Incorporation of the Green Fluorescent Protein into the Herpes Simplex Virus Type 1 Capsid (original) (raw)
Abstract
The herpes simplex virus type 1 (HSV-1) UL35 open reading frame (ORF) encodes a 12-kDa capsid protein designated VP26. VP26 is located on the outer surface of the capsid specifically on the tips of the hexons that constitute the capsid shell. The bioluminescent jellyfish (Aequorea victoria) green fluorescent protein (GFP) was fused in frame with the UL35 ORF to generate a VP26-GFP fusion protein. This fusion protein was fluorescent and localized to distinct regions within the nuclei of transfected cells following infection with wild-type virus. The VP26-GFP marker was introduced into the HSV-1 (KOS) genome resulting in recombinant plaques that were fluorescent. A virus, designated K26GFP, was isolated and purified and was shown to grow as well as the wild-type virus in cell culture. An analysis of the intranuclear capsids formed in K26GFP-infected cells revealed that the fusion protein was incorporated into A, B, and C capsids. Furthermore, the fusion protein incorporated into the virion particle was fluorescent as judged by fluorescence-activated cell sorter (FACS) analysis of infected cells in the absence of de novo protein synthesis. Cells infected with K26GFP exhibited a punctate nuclear fluorescence at early times in the replication cycle. At later times during infection a generalized cytoplasmic and nuclear fluorescence, including fluorescence at the cell membranes, was observed, confirming visually that the fusion protein was incorporated into intranuclear capsids and mature virions.
The physical structure required for transporting the herpes simplex virus type 1 (HSV-1) genome is an icosahedral proteinaeous capsid (26). Three types of capsids, termed A, B, and C can be isolated from HSV-1-infected cells (10). The A and C capsids are similar in protein content, but only the C capsid contains a genomic equivalent of DNA and matures into the infectious virion (reviewed in references 19 and 21). The B capsid comprises seven proteins. Their designations, and the open reading frames (ORF) encoding the proteins (in parentheses), are VP5 (UL19), VP19C (UL38), 21 (UL26), 22a (UL26.5), VP23 (UL18), VP24 (UL26), and VP26 (UL35) (4, 10, 11, 14).
VP26 is the smallest capsid protein (12 kDa) and is encoded by the UL35 ORF (6, 15). It is expressed late in the infectious cycle after the onset of DNA replication and has been shown to be present multiple phosphorylated forms (16). Reports that show that VP26 localizes to the infected cell nucleus (16) and requires the presence of VP5 for this localization in transfected cells (18) have been published. There are approximately 900 copies of this molecule, and they occupy the tips of the hexons but not the pentons, both of which are composed of VP5 (1, 24, 27, 28). Although it interacts with VP5, it is not required for capsid formation in a baculovirus expression system (22, 23). Studies in our laboratory have shown that VP26 is not required for growth in cell culture; however, it is important for the production of infectious virus in trigeminal ganglia (8).
The green fluorescent protein (GFP) of the bioluminescent jellyfish (Aequorea victoria) has been extensively used, both as a genetic reporter and to monitor the cellular locations of proteins fused to it (2). GFP absorbs UV or blue light and emits green fluorescence. It does not require any other cofactors or substrates or additional A. victoria gene products for this activity. Consequently, detection of GFP can be performed in heterologous systems and in living cells or tissues (reviewed in reference 3). Variants of the GFP chromophore that improve the use of the GFP reporter have been generated. One such variant, termed EGFP, contains amino acid substitutions that allow it to fluoresce 35-fold more intensely than wild-type GFP when excited with blue light, and therefore it is useful for analysis using fluorescence microscopy and flow cytometry (5). Furthermore, this variant contains several silent base changes that correspond to optimal human codon usage for better expression in eukaryotic systems. This modified version of GFP (Clontech) was used for the experiments described below.
The aim of the experiments described below was to incorporate the GFP into the HSV-1 capsid. A tagged nucleocapsid structure should be useful for the investigation of the early events in the uncoating of the virus particle and for monitoring the virus nucleocapsid during replication and transport in cell culture and in vivo. The rationale behind this approach was to utilize the VP26 polypeptide, which is located on the outer surface of the capsid shell. A fusion between the VP26 and GFP polypeptides was generated, and it was hoped that this fusion form of VP26 would still be capable of interaction with VP5 and would incorporate the GFP polypeptide onto the capsid structure. Therefore, the nucleocapsid and consequently the mature virion would be tagged with a fluorescent marker that is activated by light.
Construction of a VP26-GFP fusion protein.
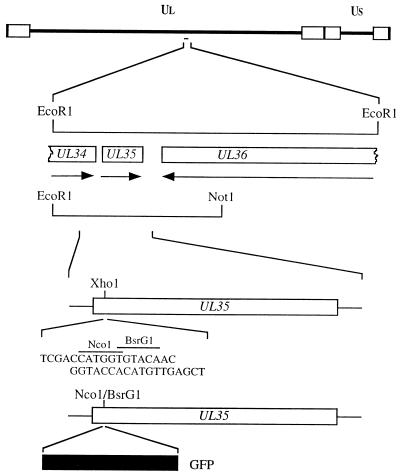
The goal of the molecular manipulations described below was to fuse the GFP ORF with that of VP26. An _Xho_I restriction enzyme site was introduced into the UL35 ORF (Fig. 1) by overlap extension PCR assays. The methodology of this assay is described in greater detail elsewhere (8, 12). The restriction site overlaps codons 5 to 7 of the UL35 ORF. The aim was to introduce the GFP ORF at this position to generate a VP26-GFP fusion construct. This was achieved by the introduction of an oligonucleotide duplex into the _Xho_I site which specified _Nco_I and _Bsr_GI restriction sites (Fig. 1). The GFP ORF was derived from pEGFP-N1 (Clontech) as an _Nco_I-to-_Bsr_GI fragment (716 bp) and cloned into the same sites in UL35. This plasmid was designated pK26GFP. The GFP ORF was fused in frame at the sequence corresponding to the VP26 N terminus to the codons for the first 4 authentic residues of VP26 and to the remaining portion of the VP26 coding sequences (108 residues) at the sequence corresponding to the C terminus. Therefore, expression of the GFP fusion polypeptide should be mediated by the UL35 promoter sequences and translation initiation should occur at the VP26 ATG codon.
FIG. 1.
Construction of a VP26-GFP fusion protein. The 5.2-kb _Eco_RI L fragment of HSV-1 strain KOS was cloned into pUC19. The _Eco_RI L fragment contains UL35 (VP26) and the C-terminal-encoding sequences of the UL34 and UL36 genes (14). A shortened version of the _Eco_RI L clone spanning the _Eco_RI and NotI restriction sites was used for subsequent manipulations. An _Xho_I restriction site which spans residues 5 and 7 of the UL35 ORF was created by overlap extension PCR assays (8, 12). This plasmid was cleaved with _Xho_I, and an oligonucleotide duplex specifying _Nco_I and _Bsr_GI restriction sites was annealed at this position. The GFP ORF derived from pEGFP-N1 (Clontech) as an _Nco_I-_Bsr_GI fragment was cloned into this plasmid to generate pK26GFP. UL, unique long; US, unique short.
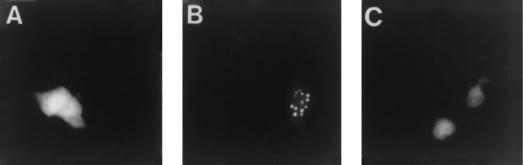
Transient transfection assays were then carried out to determine whether the GFP fusion construct was fluorescent. Briefly, Vero cell monolayers were transfected with either pK26GFP or pEGFP-N1. The latter plasmid contains the GFP sequences under the control of the constitutive human cytomegalovirus immediate-early promoter. Twenty-four hours after transfection the cells were either mock infected or infected with KOS at a multiplicity of infection (MOI) of 10 PFU per cell. Cells were then examined under a fluorescence microscope at various times after infection. Whereas fluorescence was detected in pEGFP-N1-transfected cells, in both mock- and KOS-infected samples, cells transfected with pK26GFP only displayed fluorescence upon superinfection (data not shown). Fluorescence in pK26GFP-transfected cells was observed as early as 4 h after infection and was distributed throughout the cell, as observed for the pEGFP-N1-transfected cells (Fig. 2A). However, at later times (12 h) during infection, a majority of the cells displayed a fluorescence pattern in which the fluorescence was primarily within the nucleus and in distinct locations within this structure (Fig. 2B). Therefore, the VP26-GFP fusion protein was fluorescent and was localized to the nucleus, as would be expected during the assembly of the capsid structure. The VP26-GFP fusion protein was localized to the nucleus in punctate regions only in cells infected with KOS (Fig. 2B) or the VP26-null mutant (8), KΔ26Z (data not shown), both of which assemble capsids in the nuclei of infected cells. This pattern of nuclear fluorescence was not observed when cells were infected with viruses that do not assemble capsids, such as the VP5-null mutant K5ΔZ (7) (Fig. 2C). Therefore, the VP26-GFP fusion protein was biologically functional in the context of capsid assembly.
FIG. 2.
Transfection-infection assays with pK26GFP. Vero cells seeded at 3.5 × 104 cells/cm2 in culture dishes were transfected approximately 16 h later with 5 μg each of plasmids pEGFP-N1 (A) or pK26GFP (B and C) (CellPhect; Pharmacia Upjohn). Twenty-four hours after transfection the cells were infected with either KOS (B) or K5ΔZ (C) at an MOI of 10 PFU/cell or were mock infected (A). Cells were visualized live 12 h postinfection with an Olympus BH-2 fluorescence microscope (×20 objective).
Transfer of the VP26-GFP marker into the virus genome.
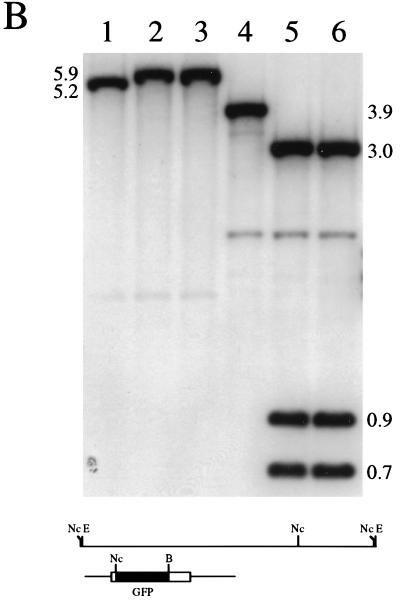
The next step was to introduce the VP26-GFP marker into the virus genome. This was carried out by standard marker transfer procedures in Vero cells. Cell monolayers were cotransfected with KOS viral DNA and linearized pK26GFP plasmid DNA. Forty-eight hours after transfection lysates were prepared and the transfection progeny were diluted and plated on Vero cell monolayers. The plaques that formed were visualized in a fluorescence microscope to detect recombinant viruses. Fluorescent plaques indicative of recombinant viruses were observed, and these were marked and picked. Two independent isolates were purified through two further cycles of limiting dilution, and the viruses were designated K26GFP-1 and K26GFP-2. When plaques of K26GFP (either isolate) were examined in a fluorescence microscope, all the plaques were found to be fluorescent, indicative of the purity of the virus stocks. A single fluorescent plaque of K26GFP-1 is shown in Fig. 3A. Southern blot analysis was performed on DNA extracted from both KOS- and K26GFP-infected cells to confirm the genotypes of the recombinant viruses (Fig. 3B). Viral DNA was either digested with _Eco_RI (lanes 1 to 3) or double digested with _Nco_I and _Bsr_GI (lanes 4 to 6), and the resulting fragments, following transfer to nitrocellulose, were hybridized to 32P-labeled pK26GFP. The probe hybridized to the 5.2-kb _Eco_RI L fragment of KOS DNA (lane 1); this fragment exhibited a decreased mobility in K26GFP-1 (lane 2) and K26GFP-2 (lane 3) DNA due to the insertion of the 716-bp GFP coding sequence. In the case of the digestion with _Nco_I and _Bsr_GI the probe hybridized to a 3.9-kb fragment in wild-type DNA (lane 4), which was cleaved in K26GFP-1 (lane 5) and K26GFP-2 (lane 6) DNA into three fragments of 3.0, 0.9, and 0.7 kb due to the introduction of _Nco_I and _Bsr_GI sites into UL35 followed by the insertion of the 0.7-kb GFP sequence. Weak hybridization of the probe to a 2.0-kb _Nco_I fragment, observed in lanes 4 to 6, was due to the presence of sequences in the probe that correspond to DNA spanning the _Nco_I and _Eco_RI sites at the 3′ end of the _Eco_RI L fragment. This analysis confirmed the introduction of the ORF for the VP26-GFP fusion construct into the virus genome. Since both isolates were phenotypically and genotypically identical, the experiments presented below were carried out with only one of the isolates, now referred to as K26GFP.
FIG. 3.
Marker rescue of VP26-GFP into the virus genome. Vero cells seeded at 3.5 × 104 cells/cm2 in culture dishes were transfected approximately 16 h later with KOS viral DNA (2 μg) and linearized pK26GFP (5 μg) (CellPhect; Pharmacia Upjohn). The transfection progeny was harvested 2 days later and plated for single plaques. Plaques were visualized in a fluorescence microscope, and the fluorescent plaques were picked and purified further. (A) One such virus, designated K26GFP-1, was plated on Vero cell monolayers, and a single plaque was photographed upon visualization in a fluorescence microscope (×10 objective). (B) Two micrograms of KOS (lanes 1 and 4), K26GFP-1 (lanes 2 and 5), and K26GFP-2 (lanes 3 and 6) was digested with _Eco_RI (lanes 1 to 3) or double digested with _Nco_I and _Bsr_GI (lanes 4 to 6), the restriction fragments were resolved by agarose gel electrophoresis and transferred to nitrocellulose (Schleicher and Schuell), and the filters were probed with 32P-labeled pK26GFP. The diagram below the blot shows the _Eco_RI L region of HSV-1 strain KOS. The probe, pK26GFP, is shown below this. Relevant restriction sites are indicated: E, _Eco_RI; Nc, _Nco_I; and B, _Bsr_GI. For details of autoradiograph scanning, see the legend for Fig. 4.
A null mutation in the UL35 ORF resulted in a twofold decrease in the virus yield from infected cells compared to that for wild-type virus, as judged by single-step growth assays (8). Similar assays using the K26GFP virus revealed that the recombinant GFP virus replicated at levels comparable to that of the wild-type virus (data not shown). Therefore, the VP26-GFP fusion protein retained biological activity during the virus replication cycle.
VP26-GFP is incorporated into the capsid shell and the mature virion.
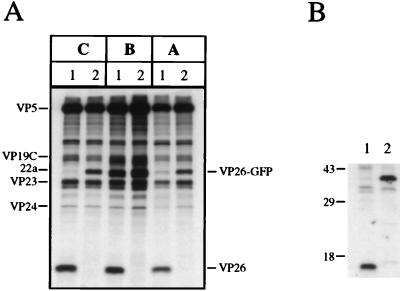
The next set of experiments was carried out to determine the composition of the capsids formed in K26GFP-infected cells. HEL cell monolayers were infected with KOS and K26GFP at an MOI of 10 PFU/cell and metabolically labeled with [35S]methionine from 8 to 24 h postinfection. Prepared nuclear lysates were sedimented through sucrose gradients, and the fractions corresponding to A, B, and C capsids were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. 4A). The VP26 and GFP polypeptides are 12 and 27 kDa in size, respectively; therefore, a VP26-GFP fusion protein would be expected to have a mobility of 39 kDa in SDS-PAGE gels. For KOS (lanes 1) capsids (A, B, or C) the 12-kDa VP26 polypeptide was evident. In the K26GFP (lanes 2) capsid fractions the 12-kDa band was absent and a novel polypeptide of approximately 39 to 40 kDa was present. This was presumably the VP26-GFP fusion protein. The VP26-GFP fusion protein was obscured in B capsids by the presence of the abundant 22a polypeptide, which is similar in size. Therefore, the VP26-GFP fusion protein was capable of interacting with the major capsid protein (VP5) and was incorporated into the capsid shell. Immunoprecipitation assays were carried out with [35S]methionine-labeled infected cell extracts and a peptide antibody specific to the C terminus of VP26. The 12-kDa VP26 polypeptide was precipitated from KOS-infected cell extracts (Fig. 4B, lane 1), and a polypeptide of approximately 40 kDa was observed in extracts of K26GFP-infected cells (lane 2). This polypeptide had the same mobility as the VP26-GFP fusion protein observed in capsids.
FIG. 4.
Sedimentation analysis of K26GFP intranuclear capsids. (A) HEL cells seeded at 6.4 × 104 cells/cm2 and infected approximately 16 h later with KOS (lanes 1) and K26GFP (lanes 2) at an MOI of 10 PFU/cell were metabolically labeled with [35S]methionine from 8 to 24 h postinfection (17). Nuclear lysates were prepared and sedimented in 20 to 50% sucrose gradients (7). The radioactivity in each fraction collected was determined by liquid scintillation counting. The peak fractions containing A (fraction 10), B (fraction 8), and C (fraction 4) capsids were precipitated with trichloroacetic acid and analyzed by SDS-PAGE (17% acrylamide). The positions of the capsid proteins are indicated on the left of the figure, and those of VP26 and VP26-GFP are indicated on the right. (B) Vero cells seeded at 1.1 × 105 cells/cm2 and infected approximately 16 h later with KOS (lane 1) and K26GFP (lane 2) at an MOI of 10 PFU/cell and labeled with [35S]methionine as described for panel A. Infected cell extracts were prepared and were precipitated with a peptide antibody to VP26, and the resulting precipitates were resolved by SDS-PAGE (17% acrylamide). Shown to the left of the figure are the relative mobilities of protein molecular weight standards. The autoradiographs in both panels were scanned on a Umax Powerlook II scanner. The images were scanned at 300 dots per inch into Adobe Photoshop 3.0 and were transported as PICT files into Microsoft Powerpoint for figure presentation and printing.
In order to determine whether the GFP tag in the virus was still fluorescent, experiments were carried out with a fluorescence-activated cell sorter (FACS), an EPICS profile analyzer (Coulter Corp.). Vero cell monolayers were infected with KOS and K26GFP at an MOI of 250 PFU/cell or were mock infected. The cells were incubated prior to and during the infection period in cycloheximide to prevent de novo protein synthesis. Cells were harvested at 4 h postinfection, resuspended in phosphate-buffered saline at 106 cells/ml, and subjected to FACS analysis using the FL1 emission channel. Background levels of fluorescence were determined with mock-infected cells. The majority of KOS-infected cells (99.1%) exhibited background levels of fluorescence, whereas only 1.7% of K26GFP-infected cells displayed background fluorescence. For the remainder of the K26GFP-infected cells, 98.3%, there was a shift in peak detected mean fluorescence. The mean detected fluorescence levels were 1.8 and 2.1 for mock- and KOS-infected cells, respectively, whereas the mean detected fluorescence level for K26GFP-infected cells was 28.4. Therefore, the VP26-GFP fusion protein incorporated into the virus capsid retained fluorescent activity.
Localization of VP26-GFP in infected cells.
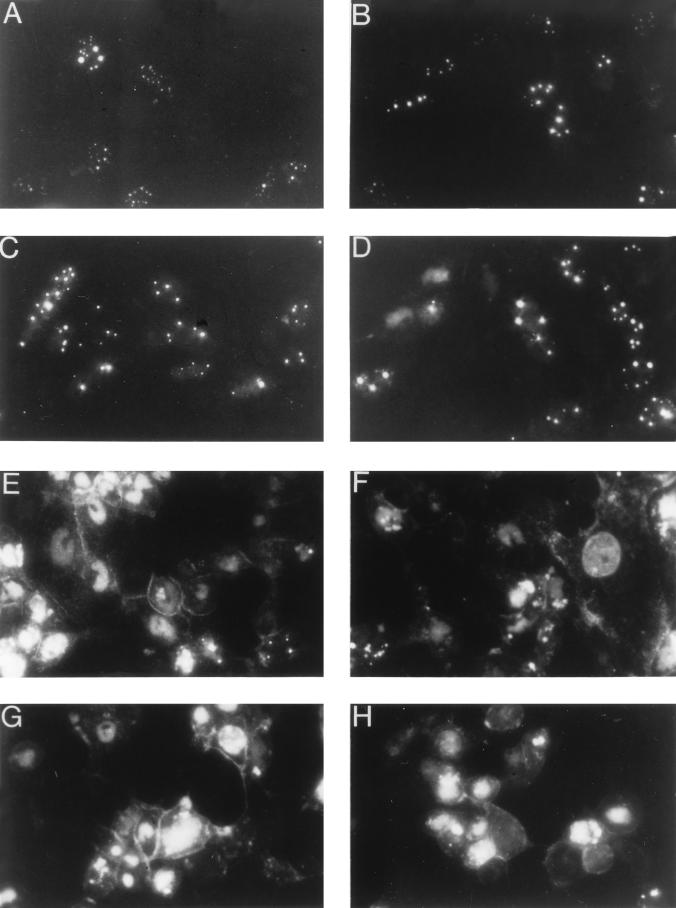
The next experiment was performed to monitor the localization of the VP26-GFP fusion protein and its incorporation into the virus capsid. Vero cells were infected with K26GFP and visualized “live” at various times after infection throughout the virus replication cycle (Fig. 5). Early in infection (6 and 8 h; Fig. 5A and B) the fusion protein localized to distinct regions within the nucleus of the infected cell. This punctate pattern was similar to that reported by others (25) and probably corresponds to compartments within the nucleus in which capsids are assembled. There was either very little or undetectable cytoplasmic fluorescence, which indicated that VP26 was sequestered in the nucleus with VP5. When the VP26-GFP marker was introduced into a virus genome that specified a null mutation in VP5 (KΔ5Z), the fluorescence observed in those infected cells was uniformly distributed throughout the cell (data not shown) and was similar to that observed in Fig. 2C. At 10 and 12 h (Fig. 5C and D) after infection the intensity of the fluorescence as well as the number of fluorescent cells increased. At later times during infection with K26GFP, the punctate nuclear fluorescence became less intense and was replaced with a more generalized nuclear fluorescence and an increase in cytoplasmic fluorescence was observed. There was also an accumulation of fluorescence at the plasma membrane. This can dramatically be seen in Fig. 5E to H. Presumably the change in the fluorescence pattern reflects virus assembly in the nucleus and its transport through the cytoplasm to the plasma membrane. Fluorescence at the plasma membrane reflects the accumulation of viruses at the cell surface. In addition to being able to trace the path of the capsid from its origin in the nuclear compartment to the cytoplasm, the accumulation of fluorescence at the plasma membrane may allow one to study the release of virus from cells, as well as the spread of virus from cell to cell.
FIG. 5.
Localization of VP26-GFP in infected cells. Vero cells seeded at 3.5 × 104 cells/cm2 in culture dishes were infected at an MOI of 10 PFU/cell. Cells were visualized live at various times after infection with an Olympus BH-2 fluorescence microscope (×40 objective). Photographs were taken at 6 (A), 8 (B), 10 (C), 12 (D), 16 (E and F), 20 (G), and 24 (H) h postinfection.
Discussion.
GFP, by virtue of its properties, is a useful tag that can be used to monitor the fate of the tagged protein. In this case, by being fused to HSV-1 capsid protein VP26, it is incorporated into the capsid structure on the outer surface of the shell and therefore allows one to follow the fate of that particle in the course of the virus life cycle. Using antibodies to the capsid proteins, immunofluorescence assays have determined the location of the capsid upon entry into the cell (20). However, problems of antibody reactivity, nonspecific cross-reactivity, and the fixation procedures involved can limit the usefulness of these assays. With GFP as a marker, entry and uncoating can be monitored in living infected cells without the need to fix cells or to rely on the availability of antibodies. GFP has been used as a reporter in recombinant pseudorabies virus (13) and herpesvirus saimiri (9); however, this is the first report of using a GFP fusion protein to incorporate this molecule into the virion structure. This is therefore a useful reagent for the analysis of HSV-1 biology, in particular, the analysis of virus replication and transport both in cell culture and in the trigeminal ganglia of mice.
An interesting outcome of this project is the ability to fuse a relatively large sequence (27 kDa) to the smallest capsid protein, VP26 (12 kDa), and still retain the ability of VP26 to interact with VP5 and become incorporated into the capsid shell. The VP26-GFP fusion protein retained the structural activity, as judged by capsid assembly, and the biological activity, as judged by infectious virus progeny, of the wild-type VP26 protein. Potentially, this technique can be utilized to tag the capsid shell with other small molecular sequences or proteins.
Acknowledgments
This work was supported by Public Health Service grant AI33077 (S.P.) from the National Institutes of Health.
We gratefully acknowledge discussions of the data with Wade Gibson, Mike Baxter, Mary-Elizabeth Harmon, and Scott Plafker. We also acknowledge Richard Hampton and Zhaohao Liao for assistance with the FACS analysis.
REFERENCES
- 1.Booy F P, Trus B L, Newcomb W W, Brown J C, Conway F F, Steven A C. Finding a needle in a haystack: detection of a small protein (the 12-kDa VP26) in a large complex (the 200-MDa capsid of herpes simplex virus) Proc Natl Acad Sci USA. 1994;91:5652–5656. doi: 10.1073/pnas.91.12.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 3.Chalfie M. Green fluorescent protein. Photochem Photobiol. 1995;62:651–656. doi: 10.1111/j.1751-1097.1995.tb08712.x. [DOI] [PubMed] [Google Scholar]
- 4.Cohen G H, Ponce de Leon M, Diggleman H, Lawrence W C, Vernon S K, Eisenberg R J. Structural analysis of the capsid polypeptides of herpes simplex virus types 1 and 2. J Virol. 1980;34:521–531. doi: 10.1128/jvi.34.2.521-531.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cormack B P, Valdivia R, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 6.Davison M D, Rixon F J, Davison A J. Identification of genes encoding two capsid proteins (VP24 and VP26) of herpes simplex virus type 1. J Gen Virol. 1992;73:2709–2713. doi: 10.1099/0022-1317-73-10-2709. [DOI] [PubMed] [Google Scholar]
- 7.Desai P, DeLuca N A, Glorioso J C, Person S. Mutations in herpes simplex virus type 1 genes encoding VP5 and VP23 abrogate capsid formation and cleavage of replicated DNA. J Virol. 1993;67:1357–1364. doi: 10.1128/jvi.67.3.1357-1364.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desai, P., N. A. DeLuca, and S. Person. Herpes simplex virus type 1 VP26 is not essential for replication in cell culture but influences production of infectious virus in the nervous system of infected mice. Virology, in press. [DOI] [PubMed]
- 9.Duboise S M, Guo J, Desrosiers R C, Jung J U. Use of virion DNA as a cloning vector for the construction of mutant and recombinant herpesviruses. Proc Natl Acad Sci USA. 1996;93:11389–11394. doi: 10.1073/pnas.93.21.11389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gibson W, Roizman B. Proteins specified by herpes simplex virus. VIII. Characterization and composition of multiple capsid forms of subtypes 1 and 2. J Virol. 1972;10:1044–1052. doi: 10.1128/jvi.10.5.1044-1052.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heilman C J, Jr, Zweig M, Stephenson J R, Hampar B. Isolation of a nucleocapsid polypeptide of herpes simplex virus types 1 and 2 possessing immunologically type-specific and cross-reactive determinants. J Virol. 1979;29:34–42. doi: 10.1128/jvi.29.1.34-42.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 13.Jons A, Mettenleiter T C. Green fluorescent protein expressed by recombinant pseudorabies virus as an in vivo marker for viral replication. J Virol Methods. 1997;66:283–292. doi: 10.1016/s0166-0934(97)00065-7. [DOI] [PubMed] [Google Scholar]
- 14.McGeoch D J, Dalrymple M A, Davison A J, Dolan A, Frame M C, McNab D, Perry L J, Scott J E, Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988;69:1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- 15.McNabb D S, Courtney R J. Identification and characterization of the herpes simplex virus type 1 virion protein encoded by the UL35 open reading frame. J Virol. 1992;66:2653–2663. doi: 10.1128/jvi.66.5.2653-2663.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McNabb D S, Courtney R J. Posttranslational modification and subcellular localization of the p12 capsid protein of herpes simplex virus type 1. J Virol. 1992;66:4839–4847. doi: 10.1128/jvi.66.8.4839-4847.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Person S, Desai P. Capsids are formed in a mutant virus blocked at the maturation site of the UL26 and UL26.5 open reading frames of HSV-1 but are not formed in a null mutant of UL38 (VP19C) Virology. 1998;242:193–203. doi: 10.1006/viro.1997.9005. [DOI] [PubMed] [Google Scholar]
- 18.Rixon F J, Addison C, McGregor A, McNab S J, Nicholson P, Preston V G, Tatman J D. Multiple interactions control the intracellular localization of the herpes simplex virus type 1 capsid proteins. J Gen Virol. 1996;77:2251–2260. doi: 10.1099/0022-1317-77-9-2251. [DOI] [PubMed] [Google Scholar]
- 19.Roizman B, Sears A. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, et al., editors. Virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2231–2295. [Google Scholar]
- 20.Sodeik B, Ebersold M W, Helenius A. Microtubule-mediated transport of incoming herpes simplex virus type 1 capsids to the nucleus. J Cell Biol. 1997;136:1007–1021. doi: 10.1083/jcb.136.5.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steven A C, Spear P G. Herpesvirus capsid assembly and envelopment. In: Burnett R, Chiu W, Garcea R, editors. Structural biology of viruses. New York, N.Y: Oxford University Press; 1996. pp. 312–351. [Google Scholar]
- 22.Tatman J D, Preston V G, Nicholson P, Elliot R M, Rixon F J. Assembly of herpes simplex virus type 1 capsids using a panel of recombinant baculoviruses. J Gen Virol. 1994;75:1101–1113. doi: 10.1099/0022-1317-75-5-1101. [DOI] [PubMed] [Google Scholar]
- 23.Thomsen D R, Roof L L, Homa F L. Assembly of herpes simplex virus (HSV) intermediate capsids in insect cells infected with recombinant baculoviruses expressing HSV capsid proteins. J Virol. 1994;68:2442–2457. doi: 10.1128/jvi.68.4.2442-2457.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trus B L, Homa F L, Booy F P, Newcomb W W, Thomsen D R, Cheng N, Brown J C, Stevens A C. Herpes simplex virus capsids assembled in insect cells infected with recombinant baculoviruses: structural authenticity and localization of VP26. J Virol. 1995;69:7362–7366. doi: 10.1128/jvi.69.11.7362-7366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ward P L, Ogle W O, Roizman B. Assemblons: nuclear structures defined by aggregation of immature capsids and some tegument proteins of herpes simplex virus type 1. J Virol. 1996;70:4623–4631. doi: 10.1128/jvi.70.7.4623-4631.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wildy P, Russell W C, Horne R W. The morphology of herpes virus. Virology. 1960;12:204–222. doi: 10.1016/0042-6822(60)90195-1. [DOI] [PubMed] [Google Scholar]
- 27.Wingfield P T, Stahl S J, Thomsen D R, Homa F L, Booy F P, Trus B L, Steven A C. Hexon-only binding of VP26 reflects differences between the hexon and penton conformations of VP5, the major capsid protein of herpes simplex virus. J Virol. 1997;71:8955–8961. doi: 10.1128/jvi.71.12.8955-8961.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Z H, He J, Jakana J, Tatman J, Rixon F J, Chiu W. Assembly of VP26 in herpes simplex virus-1 inferred from structures of wild-type and recombinant capsids. Nat Struct Biol. 1995;2:1026–1030. doi: 10.1038/nsb1195-1026. [DOI] [PubMed] [Google Scholar]