A Recombinant Human Immunodeficiency Virus Type 1 Envelope Glycoprotein Complex Stabilized by an Intermolecular Disulfide Bond between the gp120 and gp41 Subunits Is an Antigenic Mimic of the Trimeric Virion-Associated Structure (original) (raw)
Abstract
The few antibodies that can potently neutralize human immunodeficiency virus type 1 (HIV-1) recognize the limited number of envelope glycoprotein epitopes exposed on infectious virions. These native envelope glycoprotein complexes comprise three gp120 subunits noncovalently and weakly associated with three gp41 moieties. The individual subunits induce neutralizing antibodies inefficiently but raise many nonneutralizing antibodies. Consequently, recombinant envelope glycoproteins do not elicit strong antiviral antibody responses, particularly against primary HIV-1 isolates. To try to develop recombinant proteins that are better antigenic mimics of the native envelope glycoprotein complex, we have introduced a disulfide bond between the C-terminal region of gp120 and the immunodominant segment of the gp41 ectodomain. The resulting gp140 protein is processed efficiently, producing a properly folded envelope glycoprotein complex. The association of gp120 with gp41 is now stabilized by the supplementary intermolecular disulfide bond, which forms with approximately 50% efficiency. The gp140 protein has antigenic properties which resemble those of the virion-associated complex. This type of gp140 protein may be worth evaluating for immunogenicity as a component of a multivalent HIV-1 vaccine.
The urgent need for an effective vaccine against human immunodeficiency virus type 1 (HIV-1) is undoubted, for only by vaccination will the worldwide spread of AIDS be stemmed (44, 46, 62). Although there is not yet universal consensus on what components will be needed in a vaccine that is able to induce protective immunity against HIV-1 infection or disease, a popular view is that both the humoral and the cellular arms of the human immune system should be efficiently stimulated (12–14, 43, 44, 46, 57, 64, 94). To do this will probably require the creation of a multivalent vaccine that incorporates several categories of immunogen, each intended to optimally evoke different, necessary immune responses. Examples would be a live recombinant virus or a DNA vector to stimulate cellular immunity, combined with a subunit protein to generate antibody responses (4, 5, 32, 93).
There has, arguably, been more progress with evoking HIV-1-specific cellular immunity than humoral immunity in recent years, although some new concepts relating to neutralizing-antibody induction that merit continued evaluation have recently been described (18, 52, 81, 90, 103). The most widely tried method of neutralizing-antibody induction, i.e., that involving recombinant monomeric gp120 proteins, has not been successful at inducing antibodies able to neutralize heterologous primary isolates at significant titers (4, 5, 22, 40, 58, 59, 81, 111, 120). This raises serious questions about the protective efficacy of vaccines that include such proteins, either alone or in combination with other immunogens (14). One of the major obstacles to neutralizing-antibody induction is the inherent resistance of primary HIV-1 isolates to such antibodies (10, 12, 13, 58, 59, 64, 66–68, 80, 81, 102, 107, 112, 120), a feature that HIV-1 shares with other lentiviruses and one which is probably necessary for viral persistence in vivo (3, 23, 65). The native HIV-1 envelope glycoprotein complex on virions, a heterotrimer containing three gp120 proteins noncovalently associated with three gp41 moieties, is recognized poorly by antibodies that efficiently bind to the individual gp120 and gp41 subunits (51, 66, 81, 98, 102, 122).
Notwithstanding the natural defenses used by HIV-1 to resist or evade humoral immunity, proteins which faithfully represent the antigenic structure of the virion-associated envelope glycoprotein complex may be worth evaluating as vaccine immunogens. For instance, the three most potent HIV-1 neutralizing antibodies yet identified, immunoglobulin b12 (IgG1b12), 2G12, and 2F5, have a high affinity for the native trimer which is comparable to or sometimes greater than their affinity for the individual gp120 or gp41 subunits (15, 34, 77, 92, 96, 98, 102, 109). These antibodies may therefore have been raised by an immune response to virions rather than to viral debris or dissociated subunits (13, 68, 80, 81).
The lability of the noncovalent interaction between gp120 and gp41, which causes extensive gp120 dissociation from virions or virus-infected cells (38, 61, 70, 87), is a major obstacle to making stable recombinant, oligomeric envelope glycoproteins. Initial attempts at making stable oligomers therefore involved the introduction of mutations to remove or replace the gp120-gp41 cleavage recognition sequence (6, 27–29). Usually, such proteins are also truncated N-terminal to the transmembrane-spanning region of gp41, so that they are efficiently secreted as soluble proteins (the internal segment of gp41 is of limited relevance for induction of humoral immune responses). A broadly similar nonrecombinant protein was isolated from a virus-infected cell line (110). The resulting proteins (gp140s) contain the gp120 moiety linked to the 20-kDa gp41 ectodomain by a peptide bond between the C terminus of gp120 and the N terminus of gp41, which is not present in the virion-associated complex. Although these uncleaved gp140 proteins (designated gp140UNC) are oligomerized by strong, noncovalent intermolecular interactions between gp41 subunits (19, 101, 116), it is questionable whether they truly mimic the native envelope glycoprotein complex. Thus, epitopes are exposed on gp140UNC proteins that are not accessible on virions (27, 28), and there are indications that coreceptor interactions of gp140UNC proteins are inefficient (31). Together, these observations imply that a structural perturbation is caused to the gp120 component by the covalently attached, improperly associated gp41 ectodomain (31). For whatever reason, immunogenicity studies carried out to date with gp140UNC proteins have not been particularly encouraging, in that primary virus-neutralizing antibodies have not been induced (27, 90, 110).
We have therefore pursued a different approach: the expression of gp140 proteins with the natural gp120-gp41 cleavage site preserved but with a disulfide bond introduced between gp120 and the gp41 ectodomain to stabilize the association of these two subunits. We report here on the antigenic properties of such a gp140 protein.
MATERIALS AND METHODS
Cloning of gp140 and furin.
Plasmid pPPI4 is a eukaryotic shuttle vector generated at Progenics Pharmaceuticals Inc. The expression of HIV-1 envelope proteins is under the control of the cytomegalovirus major immediate-early promoter-enhancer with a tissue plasminogen activator leader and bovine growth hormone poly(A) signal (106). The vector contains the dihydroxyfolate reductase gene and a simian virus 40 SV40 origin of replication, which promotes high-level replication and transient expression of open reading frames in cells expressing the SV40 large T antigen. PCR was used to clone DNA encoding the gp140 segment of the env genes of isolates JR-FL (50), DH123 (100), HxB2 (88), GUN-1wt (104), and 89.6 (21) from the corresponding HIV-1 genomic plasmids. The primers used were 5′Kpnlenv (5′-GTCTATTATGGGGTACCTGTGTGGAAAGAAGC-3′) and 3′BstB1env (5′-CGCAGACGCAGATTCGAATTAATACCACAGCCAGTT-3′). The restriction sites are underlined. The PCR products were cloned into pPPI4 by using _Kpn_I and _Bst_B1. These plasmids are designated gp140WT (wild type) to distinguish them from the mutated forms described below. A furin-expressing plasmid, pGEMfurin, was obtained from Gary Thomas, Vollum Institute, Portland, Oreg. (63). The _Eco_RI-_Hin_dIII fragment of furin was subcloned into pcDNA3.1(−) (Invitrogen Inc.) to make pcDNA3.1furin.
Mutagenesis of gp140.
A variety of double cysteine substitutions were introduced into the gp120 and gp41 moieties of gp140WT (HIV-1 JR-FL) in pPPI4 by using Quickchange mutagenesis kits (Stratagene, La Jolla, Calif.) and verified by sequencing. Details of the positions of these substitutions are provided in Results (see Fig. 3). A similar strategy was used to make the corresponding cysteine substitutions in other HIV-1 gp140 proteins. A gp120-gp41 cleavage site mutant of JR-FL gp140UNC was generated by substitution of the sequence Lys-Arg-Arg-Val-Val-Gln-Arg-Glu-Lys-Arg-Ala-Val (the C terminus of gp120 and the first two residues of gp41) by a hexameric Leu-Arg motif. This eliminates the furin cleavage motif (underlined), as described previously (30) (see Fig. 3). The PCR primers were 3′140M (TCGAAGGCGGAGACGAAGTCGTAGCCGCAGTGCCTTGGTGGGTGCTACTCCTAATGGTTC) with 5′KpnIenv and 5′140M (5′-CTACGACTTGTCTCCGCCTTCGACTACGGGGAATAGGAGCTGTG TTCCTTGGGTTCTTG-3′) with 3′BstB1env. The PCR products of these two reactions were purified, and an overlap PCR reaction was performed to generate a full-length env gene that was cloned into pPPI4 by _Kpn_I-_Bst_BI digestion.
FIG. 3.
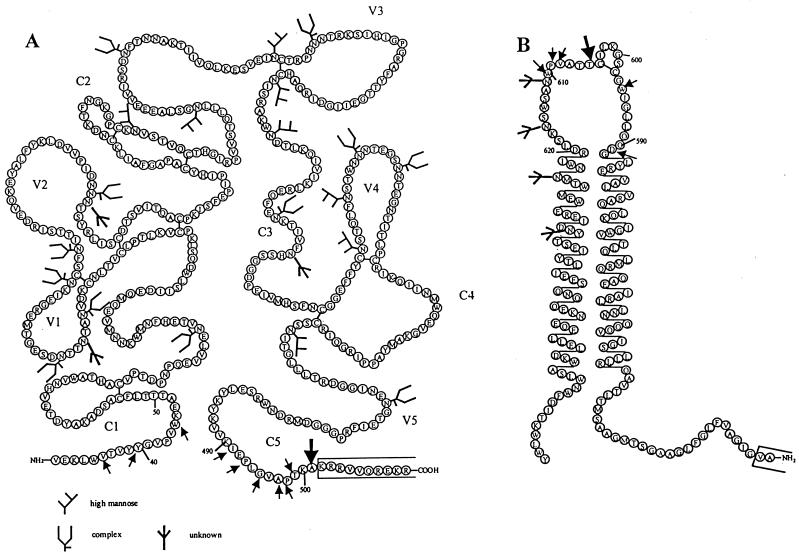
Positions of cysteine substitutions in JR-FL gp140. The various residues of the JR-FL gp140WT protein that have been mutated to cysteines in one or more mutants are indicated by arrows on the schematics of the gp120 and gp41ECTO subunits. The positions of the alanine-501 and threonine-605 residues that are both mutated to cysteine in the SOS gp140 protein are indicated by the larger arrows. (A) The depiction of JR-FL gp120, including the positioning of canonical sites for complex and high-mannose N-linked carbohydrates, is based on that of Leonard et al. (56), adjusted to reflect the sequence numbering of HIV-1 HxB2. (B) The cartoon of the JR-FL gp41-ectodomain is derived from reference 37, also adjusted to reflect the HxB2 sequence numbering. The open boxes at the C terminus of gp120 and the N terminus of gp41 indicate the regions that are mutated in the gp140UNC protein to eliminate the cleavage site between gp120 and gp41.
Note that the numbering system used to denote the positions of gp120 and gp41 residues in HIV-1 JR-FL is based on the numbering of residues in HIV-1 HxBc2, to facilitate comparison with structural information published on this envelope glycoprotein (51, 122).
Transfection, labeling, and immunoprecipitation.
Adherent 293T cells were grown in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% fetal calf serum, penicillin, streptomycin, and l-glutamine. Transient transfection of 293T cells was performed by calcium phosphate precipitation. The gp140 expression plasmids pPPI4-gp140 were transfected with or without cotransfection of the furin expression vector pcDNA3.1-furin, each at 10 μg per 10-cm2 plate. At 1 day posttransfection, the medium was changed to Dulbecco's modified Eagle's medium supplemented with 0.2% bovine serum albumin, penicillin, streptomycin and l-glutamine. For radioimmunoprecipitation analysis (RIPA), [35S]cysteine and [35S]methionine (200 μCi per plate; Amersham International PLC) were added for 24 h. The culture supernatants were then cleared of debris by centrifugation before addition of concentrated RIPA buffer to adjust the composition to 50 mM Tris-HCl–150 mM NaCl–1 mM EDTA (pH 7.2). Envelope glycoproteins were immunoprecipitated with monoclonal antibodies (MAbs) to a variety of epitopes. In some instances, the MAbs had been labeled with biotin (42). The MAbs were added in a 1-ml volume for 10 min at room temperature and then precipitated by incubation overnight at 4°C with either streptavidin-coated agarose beads (Vector Laboratories) or protein G-coated agarose beads (Pierce Inc.), as appropriate. The beads were washed three times with RIPA buffer containing 1% Nonidet P-40 detergent, and then the proteins were eluted by heating at 100°C for 5 min in 60 μl of polyacrylamide gel electrophoresis (PAGE) buffer supplemented with 2% sodium dodecyl sulfate (SDS) and, when indicated, 100 mM dithiothreitol (DTT). The immunoprecipitates were fractionated by SDS-PAGE (8% polyacrylamide) at 200 V for 1 h. Unless otherwise specified (e.g., see Fig. 2 and 10), the amounts of immunoprecipitated envelope glycoproteins loaded onto each lane were comparable, in that fixed numbers of cells were transfected with the same amount of plasmid and then a constant volume of supernatant was precipitated with a standard amount of MAb. The gels were dried and exposed to a phosphor screen. The positions of the radiolabeled proteins were determined by using a PhosphorImager with ImageQuant software (Molecular Dynamics Inc.).
FIG. 2.
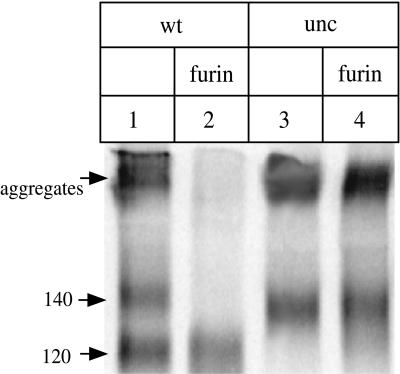
Cotransfection of furin increases the efficiency of cleavage of the peptide bond between gp120 and gp41. 293T cells were transfected with DNA expressing the HIV-1 JR-FL gp140WT or gp140UNC (gp120-gp41 cleavage site mutant) proteins, in the presence or absence of a cotransfected furin-expressing plasmid. The 35S-labeled envelope glycoproteins secreted from the cells were immunoprecipitated with the anti-gp120 MAb 2G12, boiled with SDS, and analyzed by SDS-PAGE. Lanes: 1, gp140WT (gp140/gp120 doublet); 2, gp140WT plus furin (gp120 only); 3, gp140UNC (gp140 only); 4, gp140UNC plus furin (gp140 only). The approximate molecular masses, in kilodaltons, of the major species are recorded on the left, as are higher-molecular-mass aggregates. Only one-fifth of the immunoprecipitated proteins from the transfections shown in lanes 1 and 3 were loaded onto the gel, to ensure that the amounts of envelope glycoproteins analyzed in each lane were approximately comparable.
FIG. 10.
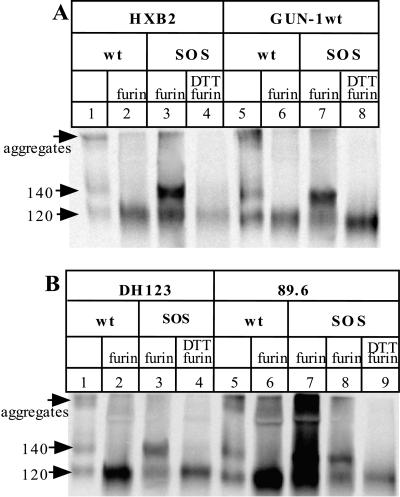
Preparation of disulfide bond-stabilized gp140 proteins from various HIV-1 isolates. 293T cells were transfected with plasmids expressing gp140 proteins from different isolates and, when indicated, a furin-expressing plasmid. The secreted, 35S-labeled glycoproteins were immunoprecipitated with the anti-gp120 MAb 2G12, boiled with SDS (and, when indicated, DTT), and analyzed by SDS-PAGE. The SOS gp140 protein from each isolate contained double cysteine substitutions at positions equivalent to alanine-501 and threonine-605 of the JR-FL gp140 protein. (A) HxB2 (lanes 1 to 4) and GUN-1wt (lanes 5 to 8). Lanes: 1 and 5, gp140WT with no cotransfected furin, producing gp120 and the gp140NON protein; 2 and 6, gp140WT plus furin, producing gp120; 3 and 7, SOS gp140 protein plus furin; 4 and 8, as lanes 3 and 7 except that the immunoprecipitates were boiled with both SDS and DTT prior to SDS-PAGE. (B) DH123 (lanes 1 to 4) and 89.6 (lanes 5 to 9). The layout of the lanes is as in panel A, except that for 89.6, lane 8 is the same as lane 7 but with only one-fifth of the immunoprecipitate loaded onto the gel and lane 9 is the same as lane 8 but with the immunoprecipitates boiled with both SDS and DTT prior to SDS-PAGE. The positions of the 120- and 140-kDa bands, and of higher-molecular-mass aggregates, are indicated on the left of each panel. Only one-fifth of the immunoprecipitated proteins from the gp140WT plus furin transfections (lanes 2 and 6) was loaded onto each gel, to approximately compensate for the increased envelope glycoprotein expression that was observed with the JR-FL gp140WT protein under these conditions.
MAbs to HIV-1 envelope glycoprotein epitopes, and sCD4.
The epitopes for, and some immunochemical properties of, anti-gp120 MAbs from various donors have been described previously (9, 71, 72). These include 19b and 83.1 to the V3 loop (74, 118), IgG1b12 and F91 to the CD4 binding site (CD4bs) (15, 72), 2G12 to a unique C3-V4 glycan-dependent epitope (108, 109), M90 to the C1 region (113), 23A and sheep antibody D7324 to the C5 region (69, 72), C11 to a discontinuous C1–C5 epitope (75), 17b to a CD4-inducible epitope (9, 47, 72, 91, 103, 105, 122, 123), A32 to a CD4-inducible C1-C4 epitope (72, 103), and G3-519 and G3-42 to C4 and C4/V3 epitopes, respectively (72, 73). MAbs to gp41 epitopes included 7B2 to epitope cluster I (a gift from James Robinson); 4D4 to cluster I (11); T4, an oligomer-specific MAb overlapping the cluster I region (28); 2.2B to cluster II (a gift from James Robinson); T15G1 to cluster II (a gift from Ab Notkins); and 2F5 to a neutralizing epitope encompassing residues 653 to 659 (11, 77, 108). The tetrameric CD4-IgG2 and monomeric soluble CD4 (sCD4) molecules, from Progenics Pharmaceuticals Inc., have also been described previously (2).
Quantitation of gp120 and gp140 proteins by ELISA.
To measure the secretion of gp120 and gp140 proteins from transfected 293T cells, we used a gp120 antigen capture enzyme-linked immunosorbent assay (ELISA) based on one that has been described previously (7, 69, 71). Briefly, envelope glycoproteins in the culture supernatants were denatured and reduced by boiling with 1% SDS and 50 mM DTT. Purified monomeric JR-FL gp120 treated in the same way was used as a reference standard for gp120 expression (106). The denatured proteins were captured on plastic by sheep antibody D7324, which was raised to the continuous sequence APTKAKRRVVQREKR at the C terminus of gp120. Bound envelope glycoproteins were detected by using a mixture of MAbs B12 and B13 against epitopes exposed on denatured gp120 (1, 71). This assay allows the efficient detection of both gp120 and any gp140 molecules in which the peptide bond between gp120 and the gp140 ectodomain is still intact (71, 106).
Sucrose velocity gradient centrifugation.
Culture supernatants from _env_-transfected 293T cells were concentrated by 100-fold on Millipore concentrators, and then a 100-μl aliquot was layered onto a 5 to 20% sucrose step gradient of 8.8 ml comprising 11 steps of 800 μl each. The gradient was overlaid to bring the volume up to 12 ml and then centrifuged for 20 h at 40,000 × g in an SW41Ti rotor at 4°C. Gradient fractions of 500 μl taken sequentially from the top were immunoprecipitated with MAb 2G12, boiled with SDS, and analyzed by SDS-PAGE.
Gel filtration analysis.
Culture supernatants from 35S-labeled _env_-transfected 293T cells were concentrated 100-fold on Millipore concentrators. A 100-μl aliquot of the concentrate was loaded onto fast protein liquid chromatography Superdex 200 HR 10/30 column (Pharmacia, Piscataway, N.J.), equilibrated with Ca2+- and Mg2+-free phosphate-buffered saline. The column was eluted at a flow rate of 0.4 ml/min, and 0.25-ml fractions were collected. Identical fractions from four runs were pooled and immunoprecipitated with MAb 2G12 as described above. Comparison of the envelope glycoprotein elution profiles with those of known protein standards allowed an approximate assessment of molecular weights.
RESULTS
Wild-type gp140 is incompletely processed by cellular proteases.
We chose to use the HIV-1 JR-FL strain (subtype B) as our template for studies of the antigenic structure of oligomerized envelope glycoproteins. Several reasons underlay this choice: (i) HIV-1 JR-FL is a primary R5 isolate with a typical neutralization resistance profile and so is representative of the most commonly transmitted HIV-1 strains (34, 107); (ii) it is a molecularly cloned virus with a well-characterized env gene (50); (iii) we have previously expressed the gp120 monomer protein from HIV-1 JR-FL (106); and (iv) we have already studied the MAb reactivity profiles of the JR-FL gp120 monomer and cell surface-expressed gp120/gp140 complex and so are familiar with their antigenic properties (34, 35, 106).
To gain experience with the gp140 form of the JR-FL envelope glycoprotein, we expressed a protein which had the natural cleavage site between gp120 and gp41 maintained intact (gp140WT). In common with all the mutants that we subsequently describe, the gp140WT protein has the gp41 moiety truncated close to the transmembrane-spanning region, so that it contains both gp120 and the gp41 ectodomain (gp41ECTO) (Fig. 1). When we expressed the gp140WT construct in 293T cells by transient transfection, we could detect envelope glycoproteins in the supernatants at between 100 and 500 ng per ml by using an antigen-capture ELISA that recognizes both gp120 and gp140 proteins after they have been deliberately denatured. When the culture supernatants were immunoprecipitated with various anti-gp120 antibodies and then subjected to denaturing SDS-PAGE analysis, two bands appeared consistently on the gels. The results of one such immunoprecipitation experiment, with the 2G12 MAb as the precipitating antibody, are shown in Fig. 2. Although higher-molecular-mass aggregates were also present, two discrete bands can be seen; one of these, which we assumed to be free gp120, migrated at 120 kDa, and the other ran at 140 kDa (Fig. 2, lane 1). This latter protein migrated identically to a gp140 protein that we had mutated in the gp120-gp41 cleavage site (gp140UNC) (lane 3).
FIG. 1.
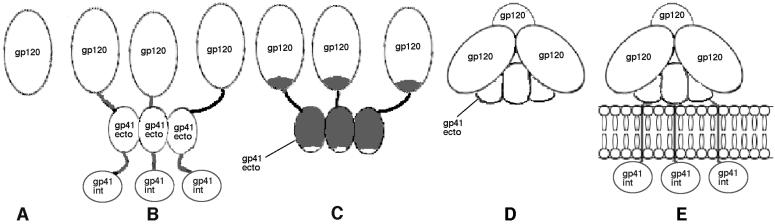
Different forms of the HIV-1 envelope glycoproteins. (A) Monomeric gp120. (B) Full-length recombinant gp160 (in practice, this protein may form higher-order aggregates in solution because of associations between various hydrophobic domains). (C) Proteolytically unprocessed gp140 trimer with the peptide bond maintained between gp120 and gp41 (gp140UNC or gp140NON). (D) The SOS gp140 protein, a proteolytically processed gp140 stabilized by an intermolecular disulfide bond. (E) Native, virion-associated gp120-gp41 trimer. The topologies of these proteins are inferred from previous reports cited in the text and from studies described in this paper. The shading of the gp140UNC protein (C) indicates the major antibody-accessible regions that are poorly or not exposed on the SOS gp140 protein or on the native gp120-gp41 trimer. The trimeric state of the SOS gp140 protein (D) has not yet been confirmed experimentally.
We reasoned that the 140-kDa band produced during expression of the gp140WT construct in transient transfections most probably arises because the host cell proteases of the furin family only incompletely cleave the scissile peptide bond between gp120 and the gp41 ectodomain. This could occur because the proteases are saturated by the large amount of gp140 expressed during a transient-transfection procedure. We therefore cotransfected a furin-encoding plasmid with the gp140WT-expressing plasmid, since such a procedure has been shown to increase the proteolytic processing of Ebola virus envelope glycoproteins (114).
In the presence of exogenous furin, the gp140WT protein was completely processed into its gp120 and gp41ECTO components (Fig. 2, lane 2). Of these, only the gp120 band is clearly visible on SDS-PAGE gels after immunoprecipitation; this is probably because the hydrophobic fusion peptide causes the 20-kDa gp41 ectodomain to self-aggregate when it is not bound to gp120. In contrast to gp140WT, the gp140UNC mutant was unaffected by the cotransfection of endogenous furin, still giving rise to only a single 140-kDa band in immunoprecipitates, because the cleavage site for furin proteases has been eliminated by mutation (lane 4). Furin cotransfection did, however, reduce by approximately fivefold the overall secretion of HIV-1 JR-FL envelope glycoproteins, as judged by the results of immunoprecipitations with polyclonal sera pooled from HIV-1-infected individuals (data not shown). This may be due to competition between the gp140- and furin-expressing plasmids for transcription or translation. We therefore adjusted the volume of supernatant used in each immunoprecipitation procedure with MAbs, to ensure that the total amounts of envelope glycoproteins present were comparable.
The above results confirm our assumption that the 140-kDa band obtained when the gp140WT protein is expressed in the absence of exogenous furin arises because of incomplete proteolytic cleavage of the peptide bond between gp120 and the gp140 ectodomain. We therefore designate this noncleaved gp140 protein gp140NON (Fig. 1). Another feature of furin cotransfection was that it eliminated the production of the high molecular weight aggregates that were visible in immunoprecipitates of both the gp140WT and gp140UNC proteins (Fig. 2, compare lane 2 with lanes 1, 3, and 4). When furin is cotransfected, the gp41 ectodomains cleaved off the gp120 subunits presumably still aggregate but are not precipitated by anti-gp120 antibodies.
Stabilization of the gp120-gp41 interaction by introduction of double cysteine substitutions.
With furin cotransfection, we could now express a soluble gp140 protein in which the gp120 and gp41ECTO components were associated only through a noncovalent linkage, mimicking what occurs in the native trimeric envelope glycoprotein complex on virions. However, the natural, noncovalent association between gp120 and gp41 is weak, leading to the gradual shedding of gp120 from virions and the surface of infected cells (38, 61, 70, 87). In practice, an unstable gp120-gp41ECTO complex is unlikely to be useful for vaccination purposes; it would, for example, be difficult to purify. We therefore sought ways to stabilize the gp120-gp41 interaction by the introduction of an intermolecular disulfide bond between the gp120 and gp41 subunits. Of note is that such bonds occur in at least a fraction of the envelope glycoprotein complexes of type C retroviruses, such as murine leukemia virus (MuLV) and human T-lymphotropic virus type 1 (HTLV-1) (25, 37, 48, 54, 55, 78, 82–86, 99).
Our mutagenesis strategy was guided by our earlier theoretical consideration of which regions of gp120 and gp41 were involved in their association (99). This analysis had, itself, been influenced by the mutational studies of Helseth et al. (45). Thus, there is strong mutagenic evidence that the first and last conserved regions of gp120 (C1 and C5 domains) are the contact sites for gp41 (45, 121). The corresponding sites on gp41 are known with less certainty. However, the positions of cysteine residues available for intermolecular disulfide bond formation in, e.g., the MuLV and HTLV-1 envelope glycoproteins strongly suggested that we should focus our attention on the central region of the gp41 ectodomain, in proximity to the intramolecular disulfide-linked loop (99). This loop is a conserved feature of retroviral envelope glycoproteins (37, 82). More recent information on the structure on the gp41 ectodomain supports this choice (8, 16, 19, 116). Precedent for the introduction of paired cysteine residues leading to the formation of intermolecular disulfide bonds has arisen from studies of HIV-1 gp41 (33) and of other viral envelope glycoproteins (39).
We therefore substituted single cysteine residues at several different positions in the C1 and C5 regions of gp120, focusing on amino acids previously shown to be important for the gp120-gp41 interaction (Fig. 3A). Simultaneously, we introduced a second cysteine substitution at several residues near the intramolecular disulfide loop of gp41 (Fig. 3B). The intent was to identify pairs of cysteine residues whose physical juxtaposition during gp140 processing was such that an intermolecular disulfide bond would form spontaneously. In all, 53 different double-cysteine substitution mutants were generated in the context of the JR-FL gp140WT protein and then coexpressed with furin by transient transfection of 293T cells (Fig. 4 and 5).
FIG. 4.
Immunoprecipitation analysis of selected double cysteine mutants of JR-FL gp140. The 35S-labeled envelope glycoproteins secreted from transfected 293T cells were immunoprecipitated with an anti-gp120 MAb, boiled with SDS, and analyzed by SDS-PAGE. The MAbs used were either 2G12 (odd-numbered lanes) or F91, which recognizes a CD4-binding site-related epitope (even-numbered lanes). The positions of the two cysteine substitutions in each protein (one in gp120, the other in gp41ECTO) are noted above the lanes. The gp140WT protein is shown in lane 15. All proteins were expressed in the presence of cotransfected furin, except for the gp140WT protein in lane 15, which serves as a reference standard for the position of 120-kDa (gp120) and 140-kDa (gp140NON) bands. Note that in this and all subsequent figures (except Fig. 10) that depict the outcome of RIPA experiments, the photographs have been cropped to show only the 120- and 140-kDa bands, since other regions of the gels were not informative.
FIG. 5.
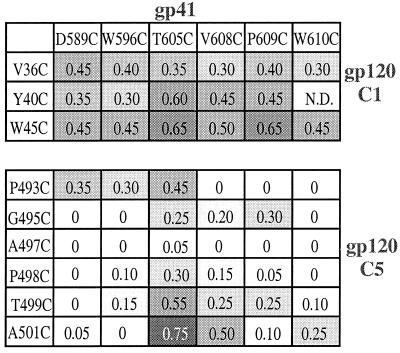
The efficiency of intermolecular disulfide bond formation is dependent upon the positions of the cysteine substitutions. The 35S-labeled envelope glycoproteins secreted from 293T cells cotransfected with furin and the various gp140 mutants were immunoprecipitated with the anti-gp120 MAb 2G12, boiled with SDS, and analyzed by SDS-PAGE. For each mutant, the intensities of the 140- and 120-kDa bands were determined by densitometry and the ratio of gp140 to gp140 + gp120 was calculated and recorded. The extent of shading is proportional to the magnitude of the ratio. The positions of the amino acid substitutions in gp41 and the C1 and C5 domains of gp120 are recorded along the top and down the sides, respectively. N.D., not done.
An initial analysis of the transfection supernatants by antigen capture ELISA indicated that all the gp140 mutants were efficiently expressed as secreted proteins, except those which contained a cysteine at residue 495 of gp120 (data not shown). We next characterized the secreted proteins by immunoprecipitation with the anti-gp120 MAbs 2G12 and F91 followed by SDS-PAGE. In this procedure, the envelope glycoproteins were eluted from the beads by boiling for 5 min in SDS-PAGE loading buffer, in the absence of any reducing agent such as DTT. In addition to the 120-kDa band (gp120), a second band of approximately 140 kDa (gp140) was precipitated by 2G12 and F91 from most of the double-cysteine mutant transfection supernatants (Fig. 4). This was not always the case, however, as exemplified by the A497C/W610C mutant, for which no 140-kDa band was visible (Fig. 4, lanes 9 and 10). There was some variation in how far the 140 kDa proteins from the different mutants migrated on the SDS-PAGE gels. For example, the V36C/W596C and T499C/T605C mutants were particularly slow moving (lanes 7 and 8 and lanes 11 and 12, respectively). The presence of diffuse bands with reduced mobility on SDS-PAGE gels is probably indicative of incomplete or improper envelope glycoprotein processing (25, 27–30, 79). High-molecular-weight aggregates similar to those in Fig. 2 were present in the immunoprecipitates of most of the double-cysteine mutants (data not shown, but see Fig. 10).
To determine which among the double-cysteine mutants was the most suitable for further analysis, we determined the relative intensities of the 140 and 120-kDa bands derived after immunoprecipitation of each mutant by MAb 2G12 followed by SDS-PAGE and densitometry (Fig. 5). We sought the mutant that produced the highest fraction of gp140 in relation to the total amount of secreted gp120 plus gp140 (i.e., the highest ratio of gp140 to gp140+gp120). Our interpretation was that such a mutant would have the most efficient formation of the intermolecular disulfide bond, while producing a 140-kDa protein that was reactive with a potently neutralizing anti-gp120 MAb.
Among the double-cysteine mutants, the one that most efficiently produced a 2G12-precipitable gp140 protein was a protein containing cysteine substitutions at alanine-501 of gp120 and threonine-605 of gp41 (A501C/T605C) (Fig. 5). Of note is that this protein migrated on SDS-PAGE gels as a discrete gp140 band with a mobility identical to that of the uncleaved gp140 protein from JR-FL (Fig. 4, compare lanes 13 and 14 with lane 15). The A501C/T605C mutant was the only one to have this property among the double cysteine mutants that we tested, a finding which suggests that a properly folded and processed gp140 protein is produced. Below, we refer to the A501C/T605C double cysteine mutant as the SOS gp140 protein.
Characterization of the SOS gp140 protein.
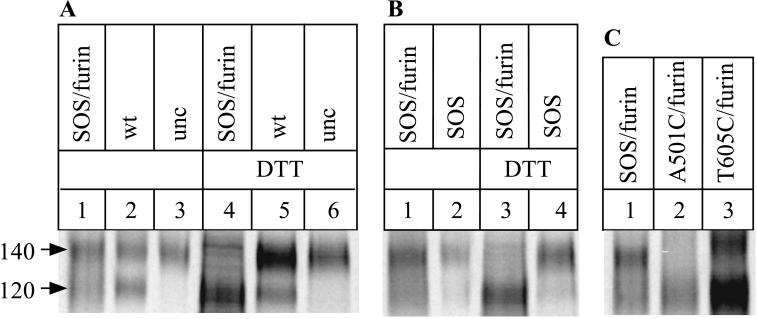
We verified that the SOS gp140 protein was indeed stabilized by an intermolecular disulfide bond by boiling the 2G12-immunoprecipitated proteins with SDS and DTT prior to gel electrophoresis; under these conditions, only a 120-kDa band was detected (Fig. 6A, lane 4, and Fig. 6B, lane 3). However, boiling with SDS alone did not eliminate the 140-kDa band (Fig. 6A and B, lanes 1). Taken together, the data imply the presence of a covalent bond between the gp120 moiety and the gp41 ectodomain of the SOS gp140 protein that is sensitive to the presence of a reducing agent, i.e., a disulfide bond. In contrast, the 140-kDa bands produced from the gp140NON (gp140WT without furin) and gp140UNC proteins were unaffected by boiling in the presence of DTT (Fig. 6A, lanes 5 and 6). In these two proteins, the gp120 and gp41ECTO subunits are attached via the uncleaved peptide bond, which is unaffected by reducing agents.
FIG. 6.
Confirmation that an intermolecular gp120-gp41 bond forms in the SOS gp140 protein. 293T cells were transfected with plasmids expressing gp140 proteins and, when indicated, a furin-expressing plasmid. The secreted, 35S-labeled glycoproteins were immunoprecipitated with the anti-gp120 MAb 2G12, boiled in the presence of SDS or, when indicated, SDS plus DTT, and analyzed by SDS-PAGE. (A) Lanes: 1 and 4, SOS gp140 protein (double cysteine mutant A501C/T605C) plus furin; 2 and 5, gp140WT protein, no furin; 3 and 6, gp140UNC protein, no furin. In lanes 1 to 3 the immunoprecipitated proteins were boiled with SDS; in lanes 4 to 6 they were boiled with SDS plus DTT. (B) Lanes: 1 and 3, SOS gp140 protein plus furin; 2 and 4, SOS gp140 protein without furin. In lanes 1 and 2 the immunoprecipitated proteins were boiled with SDS; in lanes 3 and 4 they were boiled with SDS plus DTT. (C) Lanes: 1, SOS gp140 protein (double cysteine mutant A501C/T605C) plus furin; 2, single cysteine gp140 mutant A501C plus furin; 3, single cysteine gp140 mutant T605C plus furin. The immunoprecipitated proteins in each case were boiled with SDS. High-molecular-weight aggregates were also present in immunoprecipitates of the SOS gp140 protein (data not shown but see Fig. 10).
As noted above, SDS-PAGE gels revealed that the mobility and sharpness of the 140-kDa band derived from the SOS gp140 protein was indistinguishable from those of the bands derived from the gp140NON and gp140UNC proteins (Fig. 6A, lanes 1 to 3). We also confirmed that cotransfection of furin was important for the correct formation of the SOS gp140 protein. Thus, in the absence of furin, the 140-kDa band was unaffected by boiling the immunoprecipitated proteins in the presence of DTT, suggesting that an uncleaved peptide bond still links the gp120 and gp41ECTO subunits (Fig. 6B, compare lanes 3 and 4).
Mutants containing only one of the two cysteines present in the SOS gp140 protein (gp140 mutants A501C and T605C) were also evaluated by RIPA with the 2G12 MAb (Fig. 6C). The A501C mutant produced no 140-kDa protein, and the T605C mutant produced a little, but the resulting ratio of gp140 to gp140 + gp120 was much lower with this mutant than with the SOS gp140 protein (Fig. 6C, compare lanes 1 and 3). Furthermore, the 140-kDa band derived from the T605C mutant had a lower mobility than the corresponding band from the SOS gp140 protein and probably represents a misfolded species (Fig. 6C, lane 3). Overall, this study with single cysteine substitutions provides further evidence that the 140-kDa band from the double-cysteine mutants is due to the formation of an intermolecular disulfide bond between gp120 and the gp41 ectodomain.
Attempts to improve the efficiency of disulfide bond formation in the SOS gp140 protein.
Although disulfide-stabilized gp140 proteins are secreted from cells expressing the SOS gp140 mutant, there is also significant production of gp120 monomers (Fig. 4 and 6). This implies that the disulfide bond between gp120 and the gp41 ectodomain forms with imperfect efficiency. We attempted to improve this by introducing additional amino acid substitutions near the inserted cysteines or by varying where the cysteines were positioned in gp120. We retained the gp41 cysteine at residue 605, where it is in the SOS gp140 protein, because this position seemed to be the one at which intermolecular disulfide bond formation was most favored (Fig. 5).
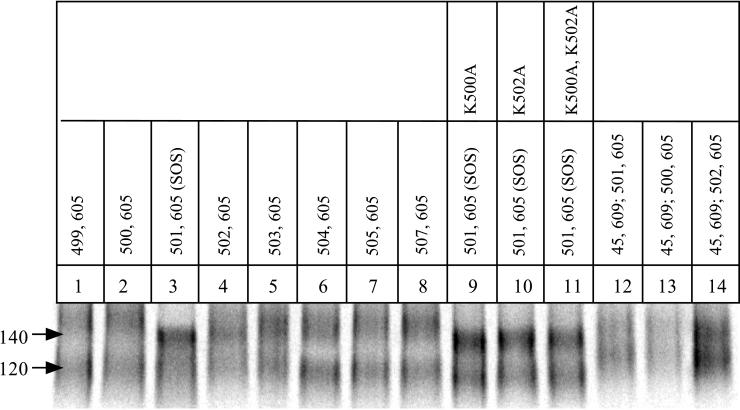
We first varied the position of the cysteine substitution in gp120, by placing it either N-terminal or C-terminal to alanine-501. The ratio of gp140 to gp140 + gp120 was not increased in any of these new mutants; it remained comparable to, or lower than, the ratio derived from the SOS gp140 protein (Fig. 7, lanes 1 to 8). Furthermore, there was a decrease in the mobility and sharpness of the gp140 band compared to that derived from the SOS gp140 protein (lanes 1 to 8). Next, we considered whether the bulky, charged side chains of the lysine residues adjacent to alanine-501 might interfere with disulfide bond formation. We therefore mutated either or both of the lysines at positions 500 and 502 to alanines in the context of the SOS gp140 protein, but these changes neither increased the ratio of gp140 to gp140 + gp120 nor affected the migration of gp140 (lanes 9 to 11). Finally, we introduced a second pair of cysteines into the SOS gp140 protein at residues 45 of gp120 and 609 of gp41, since a disulfide bond formed fairly efficiently when this cysteine pair was introduced into the wild-type protein (Fig. 5). The quadruple-cysteine mutant (W45C/A501C/T605C/P609C) was, however, poorly expressed, and the gp120 and gp140 bands that were produced both migrated unusually slowly. The same was observed with two other similar mutants (W45C/K500C/T605C/P609C) and (W45C/K502C/T605C/P609C) (Fig. 7, lanes 12 to 14). This implies that there may be protein-folding or other expression problems with quadruple-cysteine mutants of gp140.
FIG. 7.
Analysis of cysteine mutants of JR-FL gp140. The 35S-labeled envelope glycoproteins secreted from gp140-transfected 293T cells in the presence of cotransfected furin were immunoprecipitated with the anti-gp120 MAb 2G12, boiled with SDS, and analyzed by SDS-PAGE. Lanes: 1 to 8, each of the different gp140 double cysteine mutants contained the T605C substitution in gp41, in combination with a second cysteine substitution at the indicated residue in the C5 region of gp120 (the SOS gp140 protein is in lane 3); 9 to 11, gp140 proteins containing the A501C/T605C double cysteine substitutions together with the indicated lysine to alanine substitutions at residue 500 (lane 9), residue 502 (lane 10) or both residues 500 and 502 (lane 11); 12 to 14, gp140 proteins containing quadruple cysteine substitutions; each protein contained the W45C, T605C, and P609C substitutions, plus K500C (lane 12), A501C (lane 13), or K502C (lane 14).
Antigenic properties of the SOS gp140 protein.
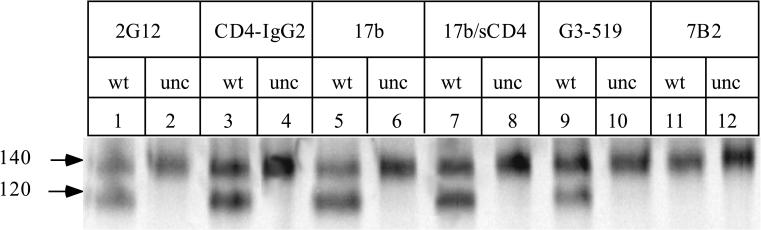
Among the JR-FL env mutants we have yet made, the efficiency of intermolecular disulfide bond formation is apparently the highest in the SOS gp140 protein (A501C/T605C). We therefore characterized the antigenic structure of this protein, by probing its topology with a panel of MAbs to a variety of gp120 and gp41 epitopes (Fig. 8). For comparison, we studied the reactivity of the same MAbs with the gp140NON protein produced when the gp140WT gene is expressed in the absence of cotransfected furin. The gp140NON protein still contains a peptide bond between the gp120 and gp140ECTO subunits (Fig. 6). Structurally, the gp140NON protein is essentially identical to the gp140UNC protein, in which the gp120-gp140 cleavage site has been deliberately replaced by mutagenesis (see Fig. 9). As an additional comparator, we used a double-cysteine mutant in which the gp120 cysteine substitution was in the C1 domain, the W45C/T605C gp140 protein (Fig. 8).
FIG. 8.
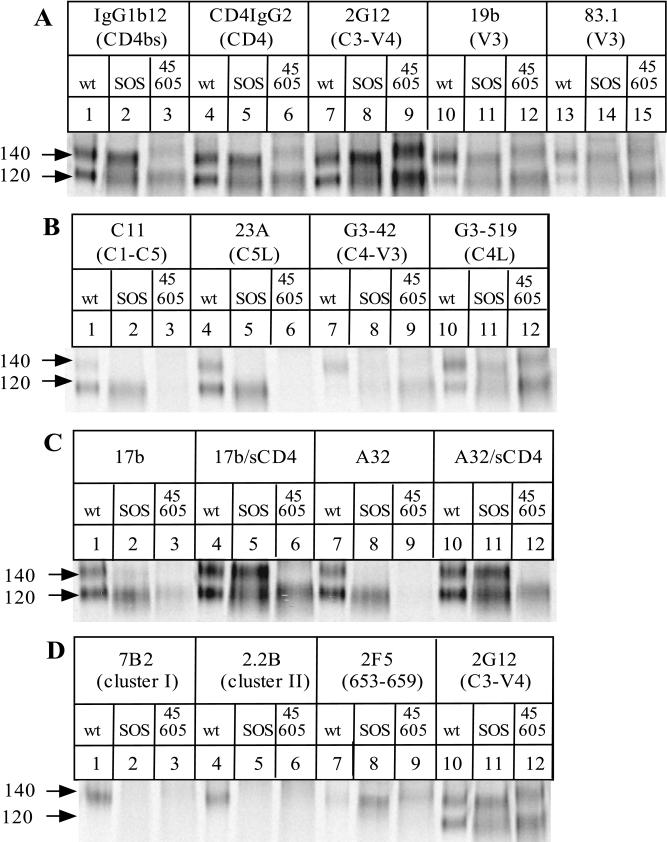
Comparison of the antigenic structures of the SOS gp140 protein, the gp140NON protein and gp120. The 35S-labeled envelope glycoproteins secreted from transfected 293T cells were immunoprecipitated with different anti-gp120 (A to C) or anti-gp41 (D) MAbs, boiled with SDS, and analyzed by SDS-PAGE. Lanes: 1, 4, 7, 10, and 13, gp140WT with no cotransfected furin, producing gp120 and the gp140NON protein; 2, 5, 8, 11, and 14, SOS gp140 protein plus cotransfected furin; 3, 6, 9, 12, and 15, gp140 protein containing the W45C/T605C double cysteine substitutions, plus co-transfected furin. Brief descriptions of the epitopes recognized by each MAb are noted above each lane; for more details, see the primary references listed in Materials and Methods. D, discontinuous epitope; L, linear epitope.
FIG. 9.
Comparison of the antigenic structures of the gp140NON and gp140UNC proteins. The 35S-labeled envelope glycoproteins secreted from transfected 293T cells were immunoprecipitated with different anti-gp120 MAbs, boiled with SDS, and analyzed by SDS-PAGE. Odd-numbered lanes contained gp140WT with no cotransfected furin, producing gp120 and the gp140NON protein. Even-numbered lanes contained gp140UNC protein with no cotransfected furin.
Compared to gp140NON, the SOS gp140 protein has several antigenic differences that we believe are desirable for a protein intended to mimic the structure of the virion-associated gp120-gp41 complex. These are summarized below.
(i) The SOS gp140 protein is efficiently recognized by the potently neutralizing antibodies IgG1b12 and 2G12 and also by the CD4-IgG2 molecule (Fig. 8A). Although the RIPA method is not sufficiently quantitative to allow a precise determination of relative affinities, the reactivities of these MAbs and of the CD4-IgG2 molecule with the SOS gp140 protein appear to be substantially greater than with the gp140NON and gp120 proteins. Clearly, the SOS gp140 protein has an intact CD4 binding site. Epitopes in the V3 loop are also accessible on the SOS gp140 protein, as shown by its reactivity with MAbs 19b and 83.1 (Fig. 8A). There is very little exposure on the SOS gp140 protein of epitopes for MAbs G3-42 and G3-519 (Fig. 8B). These MAbs bind to the C4 region of gp120, in proximity to the V3 loop and the CD4 binding site, and have neutralizing activity against T-cell-line-adapted but not primary HIV-1 isolates (67, 72, 73, 122). Each of the above MAbs also recognized the gp140NON and gp120 proteins derived from expression of gp140WT in the absence of furin (Fig. 8A). The V3 loop MAbs 19b and 83.1 and the C4-V3 MAb G3-42 bound to the gp140NON protein more strongly than to the corresponding gp120 (note the relative intensities of the 140- and 120-kDa bands in lanes 10 and 13 of Fig. 8A and lane 7 of Fig. 8B, compared to lanes 1, 4, and 7 of Fig. 8A). The V3 loop may be unusually well exposed in the uncleaved gp140 protein.
(ii) Conversely, the nonneutralizing anti-gp120 MAbs C11 and 23A did not bind detectably to the SOS gp140 protein (Fig. 8B). These MAbs recognize epitopes in the C1 and C5 domains, regions of gp120 that are involved in gp41 association and are occluded in the context of a properly formed gp120-gp41 complex (71, 121). Although the cysteine residue at position 501 of the SOS gp140 protein is located near the epitopes for MAbs C11 and 23A, it did not destroy these epitopes; thus, MAbs C11 and 23A bound efficiently to the gp120 derived from the SOS gp140 protein, which also contains the A501C substitution (Fig. 8B). In addition, MAb M90, to a discontinuous C1 epitope, bound weakly to gp120 derived from the SOS gp140 protein but not to the SOS gp140 protein itself. M90 did, however, bind to both the gp120 and gp140 components of gp140WT (data not shown). In contrast to the poor reactivity of the C1- and C5-directed MAbs with the SOS gp140 protein, these MAbs all bound to the gp140NON and gp120 proteins (Fig. 8B) and also to gp140UNC (see Fig. 9) (data not shown). This implies that the C1 and C5 regions of gp120 are abnormally accessible when a peptide bond links gp120 with the gp41 ectodomain.
(iii) The induction of the epitope for MAb 17b by the prior binding of sCD4 occurred far more efficiently on the SOS gp140 protein than on the gp140NON or gp120 protein (Fig. 8C, compare lanes 5 and 2 with lanes 4 and 1). Indeed, in the absence of sCD4, there was very little reactivity of 17b with the SOS gp140 protein (lane 2). The CD4-induced epitope for MAb 17b overlaps the coreceptor binding site on gp120 (91); it is considered that this site becomes exposed on the virion-associated gp120-gp41 complex during the conformational changes which initiate virus-cell fusion after CD4 binding (47, 91, 95, 97, 103). The induction of the 17b epitope on the SOS gp140 protein suggests that the conformation of the gp120 moieties resembles what is present on virions and is not significantly affected by the intermolecular disulfide bond with the gp41 ectodomain. The gp140NON protein bound 17b constitutively, and although there was some induction of the 17b epitope upon soluble CD4 binding, this was much less pronounced than what occurred with the SOS gp140 protein (Fig. 8C).
(iv) Another CD4-inducible epitope on gp120 is that recognized by MAb A32 (72, 103). There was negligible binding of A32 to the SOS gp140 protein in the absence of sCD4, but the epitope was strongly induced by sCD4 binding (Fig. 8C, compare lanes 11 and 8). As observed with 17b, the A32 epitope was much less efficiently induced on the gp140NON protein than on the SOS gp140 protein (compare lanes 10 and 7).
(v) Neither of the nonneutralizing anti-gp41 MAbs 7B2 and 2.2B recognized the SOS gp140 protein, whereas each bound strongly to the gp140NON protein (Fig. 8D). These anti-gp41 MAbs recognize the two major epitope clusters of the gp41 ectodomain, both of which are considered to be occluded by gp120 in the virion-associated gp120-gp41 complex (71, 98). Similar results were obtained with several other MAbs to these regions, T4, T15G1, and 4D4 (data not shown). The failure of these anti-gp41 MAbs to bind to the SOS gp140 protein is another indication that this protein adopts a configuration similar to that of the native trimer. However, we cannot exclude the possibility that the formation of an intermolecular disulfide bond involving the central region of gp41 perturbs the epitopes for several gp41 MAbs by a different mechanism. The efficient recognition of the gp140NON protein by several gp41 MAbs is consistent with the view that proteins of this type have an aberrant conformation because of the peptide bond linking gp120 with gp41 (31).
(vi) In marked contrast to what was observed with the nonneutralizing anti-gp41 MAbs, the neutralizing anti-gp41 MAb 2F5 bound efficiently to the SOS gp140 protein but not detectably to the gp140NON protein (Fig. 8D, compare lanes 11 and 10). When the experiment was repeated with a higher concentration of the gp140WT protein, some 2F5 reactivity could be observed (data not shown). However, when equivalent amounts of the gp140WT and SOS gp140 proteins were compared, it was found that 2F5 reacted more strongly with the latter (Fig. 8D). Of note is that the 2F5 epitope is the only region of gp41 thought to be well exposed in the context of native gp120-gp41 complexes (98). The ability of the SOS gp140 protein to bind 2F5 is again consistent with the view that this protein adopts a configuration similar to that of the native trimer.
We also examined whether sCD4 binding could cause the exposure of other previously occult epitopes in the C1 and C5 regions of gp120 or in several areas of the gp41 ectodomain, as happens when sCD4 induces the shedding of gp120 from gp41 on the native envelope glycoprotein complex (87, 98). However, we could not detect any increase in the exposure of any other gp41 epitopes on the SOS gp140 protein in the presence of sCD4 (data not shown). This indicates that the presence of the intermolecular disulfide bond prevents gp120 from dissociating from the gp41 ectodomain, despite the conformational changes that are induced in the gp120 moiety upon sCD4 binding.
The antigenic properties of the SOS gp140 protein were compared with those of the W45C/T605C gp140 protein. Among the set of mutants that contained a cysteine substitution within the C1 domain, this was the most efficient at gp140 formation (Fig. 5). Although the W45C/T605C gp140 protein reacted well with the 2G12 MAb (Fig. 8A, lane 9), it bound CD4-IgG2 and IgG1b12 very poorly (lanes 3 and 6). Furthermore, there was little induction of the 17b and A32 epitopes on the W45C/T605C gp140 protein by sCD4, although these epitopes were induced on the gp120 moiety from this mutant (Fig. 8C, compare lanes 6 and 12 with lanes 3 and 9). There was some reactivity of anti-gp41 MAbs with the W45C/T605C gp140 protein (Fig. 8D). The anti-gp120 MAbs C11 and 23A recognized neither the gp140 nor the gp120 form of the W45C/T605C mutant (Fig. 8B). For the C1-C5-directed MAb C11, this may be due to a direct effect of the W45C substitution on the epitope (75). Inappropriate protein folding due to the aberrant formation of disulfide bonds in the C1 and C5 regions of gp120 may be an explanation for the lack of 23A reactivity with the W45C/T605C mutant. Taken together with the fact that the gp140 bands from the W45C/T605C protein are diffuse and of relatively low mobility, these results suggest that this mutant has suboptimal antigenic properties. Indeed, the contrast between the properties of the W45C/T605C gp140 protein and the SOS gp140 protein implies that the positioning of the intermolecular disulfide bonds has a significant influence on the antigenic structure of the resulting gp140 molecule.
Comparing the antigenic structures of gp140NON and gp140UNC.
We next studied the antigenic structure of the gp140 protein produced when the cleavage site between gp120 and gp41 is replaced by mutation (gp140UNC), since this type of protein is being used in vaccine-related studies on the grounds that it is oligomeric (27, 90, 110). We compared gp140UNC with the gp140NON and gp120 proteins produced when gp140WT is expressed in the absence of cotransfected furin (Fig. 9). The gp140NON and gp140UNC proteins could not be distinguished from one another by the reactivity of any of the test MAbs; they are essentially isomorphic. The major differences in antigenic structure between the SOS gp140 protein and the gp140NON protein that were demonstrated in Fig. 8 therefore also apply to the gp140UNC protein. Of particular note is the negligible induction of the 17b epitope on the gp140UNC protein by sCD4 (Fig. 9, compare lanes 6 and 8), which may help explain why proteins of the gp140UNC type interact poorly with the CCR5 coreceptor (31). The aberrant exposure of gp41 in the gp140UNC and gp140NON proteins is also clearly revealed (compare Fig. 9, lanes 11 and 12, with Fig. 8D, lane 2).
Intersubunit disulfide bonds form in SOS gp140 proteins from other HIV-1 isolates.
To assess the generality of our observations with gp140 proteins derived from the R5 HIV-1 isolate JR-FL, we generated double-cysteine mutants of gp140s from four other HIV-1 strains. These were the R5X4 viruses GUN-1wt, 89.6, and DH123 and the T-cell-line-adapted X4 virus HxB2. In each case, the cysteines were introduced at the residues equivalent to alanine-501 and threonine-605 of HxB2. The resulting SOS gp140 proteins were precipitated with the 2G12 MAb, in comparison with the gp140WT proteins from each isolate (Fig. 10). In general, the results obtained with the GUN-1wt, 89.6, DH123, and HxB2 proteins were very similar to what was observed with JR-FL gp140s. Disulfide-stabilized gp140 proteins could be efficiently expressed from each isolate, as confirmed by the disappearance of the 140-kDa band when the immunoprecipitates were boiled with DTT before being subjected to SDS-PAGE analysis. In each case, the ratio of gp140 to gp140 + gp120 was comparable to or greater than that observed for the JR-FL SOS gp140 protein. One unexpected but advantageous observation was that furin cotransfection significantly increased the secretion of envelope glycoproteins from 89.6 gp140-transfected cells (Fig. 10B, compare lanes 6 and 7 with lane 5). This may be due to a decrease in the degradation of misfolded proteins when the scissile bond between gp120 and gp41 is correctly cleaved. We do not yet know why this should be an isolate-dependent phenomenon. To some extent, it occurs also with DH123 proteins.
Sucrose gradient analysis of the SOS gp140 and gp140UNC proteins.
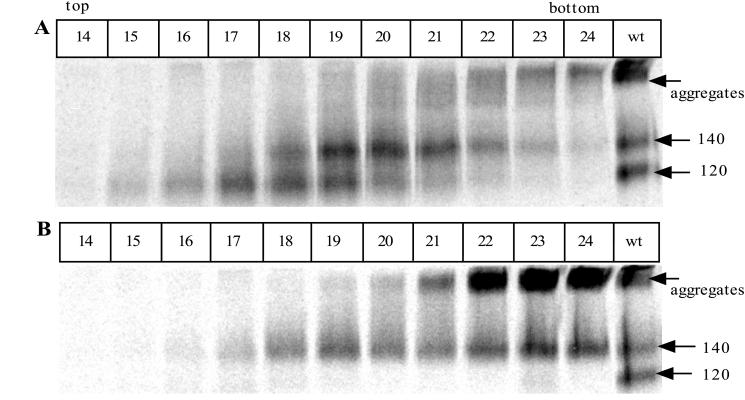
The oligomeric state of the secreted gp140 complex cannot be determined by immunoprecipitations of unfractionated supernatants, since the proteins are subsequently denatured by boiling with SDS prior to gel electrophoresis. To obtain information on the size of the gp140 protein complex under nondenaturing conditions, we performed a sucrose velocity gradient analysis on 100-fold concentrates of the proteins secreted from 293T cells transfected with SOS gp140 (JR-FL) and furin or, for comparison, with the gp140UNC mutant (Fig. 11A). To detect where various molecular species had migrated on the sucrose velocity gradient, the gradient fractions were immunoprecipitated with MAb 2G12, boiled with SDS, and analyzed by SDS-PAGE.
FIG. 11.
Sucrose gradient analysis of the JR-FL SOS gp140 and gp140UNC proteins. Envelope glycoproteins secreted from transfected 293T cells were concentrated 100-fold and then fractionated by sucrose velocity gradient centrifugation. The gradient fractions (500 μl) were immunoprecipitated with MAb 2G12, boiled with SDS, and analyzed by SDS-PAGE to detect envelope glycoproteins and determine the sizes of their denatured components. (A) JR-FL SOS gp140 protein. (B) JR-FL gp140UNC protein. The last lane in each panel shows an unconcentrated supernatant containing the JR-FL gp140WT protein expressed in the absence of furin and then immunoprecipitated with 2G12 to provide a reference standard for the positions of gp120 and gp140 proteins on the gel. These bands are marked on the right of each panel, together with the position of high-molecular-weight aggregates.
Three forms of envelope glycoproteins were detected after sucrose gradient fractionation of the SOS gp140 protein (Fig. 11A). Fractions 23 and 24 contained material of a very high molecular mass, which probably correspond to the aggregates that were noted in RIPA experiments (Fig. 2). A broad peak containing envelope glycoproteins with a subunit molecular mass of 140 kDa was centered on fraction 20. A second peak containing 120-kDa subunits was present in fractions 17 to 19, separated from the 140-kDa proteins by two fractions, or 1 ml (Fig. 11A).
When the gp140UNC protein was analyzed, the very-high-molecular-mass aggregates were again present (fractions 22 to 24), and they were more abundant than with the SOS gp140 protein (Fig. 11B). This is consistent with what was observed in the immunoprecipitation analysis shown in Fig. 2. Envelope glycoproteins with a subunit weight of 140-kDa were spread throughout fractions 18 to 24 (Fig. 11B). The 140-kDa proteins in fractions 22 to 24 were most probably derived from high-molecular-mass aggregates formed when the immunoprecipitates were boiled with SDS before being subjected to SDS-PAGE. The 140-kDa proteins in fractions 19 to 21 migrate in the same position as the 140-kDa components of the SOS gp140 preparation.
We interpret the sucrose velocity gradients to indicate that the SOS gp140 preparation contains monomeric gp120 proteins which peak in fractions 17 and 18, together with oligomeric proteins containing 140-kDa subunits which peak in fractions 19 and 20. Excluding the products of protein aggregation, only the latter proteins are present in the gp140UNC preparation. From this analysis, we cannot determine the exact molecular mass, and hence the subunit composition, of the oligomeric proteins. However, the fact that they were clearly separated from the 120-kDa gp120 monomers by an approximately 1-ml volume on a standard 5 to 20% sucrose velocity gradient of 8.8 ml (i.e., by a density difference of approximately 1.5% sucrose) indicates that they are probably of several hundred kilodaltons (a 140-kDa monomer would not be separable from a 120-kDa protein under these conditions). This would be consistent with their composition being oligomeric gp120-gp41ECTO complexes, although this cannot be proven by this type of analysis.
Gel filtration analysis of the SOS gp140 and gp140UNC proteins.
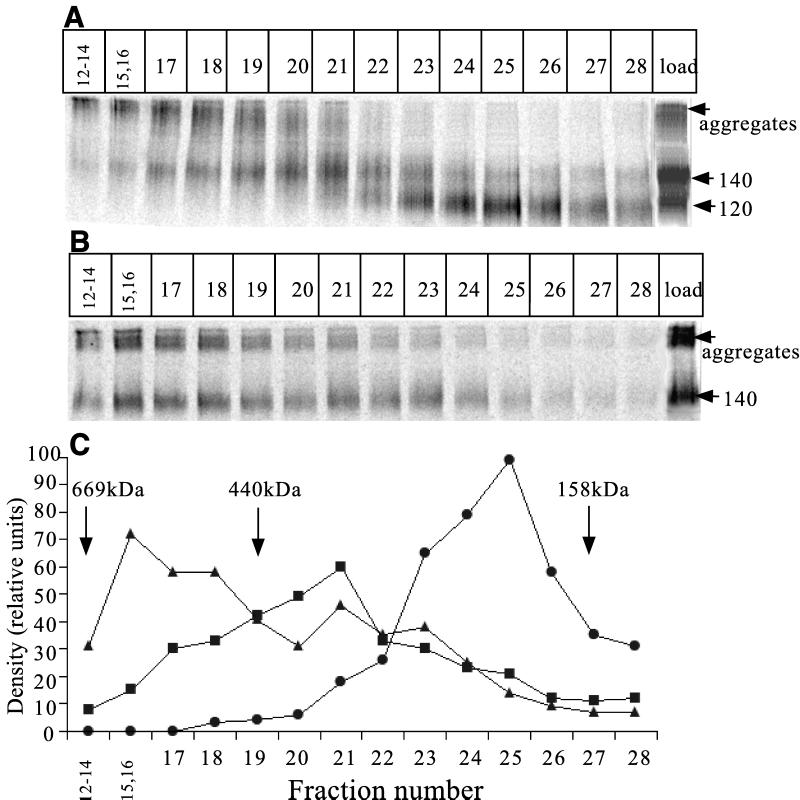
To obtain additional information on the molecular size of the gp140 protein complexes under nondenaturing conditions, we used size exclusion gel filtration chromatography (Fig. 12). This was performed on concentrates of the proteins secreted from 293T cells transfected with SOS gp140 (JR-FL) plus furin (Fig. 12A) and, for comparison, gp140UNC (Fig. 12B). To detect where various molecular species had migrated, the gradient fractions were immunoprecipitated with MAb 2G12, boiled with SDS, and analyzed by SDS-PAGE.
FIG. 12.
Gel filtration analysis of the JR-FL SOS gp140 and gp140UNC proteins. Envelope glycoproteins from transfected 293T cells were concentrated 100-fold and then fractionated by gel filtration chromatography. The fractions (250 μl) were immunoprecipitated with MAb 2G12, boiled with SDS, and analyzed by SDS-PAGE to detect envelope glycoproteins and determine the size of their constituent subunits. (A) JR-FL SOS gp140 protein. (B) JR-FL gp140UNC protein. The last lane in each panel shows an unconcentrated supernatant containing the protein under analysis and immunoprecipitated with 2G12 to provide a reference standard. These bands are marked on the right of each panel, together with the position of high-molecular-weight aggregates. (C) Densitometric analysis of the elution profile derived from the SOS gp140 protein (A and B). ▴, gp140UNC; ■, the 140-kDa component of SOS gp140; ●, the 120-kDa component of SOS gp140. The positions of molecular mass standards are indicated by arrows. These were thyroglobulin (669 kDa), ferritin (440 kDa), and aldolase (158 kDa).
Three forms of envelope glycoproteins were detected after fractionation of the SOS gp140 protein (Fig. 12A). Fractions 12 to 19 contained material of a high molecular weight, which may again correspond to the aggregates noted in RIPA experiments (Fig. 2). A broad peak containing envelope glycoproteins with a subunit molecular mass of 140 kDa was centered on fractions 20 and 21. This, we believe, is the oligomeric component of the SOS gp140 protein (see below and also Discussion). A more rapidly migrating peak containing 120-kDa subunits was found in and around fractions 24 and 25; this most probably represents the position of gp120 monomers (Fig. 12A).
When the gp140UNC protein was analyzed, the very-high-molecular-weight aggregates were again present (fractions 12 to 19), and they were more abundant than was observed with the SOS gp140 protein (Fig. 12B). This is consistent with the immunoprecipitation analysis in Fig. 2. Envelope glycoproteins with a subunit weight of 140 kDa were spread throughout fractions 15 to 23, with an apparent peak around fractions 15 and 16 (Fig. 12B). The aggregates in fractions 12 to 18 are probably derived from the unfolding of gp140 and the intermolecular association of hydrophobic gp41 moieties when the immunoprecipitates are boiled with SDS before being subjected SDS-PAGE. Of note is that the gp140 proteins in fractions 20 and 21 migrate in the same position as the 140-kDa components of the SOS gp140 preparation. The radiographs shown in Fig. 12A and B were scanned to determine the density of each band (Fig. 12C). We estimate that approximately 40 to 50% of the nonaggregated protein in the SOS gp140 protein is in the higher-molecular-weight form, with the rest being gp120.
The positions of the protein standards thyroglobulin (669 kDa), ferritin (440 kDa), and aldolase (158 kDa) are also indicated (Fig. 12C). Note that the monomeric gp120 protein migrates more rapidly, and hence appears to be of higher molecular mass, than a standard, globular protein of 158 kDa, aldolase. However, the higher-molecular-mass, oligomeric component of the SOS gp140 protein migrates more slowly, and so appears to be of lower molecular mass, than the 440-kDa ferritin standard (Fig. 12C). The unusually abundant glycosylation of the HIV-1 envelope glycoproteins is likely to affect their biophysical properties and limit the value of comparisons with traditional protein molecular weight standards.
DISCUSSION
Our goals are to make a recombinant HIV-1 envelope glycoprotein with antigenic properties mimicking those of the native, trimeric gp120/gp41 complex found on virions or virus-infected cells and then evaluate whether such a protein might be a useful component of a multivalent HIV-1 vaccine. We believe we have accomplished the initial phase in creating the SOS gp140 protein. Whether this protein will be a superior immunogen to gp120 monomers, gp140 proteins with a peptide linkage retained between the gp120 and gp41ECTO moieties, or full-length gp160, remains to be determined. We believe that the disulfide bridge in the SOS gp140 protein should provide sufficient stability for this to be a practical immunogen, considering that the 140-kDa band survives boiling and SDS treatment during PAGE analysis.
There were two technical steps necessary for the generation of the SOS gp140 protein. The first was the use of cotransfected furin to increase the efficiency with which a secreted gp140 protein is proteolytically processed into gp120 and gp41ECTO moieties. The second was the introduction of a disulfide bond, at an appropriate position, to cross-link the gp120 subunit to the gp41 ectodomain and thereby increase its stability. During the synthesis of envelope glycoproteins in HIV-1-infected cells, trimerization of the gp41 moieties in the context of the gp160 precursor precedes the cleavage of the peptide bond linking gp120 to gp41 (25, 79). The cleavage step is mediated by proteases of the furin family (25, 41, 76). This step is inefficient, but unprocessed gp160 is generally sorted intracellularly into the lysosomal pathway, and little or no uncleaved gp160 is incorporated into virions (25, 26, 60, 119). However, when the HIV-1 env gene is expressed at high levels in mammalian cells, uncleaved gp160 cleavage can be found at the cell surface, perhaps because the natural cellular complement of furin proteases is saturated or because of differences in how gp160s are routed in infected and transfected cells (60, 119; Q. J. Sattentau, personal communication). These differences may be exacerbated when soluble rather than membrane-associated proteins are expressed, as is the case with gp140s. For whatever reason, when we expressed the JR-FL gp140WT gene in 293T cells, only a fraction of the secreted gp140 proteins were properly cleaved to gp120 and gp41ECTO subunits. This problem was overcome by the exogenous supplementation of furin via transfection, a device previously used to increase the efficiency of Ebola virus envelope glycoprotein proteolytic processing (114) and one that may have general relevance for vaccine development. Furin transfection did, however, reduce the extent of envelope glycoprotein expression (except with 89.6 and perhaps DH123), perhaps because of competition between plasmids for protein translation. Careful optimization of the furin content of permanent cell lines will be required when scaling up the production of the SOS gp140 protein.
The solution to the first problem created the second: the properly processed gp120 and gp41ECTO subunits are only weakly associated by noncovalent interactions. Consequently, the gp120 moieties are rapidly shed as the complex disassembles (38, 61, 70). To overcome this, we considered whether we could modify the gp120 or gp41 primary sequences to increase the strength of the noncovalent interaction between the subunits. However, in the absence of detailed information on the structure of the gp120-gp41 interactive sites, there was no good way to predict what amino acid substitutions might work. Indeed, most of the substitutions in relevant regions of gp120 and gp41 that have been described in the literature actually weaken rather than strengthen the intersubunit association (17, 20, 45). We therefore focused on a second strategy: the stabilization of the gp120-gp41 interaction by the formation of an intersubunit disulfide bond between cysteine residues introduced into appropriate positions within gp120 and gp41.
We found that the precise positioning of the two cysteine residues introduced into gp120 and gp41 was important. Of the many double-cysteine mutants that we evaluated, the SOS gp140 protein (A501C/T605C) had the highest efficiency of disulfide bond formation, the fewest indications of poor folding, and the most favorable antigenic properties. In this protein, the cysteine substitution in gp120 is at a residue previously shown to be critical for any association of gp120 with gp41 (45). The N- and C-terminal ends of gp120 probably assume disordered, flexible conformations, a factor which provoked their deletion from the crystallized gp120 core fragment (51, 122). The flexibility of these regions may explain why so many different cysteine substitutions of residues near the gp120 N and C termini permitted at least some disulfide linking to gp140. However, several such mutants were associated with smearing of gp140 bands on SDS-PAGE gels, suggesting that an imperfectly positioned disulfide bond does have some negative effects on envelope glycoprotein folding.
The corresponding substitution in gp41 is at a location exactly equivalent to where a cysteine residue is naturally positioned in the transmembrane glycoproteins of many retroviruses, including MuLV and HTLV-1 (37, 82, 99). This cysteine is immediately C-terminal to a small loop bounded by an intramolecular disulfide bond that is a common feature of retrovirus and lentivirus transmembrane glycoproteins (37, 82). On intuitive grounds, we postulated that this region of HIV-1 gp41 was involved in gp120 binding (99); the additional cysteine present in other retroviruses probably accounts for the disulfide bond that can sometimes form between the surface and transmembrane glycoproteins (25, 54, 55, 78, 83–86). There may be a conserved mechanism of subunit association among many viral families, sometimes with the involvement of a disulfide bond and sometimes not (36, 37, 99, 124). The crystal structure of the major fragment of the gp41 ectodomain in its postfusion conformation reveals that the C-terminal helix of the gp41 trimeric coiled coil is positioned antiparallel to, and stacked on the outside of, an N-terminal trimer (19, 101, 116). This implies that the cysteine residue substituted for alanine-605 protrudes outward in the postfusion conformation of gp41. The crystal structures of the TM glycoproteins of other viruses, such as MuLV and Ebola virus, also show that the region near the intramolecular disulfide-bonded loop is solvent accessible (16, 33a, 57a, 117). At present, the conformation of the prefusion form of the gp41 ectodomain is unknown, but presumably alanine-605 must also protrude in this form of the protein since the cysteine residue substituted at this position is available for disulfide bond formation with cysteine-501 of gp120. In the correctly folded, prefusion form of the gp120-gp41 complex, these two residues must be sufficiently proximal for disulfide bond formation to be possible. If and when the prefusion form of the gp41 ectodomain is crystallized, the exact positioning of alanine-605 will be revealed.
Although we can clearly make a gp140 protein in which the gp120 and gp41ECTO moieties are stabilized by an intermolecular disulfide bond, the formation of the disulfide bond occurs with imperfect efficiency. Thus, only a fraction (perhaps 40 to 50%) of the envelope glycoprotein complexes secreted from 293T cells expressing the A501C/T605C double cysteine mutant in the presence of furin are in the form of the SOS gp140 protein (Fig. 12C). The remaining proteins are present as gp120 monomers. This reflects inefficient formation of the intermolecular disulfide bond in the transfected 293T cells, rather than a lability of this bond once it has formed; the gp120 subunit still remains attached to the gp41 ectodomain even when the SOS gp140 protein is boiled in SDS and partially denatured, indicating that the intermolecular disulfide bond is quite stable. Preliminary studies of a permanent CHO cell line show that these cells secrete essentially only disulfide-stabilized SOS gp140 proteins, with virtually no gp120 moieties being present (data not shown). The efficiency of intermolecular disulfide bond formation is probably cell type dependent.
Biophysical analyses showed that the SOS gp140 protein has a higher molecular weight than monomeric gp120. It also differs in its biophysical properties from uncleaved gp140. However, we have not yet determined whether the SOS gp140 protein contains three gp41 ectodomains, each linked to a gp120 moiety via a disulfide bond, or whether only one or two gp120s are successfully attached to trimerized gp41 subunits. A mixture of molecular species may be present. Additional studies of SOS gp140 proteins purified from a permanent cell line are necessary to address these issues.
We are, however, encouraged by the antigenic properties of the SOS gp140 protein; it has a MAb reactivity pattern that is consistent with what has been learnt from prior studies of the native trimer and of the relationship between MAb binding and HIV-1 neutralization (34, 71, 72, 98, 102, 106, 115). Thus the most commonly exposed regions on the gp120 moiety of the SOS gp140 protein correspond to neutralizing-antibody epitopes. These include areas near the CD4 binding site (e.g., the binding sites for MAb IgG1b12 and the CD4-IgG2 molecule), the C3-V4 glycan-dependent epitope for MAb 2G12, the V3 loop, and, in the presence of sCD4, the CD4-induced epitope for MAb 17b that overlaps the coreceptor binding site. For some MAbs, notably 2G12, the reactivity with the SOS gp140 protein is better than with the gp120 monomer. MAbs to nonneutralizing epitopes in the C1 and C5 domains do not bind to the SOS gp140 protein, although they recognize the uncleaved gp140 proteins quite efficiently because of the abnormal conformation conferred upon the gp120 moiety when the gp41 ectodomain is attached via a peptide bond.
Some MAbs to CD4-binding site and V3 loop epitopes (e.g., F91 and 19b) do, however, bind efficiently to the SOS gp140 protein while lacking strong neutralization activity against HIV-1 JR-FL. The binding of the nonneutralizing A32 MAb to the SOS gp140 protein in the presence of sCD4 is another example. The ability of weakly neutralizing MAbs to bind to native envelope glycoprotein complexes on the cell surface has been described previously (35). Factors such as the on-rate may be important in determining precisely which MAbs to closely related epitopes do (e.g., IgG1b12) and do not (e.g., F91) neutralize HIV-1. Kinetic parameters might not be identical in RIPA and neutralization assays, in that the slow binding of some MAbs to the native trimer might be rapid enough to be detectable in a binding assay yet too slow to be able to interfere with virus attachment and entry in a neutralization assay. There may also be differences in the rates at which neutralizing antibodies bind to soluble and membrane-associated forms of the same protein complex.
On the gp41 moiety of the SOS gp140 protein, only the epitope for the neutralizing MAb 2F5 is accessible. Nonneutralizing gp120 and gp41 antibody epitopes components are not exposed on the SOS gp140 protein, just as they are inaccessible on native trimers (71, 98). However, we cannot rule out the possibility that the occlusion of the nonneutralizing gp41 epitopes is a direct consequence of the formation of the intermolecular disulfide bond. The almost complete occlusion of the CD4-induced epitope on the SOS gp140 protein in the absence of CD4, combined with its substantial induction upon CD4 binding, is consistent with how the gp120 moieties in a native trimer are thought to be arranged (122). In this conformation, the CD4-induced epitopes are partially covered by the V1/V2 and V3 loop structures and partially occluded by interactions between the individual gp120 components of a trimer. In the gp120 monomer, the occlusion of the CD4-induced epitopes is only partial (105, 122, 123), and we observed this to be the case also in the uncleaved gp140 proteins. Thus the MAb reactivity patterns of the SOS gp140 protein are, in general, consistent with its existing as a native, oligomeric structure. We believe that the SOS gp140 protein is in a prefusion conformation, judged by the dramatic induction of the 17b epitope upon sCD4 binding.
The above properties of the SOS gp140 protein contrast markedly with the antigenic structure of gp140 proteins that retain the peptide bond between gp120 and the gp41 ectodomain. Proteins of this category have not, to date, been significantly superior to gp120 monomers as immunogens (27, 90, 110). This may be because they do not properly mimic the structure of the native trimer, as indicated by their limited ability to interact with coreceptors (31); it is probably not the ability of a protein to oligomerize that most strongly influences its immunogenicity, but its overall structure. The acid test of the value of our work to vaccine development will come from immunogenicity studies, the outcomes of which are inherently unpredictable. The preservation of the best neutralizing-antibody epitopes on the SOS gp140 protein (those for MAbs IgG1b12, 2G12, and 2F5), combined with the elimination of irrelevant epitopes, might be valuable for focusing the humoral immune response.
It may, however, be found necessary to further modify the antigenic structure of the SOS gp140 protein to improve its immunogenicity, for example by removing some of its glycan residues or variable loops (18, 89). In preliminary studies, we have found that such modifications can be made to the SOS gp140 protein without having significant effects on the efficiency of intermolecular disulfide bond formation (R. Sanders, F. Kajumo, A. Master, L. Schiffner, T. Dragic, J. P. Moore, and J. M. Binley, unpublished results). The ability of the SOS gp140 protein to bind soluble CD4 and undergo relevant conformational changes allows a further way to explore its immunogenicity, i.e., as an sCD4 complex (49). It may also be possible to make full-length, membrane-bound versions of the SOS gp140 protein by restoring the transmembrane domain. Such a protein, expressed in the context of a DNA plasmid or a live recombinant virus vector, might be useful for priming the immune system prior to boosting with a soluble version.
That the A501C/T605C double cysteine substitution works in the context not only of HIV-1 JR-FL but also with several other primary and T-cell-line-adapted subtype B isolates (Gun-1wt, DH123, 89.6, and HxB2) suggests that the method will be generally useful for generating stable trimers. We are presently making similar mutants derived from HIV-1 subtype C isolates. Thus, if the SOS gp140 mutant, or antigenic variants thereof, does induce superior neutralizing-antibody responses in small-animal models, its overall utility as a vaccine antigen could be evaluated in monkey models by using homologous and heterologous SHIVs as challenge viruses (24, 53). It may also be possible to make SOS gp140 proteins derived from SIVmac or other lentiviruses, given the likely similarity of the gp120-gp41 association among retroviruses (99). This could have useful implications for the development of vaccines against retroviruses in general and perhaps other viral families as well.
ACKNOWLEDGMENTS
We thank Gary Thomas for provision of the pGEMfurin plasmid and James Robinson and Herman Katinger for the gifts of several monoclonal antibodies. We are grateful for the sage advice of Dennis Burton and Bob Doms on the biophysical properties of HIV-1 envelope glycoproteins. We appreciate the technical assistance of Daryl Schiller.
This work was supported by RO1 grants AI 39420 and AI 45463.
REFERENCES
- 1.Abacioglu Y H, Fouts T R, Laman J D, Claassen E, Pincus S H, Moore J P, Roby C A, Kamin-Lewis R, Lewis G K. Epitope mapping and topology of baculovirus expressed HIV-1 gp160 determined with a panel of murine monoclonal antibodies. AIDS Res Hum Retroviruses. 1994;10:371–381. doi: 10.1089/aid.1994.10.371. [DOI] [PubMed] [Google Scholar]
- 2.Allaway G P, Davis-Bruno K L, Beaudry G A, Garcia E B, Wong E L, Ryder A M, Hasel K W, Gauduin M-C, Koup R A, McDougal J S, Maddon P J. Expression and characterization of CD4lgG2, a novel heterotetramer which neutralizes primary HIV-1 isolates. AIDS Res Hum Retroviruses. 1995;11:533–540. doi: 10.1089/aid.1995.11.533. [DOI] [PubMed] [Google Scholar]
- 3.Baldinotti F, Matteucci D, Mazzetti P, Giannelli C, Bandecchi P, Tozzini F, Bendinelli M. Serum neutralization of feline immunodeficiency virus is markedly dependent on passage history of the virus and host system. J Virol. 1994;68:4572–4579. doi: 10.1128/jvi.68.7.4572-4579.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnett S W, Rajasekar S, Legg H, Doe B, Fuller D H, Haynes J R, Walker C M, Steimer K S. Vaccination with HIV-1 gp120 DNA induces immune responses that are boosted by a recombinant gp120 protein subunit. Vaccine. 1997;15:869–873. doi: 10.1016/s0264-410x(96)00264-2. [DOI] [PubMed] [Google Scholar]
- 5.Belshe R B, Gorse G J, Mulligan M J, Evans T G, Keefer M C, Excler J-L, Duliege A M, Tartaglia J, Cox W I, McNamara J, Hwang K-L, Bradney A, Montefiori D C, Weinhold K J. Induction of immune responses to HIV-1 by canarypox virus (ALVAC) HIV-1 and gp120 SF-2 recombinant vaccines in uninfected volunteers. AIDS. 1998;12:2407–2415. doi: 10.1097/00002030-199818000-00009. [DOI] [PubMed] [Google Scholar]
- 6.Berman P W, Nunes W M, Haffar O K. Expression of membrane associated and secreted variants of gp160 of human immunodeficiency virus type 1 in vitro and in continuous cell lines. J Virol. 1988;62:3135–3142. doi: 10.1128/jvi.62.9.3135-3142.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Binley J M, Klasse P J, Cao Y, Jones I M, Markowitz M, Ho D D, Moore J P. Differential regulation of the antibody responses to gag and env proteins of human immunodeficiency virus type 1. J Virol. 1997;71:2799–2809. doi: 10.1128/jvi.71.4.2799-2809.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Binley J M, Moore J P. The viral mousetrap. Nature. 1997;387:346–348. doi: 10.1038/387346a0. [DOI] [PubMed] [Google Scholar]
- 9.Binley J M, Wyatt R, Desjardins E, Kwong P D, Hendrickson W, Moore J P, Sodroski J. Analysis of the interaction of antibodies with a conserved, enzymatically deglycosylated core of the HIV type 1 envelope glycoprotein 120. AIDS Res Hum Retroviruses. 1998;14:191–198. doi: 10.1089/aid.1998.14.191. [DOI] [PubMed] [Google Scholar]
- 10.Bou-Habib D C, Roderiquez G, Oravecz T, Berman P W, Lusso P, Norcross M A. Cryptic nature of envelope V3 region epitopes protects primary human immunodeficiency virus type 1 from antibody neutralization. J Virol. 1994;68:6006–6013. doi: 10.1128/jvi.68.9.6006-6013.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buchacher A, Predl R, Strutzenberger K, Steinfellner W, Trkola A, Purtscher M, Gruber G, Tauer C, Steindl F, Jungbauer A, Katinger H. Generation of human monoclonal antibodies against HIV-1 proteins: electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res Hum Retroviruses. 1994;10:359–369. doi: 10.1089/aid.1994.10.359. [DOI] [PubMed] [Google Scholar]
- 12.Burton D R. A vaccine for HIV type 1: the antibody perspective. Proc Natl Acad Sci USA. 1997;94:10018–10023. doi: 10.1073/pnas.94.19.10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burton D R, Montefiori D C. The antibody response in HIV-1 infection. AIDS. 1997;11(Suppl. A):587–598. [PubMed] [Google Scholar]
- 14.Burton D R, Moore J P. Why do we not have an HIV vaccine and how can we make one? Nat Med. 1998;4:495–498. doi: 10.1038/nm0598supp-495. [DOI] [PubMed] [Google Scholar]
- 15.Burton D R, Pyati J, Koduri R, Thornton G B, Sawyer L S W, Hendry R M, Dunlop N, Nara P L, Lamacchia M, Garratty E, Stiehm E R, Bryson Y J, Moore J P, Ho D D, Barbas C F., III Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 16.Caffrey M, Cai M, Kaufman J, Stahl S J, Wingfield P T, Covell D G, Gronenborn A M, Clore G M. Three dimensional solution structure of the 44 kDa ectodomain of SIV gp41. EMBO J. 1998;17:4572–4584. doi: 10.1093/emboj/17.16.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao J, Bergeron L, Helseth E, Thali M, Repke H, Sodroski J. Effects of amino acid changes in the extracellular domain of the HIV-1 gp41 envelope glycoprotein. J Virol. 1993;67:2747–2755. doi: 10.1128/jvi.67.5.2747-2755.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao J, Sullivan N, Desjardin E, Parolin C, Robinson J, Wyatt R, Sodroski J. Replication and neutralization of human immunodeficiency virus type 1 lacking the V1 and V2 variable loops of the gp120 envelope glycoprotein. J Virol. 1997;71:9808–9812. doi: 10.1128/jvi.71.12.9808-9812.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan D C, Fass D, Berger J M, Kim P S. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 20.Chen S L. Functional role of the leucine zipper motif region of HIV-1 transmembrane protein gp41. J Virol. 1994;68:2002–2010. doi: 10.1128/jvi.68.3.2002-2010.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collman R, Balliet J W, Gregory S A, Friedman H, Kolson D L, Nathanson N, Srinivasan A. An infectious molecular clone of an unusual macrophage-tropic and highly cytopathic strain of human immunodeficiency virus type 1. J Virol. 1992;66:7517–7521. doi: 10.1128/jvi.66.12.7517-7521.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Connor R I, Korber B T M, Graham B S, Hahn B H, Ho D D, Walker B D, Neumann A U, Vermund S H, Mestecky J, Jackson S, Fenamore E, Cao Y, Gao F, Kalams S, Kuntsman K, McDonald D, McWilliams N, Trkola A, Moore J P, Wolinsky S M. Immunological and virological analyses of persons infected by human immunodeficiency virus type 1 while participating in trials of recombinant gp120 subunit vaccines. J Virol. 1998;72:1552–1576. doi: 10.1128/jvi.72.2.1552-1576.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cook R F, Berger S L, Rushlow K E, McManus J E, Cook S J, Harrold S, Raabe M L, Montelaro R C, Issel C J. Enhanced sensitivity to neutralizing antibodies in a variant of equine infectious anemia virus is linked to amino acid substitutions in the surface unit envelope glycoprotein. J Virol. 1995;69:1493–1499. doi: 10.1128/jvi.69.3.1493-1499.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Desrosiers R C. Non-human primate models for AIDS vaccines. AIDS. 1995;9(Suppl. A):S137–S141. [PubMed] [Google Scholar]
- 25.Doms R W, Lamb R A, Rose J K, Helenius A. Folding and assembly of viral membrane proteins. Virology. 1993;193:545–562. doi: 10.1006/viro.1993.1164. [DOI] [PubMed] [Google Scholar]
- 26.Dubay J W, Dubay S R, Shin H-J, Hunter E. Analysis of the cleavage site of the human immunodeficiency virus type glycoprotein: requirement of precursor cleavage for glycoprotein incorporation. J Virol. 1995;69:4675–4682. doi: 10.1128/jvi.69.8.4675-4682.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Earl P L, Broder C C, Doms R W, Moss B. Epitope map of human immunodeficiency virus type 1 gp41 derived from 47 monoclonal antibodies produced by immunization with oligomeric envelope protein. J Virol. 1997;71:2674–2684. doi: 10.1128/jvi.71.4.2674-2684.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Earl P L, Broder C C, Long D, Lee S A, Peterson J, Chakrabarti S, Doms R W, Moss B. Native oligomeric human immunodeficiency virus type 1 envelope glycoprotein elicits diverse monoclonal antibody reactivities. J Virol. 1994;68:3015–3026. doi: 10.1128/jvi.68.5.3015-3026.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Earl P L, Doms R W, Moss B. Oligomeric structure of the human immunodeficiency virus type 1 envelope glycoprotein. Proc Natl Acad Sci USA. 1990;87:648–652. doi: 10.1073/pnas.87.2.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Earl P L, Koenig S, Moss B. Biological and immunological properties of human immunodeficiency virus type 1 envelope glycoprotein: analysis of proteins with truncations and deletions expressed by recombinant vaccinia viruses. J Virol. 1991;65:31–41. doi: 10.1128/jvi.65.1.31-41.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edinger A L, Blanpain C, Kuntsman K J, Wolinsky S M, Parmentier M, Doms R W. Functional dissection of CCR5 coreceptor function through the use of CD4-independent simian immunodeficiency virus strains. J Virol. 1999;73:4062–4073. doi: 10.1128/jvi.73.5.4062-4073.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Excler D, Plotkin S. The prime-boost concept applied to HIV preventive vaccines. AIDS. 1997;11(Suppl. A):S127–S137. [PubMed] [Google Scholar]
- 33.Farzan M, Choe H, Desjardins E, Sun Y, Kuhn J, Cao J, Archambault D, Kolchinsky P, Koch M, Wyatt R, Sodroski J. Stabilization of human immunodeficiency virus type 1 envelope glycoprotein trimers by disulfide bonds introduced into the gp41 ectodomain. J Virol. 1998;72:7620–7625. doi: 10.1128/jvi.72.9.7620-7625.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33a.Fass D, Harrison S C, Kim P S. Retrovirus envelope domain at 1.7Å resolution. Nat Struct Biol. 1996;3:465–469. doi: 10.1038/nsb0596-465. [DOI] [PubMed] [Google Scholar]
- 34.Fouts T R, Binley J M, Trkola A, Robinson J E, Moore J P. Neutralization of the HIV-1 primary isolate JR-FL by human monoclonal antibodies correlates with antibody binding to the oligomeric form of the envelope glycoprotein complex. J Virol. 1997;71:2779–2785. doi: 10.1128/jvi.71.4.2779-2785.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fouts T R, Trkola A, Fung M S, Moore J P. Interactions of polyclonal and monoclonal anti-glycoprotein 120 antibodies with oligomeric glycoprotein 120-glycoprotein 41 complexes of a primary HIV type 1 isolate: relationship to neutralization. AIDS Res Hum Retroviruses. 1998;14:591–598. doi: 10.1089/aid.1998.14.591. [DOI] [PubMed] [Google Scholar]
- 36.Gallaher W R. Similar structural models of the transmembrane proteins of Ebola and avian sarcoma viruses. Cell. 1996;85:477–478. doi: 10.1016/s0092-8674(00)81248-9. [DOI] [PubMed] [Google Scholar]
- 37.Gallaher W R, Ball J M, Garry R F, Martin-Amedec A M, Montelaro R C. A general model for the surface glycoproteins of HIV and other retroviruses. AIDS Res Hum Retroviruses. 1995;11:191–202. doi: 10.1089/aid.1995.11.191. [DOI] [PubMed] [Google Scholar]
- 38.Gelderblom H R, Reupke H, Pauli G. Loss envelope antigens of HTLV-III/LAV, a factor in AIDS pathogenesis? Lancet. 1985;ii:1016–1017. doi: 10.1016/s0140-6736(85)90570-7. [DOI] [PubMed] [Google Scholar]
- 39.Godley L, Pfeifer J, Steinhauer D, Ely B, Shaw G, Kaufmann R, Suchanek E, Pabo C, Skehel J J, Wiley D C, Wharton S. Introduction of intersubunit disulfide bonds in the membrane-distal region of the influenza hemagglutinin abolishes membrane fusion activity. Cell. 1992;68:635–645. doi: 10.1016/0092-8674(92)90140-8. [DOI] [PubMed] [Google Scholar]
- 40.Graham B S, McElrath M J, Connor R I, Schwartz D H, Gorse G J, Keefer M C, Mulligan M J, Matthews T J, Wolinsky S M, Montefiori D C, Vermund S H, Lambert J S, Corey L, Belshe R B, Dolin R, Wright P F, Korber B T, Wolff M C, Fast P E. Analysis of intercurrent human immunodeficiency virus type 1 infections in phase I and II trials of current AIDS vaccines. J Infect Dis. 1998;177:310–319. doi: 10.1086/514209. [DOI] [PubMed] [Google Scholar]
- 41.Hallenberger S, Bosch V, Angliker H, Shaw E, Klenk H D, Garten W. Inhibition of furin-mediated cleavage activation of HIV-1 glycoprotein gp160. Nature. 1992;360:358–361. doi: 10.1038/360358a0. [DOI] [PubMed] [Google Scholar]
- 42.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 43.Haynes B F, Pantaleo G, Fauci A S. Towards an understanding of the correlates of protective immunity to HIV infection. Science. 1996;271:324–328. doi: 10.1126/science.271.5247.324. [DOI] [PubMed] [Google Scholar]
- 44.Heilman C A, Baltimore D. HIV vaccines—where are we going? Nat Med. 1998;4:532–534. doi: 10.1038/nm0598supp-532. [DOI] [PubMed] [Google Scholar]
- 45.Helseth E, Olshevsky U, Furman C, Sodroski J. Human immunodeficiency virus type 1 gp120 envelope glycoprotein regions important for association with the gp41 transmembrane glycoprotein. J Virol. 1991;65:2119–2123. doi: 10.1128/jvi.65.4.2119-2123.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hilleman M R. A simplified vaccinologist's vaccinology and the pursuit of a vaccine against AIDS. Vaccine. 1998;16:778–793. doi: 10.1016/s0264-410x(97)00272-7. [DOI] [PubMed] [Google Scholar]
- 47.Hoffman T L, LaBranche C C, Zhang W, Canziani G, Robinson J, Chaiken I, Hoxie J A, Doms R W. Stable exposure of the coreceptor-binding site in a CD4-independent HIV-1 envelope protein. Proc Natl Acad Sci USA. 1999;96:6359–6364. doi: 10.1073/pnas.96.11.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hunter E, Swanstrom R. Retrovirus envelope glycoproteins. Curr Top Microbiol Immunol. 1990;157:187–253. doi: 10.1007/978-3-642-75218-6_7. [DOI] [PubMed] [Google Scholar]
- 49.Kang C-Y, Hariharan K, Nara P L, Sodroski J, Moore J P. Immunization with a soluble CD4-gp120 complex preferentially induces neutralizing anti-human immunodeficiency virus type 1 antibodies directed to conformation-dependent epitopes of gp120. J Virol. 1994;68:5854–5862. doi: 10.1128/jvi.68.9.5854-5862.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koyanagi Y, Miles S, Mitsuyasu R T, Merrill J E, Vinters H V, Chen I S. Dual infection of the central nervous system by AIDS viruses with distinct cellular tropisms. Science. 1987;236:819–822. doi: 10.1126/science.3646751. [DOI] [PubMed] [Google Scholar]
- 51.Kwong P D, Wyatt R, Robinson J, Sweet R W, Sodroski J, Hendrickson W A. Structure of an HIV gp120 envelope glycoprotein complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.LaCasse R A, Follis K E, Trahey M, Scarborough J D, Littman D R, Nunberg J H. Fusion-competent vaccines: broad neutralization of primary isolates of HIV. Science. 1999;283:357–362. doi: 10.1126/science.283.5400.357. [DOI] [PubMed] [Google Scholar]
- 53.Lamb-Wharton R J, Joag S V, Stephens E B, Narayan O. Primate models of AIDS vaccine development. AIDS. 1997;11(Suppl. A):S121–S126. [PubMed] [Google Scholar]
- 54.Leamnson R N, Halpern M S. Subunit structure of the glycoprotein complex of avian tumor virus. J Virol. 1976;18:956–968. doi: 10.1128/jvi.18.3.956-968.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leamnson R N, Shander M H M, Halpern M S. A structural protein complex in Moloney leukemia virus. Virology. 1977;76:437–439. doi: 10.1016/0042-6822(77)90318-x. [DOI] [PubMed] [Google Scholar]
- 56.Leonard C K, Spellman M W, Riddle L, Harris R J, Thomas J N, Gregory T J. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J Biol Chem. 1990;265:10373–10382. [PubMed] [Google Scholar]
- 57.Letvin N L. Progress in the development of an HIV-1 vaccine. Science. 1998;280:1875–1880. doi: 10.1126/science.280.5371.1875. [DOI] [PubMed] [Google Scholar]
- 57a.Malashkevich V N, Schneider B J, McNally M L, Milhollen M A, Pang J X, Kim P S. Core structure of the envelope glycoprotein GP2 from Ebola virus at 1.9-Å resolution. Proc Natl Acad Sci USA. 1999;96:2622–2667. doi: 10.1073/pnas.96.6.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mascola J R, Snyder S W, Weislow O S, Belay S M, Belshe R B, Schwartz D H, Clements M L, Dolin R, Graham B S, Gorse G J, Keefer M C, McElrath M J, Walker M C, Wagner K F, McNeil J G, McCutchan F E, Burke D S the National Institute of Allergy and Infectious Diseases AIDS Vaccine Evaluation Group. Immunization with envelope subunit vaccine products elicits neutralizing antibodies against laboratory adapted but not primary isolates of human immunodeficiency virus type 1. J Infect Dis. 1996;173:340–348. doi: 10.1093/infdis/173.2.340. [DOI] [PubMed] [Google Scholar]
- 59.Matthews T J. Dilemma of neutralization resistance of HIV-1 field isolates and vaccine development. AIDS Res Hum Retroviruses. 1994;10:631–632. doi: 10.1089/aid.1994.10.631. [DOI] [PubMed] [Google Scholar]
- 60.McCune J M, Rabin L B, Feinberg M B, Lieberman M, Kosek J C, Reyes G, Weissman I L. Endoproteolytic cleavage of gp160 is required for the activation of human immunodeficiency virus. Cell. 1988;53:55–67. doi: 10.1016/0092-8674(88)90487-4. [DOI] [PubMed] [Google Scholar]
- 61.McKeating J A, McKnight A, Moore J P. Differential loss of envelope glycoprotein gp120 from virions of human immunodeficiency virus type 1 isolates: effect on infectivity and neutralization. J Virol. 1991;65:852–860. doi: 10.1128/jvi.65.2.852-860.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mertens T E, Burton A. Global estimates and epidemiology of HIV-1 infection and AIDS. AIDS. 1996;10(Suppl. A):S221–S228. doi: 10.1097/00002030-199601001-00031. [DOI] [PubMed] [Google Scholar]
- 63.Molloy S S, Thomas L, VanSlyke J K, Stenberg P E, Thomas G. Intracellular trafficking and activation of the furin proprotein convertase: localization to the TGN and recycling from the cell surface. EMBO J. 1994;13:18–33. doi: 10.1002/j.1460-2075.1994.tb06231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Montefiori D C, Evans T G. Toward an HIV type 1 vaccine that generates potent, broadly cross-reactive neutralizing antibodies. AIDS Res Hum Retroviruses. 1999;15:689–698. doi: 10.1089/088922299310773. [DOI] [PubMed] [Google Scholar]
- 65.Montefiori D C, Reimann K A, Wyand M S, Manson K, Lewis M G, Collman R G, Sodroski J G, Bolognesi D P, Letvin N L. Neutralizing antibodies in sera from macaques infected with chimeric simian-human immunodeficiency virus containing the envelope glycoproteins of either a laboratory-adapted variant or a primary isolate of human immunodeficiency virus type 1. J Virol. 1998;72:3427–3431. doi: 10.1128/jvi.72.4.3427-3431.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moore J P, Binley J M. Envelope's letters boxed into shape. Nature. 1998;393:630–631. doi: 10.1038/31359. [DOI] [PubMed] [Google Scholar]
- 67.Moore J P, Cao Y, Qing L, Sattentau Q J, Pyati J, Koduri R, Robinson J, Barbas III C F, Burton D R, Ho D D. Primary isolates of human immunodeficiency virus type 1 are relatively resistant to neutralization by monoclonal antibodies to gp120 and their neutralization is not predicted by studies with monomeric gp120. J Virol. 1995;69:101–109. doi: 10.1128/jvi.69.1.101-109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moore J P, Ho D D. HIV-1 neutralization: the consequences of viral adaptation to growth on transformed T cells. AIDS. 1995;9(Suppl. A):S117–S136. [PubMed] [Google Scholar]
- 69.Moore J P, McKeating J A, Jones I M, Stephens P E, Clements G, Thomson S, Weiss R A. Characterisation of recombinant gp120 and gp160 from HIV-1: binding to monoclonal antibodies and sCD4. AIDS. 1990;4:307–315. doi: 10.1097/00002030-199004000-00004. [DOI] [PubMed] [Google Scholar]
- 70.Moore J P, McKeating J A, Weiss R A, Sattentau Q J. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science. 1990;250:1139–1142. doi: 10.1126/science.2251501. [DOI] [PubMed] [Google Scholar]
- 71.Moore J P, Sattentau Q J, Wyatt R, Sodroski J. Probing the structure of the human immunodeficiency virus surface glycoprotein gp120 with a panel of monoclonal antibodies. J Virol. 1994;68:469–484. doi: 10.1128/jvi.68.1.469-484.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moore J P, Sodroski J. Antibody cross-competition analysis of the human immunodeficiency virus type 1 exterior envelope glycoprotein. J Virol. 1996;70:1863–1872. doi: 10.1128/jvi.70.3.1863-1872.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moore J P, Thali M, Jameson B A, Vignaux F, Lewis G K, Poon S-W, Fung M S, Sun N C, Charles M, Durda P J, Åkerblom L, Wahren B, Ho D D, Sattentau Q J, Sodroski J. Immunochemical analysis of human immunodeficiency virus type 1: probing the structure of the C4 and V4 domains and the interaction of the C4 domain with the V3 loop. J Virol. 1993;67:4785–4796. doi: 10.1128/jvi.67.8.4785-4796.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moore J P, Trkola A, Korber B, Boots L J, Kessler II J A, McCutchan F E, Mascola J, Ho D D, Robinson J, Conley A J. A human monoclonal antibody to a complex epitope in the V3 region of gp120 of human immunodeficiency virus type 1 has broad reactivity within and outside clade B. J Virol. 1995;69:122–130. doi: 10.1128/jvi.69.1.122-130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Moore J P, Willey R L, Lewis G K, Robinson J, Sodroski J. Immunological evidence for interactions between the first, second and fifth conserved domains of the gp120 surface glycoprotein of human immunodeficiency virus type 1. J Virol. 1994;68:6836–6847. doi: 10.1128/jvi.68.11.6836-6847.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Morikawa Y, Barsov E, Jones I. Legitimate and illegitimate cleavage of human immunodeficiency virus glycoproteins by furin. J Virol. 1993;167:3601–3604. doi: 10.1128/jvi.67.6.3601-3604.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Muster T, Guinea R, Trkola A, Purtscher M, Klima A, Steindl F, Palese P, Katinger H. Cross-neutralizing antibodies against divergent human immunodeficiency virus type 1 isolates induced by the gp41 sequence ELDKWAS. J Virol. 1994;68:4031–4034. doi: 10.1128/jvi.68.6.4031-4034.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Opstelten D E, Wallin M, Garoff H. Moloney murine leukemia virus envelope subunits, gp70 and Pr15E, form a stable disulfide-linked complex. J Virol. 1998;72:6537–6545. doi: 10.1128/jvi.72.8.6537-6545.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Otteken A, Earl P L, Moss B. Folding, assembly, and intracellular trafficking of the HIV-1 envelope glycoprotein analyzed with monoclonal antibodies recognizing maturational intermediates. J Virol. 1996;70:3407–3415. doi: 10.1128/jvi.70.6.3407-3415.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Parren P W H I, Burton D R, Sattentau Q J. HIV-1 antibody—debris or virion? Nat Med. 1997;3:366–367. doi: 10.1038/nm0497-366d. [DOI] [PubMed] [Google Scholar]
- 81.Parren P W H I, Moore J P, Burton D R, Sattentau Q J. The neutralizing antibody response to HIV-1: viral evasion and escape from humoral immunity. AIDS. 1999;13(Suppl. A):S137–S162. [PubMed] [Google Scholar]
- 82.Patarca R, Haseltine W A. Similarities among retrovirus proteins. Nature. 1984;312:496. doi: 10.1038/312496a0. [DOI] [PubMed] [Google Scholar]
- 83.Pinter A, Fleissner E. The presence of disulfide-linked gp70 p15(E) complexes in AKR MuLV. Virology. 1977;83:417–422. doi: 10.1016/0042-6822(77)90187-8. [DOI] [PubMed] [Google Scholar]
- 84.Pinter A, Fleissner E. Characterization of oligomeric complexes of murine and feline leukemia virus envelope and core components formed upon cross-linking. J Virol. 1979;30:157–165. doi: 10.1128/jvi.30.1.157-165.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pinter A, Kopelman R, Li Z, Kayman S C, Sanders D A. Localization of the labile disulfide bond between SU and TM of the murine leukemia virus protein complex to a highly conserved CWLC motif in SU that resembles the active-site sequence of thiol-disulfide exchange enzymes. J Virol. 1997;71:8073–8077. doi: 10.1128/jvi.71.10.8073-8077.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pique C, Pham D, Tursz T, Dokhelar M C. Human T-cell leukemia virus type 1 envelope protein maturation process: requirements for syncytium formation. J Virol. 1992;66:906–913. doi: 10.1128/jvi.66.2.906-913.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Poignard P, Fouts T, Naniche D, Moore J P, Sattentau Q J. Neutralizing antibodies to human immunodeficiency virus type-1 gp120 induce envelope glycoprotein subunit dissociation. J Exp Med. 1996;183:473–484. doi: 10.1084/jem.183.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ratner L, Fisher A, Jagodzinski L L, Mitsuya H, Liou R-S, Gallo R C, Wong-Staal F. Complete nucleotide sequences of functional clones of the AIDS virus. AIDS Res Hum Retroviruses. 1987;3:57–69. doi: 10.1089/aid.1987.3.57. [DOI] [PubMed] [Google Scholar]
- 89.Reiter J N, Means R E, Desrosiers R C. A role for carbohydrates in immune evasion in AIDS. Nat Med. 1998;4:679–684. doi: 10.1038/nm0698-679. [DOI] [PubMed] [Google Scholar]
- 90.Richardson T M, Jr, Stryjewski B L, Broder C C, Hoxie J A, Mascola J R, Earl P L, Doms R W. Humoral response to oligomeric human immunodeficiency virus type 1 envelope protein. J Virol. 1996;70:753–762. doi: 10.1128/jvi.70.2.753-762.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rizzuto C D, Wyatt R, Hernandez-Ramos N, Sun Y, Kwong P D, Hendrickson W A, Sodroski J G. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 92.Roben P, Moore J P, Thali M, Sodroski J, Barbas III C F, Burton D R. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 showing differing ability to neutralize human immunodeficiency virus type 1. J Virol. 1994;68:4821–4828. doi: 10.1128/jvi.68.8.4821-4828.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Robinson H L. DNA vaccines for immunodeficiency viruses. AIDS. 1997;11(Suppl. A):S109–S119. [PubMed] [Google Scholar]
- 94.Rowland-Jones S, Tan R, McMichael A. The role of cellular immunity in protection against HIV infection. Adv Immunol. 1997;65:448–455. [PubMed] [Google Scholar]
- 95.Sattentau Q J, Moore J P. Conformational changes induced in the human immunodeficiency virus envelope glycoproteins by soluble CD4 binding. J Exp Med. 1991;174:407–415. doi: 10.1084/jem.174.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sattentau Q J, Moore J P. Human immunodeficiency virus type 1 neutralization is determined by epitope exposure on the glycoprotein gp120 oligomer. J Exp Med. 1995;182:185–196. doi: 10.1084/jem.182.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sattentau Q J, Moore J P, Vignaux F, Traincard F, Poignard P. Conformational changes induced in the envelope glycoproteins of the human and simian immunodeficiency viruses by soluble receptor binding. J Virol. 1993;67:7383–7393. doi: 10.1128/jvi.67.12.7383-7393.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sattentau Q J, Zolla-Pazner S, Poignard P. Epitope exposure on functional, oligomeric HIV-1 gp41 molecules. Virology. 1995;206:713–717. doi: 10.1016/s0042-6822(95)80094-8. [DOI] [PubMed] [Google Scholar]
- 99.Schultz T F, Jameson B A, Lopalco L, Siccardi A G, Weiss R A, Moore J P. Conserved structural features in the interaction between retroviral surface and transmembrane glycoproteins. AIDS Res Hum Retroviruses. 1992;8:1585–1594. doi: 10.1089/aid.1992.8.1571. [DOI] [PubMed] [Google Scholar]
- 100.Shibata R, Hoggan M D, Broscius C, Englund G, Theodore T S, Buckler White A, Arthur L O, Israel Z, Schultz A, Lane H C, Martin M A. Isolation and characterization of a syncytium inducing macrophage/T-cell line tropic human immunodeficiency virus type 1 isolate that readily infects chimpanzee cells in vitro and in vivo. J Virol. 1995;69:4453–4462. doi: 10.1128/jvi.69.7.4453-4462.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shu W, Ji H, Lu M. Trimerization specificity in HIV-1 gp41: analysis with a GCN4 leucine zipper model. Biochemistry. 1999;38:5378–5385. doi: 10.1021/bi990199w. [DOI] [PubMed] [Google Scholar]
- 102.Sullivan N, Sun Y, Li J, Hofmann W, Sodroski J. Replicative function and neutralization sensitivity of envelope glycoprotein from primary and T-cell-line-passaged human immunodeficiency virus type isolates. J Virol. 1995;69:4413–4422. doi: 10.1128/jvi.69.7.4413-4422.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sullivan N, Sun Y, Sattentau Q, Thali M, Wu D, Denisova G, Gershoni J, Robinson J, Moore J P, Sodroski J. CD4-induced conformational changes in the human immunodeficiency virus type 1 gp120 glycoprotein: consequences for virus entry and neutralization. J Virol. 1998;72:4694–4703. doi: 10.1128/jvi.72.6.4694-4703.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Takeuchi Y, Akutsu M, Murayama K, Shimizu N, Hoshino H. Host range mutant of human immunodeficiency virus type 1: modification of cell tropism by a single point mutation at the neutralization epitope in the env gene. J Virol. 1991;65:1710–1718. doi: 10.1128/jvi.65.4.1710-1718.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Thali M, Moore J P, Furman C, Charles M, Ho D D, Robinson J, Sodroski J. Characterization of human immunodeficiency virus type 1 (HIV-1) gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol. 1993;67:3978–3988. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Trkola A, Dragic T, Arthos J, Binley J M, Olson W C, Allaway G P, Cheng Mayer C, Robinson J, Maddon P J, Moore J P. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 107.Trkola A, Ketas T, KewalRamani V N, Endorf F, Binley J M, Katinger H, Robinson J, Littman D R, Moore J P. Neutralization sensitivity of human immunodeficiency virus type 1 primary isolates to antibodies and CD4-based reagents is independent of coreceptor usage. J Virol. 1998;72:1876–1885. doi: 10.1128/jvi.72.3.1876-1885.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Trkola A, Pomales A B, Yuan H, Korber B, Maddon P J, Allaway G P, Katinger H, Barbas III C F, Burton D R, Ho D D, Moore J P. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4 IgG. J Virol. 1995;69:6609–6617. doi: 10.1128/jvi.69.11.6609-6617.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore J P, Katinger H. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol. 1996;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.VanCott T C, Mascola J R, Kaminski R W, Kalyanraman V, Hallberg P L, Burnett P R, Ulrich J T, Rechtman D J, Birx D L. Antibodies with specificity to native gp120 and neutralization activity against primary human immunodeficiency virus type 1 isolates elicited by immunization with oligomeric gp160. J Virol. 1997;71:4319–4330. doi: 10.1128/jvi.71.6.4319-4330.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.VanCott T C, Mascola J R, Loomis-Price L D, Sinangil F, Zitomersky N, McNeil J, Robb M L, Birx D L, Barnett S. Cross-subtype neutralizing antibodies induced in baboons by a subtype E gp120 immunogen based on an R5 primary human immunodeficiency virus type 1 envelope. J Virol. 1999;73:4640–4650. doi: 10.1128/jvi.73.6.4640-4650.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Van Cott T C, Polonis V R, Loomis L D, Michael N L, Nara P L, Birx D L. Differential role of V3-specific antibodies in neutralization assays involving primary and laboratory adapted isolates of HIV-1. AIDS Res Hum Retroviruses. 1995;11:1379–1392. doi: 10.1089/aid.1995.11.1379. [DOI] [PubMed] [Google Scholar]
- 113.Veronese F D, Rahman R, Pal R, Boyer C, Romano J, Kalyanaraman V S, Nair B C, Gallo R C, Sarngadharan M G. Delineation of immunoreactive, conserved regions in the external envelope glycoprotein of the human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 1992;8:1125–1132. doi: 10.1089/aid.1992.8.1125. [DOI] [PubMed] [Google Scholar]
- 114.Volchkov V E, Feldmann H, Volchkova V A, Klenk H D. Processing of the ebola virus glycoprotein by the proprotein convertase furin. Proc Natl Acad Sci USA. 1998;95:5762–5767. doi: 10.1073/pnas.95.10.5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Watkins B A, Buge S, Aldrich K, Davis A E, Robinson J, Reitz M S, Robert-Guroff M. Resistance of human immunodeficiency virus type 1 to neutralization by natural antisera occurs through single amino acid substitutions that cause changes in antibody binding at multiple sites. J Virol. 1996;70:8431–8437. doi: 10.1128/jvi.70.12.8431-8437.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Weissenhorn W, Dessen A, Harrison S C, Shekel J J, Wiley D C. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 117.Weissenhorn W, Carfi A, Lee K H, Skehel J J, Wiley D C. Crystal structure of Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol Cell. 1998;2:605–616. doi: 10.1016/s1097-2765(00)80159-8. [DOI] [PubMed] [Google Scholar]
- 118.White-Scharf M E, Potts B J, Smith L M, Sokolowski K A, Rusche J R, Silver S. Broadly neutralizing monoclonal antibodies to the V3 region of HIV-1 can be elicited by peptide immunization. Virology. 1993;192:197–206. doi: 10.1006/viro.1993.1022. [DOI] [PubMed] [Google Scholar]
- 119.Willey R L, Bonifacino J S, Potts B J, Martin M A, Klausner R D. Biosynthesis, cleavage, and degradation of the human immunodeficiency virus type 1 envelope protein. Proc Natl Acad Sci USA. 1988;85:9580–9584. doi: 10.1073/pnas.85.24.9580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wrin T, Nunberg J H. HIV-1 MN recombinant gp120 vaccine serum, which fails to neutralize primary isolates of HIV-1, does not antagonize neutralization by antibodies from infected individuals. AIDS. 1994;8:1622–1623. doi: 10.1097/00002030-199411000-00017. [DOI] [PubMed] [Google Scholar]
- 121.Wyatt R, Desjardin E, Olshevsky U, Nixon C, Binley J, Olshevsky V, Sodroski J. Analysis of the interaction of the human immunodeficiency virus type 1 gp120 envelope glycoprotein with the gp41 transmembrane glycoprotein. J Virol. 1997;71:9722–9731. doi: 10.1128/jvi.71.12.9722-9731.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wyatt R, Kwong P D, Desjardin E, Sweet R W, Robinson J, Hendrickson W A, Sodroski J G. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 123.Wyatt R, Moore J P, Accola M, Desjardin E, Robinson J, Sodroski J. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J Virol. 1995;69:5723–5733. doi: 10.1128/jvi.69.9.5723-5733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhou N, Kay C M, Hodges R S. Disulphide bond contribution to protein stability: positional effects of substitution in the hydrophobic core of the two-stranded alpha-helical coiled-coil. Biochemistry. 1993;32:3178–3187. doi: 10.1021/bi00063a033. [DOI] [PubMed] [Google Scholar]