(98)70082-x. [DOI] [PubMed] [Google Scholar]
- 12a.Farinas, I., J. Galceran, and R. Grosschedl. Unpublished data.
- 13.Giese K, Cox J, Grosschedl R. The HMG domain of lymphoid enhancer factor 1 bends DNA and facilitates assembly of functional nucleoprotein structures. Cell. 1992;69:185–195. doi: 10.1016/0092-8674(92)90129-z. [DOI] [PubMed] [Google Scholar]
- 14.Giese K, Grosschedl R. LEF-1 contains an activation domain that stimulates transcription only in a specific context of factor-binding sites. EMBO J. 1993;12:4667–4676. doi: 10.1002/j.1460-2075.1993.tb06155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giese K, Kingsley C, Kirshner J R, Grosschedl R. Assembly and function of a TCR alpha enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein-protein interactions. Genes Dev. 1995;9:995–1008. doi: 10.1101/gad.9.8.995. [DOI] [PubMed] [Google Scholar]
- 16.Grosschedl R, Baltimore D. Cell-type specificity of immunoglobulin gene expression is regulated by at least three DNA sequence elements. Cell. 1985;41:885–897. doi: 10.1016/s0092-8674(85)80069-6. [DOI] [PubMed] [Google Scholar]
- 17.Guger K A, Gumbiner B M. Beta-catenin has Wnt-like activity and mimics the Nieuwkoop signaling center in Xenopus dorsal-ventral patterning. Dev Biol. 1995;172:115–125. doi: 10.1006/dbio.1995.0009. [DOI] [PubMed] [Google Scholar]
- 18.Gumbiner B M. Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell. 1996;84:345–357. doi: 10.1016/s0092-8674(00)81279-9. [DOI] [PubMed] [Google Scholar]
- 19.Han M. Gut reaction to Wnt signaling in worms. Cell. 1997;90:581–584. doi: 10.1016/s0092-8674(00)80517-6. [DOI] [PubMed] [Google Scholar]
- 20.He X, Saint-Jeannet J, Woodgett J, Varmus H, David I. Glycogen synthase kinase-3 and dorsoventral patterning in Xenopus embryos. Nature. 1995;374:617–622. doi: 10.1038/374617a0. [DOI] [PubMed] [Google Scholar]
- 21.Herzlinger D, Qiao J, Cohen D, Ramakrishna N, Brown A M. Induction of kidney epithelial morphogenesis by cells expressing Wnt-1. Dev Biol. 1994;166:815–818. doi: 10.1006/dbio.1994.1360. [DOI] [PubMed] [Google Scholar]
- 22.Huber O, Korn R, McLaughlin J, Ohsugi M, Herrmann B G, Kemler R. Nuclear localization of beta-catenin by interaction with transcription factor LEF-1. Mech Dev. 1996;59:3–10. doi: 10.1016/0925-4773(96)00597-7. [DOI] [PubMed] [Google Scholar]
- 23.Jou T S, Stewart D B, Stappert J, Nelson W J, Marrs J A. Genetic and biochemical dissection of protein linkages in the cadherin-catenin complex. Proc Natl Acad Sci USA. 1995;92:5067–5071. doi: 10.1073/pnas.92.11.5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kemler R. From cadherins to catenins: cytoplasmic protein interactions and regulation of cell adhesion. Trends Genet. 1993;9:317–321. doi: 10.1016/0168-9525(93)90250-l. [DOI] [PubMed] [Google Scholar]
- 25.Klingensmith J, Nusse R, Perrimon N. The Drosophila segment polarity gene dishevelled encodes a novel protein required for response to the wingless signal. Genes Dev. 1994;8:118–130. doi: 10.1101/gad.8.1.118. [DOI] [PubMed] [Google Scholar]
- 26.Korinek V, Barker N, Morin P J, van Wichen D, de Weger R, Kinzler K W, Vogelstein B, Clevers H. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 27.Korinek V, Barker N, Willert K, Molenaar M, Roose J, Wagenaar G, Markman M, Lamers W, Destree O, Clevers H. Two members of the Tcf family implicated in Wnt/β-catenin signaling during embryogenesis in the mouse. Mol Cell Biol. 1998;18:1248–1256. doi: 10.1128/mcb.18.3.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kratochwil K, Dull M, Farinas I, Galceran J, Grosschedl R. Lef1 expression is activated by BMP-4 and regulates inductive tissue interactions in tooth and hair development. Genes Dev. 1996;10:1382–1394. doi: 10.1101/gad.10.11.1382. [DOI] [PubMed] [Google Scholar]
- 29.Lin R, Thompson S, Priess J R. pop-1 encodes an HMG box protein required for the specification of a mesoderm precursor in early C. elegans embryos. Cell. 1995;83:599–609. doi: 10.1016/0092-8674(95)90100-0. [DOI] [PubMed] [Google Scholar]
- 30.Littman D R, Thomas Y, Maddon P J, Chess L, Axel R. The isolation and sequence of the gene encoding T8: a molecule defining functional classes of T lymphocytes. Cell. 1985;40:237–246. doi: 10.1016/0092-8674(85)90138-2. [DOI] [PubMed] [Google Scholar]
- 31.Love J J, Li X, Case D A, Giese K, Grosschedl R, Wright P E. Structural basis for DNA bending by the architectural transcription factor LEF-1. Nature. 1995;376:791–795. doi: 10.1038/376791a0. [DOI] [PubMed] [Google Scholar]
- 32.Mayall T P, Sheridan P L, Montminy M R, Jones K A. Distinct roles for P-CREB and LEF-1 in TCR alpha enhancer assembly and activation on chromatin templates in vitro. Genes Dev. 1997;11:887–899. doi: 10.1101/gad.11.7.887. [DOI] [PubMed] [Google Scholar]
- 33.McCrea P, Turck C, Gumbiner B. A homolog of the armadillo protein in Drosophila (plakoglobin) associated with E-cadherin. Science. 1991;254:1359–1361. doi: 10.1126/science.1962194. [DOI] [PubMed] [Google Scholar]
- 34.Mckendry R, Hsu S-C, Harland R M, Grosschedl R. LEF-1/TCF proteins mediate wnt-inducible transcription from the Xenopus nodal-related 3 promoter. Dev Biol. 1997;192:420–431. doi: 10.1006/dbio.1997.8797. [DOI] [PubMed] [Google Scholar]
- 35.Merriam J M, Rubenstein A B, Klymkowsky M W. Cytoplasmically anchored plakoglobin induces a WNT-like phenotype in Xenopus. Dev Biol. 1997;185:67–81. doi: 10.1006/dbio.1997.8550. [DOI] [PubMed] [Google Scholar]
- 36.Miller J R, Moon R T. Signal transduction through beta-catenin and specification of cell fate during embryogenesis. Genes Dev. 1996;10:2527–2539. doi: 10.1101/gad.10.20.2527. [DOI] [PubMed] [Google Scholar]
- 37.Miller J R, Moon R T. Analysis of the signaling activities of localization mutants of β-catenin during axis specification in Xenopus. J Cell Biol. 1997;139:229–243. doi: 10.1083/jcb.139.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destree O, Clevers H. XTcf-3 transcription factor mediates β-catenin-induced axis formation in Xenopus embryos. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- 39.Moon R T, Brown J D, Torres M. WNTs modulate cell fate and behavior during vertebrate development. Trends Genet. 1997;13:157–162. doi: 10.1016/s0168-9525(97)01093-7. [DOI] [PubMed] [Google Scholar]
- 40.Moon R T, Campbell R M, Christian J L, McGrew L L, Shih J, Fraser S. Xwnt-5A: a maternal Wnt that affects morphogenetic movements after overexpression in embryos of Xenopus laevis. Development. 1993;119:97–111. doi: 10.1242/dev.119.1.97. [DOI] [PubMed] [Google Scholar]
- 41.Morin P J, Sparks A B, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler K W. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 42.Munemitsu S, Albert I, Souza B, Rubinfeld B, Polakis P. Regulation of intracellular beta-catenin levels by the adenomatous polyposis coli (APC) tumor-suppressor protein. Proc Natl Acad Sci USA. 1995;92:3046–3050. doi: 10.1073/pnas.92.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Munemitsu S, Albert I, Rubinfeld B, Polakis P. Deletion of an amino-terminal sequence stabilizes β-catenin in vivo and promotes hyperphosphorylation of the adenomatous polyposis coli tumor suppressor protein. Mol Cell Biol. 1996;16:4088–4094. doi: 10.1128/mcb.16.8.4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nusse R. A versatile transcriptional effector of Wingless signaling. Cell. 1997;89:321–323. doi: 10.1016/s0092-8674(00)80210-x. [DOI] [PubMed] [Google Scholar]
- 45.Nusse R, Varmus H E. Wnt genes. Cell. 1982;69:1073–1087. doi: 10.1016/0092-8674(92)90630-u. [DOI] [PubMed] [Google Scholar]
- 46.Okamura R M, Sigvardsson M, Galceran J, Verbeek S, Clevers H, Grosschedl R. Redundant regulation of T cell differentiation and TCR alpha gene expression by the transcription factors LEF-1 and TCF-1. Immunity. 1997;8:11–20. doi: 10.1016/s1074-7613(00)80454-9. [DOI] [PubMed] [Google Scholar]
- 47.Oosterwegel M, van de Wetering M, Timmerman J, Kruisbeek A, Destree O, Meijlink F, Clevers H. Differential expression of the HMG box factors TCF-1 and LEF-1 during murine embryogenesis. Development. 1993;118:439–448. doi: 10.1242/dev.118.2.439. [DOI] [PubMed] [Google Scholar]
- 48.Orsulic S, Peifer M. An in vivo structure-function study of armadillo, the beta-catenin homologue, reveals both separate and overlapping regions of the protein required for cell adhesion and for wingless signaling. J Cell Biol. 1996;134:1283–1300. doi: 10.1083/jcb.134.5.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Papkoff J, Rubinfeld B, Schryver B, Polakis P. Wnt-1 regulates free pools of catenins and stabilizes APC-catenin complexes. Mol Cell Biol. 1996;16:2128–2134. doi: 10.1128/mcb.16.5.2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parr B A, McMahon A P. Wnt genes and vertebrate development. Curr Opin Genet Dev. 1994;4:523–528. doi: 10.1016/0959-437x(94)90067-d. [DOI] [PubMed] [Google Scholar]
- 51.Peifer M, Rauskolb C, Williams M, Riggleman B, Wieschaus E. The segment polarity gene armadillo interacts with the wingless signaling pathway in both embryonic and adult pattern formation. Development. 1991;111:1029–1043. doi: 10.1242/dev.111.4.1029. [DOI] [PubMed] [Google Scholar]
- 52.Peifer M, Pai L M, Casey M. Phosphorylation of the Drosophila adherens junction protein Armadillo: roles for wingless signal and zeste-white 3 kinase. Dev Biol. 1994;166:543–556. doi: 10.1006/dbio.1994.1336. [DOI] [PubMed] [Google Scholar]
- 53.Peifer M, Sweeton D, Casey M, Wieschaus E. Wingless signal and Zeste-white 3 kinase trigger opposing changes in the intracellular distribution of Armadillo. Development. 1994;120:369–380. doi: 10.1242/dev.120.2.369. [DOI] [PubMed] [Google Scholar]
- 54.Porfiri E, Rubinfeld B, Albert I, Hovanes K, Waterman M, Polakis P. Induction of a β-catenin-LEF-1 complex by wnt-1 transforming mutants of β-catenin. Oncogene. 1997;15:2833–2839. doi: 10.1038/sj.onc.1201462. [DOI] [PubMed] [Google Scholar]
- 55.Prieve M G, Guttridge K L, Munguia J E, Waterman M L. The nuclear localization signal of lymphoid enhancer factor-1 is recognized by two differentially expressed Srp1-nuclear localization sequence receptor proteins. J Biol Chem. 1996;271:7654–7658. doi: 10.1074/jbc.271.13.7654. [DOI] [PubMed] [Google Scholar]
- 56.Riese J, Yu X, Munnerlyn A, Eresh S, Hsu S C, Grosschedl R, Bienz M. LEF-1, a nuclear factor coordinating signaling inputs from wingless and decapentaplegic. Cell. 1997;88:777–787. doi: 10.1016/s0092-8674(00)81924-8. [DOI] [PubMed] [Google Scholar]
- 57.Rocheleau C E, Downs W D, Lin R, Wittmann C, Bei Y, Cha Y-H, Ali M, Pries J R, Mello C C. Wnt signaling and an APC-related gene specify endoderm in early C. elegans embryos. Cell. 1997;90:707–716. doi: 10.1016/s0092-8674(00)80531-0. [DOI] [PubMed] [Google Scholar]
- 58.Rubinfeld B, Albert I, Porfiri E, Fiol C, Munemitsu S, Polakis P. Binding of GSK3beta to the APC-beta-catenin complex and regulation of complex assembly. Science. 1996;272:1023–1026. doi: 10.1126/science.272.5264.1023. [DOI] [PubMed] [Google Scholar]
- 59.Rubinfeld B, Robbins P, El-Gamil M, Albert I, Porfiri E, Polakis P. Stabilization of beta-catenin by genetic defects in melanoma cell lines. Science. 1997;275:1790–1792. doi: 10.1126/science.275.5307.1790. [DOI] [PubMed] [Google Scholar]
- 60.Sanson B, White P, Vincent J P. Uncoupling cadherin-based adhesion from wingless signalling in Drosophila. Nature. 1996;383:627–630. doi: 10.1038/383627a0. [DOI] [PubMed] [Google Scholar]
- 61.Schneider S, Steinbeisser H, Warga R M, Hausen P. Beta-catenin translocation into nuclei demarcates the dorsalizing centers in frog and fish embryos. Mech Dev. 1996;57:191–198. doi: 10.1016/0925-4773(96)00546-1. [DOI] [PubMed] [Google Scholar]
- 62.Schreiber E, Matthias P, Muller M M, Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Siegfried E, Chou T B, Perrimon N. Wingless signaling acts through zeste-white 3, the Drosophila homolog of glycogen synthase kinase-3, to regulate engrailed and establish cell fate. Cell. 1992;71:1167–1179. doi: 10.1016/s0092-8674(05)80065-0. [DOI] [PubMed] [Google Scholar]
- 64.Siegfried E, Perrimon N. Drosophila wingless: a paradigm for the function and mechanism of Wnt signaling. Bioessays. 1994;16:395–404. doi: 10.1002/bies.950160607. [DOI] [PubMed] [Google Scholar]
- 65.Sokol S, Klingensmith J, Perrimon N, Itoh K. Dorsalizing and neuralizing properties of XDsh, a maternally expressed homolog of dishevelled. Development. 1995;121:3487–3497. doi: 10.1242/dev.121.10.3487. [DOI] [PubMed] [Google Scholar]
- 66.Starr D B, Matsui W, Thomas J R, Yamamoto K R. Intracellular receptors use a common mechanism to interpret signaling information at response elements. Genes Dev. 1996;10:1271–1283. doi: 10.1101/gad.10.10.1271. [DOI] [PubMed] [Google Scholar]
- 67.Thorpe C J, Schlesinger A, Carter J C, Bowerman B. Wnt signaling polarizes an early C. elegans blastomere to distinguish endoderm from mesoderm. Cell. 1997;90:695–705. doi: 10.1016/s0092-8674(00)80530-9. [DOI] [PubMed] [Google Scholar]
- 68.Travis A, Amsterdam A, Belanger C, Grosschedl R. LEF-1, a gene encoding a lymphoid-specific protein with an HMG domain, regulates T-cell receptor alpha enhancer function. Genes Dev. 1991;5:880–894. doi: 10.1101/gad.5.5.880. [DOI] [PubMed] [Google Scholar]
- 69.van de Wetering M, Cavallo R, Dooijes D, van Beest M, van Es J, Loureiro J, Ypma A, Hursh D, Jones T, Bejsovec A, Peifer M, Mortin M, Clevers H. Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell. 1997;88:789–799. doi: 10.1016/s0092-8674(00)81925-x. [DOI] [PubMed] [Google Scholar]
- 70.van de Wetering M, Oosterwegel M, Dooijes D, Clevers H. Identification and cloning of TCF-1, a T lymphocyte-specific transcription factor containing a sequence-specific HMG box. EMBO J. 1991;10:123–132. doi: 10.1002/j.1460-2075.1991.tb07928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van Genderen C, Okamura R M, Farinas I, Quo R G, Parslow T G, Bruhn L, Grosschedl R. Development of several organs that require inductive epithelial-mesenchymal interactions is impaired in LEF-1-deficient mice. Genes Dev. 1994;8:2691–2703. doi: 10.1101/gad.8.22.2691. [DOI] [PubMed] [Google Scholar]
- 72.Waterman M L, Fischer W H, Jones K A. A thymus-specific member of the HMG protein family regulates the human T cell receptor C alpha enhancer. Genes Dev. 1991;5:656–669. doi: 10.1101/gad.5.4.656. [DOI] [PubMed] [Google Scholar]
- 73.Yang-Snyder J, Miller J R, Brown J D, Lai C J, Moon R T. A frizzled homolog functions in a vertebrate Wnt signaling pathway. Curr Biol. 1996;6:1302–1306. doi: 10.1016/s0960-9822(02)70716-1. [DOI] [PubMed] [Google Scholar]
- 74.Yost C, Torres M, Miller J R, Huang E, Kimelman D, Moon R T. The axis-inducing activity, stability, and subcellular distribution of beta-catenin is regulated in Xenopus embryos by glycogen synthase kinase 3. Genes Dev. 1996;10:1443–1454. doi: 10.1101/gad.10.12.1443. [DOI] [PubMed] [Google Scholar]
- 75.Young C S, Kitamura M, Hardy S, Kitajewski J. Wnt-1 induces growth, cytosolic β-catenin, and Tcf/Lef transcriptional activation in Rat-1 fibroblasts. Mol Cell Biol. 1998;18:2474–2485. doi: 10.1128/mcb.18.5.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]