Translational Effects of Mutations and Polymorphisms in a Repressive Upstream Open Reading Frame of the Human Cytomegalovirus UL4 Gene (original) (raw)
Abstract
The human cytomegalovirus (HCMV) gpUL4 mRNA contains a 22-codon upstream open reading frame (uORF2), the peptide product of which represses downstream translation by blocking translation termination at its own stop codon and by causing ribosomes to stall on the mRNA. A distinctive feature of this unusual mechanism is its strict dependence on the uORF2 peptide sequence. To delineate sequence elements that function in the inhibitory mechanism, deletions and missense mutations affecting the previously uncharacterized amino-terminal region of uORF2 were analyzed in transient-transfection and infection assays. These experiments identified multiple codons in this region that are necessary for inhibition of downstream translation by uORF2 and, in conjunction with previous results, demonstrated that amino acids dispersed throughout the uORF2 peptide participate in the repressive mechanism. In contrast to the highly conserved carboxy terminus, the amino-terminal portion of the uORF2 peptide is polymorphic. A survey of uORF2 sequences in HCMV clinical isolates revealed that although most have uORF2 sequences that are predicted to retain the uORF2 inhibitory activity, ∼15% contain polymorphisms at codons that are essential for full inhibition by uORF2. Consistent with predictions based on analyses of engineered mutations, two viral isolates with uORF2 sequences that do not inhibit downstream translation in transfection assays expressed much more gpUL4 protein but similar levels of UL4 mRNA compared to the levels produced by the prototypic laboratory strain HCMV (Towne) and another clinical isolate with an inhibitory variant uORF2. These results demonstrate that uORF2 is polymorphic in sequence and repressive activity and suggest that the uORF2 regulatory mechanism, although prevalent among natural HCMV isolates, is not absolutely essential for viral replication.
Previous investigations into the basis for the scarcity of the human cytomegalovirus (HCMV) gpUL4 protein (gp48) at early times in infection revealed that its expression is controlled by an unusual translational mechanism (2–4, 6). The second of three short upstream open reading frames (uORFs), uORF2, within the 5′ leader of the gpUL4 mRNA encodes a 22-codon peptide that mediates inhibition of downstream translation. Ribosomes that translate uORF2 fail to cleave the peptidyl-tRNA bond linking the nascent uORF2 peptide to tRNAPro, the tRNA responsible for decoding the last uORF2 codon (3). These results suggest a model in which the nascent uORF2 peptide inhibits downstream translation by blocking translation termination at its own stop codon, thereby causing ribosomes to stall on the mRNA and to block access to the gpUL4 initiation codon by other scanning ribosomes.
Although the precise mechanism by which the uORF2 peptide inhibits translational termination is not yet known, the uORF2 peptide sequence is critical for this activity (2, 3, 6). Mutations that alter carboxy-terminal amino acids release the inhibitory effect of uORF2 on downstream translation and also eliminate both the ribosomal stalling and the blockage of peptidyl-tRNA hydrolysis that occur after translation of wild-type uORF2. In contrast, synonymous mutations of these same codons retain the inhibitory properties of wild-type uORF2. Thus, like a few uORFs in other eukaryotic genes (9, 12, 13, 15, 22), uORF2 acts in a peptide sequence-dependent manner. Comparisons among known sequence-dependent uORFs have not yet revealed any consensus sequences or shared motifs that might help to identify molecular interactions between these uORFs and the cellular translational machinery that are responsible for the inhibitory effects.
In order to gain additional insight into the uORF2-mediated inhibitory mechanism, we extended our investigations by determining the role of codons in the more amino-terminal portion of uORF2. Mutational analyses identified several codons within this region that are essential for the uORF2 inhibitory function. A survey of uORF2 sequences among clinical HCMV isolates revealed that some of these essential codons are polymorphic. Consistent with analyses of engineered mutations, certain polymorphisms eliminate the inhibitory effect of uORF2 on downstream translation, demonstrating that uORF2 is structurally and functionally polymorphic.
MATERIALS AND METHODS
Cells, virus, and viral DNA.
HCMV (Towne) was grown on primary human fibroblasts (HF) as described previously (6). Frozen suspensions of cells infected with HCMV strains isolated from bone marrow transplant recipients were obtained from the Clinical Virology Laboratory and from B. Torok-Storb of the Fred Hutchinson Cancer Research Center. Clinical isolates C1, C4, and C6 were plaque purified prior to use in infection experiments. To purify viral DNA for sequencing, a sample of ∼10 to ∼20 μl obtained by scraping a frozen virus-infected cell suspension with a wooden applicator stick was resuspended in either proteinase K buffer (10 mM Tris [pH 7.8], 5 mM EDTA, 0.5% sodium dodecyl sulfate [SDS]) or lysis buffer (50 mM KCl, 10 mM Tris-HCl [pH 8.3], 0.01% gelatin, 0.45% NP-40, 0.45% Tween 20). After proteinase K was added to a final concentration of 100 μg/ml, the reaction mixtures were incubated at 37°C for 1 h and then extracted twice with phenol-chloroform (pH 8.0). Viral DNA was precipitated with ethanol and resuspended in TE buffer (10 mM Tris [pH 8.0], 1 mM EDTA).
Plasmids and sequence analyses.
Plasmids pEQ239, pEQ325, pEQ422, pEQ429, and pEQ430 have been described previously (4, 6, 17). A series of β-galactosidase (β-Gal) expression plasmids (pEQ641 through pEQ648, pEQ700 through pEQ708, and pEQ724 through pEQ729) containing the gpUL4 (gp48) transcript leader with various mutations or deletions in the uORF2 sequence were created by PCR-based mutagenesis with plasmid pEQ422 as a template and with the 5′ primers shown in Table 1 and the 3′ primer 7 (5′CGAGGTGCTGTTTCTGGT). The amplified products were digested with _Hin_dIII and _Afl_II and inserted into pEQ422 that had been digested with _Hin_dIII and _Afl_II, thus replacing the uORF2 sequences as depicted in Table 1 and Fig. 1 through 5. The gpUL4 leader sequences in these plasmids were verified by automated sequence analysis with primer 7 and an Amplitaq FS kit (Perkin-Elmer).
TABLE 1.
uORF2 expression plasmids
| Plasmid | uORF2 peptide sequencea | Reference or 5′ PCR primerb |
|---|---|---|
| pEQ239 | MQPLVLSAKKLSSLLTCKYIPP | 17 |
| pEQ422 | -E-------------------- | 4 |
| pEQ429 | -E-------------------A | 4 |
| pEQ325 | NAc | 17 |
| pEQ643 | -E ------------------- | CTGGTTCTCTCGGCGAA |
| pEQ644 | -E --------------- | CTGGTTCTCTCGGCGAA |
| pEQ645 | -E ---------- | TCTTTGCTGACTTGC |
| pEQ641 | -EAILITGRRI----------- | GCGATTCTGATCACCGGCCGACGAATA |
| pEQ642 | -EYQEQFRVVQ----------- | TATCAGGAACAATTTCGGGTAGTACAG |
| pEQ646 | -EYQE----------------- | TATCAGGAACTCTCGGCGAAAAAACTG |
| pEQ647 | -E---QFR-------------- | CCGCTGGTTCAATTTCGGAAAAAACTG |
| pEQ648 | -E------VVQ----------- | CCGCTGGTTCTCTCGGCGGTAGTACAG |
| pEQ700 | -EY------------------- | TATCTGGTTCTCTCGGCGAAAAAACTG |
| pEQ701 | -E-Q------------------ | CCGCAGGTTCTCTCGGCGAAAAAACTG |
| pEQ702 | -E--E----------------- | CCGCTGGAACTCTCGGCGAAAAAACTG |
| pEQ703 | -E---Q---------------- | CCGCTGGTTCAATCGGCGAAAAAACTG |
| pEQ704 | -E----F--------------- | CCGCTGGTTCTCTTTGCGAAAAAACTG |
| pEQ705 | -E-----R-------------- | CCGCTGGTTCTCTCGCGGAAAAAACTG |
| pEQ706 | -E------V------------- | CCGCTGGTTCTCTCGGCGGTAAAACTG |
| pEQ707 | -E-------V------------ | CCGCTGGTTCTCTCGGCGAAAGTACTG |
| pEQ708 | -E--------Q----------- | CCGCTGGTTCTCTCGGCGAAAAAACAG |
| pEQ727 | -E-------Q------------ | CCGCTGGTTCTCTCGGCGAAACAACTG |
| pEQ726 | -E--------P----------- | CCGCTGGTTCTCTCGGCGAAAAAACTGCCGTCTTTGC |
| pEQ448 | --------EE-L---I------ | —d |
| pEQ449 | -R---F-N-------------- | — |
| pEQ450 | ------L-EE-----I------ | — |
| pEQ725 | -E----L--------------- | CCGCTGGTTCTCTTAGCGAAAAAACTG |
| pEQ729 | -E------E------------- | CCGCTGGTTCTCTCGGCGGAGAAACTG |
| pEQ728 | -E-------E------------ | CCGCTGGTTCTCTCGGCGAAACAACTG |
| pEQ724 | -E-------------I------ | CCGCTGGTTCTCTCGGCGAAAAAACTGTC |
| GTCTTTGCTGATTTGCAAATACATCC |
FIG. 1.
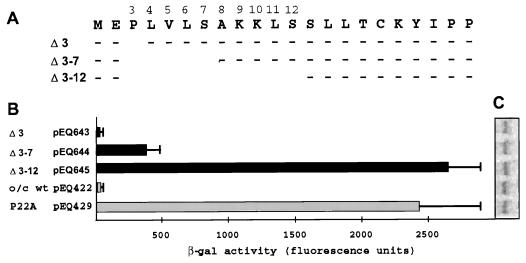
Effects of deletion of codons 3 through 12 on uORF2 inhibitory activity. (A) Plasmids containing deletions of codon 3 (Δ 3, pEQ643), codons 3 through 7 (Δ 3–7, pEQ644), and codons 3 through 12 (Δ 3–12, pEQ645) of uORF2 were transfected into triplicate dishes of HF. Control plasmids were pEQ422, which contains an optimal-context AUG codon (o/c) but otherwise wild-type uORF2 (wt), and pEQ429, which contains an optimal-context AUG codon and a missense mutation of proline to alanine at codon 22 (P22A). Subsequent to HCMV infection, β-Gal activity (B) and lacZ mRNA levels (C) were measured as described in Materials and Methods.
FIG. 5.
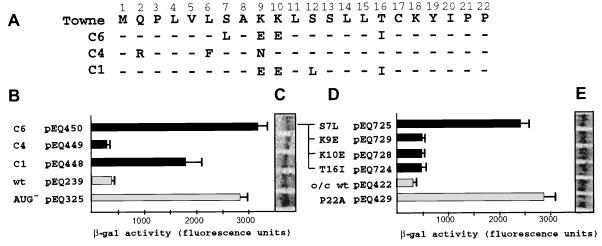
Effects of polymorphisms found in clinical isolates on uORF2 inhibitory activity. (A) Deduced amino acid sequence of clinical isolates C6, C4, and C1. Dashes indicate amino acid identity with HCMV (Towne). Plasmids containing the gpUL4 leader from these clinical isolates (B and C) or containing individual mutations found in isolate C6 (D and E) were transfected into HF. After infection with HCMV, β-Gal activity (B and D) and lacZ mRNA levels (C and E) were measured as described in Materials and Methods. o/c, optimal-context AUG codon; wt, wild-type uORF2.
DNAs from cells infected with clinical HCMV isolates were amplified by PCR with primers 19 (5′GATCAAGCTTTGACTATAAGGATCGCGACCG), 41 (5′CCCCGTAAGATGATCCTCG), or 165 (5′GATCAAGCTTAATCAGATGCCGGCCTTGT) and either 28 (5′GATCGGTACCATCATAACGATACTCTTTCAGCCTTAC) or 55 (5′CCGCCGACGGTCCCTGAG). Control PCR mixtures to which no DNA was added were included after every third experimental sample to ensure that no cross-contamination occurred. PCR products that were flanked by blank control reaction mixtures were gel purified and sequenced with primers 19 (5′GATCAAGCTTTGACTATAAGGATCGCGACCG), 28, 41, 88 (5′AGGCGTGTACGGTGGGAGGTCTAT), and/or 190 (5′CCTCGTATCACATGAGGT). The analysis was repeated, if necessary, to resolve ambiguous nucleotides.
The gpUL4 transcript leaders from clinical isolates C1 and C6 were PCR amplified with primers 18 and 28, and the resulting amplified products were inserted as _Hin_dIII/_Asp_718 fragments into pEQ176 (17). The gpUL4 leader from clinical isolate C4 was PCR amplified with primers 19 and 28, and the resulting amplified product was digested with _Nae_I and _Asp_718 and cloned into pEQ176 that had been digested with _Bgl_II, blunted with DNA polymerase (Klenow fragment), and then digested with _Asp_718. The resulting plasmids, pEQ448, pEQ449, and pEQ450 (corresponding to clinical isolates C1, C4, and C6, respectively), were sequenced with primer 7.
The gpUL4 coding region was amplified from HCMV (Towne) DNA with primers 21 (5′TTAGGACACGGTCAGATTG) and 22 (5′CCGTGGATCCATGATGCTTAGAGCGTGGAG). The product was cloned as a _Bam_HI blunt fragment into the _Bam_HI and _Sma_I sites of pBS+ to generate pEQ371.
Transfection and RNA analyses.
β-Gal expression plasmids were transfected into primary HF with DEAE-dextran as described previously (1). Twenty-four hours after transfection, cells were infected with HCMV at a multiplicity of infection of 3, and at 48 h postinfection, β-Gal expression was measured by adding the fluorogenic substrate 4-methylumbelliferyl-β-d-galactoside to the cell culture medium. The mean (plus standard deviation) fluorescence values from triplicate 60-mm-diameter dishes minus the mean value for cells transfected with control pEQ430 are plotted. Whole-cell RNA was extracted by guanidinium isothiocyanate solubilization of cells, pooling of lysates from triplicate dishes, and pelleting of RNA through CsCl (8). β-Gal mRNA was detected by Northern blot analysis as described previously (7).
Whole-cell RNA from HF infected with HCMV was isolated as described previously (18). gpUL4 mRNA was detected by Northern blot analysis with a probe derived from pEQ371.
Immunoblot assays.
HF in 100-mm-diameter dishes were infected in duplicate with HCMV (Towne) or with clinical isolate C1, C4, or C6 at a multiplicity of infection of 0.1. RNA was isolated from one of each set of duplicate plates as described above at 18 days postinfection, a time when all cells displayed viral cytopathic effects. For protein analysis, cells on the second plate were washed twice with phosphate-buffered saline and lysed in 500 μl of 2% SDS at 60°C and the DNA was sheared by multiple passages through a 27-gauge needle. Approximately 40 μg of protein per sample was separated by SDS-polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes (Micron Separations, Inc.), which were immunoblotted with a Western-Light Plus protein detection kit (Tropix). For detection of gpUL4 protein, rabbit polyclonal antibody (gp48MAPC), raised to the multiple antigenic peptide (19) (TTENSRNYYFRREDAN), was used at a 1:10,000 dilution. For ppUL44, a mouse monoclonal antibody (Virusys Corporation, Berwick, Maine) was used at a 1:20,000 dilution.
RESULTS
Effect of amino-terminal deletions of uORF2 on downstream translation.
To test whether the amino-terminal portion of uORF2 contributes to inhibition of downstream translation, we constructed plasmids that express transcripts containing mutations affecting codons 3 through 12 of uORF2 within the gpUL4 mRNA leader upstream of the β-Gal ORF. The translational impact of these uORF2 mutations was assessed by measuring β-Gal activity and β-Gal RNA levels after transfection of the expression plasmids into HF and subsequent infection with HCMV.
We first analyzed deletions that eliminate codon 3 (pEQ643), codons 3 through 7 (pEQ644), and codons 3 through 12 (pEQ645) of uORF2 (Fig. 1). In transfection analyses, the plasmid with the codon 3 deletion expressed low levels of β-Gal, similar to what occurred with pEQ422, a plasmid with an optimal context AUG codon initiating an otherwise wild-type uORF2. The plasmid with the deletion of codons 3 through 7 expressed an intermediate level of β-Gal. Deletion of codons 3 through 12 completely alleviated uORF2 peptide-mediated translational inhibition, a result similar to that produced by the missense mutation of codon 22 (P22A) present in pEQ429, which was previously shown to eliminate uORF2 inhibitory activity (4).
Northern blot analysis of whole-cell RNAs isolated from the transfected and infected cells revealed similar β-Gal RNA levels among all samples (Fig. 1C), verifying that differences in β-Gal activity were not due to differences in transcript accumulation. Also, levels of transfection efficiency were similar among samples as judged by the approximately equal levels of mRNA expression from a cotransfected plasmid expressing a catalytically inactive lacZ control (pEQ430) (data not shown). These data confirm that the observed variation in β-Gal activity resulted from translational effects of the uORF2 deletions and not from variation in levels of transfection or RNA accumulation.
Effects of amino-terminal amino acid substitutions on uORF2-mediated inhibition.
The deletion mutations described above alter both the length of the uORF2 peptide and its amino acid content. To distinguish which of these changes was responsible for the reduction of uORF2 inhibitory activity, we replaced codons 3 through 11 in toto with either conservative or nonconservative substitutions. If the length but not a particular sequence of the amino-terminal region is required for the inhibitory mechanism, then both the conservative and nonconservative mutants should retain the inhibitory activity. On the other hand, if amino acid coding sequences within the amino terminus of uORF2 are required for translational inhibition, then conservative mutations may have little or no effect while nonconservative substitutions would be more likely to disrupt uORF2 peptide-mediated inhibition.
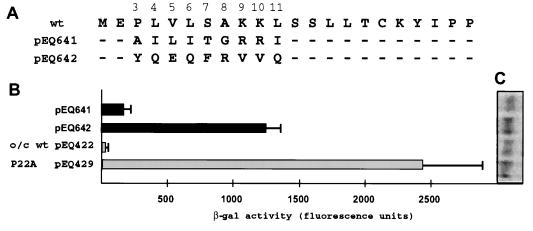
In transfection assays (Fig. 2), the mutant containing nonconservative amino acid substitutions (pEQ642) expressed high levels of β-Gal while the mutant containing conservative amino acid substitutions (pEQ641) expressed much less β-Gal. Levels of RNA expression were similar among all constructs (Fig. 2C). These data suggest that amino acids within the amino-terminal portion of the uORF2 peptide are required for its full inhibitory effect.
FIG. 2.
Missense mutations of codons 3 through 11 reduce uORF2 inhibitory activity. (A) Plasmids having conservative (pEQ641) and nonconservative (pEQ642) substitutions in uORF2 and the same control plasmids as those used for the Fig. 1 experiments were transfected into HF. After HCMV infection, β-Gal activity (B) and lacZ mRNA levels (C) were measured as described in Materials and Methods. o/c, optimal-context AUG codon; wt, wild-type uORF2.
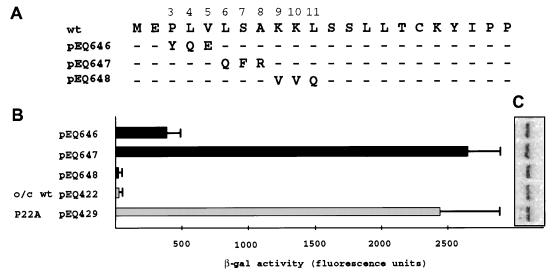
To further delineate which amino acids within the amino-terminal portion of the uORF2 peptide are required for inhibition, we created uORF2 expression constructs having mutations in codons 3 through 5, 6 through 8, or 9 through 11 (pEQ646, pEQ647, or pEQ648, respectively) (Fig. 3A). These mutations incorporated subsets of amino acid changes present in the nonconservative mutant, pEQ642 (Fig. 2A). Analysis of expression from cells transfected with these plasmids demonstrated that pEQ647 and, to a lesser extent, pEQ646 expressed high levels of β-Gal (Fig. 3B). In contrast, pEQ648 was as inhibitory as the wild-type control (pEQ422). These results, in combination with the results of the analysis of RNAs expressed from these plasmids, implicate at least one codon in positions 3 through 5 and at least one additional codon in positions 6 through 8 as being necessary for full inhibition by uORF2.
FIG. 3.
At least two codons, one in positions 3 through 5 and one in positions 6 through 8, are required for full uORF2 inhibitory activity. (A) Plasmids containing triple amino acid uORF2 substitutions derived from subsets of the nonconservative missense mutations in pEQ642 (Fig. 2) were transfected into HF. Following infection with HCMV, β-Gal activity (B) and lacZ mRNA levels (C) were measured as described in Materials and Methods. o/c, optimal-context AUG codon; wt, wild-type uORF2.
Multiple codons in the amino-terminal region of uORF2 are each necessary for inhibition of downstream translation.
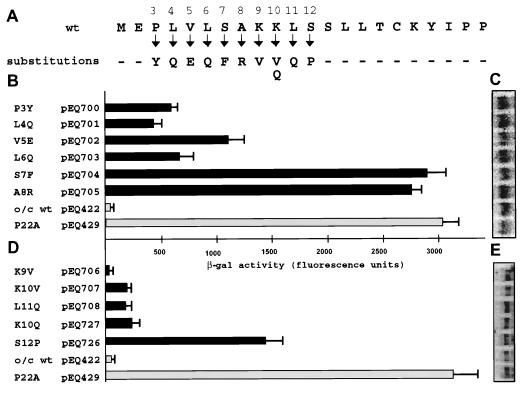
The preceding results suggest that specific amino acids within the amino-terminal portion of the uORF2 peptide are necessary for translational inhibition. To precisely define the critical codons, we analyzed expression downstream from uORF2 mutants with single missense mutations in codons 3 through 12 (Fig. 4A). Complete release of the inhibitory signal occurred when either codon 7 or 8 was mutated (pEQ704 or pEQ705, respectively) (Fig. 4B). Mutation at codon 3, 4, 5, or 6 (pEQ700, pEQ701, pEQ702, or pEQ703, respectively) released the inhibitory signal partially. The increase in β-Gal expression from the codon 3 mutant was somewhat surprising since deletion of codon 3 (pEQ700) maintained the inhibitory signal (Fig. 1). Mutation of codons 9, 10, and 11 (pEQ706, pEQ707, and pEQ708, respectively) individually maintained most of the repressive affect of uORF2 on downstream translation (Fig. 4D), consistent with the preservation of the inhibitory effect by the triple mutant with mutations in codons 9 through 11 (pEQ648) (Fig. 3B).
FIG. 4.
Individual codons within the amino-terminal portion of uORF2 are important for inhibitory activity. Plasmids containing individual missense mutations (A) were transfected into HF in two separate experiments (B and C; D and E). Following infection with HCMV, β-Gal activity (B and D) and lacZ mRNA levels (C and E) were measured as described in Materials and Methods. o/c, optimal-context AUG codon; wt, wild-type uORF2.
In a previous study (6), we identified a double mutation (K10Q and S12P) that completely eliminated uORF2-mediated translational inhibition. Because this dual mutation lies within the region investigated in this report, we examined the individual contribution of each of these mutations. Like the K10V mutation (pEQ707) (Fig. 4) and a previously reported K10E mutation (6), the isolated K10Q mutation (pEQ727) expressed low levels of β-Gal, similar to the level produced by the wild-type sequence (Fig. 4D). In contrast, the missense mutation of codon 12 (S12P; pEQ726) by itself eliminated most of the inhibitory effect of uORF2. Accumulated β-Gal mRNA levels were similar among all samples (Fig. 4C and E). These results indicate that S12P is the key mutation responsible for the loss of uORF2 inhibitory activity in the K10Q-S12P double mutant.
These transfection experiments establish that individual codons in the amino-terminal region of uORF2 are essential for full uORF2-mediated repression of downstream translation. Codons 7, 8, and 12 are most critical, codons 3 through 6 have an intermediate role, and codons 9 through 11 contribute little if any to uORF2 inhibitory activity. Although these conclusions are based on analyses of only one mutation at most positions, they provide a foundation for predicting the effects of naturally occurring polymorphisms present in other HCMV isolates.
Survey of uORF2 in HCMV clinical isolates.
We previously showed that clinical isolates of HCMV contain sequence polymorphisms in uORF2 (6). While key carboxy-terminal codons, such as proline 21 and proline 22, were consistently highly conserved, some polymorphisms located closer to the amino terminus affected some codons that, based on the results of the above-described analyses, seemed to be necessary for uORF2 function.
To examine further the frequency and diversity of uORF2 polymorphisms among clinical isolates, we determined the nucleotide sequences of the gpUL4 transcript leaders and deduced the uORF2 amino acid sequences from 19 independent clinical isolates of HCMV obtained from bone marrow transplant recipients (Table 2). uORF2 was present and was 22 codons long in each isolate. Polymorphisms were present at distinct positions within the uORF2 peptide sequence. Nine of the isolates possessed nucleotide changes that did not alter the amino acid sequence of uORF2 from that of HCMV (Towne). Two additional isolates had uORF2 amino acid sequences identical to that of another prototypic laboratory strain, AD169. Thus, ∼60% of the clinical isolates have uORF2 coding sequences identical to those found in the laboratory strains that have been shown to inhibit downstream translation (17).
TABLE 2.
uORF2 polymorphisms in clinical HCMV isolates
| Isolate | Amino acid at indicated codon that differs from indicated Towne amino acida | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | |
| M | Q | P | L | V | L | S | A | K | K | L | S | S | L | L | T | C | K | Y | I | P | P | |
| Towne-like isolates | ||||||||||||||||||||||
| S1 | — | — | — | — | — | — | — | — | * | — | — | — | — | — | — | — | — | — | — | — | — | — |
| S3 | — | — | — | — | — | — | — | — | * | — | — | — | — | — | — | — | — | — | * | — | — | — |
| S5 | — | — | — | * | — | — | — | — | * | — | — | — | — | — | — | — | — | — | — | — | — | — |
| S8 | — | — | — | * | — | — | — | — | * | — | — | — | — | — | — | — | — | — | — | — | — | — |
| S20 | — | — | — | * | — | — | — | — | * | — | — | — | — | — | — | — | — | — | — | — | — | — |
| S22 | — | — | — | * | — | — | — | — | * | — | — | — | — | — | — | — | — | — | — | — | — | — |
| S31 | — | — | — | — | — | — | — | — | * | — | — | — | — | — | — | — | — | — | — | — | — | — |
| S34 | — | — | — | * | — | — | — | — | * | — | — | — | — | — | — | — | — | — | — | — | * | — |
| S36 | — | — | — | — | — | — | — | — | * | — | — | — | — | — | — | — | — | — | — | — | — | — |
| AD169-like isolates | ||||||||||||||||||||||
| AD169 | — | — | — | — | — | — | — | — | * | — | * | — | — | * | * | A | — | — | — | — | * | — |
| S2 | — | — | — | — | — | — | — | — | * | — | * | — | — | * | * | A | — | — | — | — | * | — |
| S6 | — | — | — | — | — | — | — | — | * | — | * | — | — | * | * | A | — | — | — | — | * | — |
| S24 | — | — | — | — | — | I | — | — | * | — | * | — | — | — | * | A | — | — | — | — | * | — |
| EEIb isolates | ||||||||||||||||||||||
| S7 | — | — | — | * | — | — | * | — | E | E | * | — | — | * | * | I | — | — | — | — | * | — |
| S27 | — | — | — | — | — | — | L | — | E | E | * | — | — | * | — | I | — | — | — | — | * | — |
| S28 | — | — | — | * | — | — | * | — | E | E | * | — | — | * | * | I | — | — | — | — | * | — |
| S29 | — | — | — | * | — | — | * | — | E | E | * | — | — | * | — | I | — | — | — | — | * | — |
| S32 | — | — | — | * | — | — | * | — | E | E | * | — | — | * | * | I | — | — | — | — | * | — |
| Miscellaneous isolates | ||||||||||||||||||||||
| S21 | — | — | S | * | — | * | — | — | * | — | * | — | — | — | — | — | — | — | — | — | * | — |
| S33 | — | — | — | — | — | — | — | — | * | — | — | — | C | — | — | — | — | — | * | — | — | — |
Five of the isolates share a triple codon polymorphism, K9E, K10E, and T16I. Based on results of transfection experiments (reference 6 and Fig. 4), none of these changes is expected to alter uORF2-mediated inhibition. However, isolate S27 has an additional substitution at codon 7, a position that is critical for inhibition. Isolates S21 and S24 have substitutions at codons 3 and 6, respectively, which, based on transfection results (Fig. 4), may partially release uORF2 inhibitory activity. Isolate S33 has a change at codon 13, a position that has not been analyzed. Thus, at least ∼15% of isolates have polymorphisms predicted to eliminate, at least partially, uORF2 inhibitory activity.
Inhibitory activity of naturally occurring uORF2 variants.
To test whether polymorphisms found in clinical isolates affect uORF2 function, we cloned the gpUL4 transcript leaders from three previously sequenced clinical isolates, C1, C4, and C6 (6) into lacZ expression plasmids (pEQ448, pEQ449, and pEQ450, respectively) (Fig. 5A) and analyzed β-Gal expression in transfection and infection assays. The uORF2 from these clinical isolates had various effects on downstream translation, ranging from full inhibition with the C4 sequence to partial or full release of inhibition with the C1 and C6 uORF2 sequences (Fig. 5B). Since these plasmids contain the wild-type, rather than the optimal-context, AUG codon at the start of uORF2, we used pEQ239, which contains the gpUL4 leader from HCMV (Towne) and has the wild-type AUG context, and pEQ325, which contains a mutation of the uORF2 AUG to AAG (AUG−), as controls in this experiment.
The amino acid polymorphisms found in clinical isolate C6 were analyzed for their individual effects on uORF2 function. The four substitutions were each cloned into an expression construct with a uORF2 containing an optimal-context AUG codon. Consistent with the predictions based on previous data (Fig. 4 and reference 6), missense mutations at positions 9, 10, and 16 (pEQ729, pEQ728, and pEQ724, respectively) preserved the inhibitory effect of uORF2. In contrast, the mutation at codon 7 (pEQ725) resulted in high-level β-Gal expression, indicating that S7L was the key alteration of the quartet found in the C6 uORF2 sequence. Again, differences in β-Gal expression could not be explained by variation in RNA accumulation (Fig. 5C and E).
Expression of gpUL4 after infection with clinical isolates.
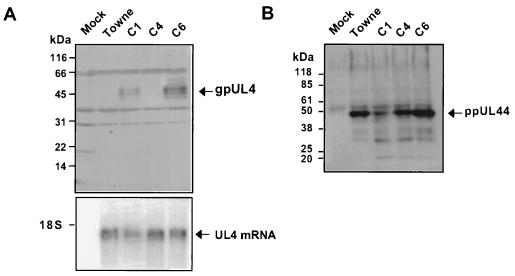
The above-described experiments demonstrated that naturally occurring polymorphisms in some clinical isolates of HCMV eliminate the inhibitory effect of uORF2 in transient-transfection assays. These results predict that gpUL4, the product of the downstream reading frame in the authentic viral mRNA, should be translated more efficiently after infection by these strains. However, since results of transient-transfection assays are not always reliable indicators of regulatory mechanisms, we tested this prediction by analyzing gene expression in cells infected with the C1, C4, C6, and Towne isolates of HCMV (Fig. 6). Substantially more gpUL4 protein was detected by immunoblot assay after C1 and C6 infection than after Towne or C4 infection (Fig. 6A), despite the similar levels of abundance of UL4 mRNA in all samples (Fig. 6A, bottom). To ensure that the differences in gpUL4 protein expression were not due to a generalized lower level of viral protein synthesis in C4- and Towne-infected cells, we also measured the expression of the viral ppUL44 DNA binding protein (Fig. 6B). An immunoblot revealed similar levels of ppUL44 protein in C4-, C6-, and Towne-infected cell extracts and slightly less in C1-infected cell extracts. The greater expression of gpUL4 protein in the C1- and C6-infected cells despite comparable levels of UL4 RNA and ppUL44 protein in all samples supports the conclusion that uORF2 exerts a major impact on gpUL4 expression during viral infection.
FIG. 6.
gpUL4 expression after infection with HCMV (Towne) and clinical isolates. HF extracts obtained 18 days after mock infection or after infection with the HCMV isolate C1, C4, C6, or Towne were separated by SDS-polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes. Immunoblots were probed with a polyclonal antiserum to gpUL4 (A) or an anti-ppUL44 monoclonal antibody (B) as described in Materials and Methods. Molecular size markers are indicated on the left of each blot. Whole-cell mRNA was examined by Northern blot analysis with a UL4 probe (A, bottom). The position of the 18S rRNA migration is indicated.
DISCUSSION
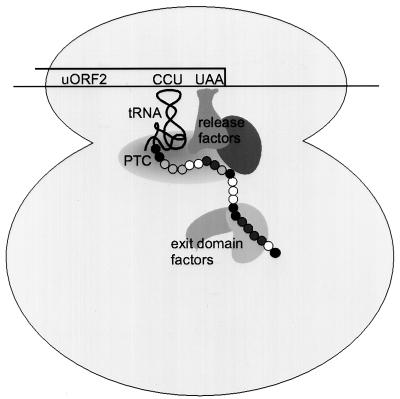
Translational repression by uORF2 in the HCMV gpUL4 mRNA occurs by a highly unusual mechanism. The nascent uORF2 peptide remains covalently attached to tRNAPro, the tRNA that decodes the final uORF2 codon (3). In addition, ribosomes stall at the uORF2 termination codon (2). These observations suggest that the nascent uORF2 peptide interferes with the peptidyl-tRNA hydrolysis reaction that normally occurs when a ribosome encounters a stop codon. Consequently, ribosomes remain at the uORF termination site on the mRNA, creating an obstacle that prevents other ribosomes from scanning to the downstream cistron (Fig. 7).
FIG. 7.
Model of uORF2 peptide interactions with ribosomes or ribosome-associated translation factors. When a ribosome reaches the uORF2 termination codon (UAA), the nascent uORF peptide (chain of circles) remains linked to the tRNA decoding the final uORF2 proline codon, CCU. The ribosome stalls at this site, creating a roadblock that obstructs other ribosomes from scanning to the downstream cistron. Changes in uORF2 amino acids that fully (black circles) or partially (dark-gray circles) alleviate uORF2 inhibitory function, do not affect it (white circles), or for which insufficient data are available (light-gray circles) are shown. The repressive effects of uORF2 may be mediated through its interactions with ribosomal components, such as the peptidyl transferase center (PTC) and the peptide exit domain, or translation factors involved in peptidyl-tRNA hydrolysis, such as eukaryotic release factors.
A distinctive feature of this mechanism is its strict dependence on the uORF2 peptide sequence for inhibition of peptidyl-tRNA hydrolysis, ribosomal stalling, and inhibition of downstream translation (2, 3, 6). In the present study, we discovered that sequences located in the amino-terminal region, like those at the carboxy terminus, are also required for translational inhibition. Mutation of certain amino acid positions (e.g., codons 7, 8, and 12) alleviated much or all of the uORF2 repressive activity. Mutations at other positions had less (e.g., codons 3 through 6) or no (e.g., codons 9 through 11) effect on uORF2 inhibitory activity. Since only one amino acid substitution was analyzed for most codons in uORF2, we cannot exclude the possibility that other amino acid substitutions might have different effects. In fact, nonconservative mutations at codons 7 and 8 (Fig. 4) completely eliminated the inhibitory effect of uORF2 while conservative mutations affecting codons 7 and 8 along with codons 3 through 11 (Fig. 2) only partially reduced the uORF2 inhibitory effect. On the other hand, multiple mutations at codons 9 (K9V, K9Q, and K9E), 10 (K10V and K10E), 11 (L11M and L11Q), 16 (T16A and T16I), 21 (P21A and P21S), and 22 (P22A and P22T) (reference 6 and Fig. 4) have been tested and in all these cases alternative mutations affected uORF2 function in a concordant manner. Thus, these transfection data provide a provisional guide to uORF2 codons that are essential and dispensable for inhibition of downstream translation.
Only a few other eukaryotic genes contain uORFs that have been shown to function in a peptide sequence-dependent manner like uORF2. Inhibition of mammalian _S_-adenosylmethionine decarboxylase translation requires specific amino acids at the three carboxy-terminal codons of the 6-amino-acid uORF (9), while codons 2 and 3 can be altered without affecting the inhibitory effect. Analyses of the uORF sequence requirements in the cases of Saccharomyces cerevisiae CPA1 and its Neurospora crassa homologue arg-2 (12, 22), the mammalian β2-adrenergic receptor (13), and the mammalian retinoic acid receptor β2 (15) are less complete, but at least some codons located near the middle regions of these uORFs are essential for translational inhibition. We now have evidence that codons dispersed throughout uORF2 in the gpUL4 mRNA are required for the inhibitory mechanism. Comparisons of the uORF sequences among these few examples of sequence-dependent uORFs and those shown to act in a sequence-independent manner (5, 10, 14, 21) do not reveal any distinguishing motifs. Thus, sequence data alone are insufficient for accurately predicting which uORFs, among those found in 5 to 10% of cellular genes and in a strikingly high percentage of some subsets such as oncogenes (11), act in a sequence-dependent manner.
Prompted by the finding that some codons that are critical for uORF2 function are ones that are polymorphic among clinical isolates of HCMV, we investigated the structural and functional diversity of uORF2. In the majority of the 19 clinical HCMV isolates studied here, the uORF2 peptide sequence was the same as that found in the laboratory strains Towne and AD169, both of which preserve the inhibitory effect on downstream translation (17). However, in ∼15% of the isolates surveyed, polymorphisms that affect critical codons in uORF2 were present, and thus these strains may express gpUL4 at higher levels. Consistent with these predictions, polymorphisms present in C1 and C6 both allowed high levels of β-Gal expression while those present in C4 did not (Fig. 5B). Further, with C6, of the four codons that differ from those of uORF2 in the prototypic strain Towne, the S7L polymorphism accounted for the loss of repression by the C6 uORF2 (Fig. 5D) as predicted by analyses of engineered mutations (Fig. 4).
The transfection results led us to evaluate whether gpUL4 translation is increased in cells infected with isolates having noninhibitory uORF2 variants. Indeed, gpUL4 protein is more abundant after infection with C1 and C6 than after infection with HCMV (Towne) or C4 despite similar levels of UL4 RNA (Fig. 6). We cannot exclude the possibility that other genetic differences among isolates somehow contributed to the differences in gpUL4 expression. However, these findings strongly support the hypothesis that uORF2 polymorphisms are major determinants of gpUL4 expression. Studies of isogenic viruses with and without uORF2 will be needed to definitively establish the role of uORF2 in controlling gpUL4 expression during viral infection.
The observed high-level expression of gpUL4 after C1 and C6 infection of cells in culture demonstrates that the uORF2 repressive mechanism is not essential for viral growth in cell culture. Since these and several other isolates predicted to contain uORF2 sequences that allow efficient downstream translation are low-number-passage clinical isolates, it seems likely that the uORF2 inhibitory mechanism is also not essential for viral replication in humans. However, it is possible that the uORF2 polymorphisms that are inactive in HF in cell culture remain active in other cell types in infected patients. In addition, although the isolates were passaged only a few times in culture, some of these polymorphisms may have arisen ex vivo. Determining whether polymorphisms in uORF2 sequence and function affect the pathogenesis or replication efficiency of HCMV during natural infections will require additional studies. The polymorphisms identified here may be useful for typing of HCMV isolates in efforts to discover linkages between viral genetic loci and disease manifestations (16, 20).
Although we do not know the precise molecular interactions between the uORF2 peptide and the cellular translational machinery that repress translation termination, our experiments support the model depicted in Fig. 7. The uORF2 peptide prevents the peptidyl-tRNA hydrolysis reaction during translation termination, resulting in ribosomal stalling and inhibition of downstream translation. Thus, we hypothesize that the nascent uORF2 peptide interacts with a ribosomal constituent or a translation factor involved in the translation termination reaction. The identification of amino acids dispersed throughout the 22 codons of uORF2 that are necessary for inhibiting translation should be useful in guiding attempts to identify the factors with which the nascent uORF2 peptide interacts.
ACKNOWLEDGMENTS
We thank Beverley Torok-Storb and the Clinical Virology Laboratory of the Fred Hutchinson Cancer Research Center for providing clinical isolates of HCMV; Mark Stinski for providing gpUL4 antiserum; Stephanie Child for technical assistance; Ed Mocarski, Catherine Degnin, and Mark Schleiss for providing plasmids; and Michael Bates, Wei-Chun Goh, and Michael Katze for helpful comments on the manuscript. We also thank the Biotechnology, Biocomputing, and Image Analysis Resources of the Fred Hutchinson Cancer Research Center for technical assistance.
This work was support by Public Health Service grant AI-26672 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Biegalke B J, Geballe A P. Translational inhibition by cytomegalovirus transcript leaders. Virology. 1990;177:657–667. doi: 10.1016/0042-6822(90)90531-u. [DOI] [PubMed] [Google Scholar]
- 2.Cao J, Geballe A P. Coding sequence-dependent ribosomal arrest at termination of translation. Mol Cell Biol. 1996;16:603–608. doi: 10.1128/mcb.16.2.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cao J, Geballe A P. Inhibition of nascent-peptide release at translation termination. Mol Cell Biol. 1996;16:7109–7114. doi: 10.1128/mcb.16.12.7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao J, Geballe A P. Translational inhibition by a human cytomegalovirus upstream open reading frame despite inefficient utilization of its AUG codon. J Virol. 1995;69:1030–1036. doi: 10.1128/jvi.69.2.1030-1036.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Child, S. J., M. K. Miller, and A. P. Geballe. Translational control by an upstream open reading frame in the HER-2/neu transcript. J. Biol. Chem., in press. [DOI] [PubMed]
- 6.Degnin C R, Schleiss M R, Cao J, Geballe A P. Translational inhibition mediated by a short upstream open reading frame in the human cytomegalovirus gpUL4 (gp48) transcript. J Virol. 1993;67:5514–5521. doi: 10.1128/jvi.67.9.5514-5521.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geballe A P, Leach F S, Mocarski E S. Regulation of cytomegalovirus late gene expression: gamma genes are controlled by posttranscriptional events. J Virol. 1986;57:864–874. doi: 10.1128/jvi.57.3.864-874.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geballe A P, Spaete R R, Mocarski E S. A cis-acting element within the 5′ leader of a cytomegalovirus beta transcript determines kinetic class. Cell. 1986;46:865–872. doi: 10.1016/0092-8674(86)90068-1. [DOI] [PubMed] [Google Scholar]
- 9.Hill J R, Morris D R. Cell-specific translational regulation of S-adenosylmethionine decarboxylase mRNA. Dependence on translation and coding capacity of the cis-acting upstream open reading frame. J Biol Chem. 1993;268:726–731. [PubMed] [Google Scholar]
- 10.Hinnebusch A G. Translational control of GCN4: gene specific regulation by phosphorylation of eIF2. In: Hershey J W B, Matthews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 199–244. [Google Scholar]
- 11.Kozak M. An analysis of vertebrate mRNA sequences: intimations of translational control. J Cell Biol. 1991;115:887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo Z, Sachs M S. Role of an upstream open reading frame in mediating arginine-specific translational control in Neurospora crassa. J Bacteriol. 1996;178:2172–2177. doi: 10.1128/jb.178.8.2172-2177.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parola A L, Kobilka B K. The peptide product of a 5′ leader cistron in the beta 2 adrenergic receptor mRNA inhibits receptor synthesis. J Biol Chem. 1994;269:4497–4505. [PubMed] [Google Scholar]
- 14.Polymenis M, Schmidt E V. Coupling of cell division to cell growth by translational control of the G1 cyclin CLN3 in yeast. Genes Dev. 1997;11:2522–2531. doi: 10.1101/gad.11.19.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reynolds K, Zimmer A M, Zimmer A. Regulation of RAR beta 2 mRNA expression: evidence for an inhibitory peptide encoded in the 5′-untranslated region. J Cell Biol. 1996;134:827–835. doi: 10.1083/jcb.134.4.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosen H R, Corless C L, Rabkin J, Chou S. Association of cytomegalovirus genotype with graft rejection after liver transplantation. Transplantation. 1998;66:1627–1631. doi: 10.1097/00007890-199812270-00010. [DOI] [PubMed] [Google Scholar]
- 17.Schleiss M R, Degnin C R, Geballe A P. Translational control of human cytomegalovirus gp48 expression. J Virol. 1991;65:6782–6789. doi: 10.1128/jvi.65.12.6782-6789.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siebert P D, Chenchik A. Modified acid guanidinium thiocyanate-phenol-chloroform RNA extraction method which greatly reduces DNA contamination. Nucleic Acids Res. 1993;21:2019–2020. doi: 10.1093/nar/21.8.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tam J P. Synthetic peptide vaccine design: synthesis and properties of a high-density multiple antigenic peptide system. Proc Natl Acad Sci USA. 1988;85:5409–5413. doi: 10.1073/pnas.85.15.5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Torok-Storb B, Boeckh M, Hoy C, Leisenring W, Myerson D, Gooley T. Association of specific cytomegalovirus genotypes with death from myelosuppression after marrow transplantation. Blood. 1997;90:2097–2102. [PubMed] [Google Scholar]
- 21.Wang L, Wessler S R. Inefficient reinitiation is responsible for upstream open reading frame-mediated translational repression of the maize R gene. Plant Cell. 1998;10:1733–1746. doi: 10.1105/tpc.10.10.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Werner M, Feller A, Messenguy F, Pierard A. The leader peptide of yeast gene CPA1 is essential for the translational repression of its expression. Cell. 1987;49:805–813. doi: 10.1016/0092-8674(87)90618-0. [DOI] [PubMed] [Google Scholar]