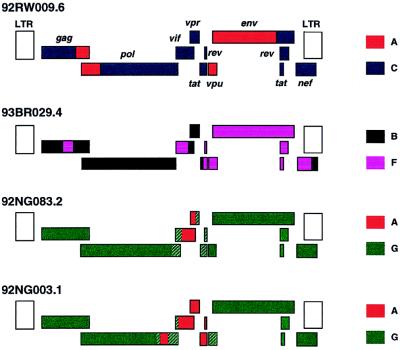
A Comprehensive Panel of Near-Full-Length Clones and Reference Sequences for Non-Subtype B Isolates of Human Immunodeficiency Virus Type 1 (original) (raw)
Abstract
Non-subtype B viruses cause the vast majority of new human immunodeficiency virus type 1 (HIV-1) infections worldwide and are thus the major focus of international vaccine efforts. Although their geographic dissemination is carefully monitored, their immunogenic and biological properties remain largely unknown, in part because well-characterized virological reference reagents are lacking. In particular, full-length clones and sequences are rare, since subtype classification is frequently based on small PCR-derived viral fragments. There are only five proviral clones available for viruses other than subtype B, and these represent only 3 of the 10 proposed (group M) sequence subtypes. This lack of reference sequences also confounds the identification and analysis of mosaic (recombinant) genomes, which appear to be arising with increasing frequency in areas where multiple sequence subtypes cocirculate. To generate a more representative panel of non-subtype B reference reagents, we have cloned (by long PCR or lambda phage techniques) and sequenced 10 near-full-length HIV-1 genomes (lacking less than 80 bp of long terminal repeat sequences) from primary isolates collected at major epicenters of the global AIDS pandemic. Detailed phylogenetic analyses identified six that represented nonrecombinant members of HIV-1 subtypes A (92UG037.1), C (92BR025.8), D (84ZR085.1 and 94UG114.1), F (93BR020.1), and H (90CF056.1), the last two comprising the first full-length examples of these subtypes. Four others were found to be complex mosaics of subtypes A and C (92RW009.6), A and G (92NG083.2 and 92NG003.1), and B and F (93BR029.4), again emphasizing the impact of intersubtype recombination on global HIV-1 diversification. Although a number of clones had frameshift mutations or translational stop codons in major open reading frames, all the genomes contained a complete set of genes and three had intact genomic organizations without inactivating mutations. Reconstruction of one of these (94UG114.1) yielded replication-competent virus that grew to high titers in normal donor peripheral blood mononuclear cell cultures. This panel of non-subtype B reference genomes should prove valuable for structure-function studies of genetically diverse viral gene products, the generation of subtype-specific immunological reagents, and the production of DNA- and protein-based subunit vaccines directed against a broader spectrum of viruses.
One critical question facing current AIDS vaccine development efforts is to what extent human immunodeficiency virus type 1 (HIV-1) genetic variation has to be considered in the design of candidate vaccines (11, 21, 41, 72). Phylogenetic analyses of globally circulating viral strains have identified two distinct groups of HIV-1 (M and O) (33, 45, 61, 62), and 10 sequence subtypes (A to J) have been proposed within the major group (M) (29, 30, 45, 72). Sequence variation among viruses belonging to these different lineages is extensive, with envelope amino acid sequence variation ranging from 24% between different subtypes to 47% between the two different groups. Given this extent of diversity, the question has been raised whether immunogens based on a single virus strain can be expected to elicit immune responses effective against a broad spectrum of viruses or whether vaccine preparations should include mixtures of genetically divergent antigens and/or be tailored toward locally circulating strains (11, 21, 41, 72). This is of particular concern in developing countries, where multiple subtypes of HIV-1 are known to cocirculate and where subtype B viruses (which have been the source of most current candidate vaccine preparations [10, 21]) are rare or nonexistent (5, 24, 40, 72).
Although the extent of global HIV-1 variation is well defined, little is known about the biological consequences of this genetic diversity and its impact on cellular and humoral immune responses in the infected host. In particular, it remains unknown whether subtype-specific differences in virus biology exist that have to be considered for vaccine design. Thus far, such differences have not been identified. For example, several studies have shown that there is no correlation between HIV-1 genetic subtypes and neutralization serotypes (38, 42, 46, 68). Some viruses are readily neutralized, while most are relatively neutralization resistant (42). Although the reasons for these different susceptibilities remain unknown, it is clear that neutralization is not a function of the viral genotype (38, 42, 46, 68). Similarly, recent studies have identified vigorous cross-clade cytotoxic T-lymphocyte (CTL) reactivities in individuals infected with viruses from several different clades (3, 6), as well as in recipients of a clade B vaccine (15). These results are very encouraging, since they suggest that CTL cross-recognition among HIV-1 clades is much more prevalent than previously anticipated and that immunogens based on a limited number of variants may be able to elicit a broad CTL response (6). Nevertheless, it would be premature to conclude that HIV-1 variation poses no problem for AIDS vaccine design. Only a comprehensive analysis of genetically defined representatives of the various groups and subtypes will allow us to judge whether certain variants differ in fundamental viral properties and whether such differences will have to be incorporated into vaccine strategies. Obviously, such studies require well-characterized reference reagents, in particular full-length and replication-competent molecular clones that can be used for functional and biological studies.
Full-length reference sequences representing the various subtypes are also urgently needed for phylogenetic comparisons. Recent analyses of subgenomic (23, 52, 54, 58) as well as full-length (7, 18, 53, 60) HIV-1 sequences identified a surprising number of HIV-1 strains which clustered in different subtypes in different parts of their genome. All of these originated from geographic regions where multiple subtypes cocirculated and are the results of coinfections with highly divergent viruses (52, 60, 62). Detailed phylogenetic characterization revealed that most of them have a complex genome structure with multiple points of crossover (7, 18, 53, 60). Some recombinants, like the “subtype E” viruses, which are in fact A/E recombinants (7, 18), have a widespread geographic dissemination and are responsible for much of the Asian HIV-1 epidemic (69, 70). In other areas, recombinants appear to be generated with increasing frequencies since many randomly chosen isolates exhibit evidence of mosaicism (4, 8, 31, 66, 71). Since recombination provides the opportunity for evolutionary leaps with genetic consequences that are far greater than those of the steady accumulation of individual mutations, the impact of recombination on viral properties must be monitored. We therefore need full-length nonrecombinant reference sequences for all major HIV-1 groups and subtypes before we can map and characterize the extent of intersubtype recombination.
The number of molecular reagents for non-subtype B viruses is very limited. There are currently only five full-length, nonrecombinant molecular clones available for viruses other than subtype B (45), and these represent only three of the proposed (group M) subtypes (A, C, and D). Moreover, only three clones (all derived from subtype D viruses) are replication competent and thus useful for studies requiring functional gene products (45, 48, 65). Given the unknown impact of genetic variation on correlates of immune protection, subtype-specific reagents are critically needed for phylogenetic, immunological, and biological studies. In this paper, we report the cloning (by long PCR and lambda techniques) of 10 near-full-length HIV-1 genomes from isolates previously classified as non-subtype B viruses. Detailed phylogenetic analysis showed that six comprise nonmosaic representatives of five major subtypes, including two for which full-length representatives have not been reported. Four others were identified as complex intersubtype recombinants, again emphasizing the prevalence of hybrid genomes among globally circulating HIV-1 strains. We also describe a strategy for the biological evaluation of long-PCR-derived genomes and report the generation of a replication-competent provirus by this approach. The effect of these reagents on vaccine development is discussed.
MATERIALS AND METHODS
Virus isolates.
All viruses used in this study were propagated in normal donor peripheral blood mononuclear cells (PBMCs) and thus represent primary isolates. Their biological phenotype (SI/NSI), year of isolation, relevant epidemiological and clinical information, and appropriate references are summarized in Table 1. For consistency, isolates are labelled according to World Health Organization (WHO) nomenclature (28); some isolates have previously been reported under different names (1, 43), which are listed in parentheses. Preliminary subtype classification was made on the basis of partial env and/or gag gene sequences (1, 17, 19, 43).
TABLE 1.
Epidemiological and clinical information for study isolates
| Isolatea | Sexb | Age (yr) | City | Country | Risk factorc | Disease statusd | Antiviral therapy | Yr of isolation | Sourcee | Biological phenotypef | Preliminary subtype assignment | Reference(s) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 92UG037 | F | 31 | Entebbe | Uganda | Het | AS | No | 1992 | WHO | NSI | A | 19, 72 |
| 92BR025 | M | 23 | Porto Alegre | Brazil | Hemo | AS | No | 1992 | WHO | NSI | C | 19, 72 |
| 94UG114 | M | 31 | Butuku | Uganda | Het | AS | No | 1994 | WHO | NSI | NA | |
| 84ZR085 (H85) | NAg | NA | NA | Zaire | NA | AIDS | No | 1984 | TJU | NA | NA | |
| 93BR020 | M | 52 | Rio de Janeiro | Brazil | Bi | AS | No | 1993 | WHO | SI | F | 19, 72 |
| 90CF056 (U4056) | M | NA | Bangui | CAR | Het | AS | No | 1990 | PIB | NSI | U | 43 |
| 92RW009 | F | 24 | Kigali | Rwanda | Het | AS | No | 1992 | WHO | NSI | Ah | 17, 72 |
| 93BR029 | M | 17 | Sao Paulo | Brazil | NA | AS | No | 1993 | WHO | NSI | Fh | 19, 72 |
| 92NG083 (JV1083) | F | 27 | Jos | Nigeria | NA | AIDS | No | 1992 | IHV | NSI | Gh | 1 |
| 92NG003 (G3) | F | 24 | Jos | Nigeria | Het | AS | NA | 1992 | IHV | NSI | Gh | 1 |
Amplification of near-complete HIV-1 genomes by using long-PCR methods.
Near-full-length HIV-1 genomes were amplified from DNA of short-term-cultured PBMCs essentially as described previously (18, 56) with the GeneAmp XL kit (Perkin-Elmer Cetus, Foster City, Calif.) and primers spanning the tRNA primer binding site (upstream primer UP1A: 5′-AGTGGCGCCCGAACAGG-3′) and the R/U5 junction in the 3′ long terminal repeat (LTR) (downstream primer Low2: 5′-TGAGGCTTAAGCAGTGGGTTTC-3′). Some isolates were amplified with primers containing _Mlu_I restriction enzyme sites to facilitate subsequent subcloning into plasmid vectors (upstream primer UP1AMlu1: 5′-TCTCTacgcgtGGCGCCCGAACAGGGAC-3′; downstream primer Low1Mlu1: 5′-ACCAGacgcgtACAACAGACGGGCACACACTACTT-3′ [lowercase letters indicate the _Mlu_I restriction site]). Whenever possible, PBMC DNAs were diluted before PCR analysis to attempt amplification from single proviral templates. Cycling conditions included a hot start (94°C for 2 min), followed by 20 cycles of denaturation (94°C for 30 s) and extension (68°C for 10 min), followed by 17 cycles of denaturation (94°C for 30 s) and extension (68°C for 10 min) with 15-s increments per cycle. PCR products were visualized by agarose gel electrophoresis and subcloned into pCRII by T/A overhang (92UG037.1, 92BR025.8, 93BR020.1, and 90CF056.1) or following cleavage with _Mlu_I into a modified pTZ18 vector (pTZ18Mlu1) containing a unique _Mlu_I site in its polylinker (94UG114.1, 92RW009.6, 93BR029.4, 92NG083.2, and 92NG003.1). Transformations were performed in INVαF′ cells (OneShot kit; Invitrogen, San Diego, Calif.), and colonies were screened by restriction enzyme digestion for full-length inserts (transformation efficiencies were generally poor, yielding only a few recombinant colonies; however, once subcloned, full-length genomes were stable in their respective vectors). One full-length clone per isolate was randomly chosen for subsequent sequence analysis.
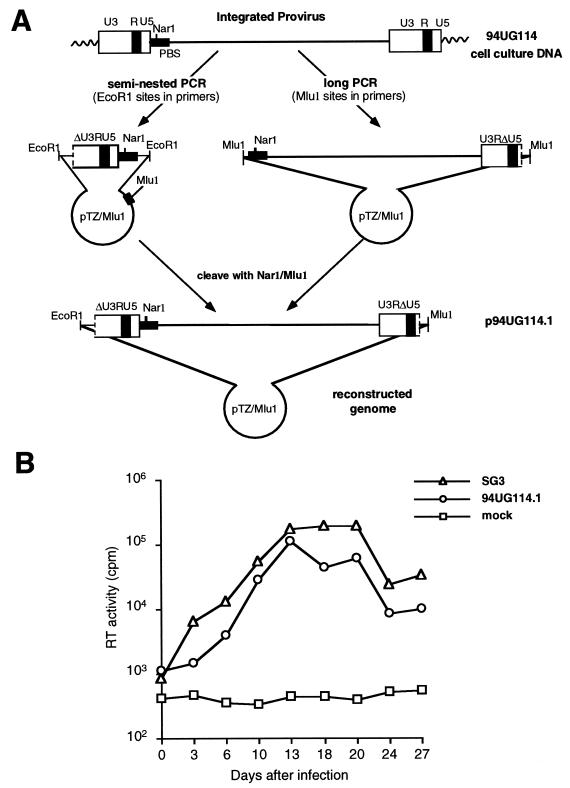
Construction of a full-length and infectious molecular clone of 94UG114.1.
A 674-bp fragment spanning most of the viral LTR (lacking positions 1 to 92 of U3 sequences), as well as the untranslated leader sequence preceding gag, was amplified from 94UG114 PBMC DNA by using primers and conditions described previously (18). After sequence confirmation, this LTR fragment was cloned into the pTZ18Mlu1 vector, which was subsequently cleaved with _Nar_I (in the primer binding site) and _Mlu_I (in the polylinker) to allow the insertion of the 94UG114.1 long-PCR product cleaved with the same restriction enzymes. The resulting plasmid clone comprised a full-length 94UG114.1 genome with 3′ and 5′ LTR fragments containing all regulatory elements necessary for viral replication.
Lambda phage cloning.
The 84ZR085.1 genome was cloned by lambda phage methods as previously described (36). Briefly, high-molecular-weight DNA from a primary PBMC culture was digested with _Sac_I (an enzyme that cleaves the viral LTR), fractionated by sucrose gradient centrifugation to enrich for fragments 9 to 15 kb in length, and ligated into purified arms of λgtWes.λB. Ligation products were packaged in vitro, subjected to titer determination, and plated on LE392 cells. Recombinant phage plaques were screened with a full-length HIV-1 probe (BH10) (22). One positive phage recombinant was plaque purified, and its restriction map was determined by multiple enzyme digestions. The viral insert was released by digestion with _Sac_I and subcloned into pUC19.
Sequence analysis of HIV-1 genomes.
92UG037.1, 92BR025.8, 84ZR085.1, 93BR020.1, 90CF056.1, 92RW009.6, and 93BR029.4 were sequenced by the shotgun sequencing approach (37). Briefly, viral genomes were released from their respective plasmid vectors by cleavage with the appropriate restriction enzymes, purified by gel electrophoresis, and sonicated (model XL2020 sonicator; Heat System Inc., Farmingdale, N.Y.) to generate randomly sheared DNA fragments of 600 to 1,000 bp. Following purification by gel electrophoresis, fragments were end repaired with T4 DNA polymerase and Klenow enzyme and ligated into _Sma_I-digested and dephosphorylated M13 or pTZ18 vectors. Approximately 200 shotgun clones were sequenced for each viral genome by using cycle-sequencing and dye terminator methods on an automated DNA Sequenator (model 377A; Applied Biosystems, Inc.). Sequences were determined for both strands of DNA. 92UG114.1, 92NG083.2, and 92NG003.1 were sequenced directly by the primer-walking approach (primers were designed approximately every 300 bp along the genome for both strands). Proviral contigs were assembled from individual sequences with the Sequencher program (Gene Codes Corp., Ann Arbor, Mich.). Sequences were analyzed with Eugene (Baylor College of Medicine, Houston, Tex.) and MASE (12).
Phylogenetic tree analysis.
Phylogenetic relationships of the newly derived viruses were estimated from sequence comparisons with previously reported representatives of HIV-1 group M (45). Multiple gag and env sequence alignments were obtained from the Los Alamos sequence database (http://hiv- web .lanl .gov/HTML/alignments.html). Newly derived gag and env sequences were added to these alignments by using the CLUSTAL W profile alignment option (67) and adjusted manually with the alignment editor MASE (12). All partial sequences were removed from these alignments. Sites where there was a gap in any of the remaining sequences, as well as areas of uncertain alignment, were excluded from all sequence comparisons. Pairwise evolutionary distances were estimated by Kimura’s two parameter method to correct for superimposed substitutions (26). Phylogenetic trees were constructed by the neighbor-joining method (55), and the reliability of topologies was estimated by performing bootstrap analysis with 1,000 replicates (13). NJPLOT was used to draw trees for illustrations (49). Phylogenetic relationships were also determined by using maximum-parsimony (with repeated randomized input orders; 10 iterations) and maximum-likelihood approaches, implemented with the programs DNAPARS and DNAML from the PHYLIP package (14).
Complete genome alignment.
All newly derived HIV-1 genome sequences were aligned with previously reported (45) full-length representatives of HIV-1 subtype A (U445), B (LAI, RF, OYI, MN, SF2), C (C2220), D (ELI, NDK, Z2Z6), and “E” (90CF402.1, 93TH253.3, CM240), as well as SIVcpzGAB as an outgroup, by using the CLUSTAL W (67) profile alignment option (the alignment includes the untranslated leader sequence, gag, pol, vif, vpr, tat, rev, vpu, env, nef, and available 3′ LTR sequences). Sequences that had to be excluded from any particular analysis were removed only after gap tossing was performed on the complete alignment containing all sequences. This ensured that all positions were comparable in different runs with different sequences. The complete genome alignment is available upon request.
Diversity plots.
The percent diversity between selected pairs of sequences was determined by moving a window of 500 bp along the genome alignment in 10-bp increments. The divergence values for each pairwise comparison were plotted at the midpoint of the 500-bp segment.
Bootstrap plots.
Bootscanning was performed on neighbor-joining trees by using SEQBOOT, DNADIST (with Kimura’s correction), NEIGHBOR, and CONSENSUS from the PHYLIP package (14) for a window of 500 bp moving along the alignment in increments of 10 bp. We evaluated 1,000 replicates for each phylogeny. The program ANALYZE from the bootscanning package (57) was used to examine the clustering of the putative hybrid with representatives of the subtypes presumed to have been involved in the recombination event. The bootstrap values for these sequences were plotted at the midpoint of each window.
Exploratory tree analysis.
Exploratory tree analysis was performed by the bootstrap plot approach described above, except in this case an increment of 100 bp was used and each neighbor-joining tree was viewed with DRAWTREE from the PHYLIP package (14). In addition, all full-length sequences (except known recombinants) were included in the analysis.
Informative site analysis.
To estimate the location and significance of crossovers, each putative hybrid sequence was compared with a representative of each of the two subtypes inferred to have been involved in the recombination event and an appropriate outgroup. Recombination breakpoints were mapped by examining the linear distribution of phylogenetically informative sites supporting the clustering of the hybrid with each of the two “parental” subtypes, essentially as described previously (52, 53). Potential breakpoints were inserted between each pair of adjacent informative sites, and the extent of heterogeneity between the two sides of the breakpoint, with respect to numbers of the two kinds of informative site, was calculated as a 2 × 2 chi-square value; the likely breakpoint was identified as that which gave the maximal chi-square value. Since the alignments contained more than one putative crossover, this analysis was performed by looking for one and two breakpoints at a time and repeated on subsections of the alignment defined by breakpoints that had already been identified. To assess the probability of obtaining (by chance) chi-square values as high as those observed, 10,000 random permutations of the informative sites were examined.
DNA transfection and viral infectivity studies.
Ten micrograms of the reconstructed 94UG114.1 plasmid subclone was transfected into 293 T cells by a calcium phosphate precipitation method (2). Two days after infection, cultured supernatants were analyzed for reverse transcriptase (RT) activity and used to infect phytohemagglutinin (PHA)-stimulated normal donor PBMCs (20). Cultures were monitored for virus replication every 3 to 4 days.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the near-full length HIV-1 proviral sequences reported in this study are listed in Table 2.
TABLE 2.
Inactivating mutations in near-complete HIV-1 genomes
| Clone | Defective gene(s) | In-frame stop codona | Frameshift mutationa | Altered initiation codona | Plasmid vectord | GenBank accession no. |
|---|---|---|---|---|---|---|
| 92UG037.1 | pol | 3144 | pCRII | U51190 | ||
| 92BR025.8 | pol | 2141, 3115 | 4131 | pCRII | U52953 | |
| 94UG114.1 | None | pTZ18Mlu1 | U88824 | |||
| 84ZR085.1 | gag/pol | 1692 | pUC19 | U88822 | ||
| 93BR020.1 | None | pCR2.1 | AF005494 | |||
| 90CF056.1 | None | pCR2.1 | AF005496 | |||
| 92RW009.6 | gag | 213 | pTZ18Mlu1 | U88823 | ||
| 93BR029.4 | gag | 260, 472 | pTZ18Mlu1 | AF005495 | ||
| 92NG083.2 | gag, vpu | 360 | 5462b | 157 | pTZ18Mlu1 | U88826 |
| 92NG003.1c | vpr, vpu, nef | 5024b, 5485b | 8113 | pTZ18Mlu1 | U88825 |
RESULTS
Molecular cloning of non-subtype B HIV-1 isolates.
The purpose of this study was to (i) molecularly clone a panel of near-full-length reference genomes for non-subtype B isolates of HIV-1, (ii) determine their nucleotide sequence and phylogenetic relationships, and (iii) generate proviral constructs for biological and functional studies. To accomplish this, we selected 10 geographically diverse HIV-1 isolates, 7 of which had previously been classified as members of (group M) subtypes A (92UG037 and 92RW009), C (92BR025), F (93BR020 and 93BR029), and G (92NG003 and 92NG083) on the basis of env (17, 19) and/or gag sequences (1). The remaining three (84ZR085, 90CF056, and 94UG114) were chosen because they originated from major epicenters of the African AIDS epidemic, including a potential vaccine evaluation site (94UG114). In addition, 90CF056 was of interest because it did not fall into any known subtype at the time of its first genetic characterization (43). Table 1 summarizes available demographic and clinical information, as well as biological data concerning the isolate phenotype (SI/NSI). Only viruses grown in normal donor PBMCs were selected for analysis.
Of the 10 viral genomes, 9 were cloned by long-PCR methods with primers homologous to the tRNA primer binding site (upstream primer) and the polyadenylation signal in the 3′ LTR (downstream primer). This amplification strategy generated near-full-length genomes containing all coding and regulatory regions, except for 70 to 80 bp of 5′ unique LTR sequences (U5). All isolates, regardless of subtype classification, yielded long-PCR products with the same set of primer pairs. In some instances, genomes were amplified with primers containing _Mlu_I restriction enzyme sites. This greatly facilitated subsequent subcloning into a plasmid vector (Table 2). One provirus (84ZR085.1) was cloned by standard lambda phage techniques (36) with _Sac_I sites in the viral LTRs as the cloning enzymes.
Sequence analysis of near-full-length HIV-1 genomes.
All 10 HIV-1 genomes were sequenced in their entirety by either shotgun sequencing or primer-walking approaches. The long-PCR-derived clones ranged in size from 8,952 to 8,999 bp and spanned the genome from the primer binding site to the R/U5 junction of the 3′ LTR. The lambda phage-derived 84ZR085.1 genome was 8,975 bp in length and ranged from the 5′ TAR domain to the 3′ U3 region (unlike most other HIV-1 strains, 84ZR085.1 contains two _Sac_I sites in the LTR). Inspection of potential coding regions revealed that all clones contained the expected reading frames for gag, pol, vif, vpr, tat, rev, vpu, env, and nef. In addition, all major regulatory sequences, including promoter and enhancer elements in the LTR, the packaging signal, and splice sites, appeared to be intact. None of the genomes had major deletions or rearrangements, although inspection of the deduced protein sequences identified inactivating mutations in 7 of the 10 clones (Table 2). However, most of these were limited to point mutations in single genes and were thus amenable to repair. Only two genomes (92NG003.1 and 92NG083.2) contained stop codons, small deletions, and frameshift mutations in several genes, rendering them multiply defective. Importantly, no inactivating mutations were identified in 94UG114.1, 93BR020.1, and 90CF056.1, suggesting that these clones encoded biologically active genomes (Table 2).
Phylogenetic analyses in gag and env regions.
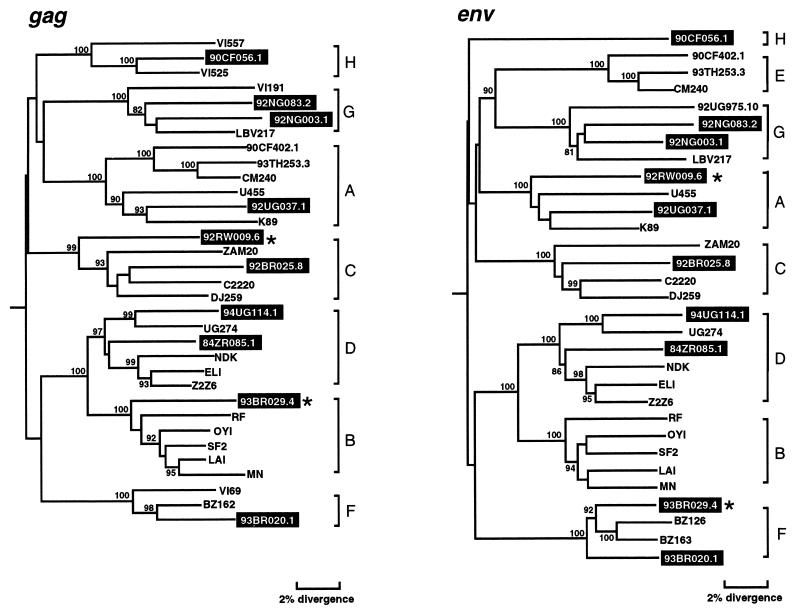
To determine the phylogenetic relationships of the newly characterized viruses, we first constructed evolutionary trees from full-length gag and env sequences. This was done to confirm the authenticity of previously characterized strains, classify the new viruses, and compare viral branching orders in trees from two genomic regions. The results confirmed a broad subtype representation among the selected viruses (Fig. 1). Strains fell into six of the seven major (non-B) clades, including three for which full-length sequences are not available (i.e., F, G, and H). However, comparison of the gag and env topologies also identified two strains with discordant branching orders. 92RW009.6 grouped with subtype C viruses in gag but with subtype A viruses in env. Similarly, 93BR029.4 clustered with subtype B viruses in gag but with subtype F viruses in env. These different phylogenetic positions were supported by high bootstrap values and thus indicated that these two strains were intersubtype recombinants.
FIG. 1.
Phylogenetic relationships of the newly characterized viruses (highlighted) to representatives of all major HIV-1 (group M) subtypes in gag and env regions. Trees were constructed from full-length gag and env nucleotide sequences by using the neighbor-joining method (see the text for details of the method). Horizontal branch lengths are drawn to scale (the scale bar represents 0.02 nucleotide substitution per site); vertical separation is for clarity only. Values at the nodes indicate the percent bootstraps in which the cluster to the right was supported (bootstrap values of 75% and higher are shown). Asterisks denote two hybrid genomes with discordant branching orders in gag and env trees. Brackets on the right represent the major sequence subtypes of HIV-1 group M. Trees were rooted by using SIVcpzGAB as an outgroup.
Diversity plots.
To characterize the two putative recombinants as well as the other eight strains in regions outside gag and env, we performed pairwise sequence comparisons with available full-length sequences from the database. A multiple genome alignment was generated which included the new sequences as well as U455 (subtype A); LAI, RF, OYI, MN, and SF2 (subtype B); C2220 (subtype C); ELI, NDK, and Z2Z6 (subtype D); and 90CF402.1, 93TH253.3, and CM240 (“subtype E”). The percent nucleotide sequence diversity between sequence pairs was then calculated for a window of 500 bp moved in steps of 10 bp along the alignment. Importantly, distance values were calculated only after all sites with a gap in any of the sequences were removed from the alignment. This ensured that all comparisons were made across the same sites.
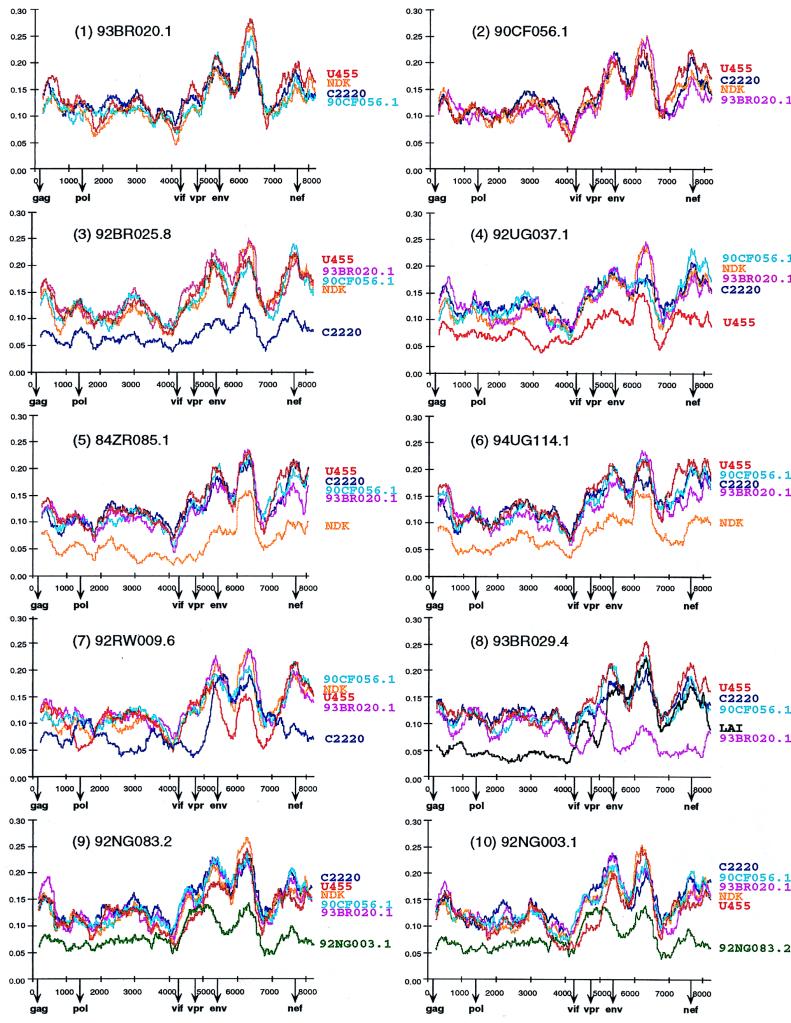
Figure 2 depicts selected distance plots for the newly characterized viruses. For example, in panel 1, 93BR020.1 (putative subtype F) is compared to U455 (subtype A), NDK (subtype D), C2220 (subtype C), and 90CF056.1 (putative subtype H). The resulting plots all exhibit very similar diversity profiles characterized by alternating regions of sequence variability and conservation (values range from 7% divergence near the 5′ and 3′ ends of pol to 30% in the segment of env encoding the V3 region). Moreover, the four plots are virtually superimposable, indicating that 93BR020.1 is roughly equidistant from U455, NDK, C2220, and 90CF056.1 over the entire length of its genome. A very similar set of distance curves was also obtained from comparisons of 90CF056.1 with 93BR020.1, U455, NDK, and C2220 (panel 2) and from comparisons of both 93BR020.1 and 90CF056.1 with representatives of subtype B and “E” (data not shown). These results indicating that 93BR020.1 and 90CF056.1 are equidistant from each other as well as from members of subtypes A, B, C, D, and “E,” together with the gag and env phylogenetic trees (Fig. 1), suggest that 93BR020.1 and 90CF056.1 represent nonrecombinant members of subtypes F and H, respectively.
FIG. 2.
Diversity plots comparing the sequence relationships of the newly characterized viruses to each other and to reference sequences from the database. In each panel, the sequence named above the plots is compared to the sequences listed on the right (sequences are color coded). U455, LAI, C2220, and NDK are published reference sequences for subtypes A, B, C, and D, respectively (45). Distance values were calculated for a window of 500 bp moving in steps of 10 nucleotides. The x axis indicates the nucleotide positions along the alignment (gaps were stripped and removed from the alignment). The positions of the start codons of the gag, pol, vif, vpr, env, and nef genes are shown. The y axis denotes the distance between the viruses compared (0.05 = 5% divergence).
Very similar data were also obtained when 92BR025.8, 92UG037.1, 84ZR085.1, and 94UG114.1 were subjected to diversity plot analysis with the same set of reference sequences (Fig. 2, panels 3 to 6). Again, distance curves exhibited very similar profiles indicating approximate equidistance among the strains analyzed, except when viruses from the same subtype were compared. For example, in panel 3, distances between 92BR025.8 (putative subtype C) and U455, 93BR020.1, 90CF056.1, NDK, and C2220 are depicted. As expected, the C2220 plot falls clearly below all others, indicating the lower level of sequence divergence between viruses from the same subtype (ranging from 4% in pol to 12% in env). Importantly, however, inter- and intradiversity plots follow each other very closely; i.e., the same genomic regions exhibit proportionally higher and lower levels of divergence (also see panels 4 to 6). Thus, at the level of both inter- and intrasubtype comparisons, there was no evidence of mosaicism in the genomes of these four viruses. Together with the results in Fig. 1, this suggests that these strains represent nonmosaic members of subtypes A (92UG037.1), C (92BR025.8), and D (84ZR085.1 and 94UG114.1), respectively.
By contrast, the diversity plots of the putative recombinants 92RW009.6 and 93BR029.4 exhibited disproportionate levels of sequence divergence from different subtypes along their genome, consistent with their discordant branching orders in gag and env trees. As shown in Fig. 2, panel 7, 92RW009.6 is most similar to the subtype C strain C2220 in the 5′ half of gag, most of pol, vif, vpr, as well as nef (the dark blue curve falls below all others). However, in the 3′ end of gag, the 5′ end of pol, and most of env, 92RW009.6 is most similar to the subtype A strain U455 (the red curve falls below all the others). Similarly in panel 8, 93BR029.4 is most similar to the subtype B strain LAI (black curve) in gag, pol, and vpr, while it is most similar to the putative subtype F strain 93BR020.1 (magenta curve) in the vif, env, and nef regions. In each case, the magnitude of the difference between the new sequence and the most similar subtype was no greater than the diversity seen within subtypes. Thus, these data suggest that 92RW009.6 and 93BR029.1 represent mosaics, comprised of subtypes A/C and B/F, respectively. In each case, the plots suggested several (at least four) crossovers; these are the minimum number of recombination breakpoints, since the window size used makes it unlikely that recombinant regions shorter than 500 bp would be detected.
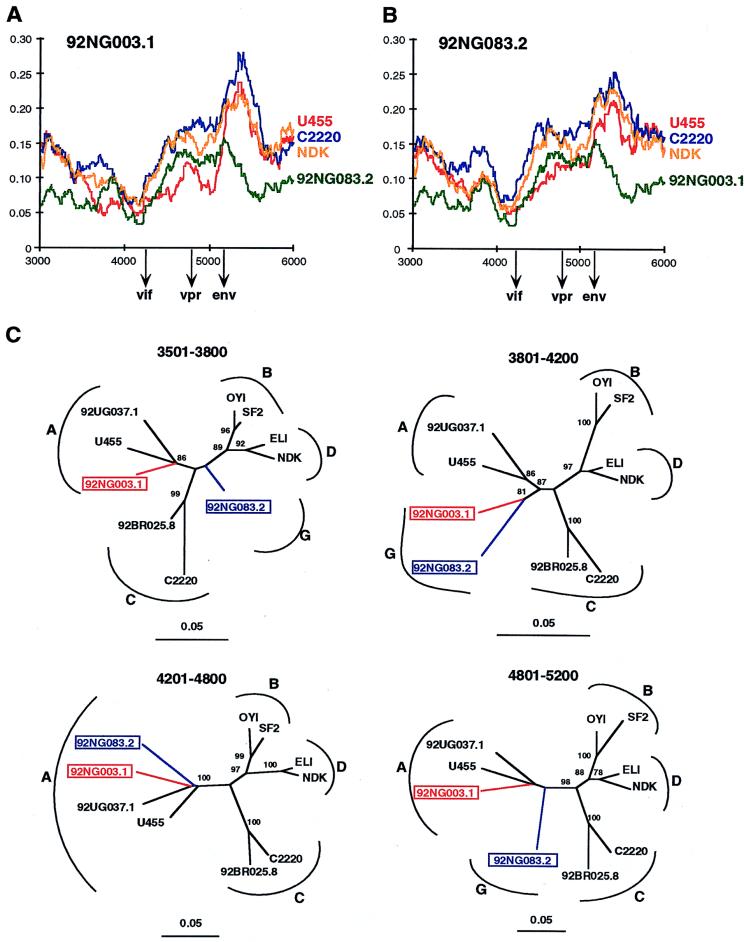
Finally, inspection of the diversity plots for 92NG003.1 and 92NG083.2 also revealed disproportionate levels of sequence variation, although not as pronounced as for 92RW009.6 and 93BR029.4. As shown in Fig. 2, panels 9 and 10, 92NG003.1 and 92NG083.2 are equidistant from members of subtypes A, C, D, F, and H (as well as B and “E” [data not shown]) for most of their genome, suggesting that they represent an independent subtype, i.e., subtype G. However, in the vif/vpr region, the U455 distance plot falls below all others (including the 92NG003.1/92NG083.2 distance plot depicted in green in panels 9 and 10), suggesting a disproportionately closer relationship to subtype A. Assuming that U455 is nonmosaic, these results suggest that both 92NG003.1 and 92NG083.2 contain short fragments of subtype A sequence in the central region of their genome.
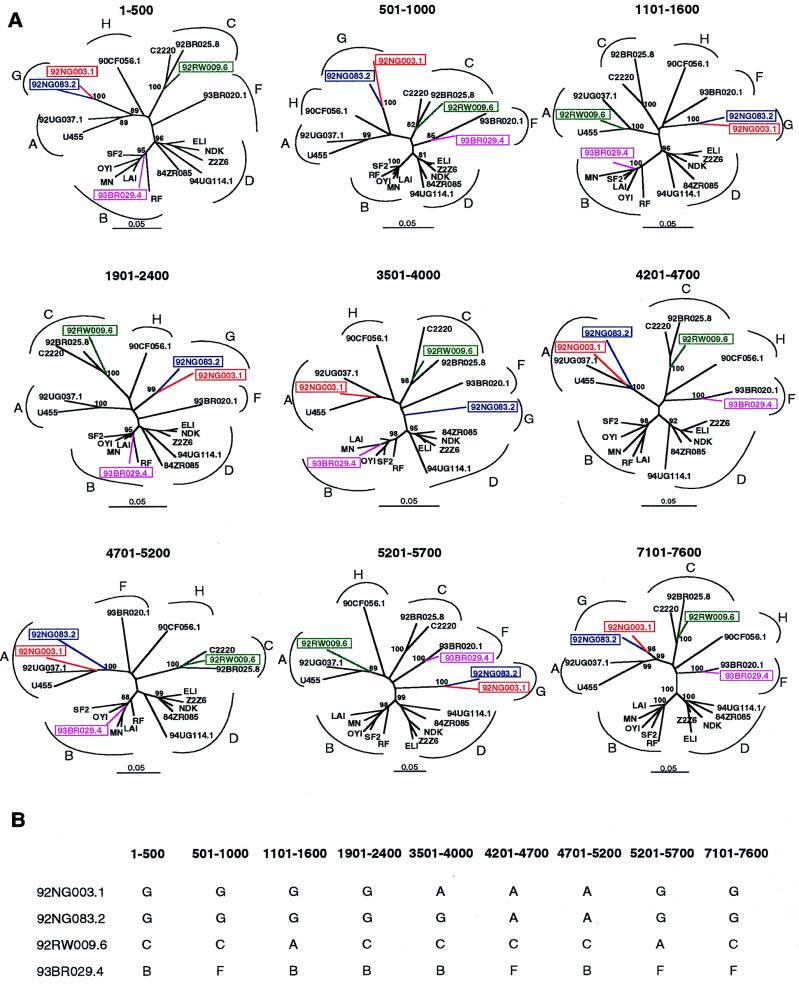
Exploratory tree analyses.
To examine the phylogenetic position of the newly derived strains relative to each other and to the reference sequences over the entire genome, we performed exploratory tree analyses by using the same multiple genome alignment generated for the diversity plots (Fig. 3). A total of 79 trees were constructed for overlapping fragments of 500 bp, moving in 100-bp increments along the alignment. As expected, four genomes that clustered in different subtypes in different parts of their genome were identified (representative trees are depicted in Fig. 3A). These included 93BR029.4, which alternated between subtypes F and B, 92RW009.6, which alternated between subtypes A and C, and 92NG083.2 and 92NG003.1, which grouped either independently or within subtype A. Interestingly, the last two strains exhibited distinct patterns of mosaicism. In trees spanning the region from 3501 to 4000, 92NG003.1 clustered within subtype A while 92NG083.2 clustered independently, presumably representing subtype G (Fig. 3B). In contrast to these strains, there was no evidence for a hybrid genome structure in 92UG037.1, 92BR025.8, 94UG114.1, 84ZR085.1, 93BR020.1, or 90CF056.1. As shown in Fig. 3A, these viruses branched consistently in all regions analyzed. Based on these findings and the results of the diversity plots, we thus concluded that 6 of the 10 selected HIV-1 strains represent nonrecombinant reference strains for subtypes A (92UG037.1), C (92BR025.8), D (94UG114.1 and 84ZR085.1), F (93BR020.1), and H (90CF056.1), respectively, while four are intersubtype recombinants.
FIG. 3.
Exploratory tree analysis. (A) Neighbor-joining trees were constructed for a 500-bp window moving in increments of 100 bp along the multiple genome alignment. Trees depicting discordant branching orders among the newly determined sequences are shown (hybrid sequences are boxed and color coded). The position of each tree in the alignment is indicated; subtypes are identified by curved brackets. Numbers at the nodes indicate the percentage of bootstrap values with which the adjacent cluster is supported (only values above 80% are shown). Branch lengths are drawn to scale. (B) Summary of the subtype assignments of the four recombinants illustrated in panel A.
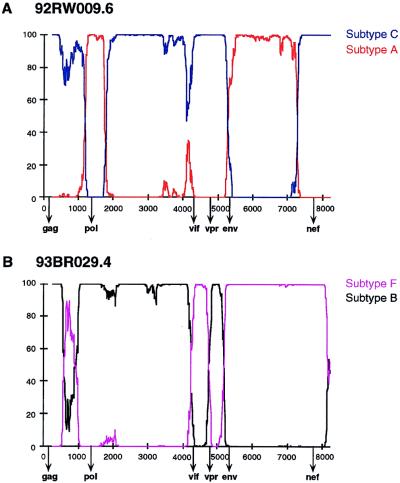
Recombination breakpoint analysis in 92RW009.6 and 93BR029.4.
To map the location of the recombination breakpoints in 92RW009.6 and 93BR029.4, we used bootstrap plots and informative site analyses (18, 52, 53). Unrooted trees which included U455, 92UG037.1, LAI, MN, OYI, SF2, RF, C2220, 92BR025.1, NDK, ELI, Z2Z6, 93BR020.1, and 90CF056.1 were constructed; then the magnitudes of the bootstrap values supporting (i) the clustering of 92RW009.6 with members of subtype A (U455 and 92UG037.1) or C (2220 and 92BR025.8) and (ii) the clustering of 93BR029.4 with members of subtype B (LAI, MN, OYI, MN, and RF) or F (92BR020.1) were determined (in the latter case, subtype D viruses were excluded because of their known close relationship to subtype B viruses). Figure 4 depicts the results of 797 such phylogenetic analyses generated for each genome, performed on a window of 500 nucleotides and moving in steps of 10 nucleotides. Very high bootstrap values (>80%) supporting the clustering of 92RW009.6 with subtype C were apparent in gag, the 3′ two-thirds of pol, and nef. By contrast, significant branching of 92RW009.6 with subtype A was apparent in the gag/pol overlap and the env region. In a small region (positions 4000 to 4200) in the middle of the genome, 92RW009.6 appeared not to cluster significantly with either subtype, but further inspection revealed that this was due to a small number of informative sites. These data thus indicated four points of recombination crossovers between subtypes A and C (Fig. 4A). A similar analysis identified six recombination breakpoints between subtypes B and F in 93BR029.4 (Fig. 4B). These included two more (in gag) than were apparent from the diversity-plot analysis (compare Fig. 2), indicating a greater sensitivity of this approach.
FIG. 4.
Recombination breakpoint analysis for 92RW009.6 and 93BR029.4. (A) Bootstrap plots depicting the relationship of 92RW009.6 to representatives of subtype A (red) and C (blue), respectively. Trees were constructed from the multiple genome alignment, and the magnitude of the bootstrap value supporting the clustering of 92RW009.6 with U455 and 92UG037.1 (subtype A) or with C2220 and 92BR025.8 (subtype C), respectively, was plotted for a window of 500 bp moving in increments of 10 bp along the alignment. Regions of subtype A or C origin are identified by very high bootstrap values (>90%). Points of crossover of the two curves indicate recombination breakpoints. The beginnings of gag, pol, vif, vpr, env, and nef open reading frames are shown. The y axis indicates the percentage of bootstrap replicates which support the clustering of 92RW009.6 with representatives of the respective subtypes. (B) Bootstrap plots depicting the relationship of 93BR029.4 to representatives of subtypes B (black) and F (magenta), respectively. Analyses are as in panel A, except that the bootstrap values supporting the clustering of 93BR029.4 with SF2, OYI, MN, LAI, and RF (subtype B) or with 93BR020.1 (subtype F), respectively, were plotted. Subtype D viruses were excluded from this analysis because of their known close relationship to subtype B viruses.
To map the recombination crossover points in 92RW009.6 and 93BR029.1 more precisely, we examined the distribution of phylogenetically informative sites supporting alternative tree topologies (52, 53). Briefly, this was done in a four-sequence alignment which included the query sequence, a representative of each of the two subtypes presumed to have been involved in the recombination event, and an outgroup. Breakpoints were identified by looking for statistically significant differences in the ratios of sites supporting one topology over another. Consistent with the bootscanning data, this analysis identified four breakpoints in 92RW009.6 (Table 3) and six in 93BR029.4 (Table 4). A schematic representation of the mosaic genomes of 92RW009.6 and 93BR029.4 is depicted in Fig. 6 (below).
TABLE 3.
Informative-site analysis of 92RW009.6
| Regiona | Subtype | No. of informative sites in: | ||
|---|---|---|---|---|
| Subtype A (U455) | Subtype C (C2220) | Outgroup (NDK) | ||
| 1–1037 | C | 8 | 32 | 8 |
| 1085–1940 | A | 17 | 5 | 4 |
| 1986–5288 | C | 18 | 99 | 27 |
| 5293–7238 | A | 60 | 9 | 13 |
| 7254–8431 | C | 12 | 55 | 12 |
TABLE 4.
Informative-site analysis of 93BR029.4
| Regiona | Subtype | No. of informative sites in: | ||
|---|---|---|---|---|
| Subtype B (LAI) | Subtype F (93BR020.1) | Outgroup (C2220) | ||
| 1–735 | B | 18 | 6 | 3 |
| 755–896 | F | 1 | 10 | 0 |
| 930–4247 | B | 99 | 10 | 14 |
| 4340–4668 | F | 2 | 15 | 1 |
| 4787–5166 | B | 15 | 0 | 5 |
| 5244–8242 | F | 15 | 139 | 13 |
| 8250–8429 | B | 13 | 0 | 0 |
FIG. 6.
Inferred structures of the four recombinant genomes characterized in this study. Regions of different subtype origin are color coded. Uncertain breakpoints are hatched. LTR sequences were not analyzed and are shown as open boxes.
Recombination breakpoint analysis in 92NG003.1 and 92NG083.2.
Because of the lack of a full-length subtype G reference sequence, recombination breakpoint analysis of 92NG003.1 and 92NG083.2 required a different approach. The analyses, summarized in Fig. 2 and 3, suggested that these two viruses contained subtype A sequences in the middle of their genome. To attempt to confirm this and to define the extent of these putative subtype A fragments, we performed a more detailed diversity plot analysis of the viral middle region (between positions 3000 and 6000) by using different viral strains and window sizes (ranging from 200 to 400 bp) to examine the extent of sequence divergence of 92NG083.2 and 92NG003.1 from members of other subtypes, including subtype A. Figures 5A and B depict representative results (with a window size of 300 bp moving in steps of 10 bp along the alignment). Similar to the data shown in Fig. 2, the two “subtype G” viruses are roughly equidistantly related to members of subtypes A (U455), C (C2220), and D (NDK), except for two regions in 92NG003.1 and one region in 92NG083.2, where both viruses are disproportionately more closely related to U455 than they are to each other (the red line drops below the green line). By noting the points at which the “G”-A distance increases or decreases relative to the others, we could tentatively identify recombination breakpoints. For example, at position 3400 in Fig. 5A, the U455 plot (red) falls whereas the C2220 (blue), NDK (yellow), and 92NG083.2 (green) plots do not, and around position 3600, the U455 plot crosses the 92NG083.2 plot. Bearing in mind the window size of 300 nucleotides, this finding suggested that a recombination crossover occurred around position 3500. Similar “G”-A plot crossings around positions 3800, 4200, and 5200 in Fig. 5A and around positions 4200 and 4800 in Fig. 5B suggested additional recombination breakpoints.
FIG. 5.
Recombination breakpoint analysis of 92NG083.2 and 92NG003.1. (A and B) Diversity plots comparing the sequence relationships of 92NG003.1 and 92NG083.2 to each other and to reference sequences from the database. In both panels, the sequence named above the plots is compared to the sequences listed on the right (sequences are color coded). U455, C2220, and NDK are published reference sequences for subtypes A, C, and D, respectively (45). Distance values were calculated for a window of 300 bp moving in steps of 10 nucleotides. The x axis indicates the nucleotide positions along the alignment (gaps were stripped and removed from the alignment). The positions of the start codons of the vif, vpr, and env genes are shown. The y axis denotes the distance between the viruses compared (0.05 = 5% divergence). (C) Neighbor-joining trees depicting discordant branching orders of 92NG003.1 and 92NG083.2 in regions delineated by breakpoints identified in panels A and B (hybrid sequences are boxed and color coded). The position of each tree in the alignment is indicated; subtypes are identified by curved brackets. Numbers at the nodes indicate the percentage of bootstrap values with which the adjacent cluster is supported (only values above 80% are shown). Branch lengths are drawn to scale.
We then constructed phylogenetic trees by using the regions of sequence defined by these putative breakpoints (Fig. 5C). This analysis generally supported the conclusions drawn from the diversity plots (i.e., 92NG003.1 clustered with subtype A viruses in the region between 3501 and 3800, whereas 92NG083.2 did not; and both 92NG003.1 and 92NG083.2 clustered with subtype A viruses in the region 4201 and 4800). However, neither the diversity plot nor the tree analysis allowed us to define the boundaries of the subtype A fragments with certainty. Nevertheless, the data indicated that (i) both 92NG083.2 and 92NG003.1 represent G/A recombinants, (ii) they are the result of different recombination events because some of their breakpoints are clearly different, and (iii) 92NG083.2 probably encodes a nonrecombinant pol gene. A schematic representation of the mosaic genomes of 92NG083.2 and 92NG003.1 is shown in Fig. 6, with shaded areas indicating regions of uncertain subtype assignment.
Reevaluation of the phylogenetic position of subtype G viruses in the gp41 region.
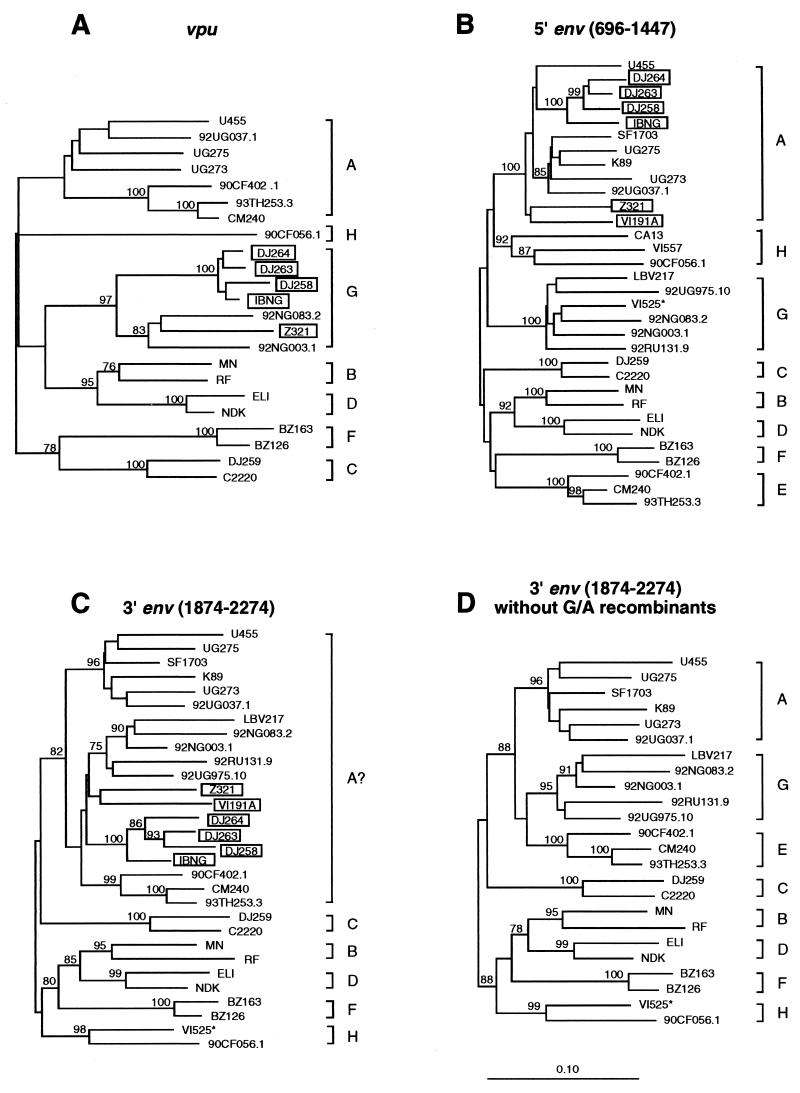
We (19) and others (40) previously reported that the env genes of subtype “G” viruses are chimeric, with sequences encoding the intracellular portion of gp41 clustering in subtype A. We were therefore surprised that neither the diversity plot nor the exploratory tree analysis provided evidence for a closer relationship of 92NG003.1 and 92NG083.2 to U455 and 92UG037.1 in this region. To investigate this further, we performed extensive tree analyses in the vpu/env region, including as many reference sequences for the various group M subtypes as were available (Fig. 7; for subtypes B and “E,” only a few representatives are shown). The results revealed that a number of viruses previously classified as subtype A in the extracellular domain of env (gp120) fell into subtype G in the vpu region (boxed viruses in Fig. 7A and B). Exclusion of these obvious recombinants from gp41 tree analyses changed the grouping of 92NG003.1 and 92NG083.3 as well as that of all other subtype G viruses. Instead of falling into a larger “subtype A cluster” (labelled “A?” in Fig. 7C), they grouped independently from both subtype A and E viruses, i.e., as subtype G, with high bootstrap values (Fig. 7D; also note that VI525 clusters in subtype H in the intracellular region of gp41, and not in subtype G, as assumed in reference 19). The inadvertent inclusion of recombinants was thus responsible for our previous erroneous classification of subtype G viruses as “A” at the 3′ end of gp41.
FIG. 7.
Phylogenetic relationships of subtype G (and “E”) viruses in vpu and env regions. Trees were constructed for the vpu (A), 5′ env (B), and 3′ env (C and D) regions to reexamine the subtype associations of previously classified subtype A, G, and “E” viruses (19). Several strains (boxed) previously thought to represent subtype A (panel B) were found to cluster in subtype G viruses in the vpu region (panel A). Exclusion of these G/A recombinants changed the topology of trees derived from the intracellular gp41 domain (panels C and D). VI525 (highlighted by an asterisk) was identified as a G/H recombinant, clustering in subtype G and H in the extracellular and intracellular portions of env, respectively. All known representatives for the different subtypes were included in the analysis, and only a few representatives for subtypes B and “E” are shown.
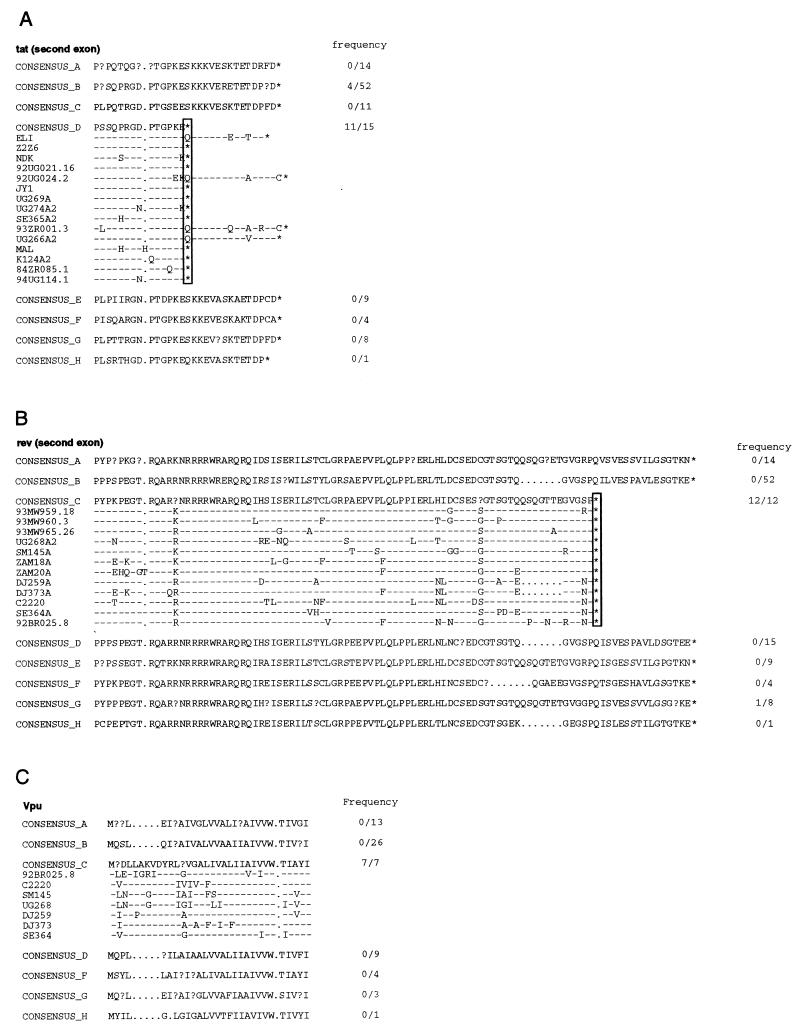
Subtype-specific genome features.
Having classified the 10 new viruses with respect to their subtype assignments, we examined their sequences for clade-specific signature sequences. Comparing deduced amino acid sequences gene by gene, we found several subtype-specific features (Fig. 8). For example, most subtype D viruses (including 84ZR085.1 and 94UG114.1) contain an in-frame stop codon in the second exon of tat, which removes 13 to 16 amino acids from the carboxy terminus of the Tat protein (Fig. 8A). Similarly, all subtype C viruses (including 92BR025.8) contain a stop codon in the second exon of rev, which would be predicted to shorten this protein by 16 amino acids (Fig. 8B). Subtype C viruses also contain a 15-bp insertion at the 5′ end of the vpu gene (Fig. 8C), which extends the putative membrane-spanning domain of the Vpu protein by 5 amino acids (data not shown). Although these changes are unlikely to alter the function of the respective gene products in a major way (e.g., the known functional domains of both Tat and Rev proteins are not affected by these changes), it is possible that they could influence their mechanism of action in a subtle (but nevertheless biologically important) manner. However, direct experimentation is necessary to examine this possibility.
FIG. 8.
Subtype-specific genome features. (A) Alignment of deduced Tat (region encoded by second exon) amino acid sequences. Consensus sequences were generated for available representatives of all major subtypes (question marks indicate sites at which fewer than 50% of the viruses contain the same amino acid residue). Dashes denote sequence identity with the consensus sequence, while dots represent gaps introduced to optimize the alignments. A vertical box highlights a premature Tat protein truncation (asterisk) which is present in 11 of 15 subtype D and 4 of 52 subtype B viruses (frequencies are listed in the column on the right). (B) Alignment of deduced Rev (region encoded by the second exon) protein sequences. (C) Alignment of deduced Vpu protein sequences.
Inspection of the sequences also revealed the lack of a previously identified signature sequence in one of the newly characterized viruses. 92BR025.8 was found to encode only two potential NF-κB binding sites in its core enhancer region (data not shown). By contrast, all other subtype C viruses, including several African isolates from Ethiopia, Zambia, and Malawi (59), as well as two additional isolates from Brazil and two from India (16), encode three NF-κB binding sites.
Construction of a replication-competent 94UG114.1 provirus.
Long-PCR approaches generally fail to generate replication-competent clones of HIV-1 because of sequence redundancies in the LTRs. Portions of the LTRs have to be added in additional cloning steps to generate a complete set of regulatory sequences required for viral DNA synthesis and reverse transcription. Although LTR sequences from any subtype (e.g., subtype B) would probably restore functionality, such chimeric proviruses could differ in their biological properties (56). To generate genomes that represent more faithfully their corresponding isolates, we have devised an amplification and cloning strategy that allows the construction of a replication-competent provirus in a two-step process (Fig. 9A). Briefly, both the 5′ LTR and a fragment containing the remainder of the genome are amplified from the same isolate DNA by regular PCR and long-PCR approaches, respectively. Both products are then subcloned into a plasmid which contains restriction enzyme sites suitable for the subsequent joining of the two fragments into a single vector. For 94UG114.1, we used _Nar_I, a unique enzyme site present in the primer binding site of all known group M and O strains of HIV-1 (45), in combination with _Mlu_I, a non-cutter of almost all HIV-1 genomes (53 of 55 complete HIV-1 sequences in the database are not cleaved by _Mlu_I [45]). The latter enzyme site was introduced via the PCR primers (Fig. 9A).
FIG. 9.
Generation of replication-competent proviral clones from long-PCR products. (A) Construction of a replication-competent 94UG114.1 provirus from two separately amplified genomic regions (see the text for details). (B) Replication potential of 94UG114.1 in primary PBMC cultures. Normal donor PBMCs were isolated, PHA stimulated and then infected with equal amounts (based on p24 antigen content) of 94UG114.1 and SG3 viruses derived from 293T transfections of proviral DNA. Virus production was monitored by measuring supernatant RT activity at 3-day intervals as described previously (20). Supernatants from a mock-transfected culture served as a negative control.
Following reconstruction, the 94UG114.1 full-length clone was transfected into 293T cells, together with positive (SG3 [20]) and negative (plasmid) control constructs. Analysis of culture supernatants revealed positive RT and p24 activity, consistent with the expression of functional gag, tat, rev, and pol gene products. Subsequent cell-free transmission of culture fluids to PHA-stimulated normal donor PBMCs established that 94UG114.1 was infectious for and grew well in natural target cells (Fig. 9B). Moreover, its replication profile was comparable to that of the highly cytopathic SG3 strain (20), indicating efficient _env_-mediated fusion and spread in the culture. These results thus document that the long-PCR-derived 94UG114.1 genome encodes functional gene products and represents a replication competent proviral clone (reconstruction of some of the other clones is under way).
DISCUSSION
Non-subtype B viruses cause the vast majority of new HIV-1 infections worldwide, yet they are only infrequently studied with respect to their biological, immunogenic, and pathogenic properties, in part because well-characterized virological reference reagents are still lacking. In this study, we selected 10 non-subtype B isolates from various geographic locations and cloned their genomes by using long-PCR or lambda phage techniques. All the genomic clones were derived from primary (PBMC-derived) isolates and thus represent biologically relevant viruses. Detailed phylogenetic analysis identified six of these viruses as nonrecombinant members of subtypes A, C, D (two), F, and H, which more than doubles the number of non-subtype B reference strains available (Table 5). Among these, the near-full-length genomes of 93BR020.1 and 90CF056.1 represent the first such strains for subtypes F and H, respectively. The four other viruses were found to represent complex mosaics of subtypes A and C, A and G (two), and B and F. Both A/G recombinants originated from Nigeria but must have arisen from independent recombination events since they are not closely related and differ in their patterns of mosaicism. One of these (92NG083.2) appears to contain only a single short (perhaps 600-bp) segment of subtype A origin in the vif/vpr region, and in the absence of (as yet) any full-length subtype G virus, it thus serves as a (nonmosaic) subtype G representative for the gag, pol, env, and nef regions. Importantly, 9 of the 10 genomes were generated in such a way that they can be tested for biological activity following a simple reconstruction step. An example of such a reconstructed genome giving rise to replication competent virus (94UG114.1) demonstrates that this approach is feasible.
TABLE 5.
Full-length group M HIV-1 sequences in the database
| Subtype | Clonea | Source | Cloning method | Replication competence | Defective gene(s) | GenBank accession no. |
|---|---|---|---|---|---|---|
| A | U455 | U937 | Phage | − | vpr, vpu, env | M62320 |
| 92UG037.1 | PBMC | PCR | − | pol | U51190 | |
| B | LAI/BRU | PBMC | Phage | +c | None | K02013 |
| BH10 | H9 | Phage | +c | vpr, nef | X01762 | |
| PV22 | H9 | Phage | NA | vpr | K02083 | |
| PM213 | NAe | NA | NA | vpr, vpud | D86069 | |
| MCK1 | NA | NA | NA | vpr | D86068 | |
| LW12-3 | H9 | Phage | +c | vif, vpr | U12055 | |
| HXB2 | H9 | Phage | + | nef, vpr, vpud | M38432 | |
| TH475b | TH4-7-5 | PCR | − | vpr | L31963 | |
| NY5 | A3.01 | Phage | − | vpud | M38431 | |
| SF2 | HUT-78 | Phage | + | vpu | K02007 | |
| P89.6 | PBMC | Phage | +c | None | U39362 | |
| RF | H9 | Phage | − | gag, vpu | M17451 | |
| ACH320.2A.1.2 | PBMC | Phage | + | None | U34603 | |
| ACH320.2A.2.1 | PBMC | Phage | + | None | U34604 | |
| AD8 | PBMC | Phage | +c | vpud | AF004394 | |
| D31 | NA | NA | NA | None | U43096 | |
| CAM1 | NA | NA | NA | vpu | D10112 | |
| F12 | HUT-78 | NA | NA | vpr | Z11530 | |
| SG3 | HUT-78 | Phage | + | vpu | L02317 | |
| WEAU | H9 | Phage | + | nef | U21135 | |
| YU2 | Brain tissue | Phage | +c | vpud | M93258 | |
| YU10 | Brain tissue | Phage | − | pol | M93259 | |
| MN | H9 | Phage | − | pol, nef, vpu | M17449 | |
| HAN2/3 | MT2 | Phage | − | env | U43141 | |
| JRCSF | PBMC | Phage | +c | None | M38429 | |
| JRFLb | PBMC | Phage | +c | None | U63632 | |
| OYIb | PBMC | Phage | − | vpud | M26727 | |
| C18MBCb | PBMC | PCR | − | nef | U37270 | |
| MANCb | Kidney tissue | PCR | − | None | U23487 | |
| WR27 | PBMC | PCR | − | NA | U26546 | |
| C | C2220 | PBMC | PCR | − | tat | U46016 |
| 92BR025.8 | PBMC | PCR | − | pol | U52953 | |
| D | NDK | CEM | Phage | + | None | M27323 |
| Z2Z6 | A3.01 | Phage | +c | None | M22639 | |
| ELI | PBMC | Phage | +c | None | X04414 | |
| 84ZR085.1 | PBMC | Phage | − | gag, pol | U88822 | |
| 94UG114.1 | PBMC | PCR | + c | None | U88824 | |
| F | 93BR020.1 | PBMC | PCR | − | None | AF005494 |
| H | 90CF056.1 | PBMC | PCR | − | None | AF005496 |
| R | 90CF402.1 A/E | PBMC | Phage | +c | vif, vpud | U51188 |
| 93TH253.3 A/E | H9 | Phage | − | env, vpud | U51189 | |
| CM240 A/E | PBMC | PCR | − | vpud | U54771 | |
| MAL A/D/I/? | PBMC | Phage | +c | vpud | K03456 | |
| ZAM184 A/C | PBMC | PCR | − | None | U86780 | |
| 92RW009.6 A/C | PBMC | PCR | − | gag | U88823 | |
| 92NG003.1 A/G | PBMC | PCR | − | vpr, vpu, env, nef | U88825 | |
| 92NG083.2 A/C | PBMC | PCR | − | gag, vpu | U88826 | |
| 93BR029.4 B/F | PBMC | PCR | − | gag | AF005495 | |
| IBNGb A/G | PBMC | PCR | − | tat | L39106 | |
| Z321Bb A/G/I/? | CEM3/HUT-78 | PCR | − | vpr, vpu | U76035 |
HIV-1 group M subtypes.
The presence of subtypes within the M group of HIV-1 was first suggested in 1992 on the basis of phylogenetic analysis of env gene sequences, which revealed five approximately equidistant clades within the HIV-1 tree (44). With the determination of additional HIV-1 sequences of diverse origins, 10 subtypes (A to J) have now been described (29, 30, 35, 45), although full-length env sequences are not yet available for subtypes I and J (29, 30). Phylogenetic analysis of gag gene sequences yielded very similar overall results (34), although for some viruses a comparison of their phylogenetic positions in the different trees revealed that they were recombinants (52, 53). Sequences for the third major retrovirus gene, pol, have thus far been available only for representatives of four subtypes (45). The data presented in this study thus allow the first estimate of a phylogeny for full-length pol gene sequences based on the sequence information of seven subtypes. The results shown in Fig. 10 are remarkably consistent with those of trees from gag and env regions (compare Fig. 1), demonstrating that the phylogenetic structure implied by the current subtype classification scheme is a real phenomenon.
FIG. 10.
Phylogeny of full-length pol sequences of seven major HIV-1 group M subtypes. The sequences determined in this study are highlighted. Horizontal branch lengths are drawn to scale (the scale bar represents 0.02 nucleotide substitution per site). Vertical separation is for clarity only. Values at the nodes indicate the percentage of bootstraps in which the cluster to the right was supported (bootstrap values of 80% and higher only are shown). Brackets on the right represent the major sequence subtypes of HIV-1 group M. Trees were rooted by using SIVcpzGAB as an outgroup.
HIV-1 intersubtype recombinants.
While the majority of HIV-1 group M sequences fall neatly into the various subtypes discussed above, a substantial minority do not. That is, the phylogenetic position of many viruses differs depending on the genomic region analyzed, indicating that they are mosaics generated by recombination. In our study, 4 of 10 geographically diverse isolates were found to represent intersubtype recombinants. Similarly, 7 of 12 full-length non-subtype B sequences in the database represent recombinants (Table 5). These numbers do not necessarily indicate the actual prevalence of mosaic viruses, because the viruses were not systematically sampled; for example, three of the recombinants in the database are “subtype E” viruses, all descended from a common ancestral recombinant virus and selected for study because of specific interest in their role in the Thai AIDS epidemic (7, 18). However, numerous subgenomic sequences have been identified as mosaic (4, 8, 31, 52, 54, 66, 71). In our initial study (52), about 10% of the database sequences appeared to be intersubtype recombinants, and more recent surveys suggest that this proportion may be increasing (8, 66, 71).
Given the apparent prevalence of mosaic viruses, it is clear that subtype-specific reference strains can be defined as such only after comprehensive recombination analysis. Small subgenomic fragments or even full-length gag and env sequences are not sufficient to identify all hybrid genomes. Although multiple crossovers are a characteristic feature of retroviral recombination and have been found in many of the mosaic HIV-1 genomes examined (7, 19, 53, 60, 62), the examples of 92NG003.1 and 92NG083.2 demonstrate that crossovers may be confined to regions outside of gag and env. Thus, elimination of the possibility that a virus is recombinant requires the determination of substantial (if not all) portions of its genome. As a consequence, subtype-specific reference reagents, such as immunogens for cross-clade CTL and neutralization assays, should be derived only from viral isolates for which a complete genome has been characterized.
These considerations emphasize the need for detailed analyses involving reliable methods for identification of recombinant viral sequences. We have found that diversity plots, depicting the distance between a query sequence and a set of reference sequences in moving windows along the genome, represent an excellent initial screening tool. The extent of sequence divergence (between any pair of viruses) varies along the genome, but since all plots are shown in the same graph, particular regions where the query sequence is anomalously highly similar to (or divergent from) other sequences can be readily identified. For example, this approach uncovered the subtype A-like regions in the middle of the putative “subtype G” genomes 92NG003.1 and 92NG083.2 (Fig. 2, panels 9 and 10; Fig. 5A and B). (An alternative program available from the database [termed RIP] [63] uses a similar approach. RIP identifies windows of sequence in which the query sequence is significantly more similar to the consensus sequence of one particular subtype; if the most similar subtype varies along the sequence, this is a sign that the query sequence is probably a recombinant.) However, the results of such analyses relying only on extents of sequence divergence must be treated with some caution, because they are susceptible to variation in evolutionary rate in different lineages. Once suspicious regions have been identified, phylogenetic analyses of windows of sequence around these regions can be used to look for discordant branching orders and to identify the subtypes likely to have been involved in the recombination event. The bootstrap value supporting the clustering of the query sequence with sequences of the supposed “parental” subtypes can be examined, again in moving windows along the genome. (The bootscanning approach of Salminen et al. [57] is very similar to this.) Finally, informative site analysis can be used to map as precisely as possible the breakpoints of the putative recombination events (52, 53).
Clearly, recombination analysis relies on the availability of accurately defined nonmosaic reference sequences. Thus, location of the breakpoints in the two G/A recombinant viruses identified here must remain tentative because of the lack of such reference sequences for subtype G. The precise positions of breakpoints in the recently characterized Thai and Central African Republic “subtype E” viruses are similarly uncertain (7, 18), in this case because of the lack of a complete nonmosaic subtype E reference sequence. It should also be emphasized that currently designated reference sequences may require revision in the future. For example, the inadvertent inclusion of recombinant “reference” sequences in previous tree analyses (19, 40) led to an incorrect subtype assignment of subtype G gp41 sequences (Fig. 7). It is therefore possible that as more sequences become available, one or more of the viral sequences currently classified as nonrecombinant may be identified as a hybrid.
Relevance of the HIV-1 subtype nomenclature.
The various subtypes differ in their geographic dissemination, and so the subtype designations have been powerful molecular epidemiological markers for tracking the course of the global pandemic (5, 24, 72). For example, the AIDS epidemic in Thailand was initially believed to have resulted from a single introduction of HIV-1. However, genetic analysis revealed that there were in fact two distinct epidemics of different origins: intravenous drug users were infected with subtype B viruses prevalent in the United States and Europe, while commercial sex workers and their contacts harbored (recombinant) “subtype E” viruses common only in Africa (7, 18, 25, 39, 43, 47). These, and other examples (5), have demonstrated the utility of subtyping as a tool to monitor the geographic distribution, prevalence, and intermixing of HIV-1 variants. Nevertheless, some aspects of the current subtype nomenclature are clearly arbitrary and are based on historical facts rather than the application of consistent nomenclature rules. For example, subtype B viruses consistently cluster with subtype D viruses in phylogenetic trees of different genes (61) (Fig. 1 and 10), and the divergence between these two subtypes is hardly any greater than the diversity seen within some other subtypes (e.g., subtype A). This suggests that the HIV-1 epidemic in North America was initiated by a virus that could have been classified as subtype D. Instead, subtype B viruses were designated as a separate subtype, because they happened to be the sole initial focus of attention. Moreover, subtypes are not the only appropriate level of classification in epidemiological tracking. Other (chance?) epidemiological events have led to identifiable geographic and phylogenetic subclusters within subtypes, such as the Thai B clade (frequently referred to as B′) or subclusters with subtype A. Nevertheless, the current subtype classification is likely to remain useful in the molecular epidemiological context.
The subtype classification would be of even greater interest if members of the different subtypes were found to differ in their biological properties. The average values for protein sequence diversity among subtypes for Gag, Pol, and Env are 15, 10, and 24%, respectively (subtype B versus D comparisons were excluded from these calculations for the reasons given above). The neutral theory of molecular evolution (27) notwithstanding, it would be surprising if proteins whose sequences differ by such an extent did not exhibit at least some variation in their biological properties. However, no subtype-specific differences in virus biology have yet been identified. Extensive studies have shown that subtypes do not correlate with neutralization serotypes (38, 42, 46, 68), and even T-cell immune responses appear to be largely independent of genetic subtypes (3, 6, 15). Members of the various subtypes have also not been found to differ in second-receptor usage (73, 74), and a proposed preferential tropism of “subtype E” viruses for skin-derived Langerhans cells (64) has not been confirmed in subsequent investigations (9, 50, 51). Thus, current data have failed to identify simple correlations between phylogenetic lineages and biological phenotypes.
Further consideration of the phylogenetic relationships within the HIV-1 M group (Fig. 1 and 10) yields some insight into the apparent lack of phenotypic correlates at the subtype level. Any subtype-specific property, i.e., a phenotype common among all members of one subtype but not found among members of other subtypes, would have to be due to sequence changes occurring on the “presubtype” branch for that subtype (here we define a presubtype branch as that connecting the common ancestor of a subtype to the common ancestor of the entire M group). These presubtype branches comprise only a fraction of the total divergence between contemporary viruses representing different subtypes. The chances of finding subtype-specific biological properties are thus similarly small, because the genetic changes responsible for these differences would have to occur on these presubtype branches. In fact, biologically meaningful sequence changes can occur at any point in the tree and certainly would not be expected to occur only (or preferentially) on presubtype branches. An expectation of biological differences along strict (and all) subtype lines is thus overly simplistic.
Nevertheless, it would be premature to conclude that there are no subtype-specific differences in virus biology. A relatively small number of viral phenotypes have been examined, and available in vitro assays may be too insensitive to identify subtle (yet important) differences in viral growth and cell tropism. Moreover, there are some sequence changes that appear to have arisen on the presubtype branches, and certain of these subtype-specific variations occur within genomic regions of known regulatory function. For example, subtype C viruses (which comprise about 36% of all globally circulating HIV-1 group M viruses based on the latest WHO estimates) are characterized by a premature truncation of their rev open reading frame (Fig. 8), an enlarged Vpu protein (Fig. 8), and three (instead of the common two) copies of a consensus NF-κB domain (59). Similarly, “subtype E” viruses (which are spreading with increasing rapidity in Asia) differ from other subtypes in having only one consensus NF-κB site (18). Such changes in enhancer copy numbers and regulatory proteins may manifest themselves only after multiple rounds of replication in vivo. Thus, subtype-specific biological differences may become apparent only in broad-based natural history studies.
Utility of subtype-specific reference reagents.
The availability of near-full-length representatives for five non-B HIV-1 group M clades, including a reconstructed replication-competent molecular clone of a subtype D isolate, should greatly facilitate efforts aimed at determining the biological consequences of HIV-1 genetic diversity and its impact on cellular and humoral immune responses in the infected host. Clones and sequences will be useful for identifying cross-clade CTL epitopes and for generating subtype-specific CTL targets. The clones will also be useful for the preparation of DNA- or protein-based subunit vaccines, including cocktails of genetically diverse immunogens. In this context, it should be noted that the representatives of subtypes F and H both contain uninterrupted reading frames. Finally, the full-length sequences are critically needed for phylogenetic studies, particularly of genomic regions other than gag and env. In collaboration with the Los Alamos database, we have compiled a list of nonmosaic reference sequences for all major HIV-1 genes (32), which is available at the Los Alamos web site (http://hiv -web.lanl.gov/subtype/subtypes.html). A similar compilation of documented intersubtype recombinants is in preparation. These listings should help investigators interested in subtyping new sequences to avoid the inclusion of mosaic sequences into phylogenetic trees.
All clones have been submitted to the National Institutes of Health Research and Reagent Program, Bethesda, Md., and all sequences have been recorded in GenBank and are available on-line through the Los Alamos HIV database. These reagents are thus available to investigators and manufacturers interested in the development and testing of HIV vaccines.
ACKNOWLEDGMENTS
We thank the NIH AIDS Research and Reference Reagent Program and Quality Biologicals Inc. for providing expanded PBMC cultures of HIV-1 isolates; the members of the WHO and NIAID Networks of HIV Isolation and Characterization for continuing collaborative interactions; and W. L. Abbott for artwork and preparation of the manuscript.
This work was supported by grants from the National Institutes of Health (N01 AI 35170, R01 AI 25291, and U01 AI 41530), by shared facilities of the UAB Center for AIDS Research (DNA Sequencing Core; P30 AI27767), and by the Birmingham Veterans Administration Medical Center.
REFERENCES
- 1.Abimiku A G, Stern T L, Zwandor A, Markham P D, Calef C, Kyari S, Saxinger W C, Gallo R C, Robert-Guroff M, Reitz M S. Subgroup G HIV type 1 isolates from Nigeria. AIDS Res Hum Retroviruses. 1994;10:1581–1583. doi: 10.1089/aid.1994.10.1581. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 3.Betts M R, Krowka J, Santamaria C, Balsamo K, Gao F, Mulundu G, Luo C, N’gandu N, Sheppard H, Hahn B H, Allen S, Frelinger J A. Cross-clade HIV-specific cytotoxic T-lymphocyte responses in HIV-infected Zambians. J Virol. 1997;71:8908–8911. doi: 10.1128/jvi.71.11.8908-8911.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bobkov A, Cheingsong-Popov R, Salminen M, McCutchan F, Louwagie J, Ariyoshi K, Whittle H, Weber J. Complex mosaic structure of the partial envelope sequence from a Gambian HIV type 1 isolate. AIDS Res Hum Retroviruses. 1996;12:169–171. doi: 10.1089/aid.1996.12.169. [DOI] [PubMed] [Google Scholar]
- 5.Brodine S K, Mascola J R, McCutchan F E. Genotypic variation and molecular epidemiology of HIV. Infect Med. 1997;14:739–748. [Google Scholar]
- 6.Cao H, Kanki P, Sankale J-L, Dieng-Sarr A, Mazzara G P, Kalams S A, Korber B, Mboup S, Walker W B. Cytotoxic T-lymphocyte cross-reactivity among different human immunodeficiency virus type 1 clades: implications for vaccine development. J Virol. 1997;71:8615–8623. doi: 10.1128/jvi.71.11.8615-8623.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carr J K, Salminen M O, Koch C, Gotte D, Artenstein A W, Hegerich P A, St. Louis D, Burke D S, McCutchan F E. Full-length sequence and mosaic structure of a human immunodeficiency virus type 1 isolate from Thailand. J Virol. 1996;70:5935–5943. doi: 10.1128/jvi.70.9.5935-5943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cornelissen M, Kampinga G, Zorgdrager F, Goudsmit J the UNAIDS Network for HIV Isolation and Characterization. Human immunodeficiency virus type 1 subtypes defined by env show high frequency of recombinant gag genes. J Virol. 1996;70:8209–8212. doi: 10.1128/jvi.70.11.8209-8212.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dittmar M T, Simmons G, Hibbitts S, O’Hare M, Louisirirotchanakul S, Beddows S, Weber J, Clapham P R, Weiss R A. Langerhans cell tropism of human immunodeficiency virus type 1 subtype A through F isolates derived from different transmission groups. J Virol. 1997;71:8008–8013. doi: 10.1128/jvi.71.10.8008-8013.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dolin R. Human studies in the development of human immunodeficiency virus vaccines. J Infect Dis. 1995;172:1175–1183. doi: 10.1093/infdis/172.5.1175. [DOI] [PubMed] [Google Scholar]
- 11.Esparaza J, Osmanov S, Heyward W. HIV preventive vaccines. Drugs. 1995;50:792–804. doi: 10.2165/00003495-199550050-00003. [DOI] [PubMed] [Google Scholar]
- 12.Faulkner D M, Jurka J. Multiple aligned sequence editor (MASE) Trends Biochem Sci. 1988;13:321–322. doi: 10.1016/0968-0004(88)90129-6. [DOI] [PubMed] [Google Scholar]
- 13.Felsenstein J. Confidence limits on phylogenies: an approach using the boostrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 14.Felsenstein J. PHYLIP (Phylogeny Inference Package), version 3.5c. Seattle: Department of Genetics, University of Washington; 1992. [Google Scholar]
- 15.Ferrari G, Humphrey W, McElrath M J, Excler J-L, Duliege A-M, Clements M L, Corey L C, Bolognesi D P, Weinhold K T. Clade B-based HIV-1 vaccines elicit cross-clade cytotoxic T lymphocyte reactivities in uninfected volunteers. Proc Natl Acad Sci USA. 1997;94:1396–1401. doi: 10.1073/pnas.94.4.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao, F., and B. H. Hahn. Unpublished data.
- 17.Gao F, Yue L, Craig S, Thornton C L, Robertson D L, McCutchan F E, Bradac J A, Sharp P M, Hahn B H. Genetic variation of HIV type 1 in four World Health Organization-sponsored vaccine evaluation sites: generation of functional envelope (glycoprotein 160) clones representative of sequence subtypes A, B, C, and E. AIDS Res Hum Retroviruses. 1994;10:1359–1368. doi: 10.1089/aid.1994.10.1359. [DOI] [PubMed] [Google Scholar]
- 18.Gao F, Robertson D L, Morrison S G, Hui H, Craig S, Decker J, Fultz P N, Girard M, Shaw G M, Hahn B H, Sharp P M. The heterosexual human immunodeficiency virus type 1 epidemic in Thailand is caused by an intersubtype (A/E) recombinant of African origin. J Virol. 1996;70:7013–7029. doi: 10.1128/jvi.70.10.7013-7029.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao F, Morrison S G, Robertson D L, Thornton C L, Craig S, Karlsson G, Sodroski J, Morgado M, Galvao-Castro B, von Briesen H, Beddows S, Weber J, Sharp P M, Shaw G M, Hahn B H the WHO and NIAID Networks for HIV Isolation and Characterization. Molecular cloning and analysis of functional envelope genes from HIV-1 sequence subtypes A through G. J Virol. 1996;70:1651–1667. doi: 10.1128/jvi.70.3.1651-1667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghosh S K, Fultz P N, Keddie E, Saag M S, Sharp P M, Hahn B H, Shaw G M. A molecular clone of HIV-1 tropic and cytopathic for human and chimpanzee lymphocytes. Virology. 1993;194:858–864. doi: 10.1006/viro.1993.1331. [DOI] [PubMed] [Google Scholar]
- 21.Graham B S, Wright P F. Candidate AIDS vaccines. N Engl J Med. 1995;333:1331–1339. doi: 10.1056/NEJM199511163332007. [DOI] [PubMed] [Google Scholar]
- 22.Hahn B H, Shaw G M, Arya S K, Popovic M, Gallo R C, Wong-Staal F. Molecular cloning and characterization of the HTLV-III virus associated with AIDS. Nature. 1984;312:166–169. doi: 10.1038/312166a0. [DOI] [PubMed] [Google Scholar]
- 23.Hahn B H, Robertson D L, Sharp P M. Intersubtype recombination in HIV-1 and HIV-2. In: Myers G, Korber B, Wain-Hobson S, Jeang K-T, Henderson L E, Pavlakis G N, editors. Human retroviruses and AIDS 1995: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; 1995. pp. III-22–III-29. [Google Scholar]
- 24.Hu D J, Dondero T J, Reyfield M A, George J R, Schochetman G, Jaffe H W, Luo C-C, Kalish M L, Weniger B G, Pau C-P, Schable C A, Curran J W. The emerging diversity of HIV: the importance of global surveillance for diagnostics, research and prevention. JAMA. 1996;275:210–216. [PubMed] [Google Scholar]
- 25.Kalish M L, Baldwin A, Raktham S, Wasi C, Luo C-C, Schochetman G, Mastro T D, Young N, Vanichseni S, Rubsamen-Waigmann H, von Briesen H, Mullins J I, Delwart E, Herring B, Esparaza J, Heyward W L, Osmanov S. The evolving molecular epidemiology of HIV-1 envelope subtypes in injecting drug users in Bangkok, Thailand: implications for HIV vaccine trials. AIDS. 1995;9:851–857. doi: 10.1097/00002030-199508000-00004. [DOI] [PubMed] [Google Scholar]
- 26.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 27.Kimura M. The neutral theory of molecular evolution. Cambridge, United Kingdom: Cambridge University Press; 1983. [Google Scholar]
- 28.Korber B T M, Osmanov S, Esparza J, Myers G WHO Network for HIV Isolation and Characterization. The World Health Organization Global Programme on AIDS Proposal for Standardization of HIV Sequence Nomenclature. AIDS Res Hum Retroviruses. 1994;10:1355–1358. doi: 10.1089/aid.1994.10.1355. [DOI] [PubMed] [Google Scholar]
- 29.Kostrikis L G, Bagdades E, Cao Y, Zhang L, Dimitriou D, Ho D D. Genetic analysis of human immunodeficiency virus type 1 strains from patients in Cyprus: identification of a new subtype designated subtype I. J Virol. 1995;69:6122–6130. doi: 10.1128/jvi.69.10.6122-6130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leitner T, Albert J. A new genetic subtype of HIV-1. In: Myers G, Korber B, Wain-Hobson S, Jeang K-T, Mellors J W, McCutchan F E, Henderson L E, Pavlakis G N, editors. Human retroviruses and AIDS 1995: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; 1995. pp. III-147–III-150. [Google Scholar]
- 31.Leitner T, Escanilla D, Marquina S, Wahlberg J, Brotstrom C, Hansson H B, Uhlen M, Albert J. Biological and molecular characterization of subtype D, G, and A/D recombinant HIV-1 transmissions in Sweden. Virology. 1995;209:136–146. doi: 10.1006/viro.1995.1237. [DOI] [PubMed] [Google Scholar]
- 32.Leitner T, Korber B T M, Robertson D L, Gao F, Hahn B H. Updated proposal of reference sequences of HIV-1 genetic subtypes. In: Korber B, Foley B, Kuiken C, Leitner T, McCutchan F, Mellors J W, Hahn B H, editors. Human retroviruses and AIDS 1997: a compilation and analysis of nucleic acid and amino acid sequence. Los Alamos, N.Mex: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; 1997. pp. III-19–III-24. [Google Scholar]
- 33.Loussert-Ajaka I, Chaic M-L, Korber B T M, Letournearu F, Gomas E, Allen E, Ly T-D, Brun-Vezinet F, Simon F, Saragosti S. Variability of human immunodeficiency virus type 1 group O strains isolated from Cameroonian patients living in France. J Virol. 1995;69:5640–5649. doi: 10.1128/jvi.69.9.5640-5649.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Louwagie J, McCutchan F E, Peeters M, Brennan T P, Sanders-Buell E, Eddy G A, van der Groen G, Fransen K, Gershy-Damet G-M, Deleys R, Burke D S. Phylogenetic analysis of gag genes from 70 international HIV-1 isolates provides evidence for multiple genotypes. AIDS. 1993;7:769–780. doi: 10.1097/00002030-199306000-00003. [DOI] [PubMed] [Google Scholar]
- 35.Louwagie J, Janssens W, Mascola J, Heyndrickx L, Hegerich P, van der Groen G, McCutchan F E, Burke D S. Genetic diversity of the envelope glycoprotein from human immunodeficiency virus type 1 isolates of African origin. J Virol. 1995;69:263–271. doi: 10.1128/jvi.69.1.263-271.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. pp. 269–295. [Google Scholar]
- 37.Martin-Gallardo A, Lamerdin J, Carrano A. Shotgun sequencing. In: Adams M D, Fields C, Venter J C, editors. Automated DNA sequencing and analysis. London, United Kingdom: Academic Press, Ltd.; 1994. pp. 37–41. [Google Scholar]
- 38.Mascola J R, Louder M K, Surman S R, VanCott T C, Yu X F, Bradac J, Porter K R, Nelson K E, Girard M, McNeil J G, McCutchan F E, Birx D L, Burke D S. Human immunodeficiency virus type 1 neutralizing antibody serotyping using serum pools and an infectivity reduction assay. AIDS Res Hum Retroviruses. 1996;12:1319–1328. doi: 10.1089/aid.1996.12.1319. [DOI] [PubMed] [Google Scholar]
- 39.McCutchan F E, Hegerich P A, Brennan T P, Phanuphak P, Singharaj P, Jugsudee A, Berman P W, Gray A M, Fowler A K, Burke D S. Genetic variants of HIV-1 in Thailand. AIDS Res Hum Retroviruses. 1992;8:1887–1895. doi: 10.1089/aid.1992.8.1887. [DOI] [PubMed] [Google Scholar]
- 40.McCutchan, F. E., M. O. Salminen, J. K. Carr, and D. S. Burke. 1996. HIV-1 genetic diversity. AIDS 10(Suppl. 3)**:**S13–S20. [PubMed]
- 41.Moore J, Trkola A. HIV type 1 coreceptors, neutralization serotypes, and vaccine development. AIDS Res Hum Retroviruses. 1997;13:733–736. doi: 10.1089/aid.1997.13.733. [DOI] [PubMed] [Google Scholar]
- 42.Moore J P, Cao Y, Leu J, Qin L, Korber B, Ho D D. Inter- and intrasubtype neutralization of human immunodeficiency virus type 1: the genetic subtypes do not correspond to neutralization serotypes but partially correspond to gp120 antigenic serotypes. J Virol. 1996;70:427–444. doi: 10.1128/jvi.70.1.427-444.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murphy E, Korber B, Georges-Courbot M-C, You B, Pinter A, Cook D, Kieny M-P, Georges A, Mathiot C, Barre-Sinoussi F, Girard M. Diversity of V3 region sequences of human immunodeficiency viruses type 1 from the Central African Republic. AIDS Res Hum Retroviruses. 1993;9:997–1007. doi: 10.1089/aid.1993.9.997. [DOI] [PubMed] [Google Scholar]
- 44.Myers G, Korber B, Berzofsky J A, Smith R F, Pavlakis G N. Human retroviruses and AIDS: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; 1992. [Google Scholar]
- 45.Myers G, Korber B, Foley B, Jeang K-T, Mellors J W, Wain-Hobson S. Human retroviruses and AIDS: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; 1996. [Google Scholar]
- 46.Nyambi P N, Nkengasong J, Lewi P, Koen A, Janssens W, Heyndrickx F K L, Piot P, van der Groen G. Multivariate analysis of human immunodeficiency virus type 1 neutralization data. J Virol. 1996;70:445–458. doi: 10.1128/jvi.70.9.6235-6243.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ou C-Y, Takebe Y, Weniger B G, Luo C-C, Kalish M L, Auwanit W, Yamazaki S, Gayle H D, Young N L, Schochetman G. Independent introductions of two major HIV-1 genotypes into distinct high-risk populations in Thailand. Lancet. 1993;341:1171–1174. doi: 10.1016/0140-6736(93)91001-3. [DOI] [PubMed] [Google Scholar]
- 48.Peden K, Emerman M, Montagnier L. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology. 1997;185:661–672. doi: 10.1016/0042-6822(91)90537-l. [DOI] [PubMed] [Google Scholar]
- 49.Perrière G, Gouy M. WWW-Query: an on-line retrieval system for biological sequence banks. Biochimie. 1996;78:364–369. doi: 10.1016/0300-9084(96)84768-7. [DOI] [PubMed] [Google Scholar]
- 50.Pope M, Frankel S S, Mascola J R, Trkola A, Isdell F, Birx D L, Ho D, Moore J P. HIV-1 strains from subtypes B and E replicate in cutaneous dendritic cell–T-cell mixtures without displaying subtype-specific tropism. J Virol. 1997;71:8001–8007. doi: 10.1128/jvi.71.10.8001-8007.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pope M, Ho D D, Moore J P, Weber J, Dittmar M T, Weiss R A. Different subtypes of HIV-1 and cutaneous dendritic cells. Science. 1997;278:786–787. doi: 10.1126/science.278.5339.785c. [DOI] [PubMed] [Google Scholar]
- 52.Robertson D L, Sharp P M, McCutchan F E, Hahn B H. Recombination in HIV-1. Nature. 1995;374:124–126. doi: 10.1038/374124b0. [DOI] [PubMed] [Google Scholar]
- 53.Robertson D L, Hahn B H, Sharp P M. Recombination in AIDS viruses. J Mol Evol. 1995;40:249–259. doi: 10.1007/BF00163230. [DOI] [PubMed] [Google Scholar]
- 54.Sabino E C, Shpaer E G, Morgado M G, Korber B T M, Diaz R S, Bongertz V, Cavalcante S, Galvão-Castro B, Mullins J I, Mayer A. Identification of human immunodeficiency virus type 1 envelope genes recombinant between subtypes B and F in two epidemiologically linked individuals from Brazil. J Virol. 1994;68:6340–6346. doi: 10.1128/jvi.68.10.6340-6346.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 56.Salminen M O, Koch C, Sanders-Buell E, Ehrenberg P K, Michael N L, Carr J K, Burke D S, McCutchan F E. Recovery of virtually full length HIV-1 provirus of diverse subtypes from primary virus cultures using the polymerase chain reaction. Virology. 1995;213:80–86. doi: 10.1006/viro.1995.1548. [DOI] [PubMed] [Google Scholar]
- 57.Salminen M O, Carr J K, Burke D S, McCutchan F E. Identification of breakpoints in intergenotypic recombinants of HIV-1 by bootscanning. AIDS Res Hum Retroviruses. 1995;11:1423–1425. doi: 10.1089/aid.1995.11.1423. [DOI] [PubMed] [Google Scholar]
- 58.Salminen M O, Carr J K, Burke D S, McCutchan F E. Genotyping of HIV-1. In: Myers G, Korber B, Wain-Hobson S, Jeang K-T, Henderson L E, Pavlakis G N, editors. Human retroviruses and AIDS 1995: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; 1995. pp. III-30–III-34. [Google Scholar]
- 59.Salminen M O, Johansson B, Sonerborg A, Ayehunie S, Gotte D, Leinikki P, Burke D S, McCutchan F E. Full length sequence of an Ethiopian human immunodeficiency virus type 1 (HIV-1) isolate of genetic subtype C. AIDS Res Hum Retroviruses. 1996;12:1329–1339. doi: 10.1089/aid.1996.12.1329. [DOI] [PubMed] [Google Scholar]
- 60.Salminen M O, Carr J K, Robertson D L, Hegerich P, Gotte D, Koch C, Sanders-Buell E, Gao F, Sharp P M, Hahn B H, Burke D S, McCutchan F E. Evolution and probable transmission of intersubtype recombinant human immunodeficiency virus type 1 in a Zambian couple. J Virol. 1997;71:2647–2655. doi: 10.1128/jvi.71.4.2647-2655.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sharp P M, Robertson D L, Gao F, Hahn B H. Origins and diversity of human immunodeficiency viruses. AIDS. 1994;8:S27–S42. [Google Scholar]
- 62.Sharp P M, Robertson D L, Hahn B H. Cross-species transmission and recombination of AIDS viruses. Philos Trans R Soc London Ser B. 1995;349:41–47. doi: 10.1098/rstb.1995.0089. [DOI] [PubMed] [Google Scholar]
- 63.Siepel A C, Korber B T. Scanning the database for recombinant HIV-1 genomes. In: Myers G, Korber B, Wain-Hobson S, Jeang K-T, Henderson L E, Pavlakis G N, editors. Human retroviruses and AIDS 1995: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; 1995. pp. III-35–III-60. [Google Scholar]
- 64.Soto-Ramirez L E, Renjifo B, McLane M F, Marlink R, O’Hara C, Sutthent R, Wasi C, Vithayasai P, Vithayasai V, Apichartpiyakul C, Auewarakul P, Pena Cruz V, Chui D-S, Osathanondh R, Mayer K, Lee T-H, Essex M. HIV-1 Langerhans’ cell tropism A associated with heterosexual transmission of HIV. Science. 1996;271:1291–1293. doi: 10.1126/science.271.5253.1291. [DOI] [PubMed] [Google Scholar]
- 65.Spire B, Sire J, Zachar V, Rey F, Barré-Sinoussi F, Galibert F, Hampe A, Chermann J-C. Nucleotide sequence of HIV1-NDK: a highly cytopathic strain of the human immunodeficiency virus. Gene. 1989;81:275–284. doi: 10.1016/0378-1119(89)90188-1. [DOI] [PubMed] [Google Scholar]
- 66.Takehisa J, Oseikwasi M, Ayisi N K, Hishida O, Miura T, Igarashi T, Brandful J, Ampofo W, Netty V B A, Mensah M, Yamashita M, Ido E, Hayami M. Phylogenetic analysis of human immunodeficiency virus 1 in Ghana. Acta Virol. 1997;41:51–54. [PubMed] [Google Scholar]
- 67.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W—improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weber J, Fenyö E-M, Beddows S, Kaleebu P, Björndal Å the WHO Network for HIV Isolation and Characterization. Neutralization serotypes of human immunodeficiency virus type 1 field isolates are not predicted by genetic subtype. J Virol. 1996;70:7827–7832. doi: 10.1128/jvi.70.11.7827-7832.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weniger, B. G., K. Limpakarnjanarat, K. Ungchusak, S. Thanprasertsuk, K. Choopanya, S. Vanichseni, T. Uneklabh, P. Thongcharoen, and C. Wasi. 1991. The epidemiology of HIV infection in AIDS in Thailand. AIDS 5(Suppl. 2)**:**S71–S85. [DOI] [PubMed]
- 70.Weniger, B. G., Y. Takebe, C.-Y. Ou, and S. Yamazaki. 1994. The molecular epidemiology of HIV in Asia. AIDS 8(Suppl. 2)**:**S13–S28. [PubMed]
- 71.Wieland U, Seelhoff A, Hofmann A, Kuhn J E, Eggers H J, Mugyenyi P, Schwander S. Diversity of the vif gene of human immunodeficiency virus type 1 in Uganda. J Gen Virol. 1997;78:393–400. doi: 10.1099/0022-1317-78-2-393. [DOI] [PubMed] [Google Scholar]
- 72.World Health Organization Network for HIV Isolation and Characterization. HIV-1 variation in WHO-sponsored vaccine-evaluation sites: genetic screening, sequence analysis and preliminary biological characterization of selected viral strains. AIDS Res Hum Retroviruses. 1994;10:1327–1344. doi: 10.1089/aid.1994.10.1327. [DOI] [PubMed] [Google Scholar]
- 73.Zhang L, Huang Y, He T, Cao Y, Ho D D. HIV-1 subtype and second-receptor use. Nature (London) 1996;383:768. doi: 10.1038/383768a0. [DOI] [PubMed] [Google Scholar]
- 74.Zhang L, Carruthers C D, He T, Huany Y, Cao Y, Wang G, Hahn B, Ho D D. HIV-1 subtypes, co-receptor usage, and CCR5 polymorphism. AIDS Res Hum Retroviruses. 1997;13:1357–1366. doi: 10.1089/aid.1997.13.1357. [DOI] [PubMed] [Google Scholar]