Quasispecies of TT Virus (TTV) with Sequence Divergence in Hypervariable Regions of the Capsid Protein in Chronic TTV Infection (original) (raw)
Abstract
Three hypervariable regions were identified in a central portion of open reading frame 1 of TT virus DNA, which codes for a putative capsid protein of 770 amino acids. TT virus circulates as quasispecies, with many amino acid substitutions in hypervariable regions, to evade immune surveillance of the hosts and to establish a persistent infection.
Late in 1997, a nonenveloped, single-stranded DNA virus was recovered from a patient, who developed posttransfusion hepatitis not related to any of the known hepatitis viruses (non-A through non-G), and named TT virus (TTV) (14, 17). Because TTV has a circular genomic structure (9, 10, 20), it is classified tentatively in the Circoviridae family (6). Despite being a DNA virus, TTV has an extremely wide sequence divergence which causes it to fall into at least 16 genotypes separated by an evolutionary distance >0.30 (4, 10, 11, 13, 17–20, 22, 25–27).
TTV may have a hypervariable region (HVR), because amino acid substitutions among distinct TTV strains of the same genotype are found more frequently in a central portion of open reading frame 1 (ORF1) than in the other genomic regions (10, 24). For further defining the HVR of TTV, the amino acid sequences of the two ORFs (ORF1 and ORF2) of nine TTV isolates of genotype 1a, including three newly sequenced and six previously reported (10, 17, 24), were compared. Furthermore, HVR sequences were compared among the many TTV clones recovered from the sera of five individuals with chronic or acute TTV infection. The quasispecies nature of circulating TTV may help it evade immune surveillance and establish persistent infection, as is seen for hepatitis C virus (HCV) and human immunodeficiency virus type 1 (2, 7, 8, 16, 29).
HVRs in ORF1 of TTV.
Two patients (TRM1, with HCV-associated hepatocellular carcinoma, and TK16, with non-A through non-G acute hepatitis) and one blood donor (TP1-3) were infected with TTV of genotype 1a. TTV DNAs in their plasma were sequenced (3,308 nucleotides [nt] corresponding to >85% of the genomic sequence) by a long-distance PCR method for amplifying 3.4 kb (19). The sequences were compared with the sequences of six reported isolates of the same genotype (10, 17, 24) within various genomic regions.
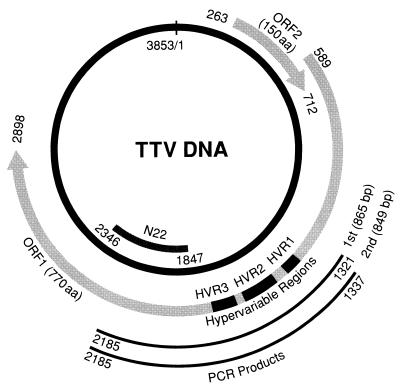
There were three regions with marked sequence divergence located in the same positions on the genome of nine isolates, including the three obtained in the present study. They were in a central portion of ORF1 and spanned amino acids (aa) 275 to 296, 314 to 360, and 372 to 402 (Fig. 1) and were designated HVR1, HVR2, and HVR3, respectively. The TTV HVRs were located outside the N22 region (14), the sequence of which is used for detecting TTV DNA and classifying various genotypes (4, 17–20, 22, 25–27) (Fig. 1).
FIG. 1.
Genomic organization of TTV and positions of the three HVRs in ORF1. The genomic organization of the prototype TTV isolate of genotype 1a (TA278), consisting of 3,853 nt (17, 20), is shown, with the positions of the two ORFs indicated by arrows. The positions of HVR1 (nt 1411 to 1476 [aa 275 to 296]), HVR2 (nt 1528 to 1668 [aa 314 to 360]), and HVR3 (nt 1702 to 1794 [aa 372 to 402]) are indicated by black bars in ORF1. A nucleotide sequence including the HVRs was amplified by PCR with heminested primers with products indicated at the bottom. The position of the N22 clone (14), from which primers for detection and genotyping of TTV DNA are deduced (18, 19), is indicated.
Table 1 compares the nucleotide and amino acid sequences of ORF1 among the nine isolates. TRM1 was used as the standard, because comparison with the other eight isolates gave the lowest amount of sequence divergence. The eight isolates were different from TRM1 at 1.7 to 7.2% (mean, 2.9%) of the 2,310 nt in ORF1. Divergent nucleotides clustered in HVRs at a rate of 31.9 to 68.1%. Within the three HVRs, totalling 300 nt, 15 to 53 nt (mean, 31.6 nt) were divergent, corresponding to 5 to 18% (mean, 10.5%) of the 300 nt. Sequences outside the three HVRs in ORF1 (total, 2,010 nt) were much conserved, by contrast, showing a divergence of only 0.7 to 5.6% (mean, 1.7%). Nucleotide sequence divergence occurred most frequently in the third letter of a codon and accounted for a mean of 68.7%, which was much more frequent than the 39.2% occurrence in HVRs. Of the 770 aa coded for by ORF1, 20 to 32 aa (mean, 28.2 aa) were divergent, corresponding to 2.6 to 4.2% (mean, 3.7%) of the 770 aa. Of the divergent amino acids, 46 to 86% occurred in HVRs. Of the 100 aa representing the three HVRs, 12 to 25 aa (mean, 18.9 aa) were divergent, accounting for 12 to 25% (mean, 18.9%) of the 100 aa.
TABLE 1.
Comparison of nucleotide and amino acid sequences within the HVRs and the remaining regions in ORF1 between the TRM1 isolate and the other eight isolates of the same genotype (1a)
| Isolate | Accession no. | ORF1 (2,310 nt) | No. of nucleotide substitutions (%) in: | No. of amino acid substitutions (%) in: | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HVR (300 nt)a | Non-HVR (2,010 nt)b | ORF1 (770 aa) | HVRa (100 aa) | Non-HVRb (670 aa) | |||||||||
| Subtotal | Codon position | Subtotal | Codon position | ||||||||||
| 1st | 2nd | 3rd | 1st | 2nd | 3rd | ||||||||
| TA278 | AB008394 | 47 (2.0) | 32 (11) | 11 | 12 | 9 | 15 (0.7) | 3 | 3 | 9 | 29 (3.8) | 25 (25) | 4 (0.6) |
| TK16 | AB026346 | 54 (2.3) | 32 (11) | 11 | 10 | 11 | 22 (1.1) | 4 | 4 | 14 | 32 (4.2) | 22 (22) | 10 (1.5) |
| TP1-3 | AB026347 | 62 (2.7) | 37 (12) | 14 | 10 | 13 | 25 (1.2) | 4 | 6 | 15 | 32 (4.2) | 23 (23) | 9 (1.3) |
| G97801 | AB011486 | 52 (2.3) | 28 (9) | 11 | 8 | 9 | 24 (1.2) | 5 | 4 | 15 | 27 (3.5) | 19 (19) | 8 (1.2) |
| G103301 | AB011487 | 39 (1.7) | 15 (5) | 6 | 5 | 4 | 24 (1.2) | 6 | 2 | 16 | 20 (2.6) | 12 (12) | 8 (1.2) |
| G104901 | AB011489 | 51 (2.2) | 32 (11) | 11 | 14 | 7 | 19 (0.9) | 5 | 2 | 12 | 28 (3.6) | 15 (15) | 13 (1.9) |
| TTVCHN1 | AF079173 | 56 (2.4) | 24 (8) | 5 | 6 | 13 | 32 (1.6) | 8 | 7 | 17 | 28 (3.6) | 13 (13) | 15 (2.2) |
| GH1 | AF122913 | 166 (7.2) | 53 (18) | 20 | 6 | 37 | 113 (5.6) | 18 | 5 | 90 | 29 (3.8) | 22 (22) | 7 (1.0) |
| Mean no. | 65.9 | 31.6 | 11.1 | 8.9 | 12.9 | 34.3 | 6.6 | 4.1 | 23.5 | 28.2 | 18.9 | 9.3 | |
| Mean % | 2.9 | 10.5 | 39.2 | 1.7 | 68.7 | 3.7 | 18.9 | 1.4 |
Sequence divergence in HVRs of TTV clones from patients with chronic TTV infection (C1 and C2) and its evolution with time.
Sequential sera were obtained from two patients with non-A through non-G chronic hepatitis complicated by cirrhosis (C1 and C2), both of whom were persistently infected with TTV. C1 had received transfusions during a hysterectomy at the age of 44, and was diagnosed with cirrhosis at 52 years of age. TTV DNA was recovered from serum samples taken at 60, 64, and 68 years of age. C2 was diagnosed with cirrhosis at the age of 74. TTV DNA was recovered from serum samples taken at 79 and 81 years of age.
An 849-bp sequence in ORF1 bearing HVRs (Fig. 1) was amplified by PCR in the presence of TaKaRa Ex Taq (TaKaRa Shuzo Co., Shiga, Japan) with heminested primers. The first-round PCR was performed with NG161 (sense, 5′-GCA ACC GCA GCG GAT ATG CAA TAT CCG TTC-3′ [nt 1321 to 1350]) and NG063 (antisense, 5′-CTG GCA TTT TAC CAT TTC CAA AGT T-3′ [nt 2161 to 2185]) for 35 cycles, and the second-round PCR was done for 25 cycles with NG152 (sense, 5′-TGC AAT ATC CGT TCG GCT CAC CAC-3′ [nt 1337 to 1360]) and NG063. The conditions for both PCR methods were: 94°C, 30 s; 60°C, 30 s; 72°C, 60 s (with an additional 7 min for the last cycle). The amplification products by the first-round PCR measured 865 bp, and those by the second-round PCR measured 849 bp.
Products of the second-round PCR were inserted into pT7BlueT-Vector, and the independent TTV clones thus obtained were confirmed to belong to genotype 1a by PCR with primers specific for this genotype, i.e., NG164 (sense, 5′-GGA TAT GTA GAA TTT TGT GCA-3′ [nt 2020 to 2040]) and NG165 (antisense, 5′-AAA GCC TTT TGT GGG GTC TG-3′ [nt 2126 to 2145]). The obtained clones were sequenced with the BigDye Terminator Cycle Sequence Ready Reaction Kit (PE Applied Biosystems, Foster City, Calif.). Sequence analysis of TTV clones was performed with Genetyx-Mac version 10.1 (Software Development Co., Tokyo, Japan) and ODEN program version 1.1.1 (5) of DDBJ (DNA Data Bank of Japan, National Institute of Genetics, Mishima, Japan).
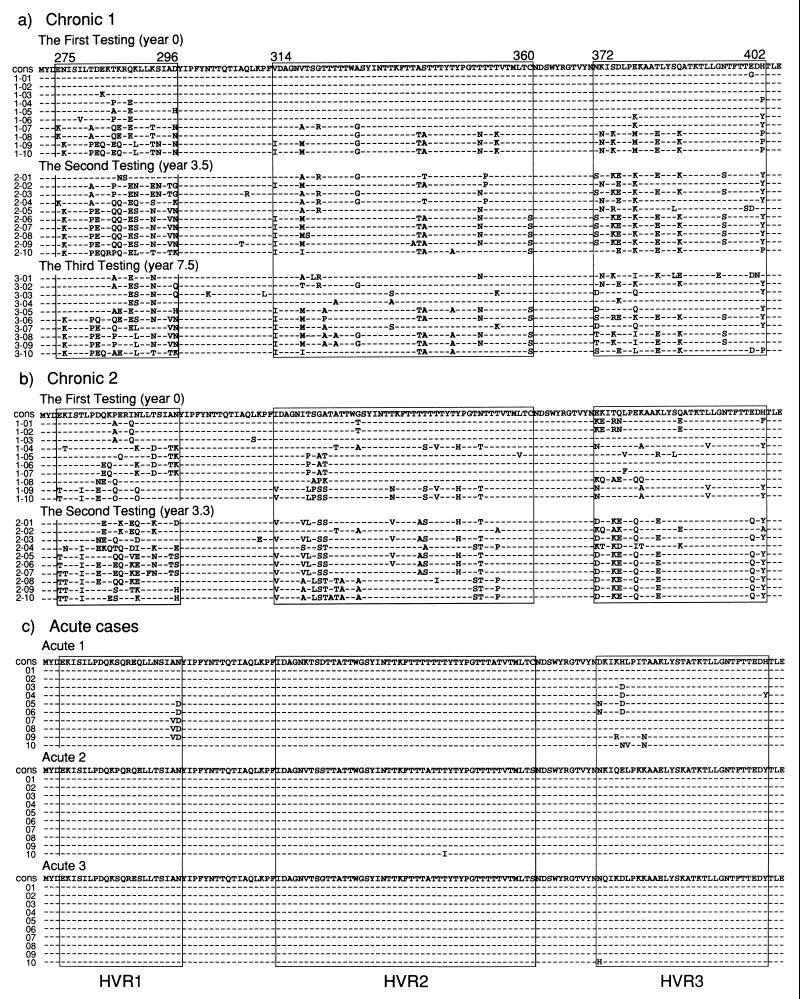
Ten TTV clones from C1 at year 0 revealed quasispecies with marked sequence divergence in the HVRs, similar to those at years 3.5 and 7.5 (Fig. 2a). The sequences of TTV clones at year 0 was somewhat similar to those at year 3.5; such a similarity was observed also between TTV clones at years 3.5 and 7.5. Ten TTV clones from C2 also showed remarkable heterogeneity in HVR sequences from both testings (Fig. 2b). There was a close relationship between clones obtained at years 0 and 3.3 from C2, which would reflect continuous evolution in the HVR sequence.
FIG. 2.
Amino acid sequences of the three HVRs of TTV clones in five patients with chronic (C1 and C2) or acute (A1, A2, and A3) infection. (a and b) Sequences of HVR1, HVR2, and HVR3 are shown for 10 clones, each obtained from sequential sera of two patients with chronic infection. The consensus sequence (cons) in 10 clones at year 0 is indicated at the top. (c) Sequences of 10 clones each from three patients with acute infection. Dashes indicate the same amino acids as those in the consensus sequence (cons).
Sequence of HVRs in TTV clones from three patients with acute TTV infection (A1, A2, and A3).
Sera were obtained from two patients with posttransfusion non-A through non-G hepatitis associated with acute TTV infection (14), 10 and 8 weeks after they received transfusions, respectively. An additional serum sample was obtained from one patient with HCV-associated hepatocellular carcinoma 6 weeks after he contracted sporadic TTV infection. His transaminase levels did not change during the 8 weeks that TTV DNA was detected in his serum. All three patients with acute infection lost TTV DNA from their serum by 2 to 7 weeks after the time of testing.
The sequences of the HVRs of 10 TTV clones obtained from each of the patients’ sera are shown in Fig. 2c; they were recovered from unfractionated sera (Table 2). In sharp contrast to the marked sequence divergence in TTV clones from the patients with chronic infection, the 10 clones from the three patients with acute infection were strikingly homogeneous in the HVR amino acid sequences.
TABLE 2.
Profiles of individuals with chronic or acute TTV infection
| Individual | Age (yr) and sexa | Time of sampling | Clinical diagnosis (etiology) | Amt of TTV DNAb (copies/ml) | Immunoprecipitationc |
|---|---|---|---|---|---|
| Sup Ppt | |||||
| C1 | 60F | Yr 0 | Liver cirrhosis (non-A to G) | 104 | 0 < 10.0 |
| 64 | Yr 3.5 | 103 | 0 < 5.4 | ||
| 68 | Yr 7.5 | 104 | 0 < 11.6 | ||
| C2 | 79F | Yr 0 | Liver cirrhosis (non-A to G) | 104 | 0 < 13.6 |
| 81 | Yr 3.3 | 104 | 0 < 13.2 | ||
| A1 | 58M | Wk 10d | Posttransfusion hepatitis (non-A to G) | 103 | 0.9 < 2.8 |
| A2 | 70F | Wk 8d | Posttransfusion hepatitis (non-A to G) | 102 | 1.5 > 0.1 |
| A3 | 70M | Wk 6e | Sporadic TTV infection | 104 | 1.7 < 3.0 |
TTV complexed with IgG in sera from patients with chronic infection.
TTV particles in sera were immunoprecipitated with ICN/CAPPEL goat antiserum to human immunoglobulin G (IgG) (whole molecule) (ICN Biomedicals, Aurora, Ohio), and supernatant was separated from precipitate (28). They were tested for TTV DNA by PCR (Table 2). Samples were judged to contain TTV complexed with IgG when precipitate generated a higher density of amplification products than supernatant after treatment with goat antiserum to human IgG and when no significant amplification signals were observed in the precipitate fraction after treatment with normal goat serum (ICN Biochemicals).
TTV DNA was detected only in the precipitate fraction of sera from two patients with chronic infection taken at different time points (Table 2). In view of the sensitivity of this experiment, TTV particles in the supernatant were deduced to be less than 1/10 to 1/100 of those in the precipitate for these two patients, with serum titers at 103 to 104 copies/ml.
By contrast, TTV particles were detected in supernatant fractions from all three patients with acute infection. Two patients (A1 and A3) were tested when they had peak TTV DNA titers, and their sera had higher TTV DNA titers in precipitate than supernatant. Serum of the remaining patient (A2) taken 3 weeks earlier than the peak TTV DNA level, however, possessed a much higher TTV DNA titer in supernatant than precipitate.
Discussion.
The Circoviridae family consists of nonenveloped, single-stranded circular DNA viruses (6); there have been only three animal circoviruses known, i.e., chicken anemia virus, beak and feather disease virus of parrots, and porcine circovirus (1, 6, 12, 15). Due to its circular genomic structure (9, 10, 20), TTV may qualify as a fourth animal circovirus, although its genomic size (3,818 to 3,853 nt, depending on genotype) is much larger than those of the other three (1,758 to 2,319 nt) (1, 12, 15, 20).
In spite of being a DNA virus, TTV has an extremely high level of sequence divergence, ranging to 60.5% for the entire genome (20); at least 16 genotypes that differ by >30% from one another have been distinguished (19). The reason for the outstanding genetic heterogeneity of TTV is not clear. The replication of TTV might involve reverse transcription, making for an accelerated mutation rate as is the case for hepatitis B virus (23), a double-stranded DNA virus encoding a reverse transcriptase; a sequence motif for reverse transcriptase has not been identified in the TTV genome, however.
Nucleotide sequences vary considerably even among TTV isolates of the same genotype. Sequence divergence resulting in amino acid conversions is the highest in a central portion of ORF1 (10, 24), the translation product of which has an arginine-rich sequence in the N terminus. By analogy with VP1 of chicken anemia virus, which also has this sequence (1, 15), the ORF1 in TTV may encode a capsid protein. Comparison of nine TTV isolates of genotype 1a highlighted three regions in ORF1 that had a markedly divergent amino acid sequence (Fig. 1). They were designated HVR1, HVR2, and HVR3 and coded for 22, 47, and 31 aa, respectively.
The nine TTV isolates of genotypes 1a were 92 to 96% similar in the amino acid sequence of the ORF1 product. Most amino acid substitutions in ORF1 (47 to 86%) clustered in HVRs that spanned 100 aa altogether; regions outside HVRs in ORF1 were well conserved, with a similarity of ≥98%. HVRs in TTV are comparable to the HVR in chicken anemia virus, which stretches for 13 aa (position, 139 to 151) in the 450-aa VP1 (21).
In individuals with chronic infection, TTV circulates as quasispecies, with sequence divergence in the HVR. TTV evolves in hosts by changing its HVR sequences to escape immune surveillance. The adaptability of the HVR for viral persistence has been well established for HCV and human immunodeficiency virus type 1 (2, 7, 8, 16, 29). Quasispecies of TTV due to variation in the HVR were found restricted to hosts with chronic infection. Circulating TTV in hosts with acute resolving infection, rarely, if ever, showed sequence divergence in the HVR (Fig. 2).
Should variation in HVR reflect a strategy of TTV to evade immune surveillance of hosts, humoral antibodies to HVR sequences may well be present in the circulation. Antibodies to HVR sequences would bind with circulating TTV to form immune complexes, as is seen in chronic HCV infection (3). This was actually the case for TTV (Table 2). In individuals with chronic TTV infection, by far the most TTV particles in serum were precipitated with goat anti-human IgG, thereby indicating that they would form immune complexes in the circulation. In individuals with acute resolving infection, by contrast, free TTV particles uncomplexed with IgG always occurred, in amounts exceeding those of complexed TTV in some instances.
Nucleotide sequence accession numbers.
The nucleotide sequence data in this report have been deposited in the DDBJ/EMBL/GenBank nucleotide sequence databases under accession no. AB026345 to AB026347 for 3,308 nt of TRM1, TK16, and TP1-3 and AB026348 to AB026427 for 80 TTV isolates consisting of 800 nt covering the HVRs.
REFERENCES
- 1.Claessens J A, Schrier C C, Mockett A P, Jagt E H, Sondermeijer P J. Molecular cloning and sequence analysis of the genome of chicken anaemia agent. J Gen Virol. 1991;72:2003–2006. doi: 10.1099/0022-1317-72-8-2003. [DOI] [PubMed] [Google Scholar]
- 2.Hijikata M, Kato N, Ootsuyama Y, Nakagawa M, Ohkoshi S, Shimotohno K. Hypervariable regions in the putative glycoprotein of hepatitis C virus. Biochem Biophys Res Commun. 1991;175:220–228. doi: 10.1016/s0006-291x(05)81223-9. [DOI] [PubMed] [Google Scholar]
- 3.Hijikata M, Shimizu Y K, Kato H, Iwamoto A, Shih J W, Alter H J, Purcell R H, Yoshikura H. Equilibrium centrifugation studies of hepatitis C virus: evidence for circulating immune complexes. J Virol. 1993;67:1953–1958. doi: 10.1128/jvi.67.4.1953-1958.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hohne M, Berg T, Muller A R, Schreier E. Detection of sequences of TT virus, a novel DNA virus, in German patients. J Gen Virol. 1998;79:2761–2764. doi: 10.1099/0022-1317-79-11-2761. [DOI] [PubMed] [Google Scholar]
- 5.Ina Y. ODEN: a program package for molecular evolutionary analysis and database search of DNA and amino acid sequences. Comput Appl Biosci. 1994;10:11–12. doi: 10.1093/bioinformatics/10.1.11. [DOI] [PubMed] [Google Scholar]
- 6.Lukert P D, de Boer G F, Dale J L, Keese P, McNulty M S, Randers J W, Tisher I. Family Circoviridae. In: Murphy F A, Fauquet C M, Bishop D H L, Ghabrial S A, Jarvis A W, Martelli G P, Mayo M A, Summers M D, editors. Virus taxonomy. Classification and nomenclature of viruses, sixth report of the International Committee on Taxonomy of Viruses. New York, N.Y: Springer; 1995. pp. 166–168. [Google Scholar]
- 7.Martell M, Esteban J I, Quer J, Genesca J, Weiner A, Esteban R, Guardia J, Gomez J. Hepatitis C virus (HCV) circulates as a population of different but closely related genomes: quasispecies nature of HCV genome distribution. J Virol. 1992;66:3225–3229. doi: 10.1128/jvi.66.5.3225-3229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meyerhans A, Cheynier R, Albert J, Seth M, Kwok S, Sninsky J, Morfeldt-Manson L, Asjo B, Wain-Hobson S. Temporal fluctuations in HIV quasispecies in vivo are not reflected by sequential HIV isolations. Cell. 1989;58:901–910. doi: 10.1016/0092-8674(89)90942-2. [DOI] [PubMed] [Google Scholar]
- 9.Miyata H, Tsunoda H, Kazi A, Yamada A, Khan M A, Murakami J, Kamahora T, Shiraki K, Hino S. Identification of a novel GC-rich 113-nucleotide region to complete the circular, single-stranded DNA genome of TT virus, the first human circovirus. J Virol. 1999;73:3582–3586. doi: 10.1128/jvi.73.5.3582-3586.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mushahwar I K, Erker J C, Muerhoff A S, Leary T P, Simons J N, Birkenmeyer L G, Chalmers M L, Pilot-Matias T J, Dexai S M. Molecular and biophysical characterization of TT virus: evidence for a new virus family infecting humans. Proc Natl Acad Sci USA. 1999;96:3177–3182. doi: 10.1073/pnas.96.6.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naoumov N V, Petrova E P, Thomas M G, Williams R. Presence of a newly described human DNA virus (TTV) in patients with liver disease. Lancet. 1998;352:195–197. doi: 10.1016/S0140-6736(98)04069-0. [DOI] [PubMed] [Google Scholar]
- 12.Niagro F D, Forsthoefel A N, Lawther R P, Kamalanathan L, Ritchie B W, Latimer K S, Lukert P D. Beak and feather disease virus and porcine circovirus genomes: intermediates between the geminiviruses and plant circoviruses. Arch Virol. 1998;143:1723–1744. doi: 10.1007/s007050050412. [DOI] [PubMed] [Google Scholar]
- 13.Niel C, de Oliveira J M, Ross R S, Gomes S A, Roggendorf M, Viazov S. High prevalence of TT virus infection in Brazilian blood donors. J Med Virol. 1999;57:259–263. [PubMed] [Google Scholar]
- 14.Nishizawa T, Okamoto H, Konishi K, Yoshizawa H, Miyakawa Y, Mayumi M. A novel DNA virus (TTV) associated with elevated transaminase levels in posttransfusion hepatitis of unknown etiology. Biochem Biophys Res Commun. 1997;241:92–97. doi: 10.1006/bbrc.1997.7765. [DOI] [PubMed] [Google Scholar]
- 15.Noteborn M H, de Boer G F, van Roozelaar D J, Karreman C, Kranenburg O, Vos J G, Jeurissen S H, Hoeben R C, Zantema A, Koch G, van Ormondt H, van der Eb A J. Characterization of cloned chicken anemia virus DNA that contains all elements for the infectious replication cycle. J Virol. 1991;65:3131–3139. doi: 10.1128/jvi.65.6.3131-3139.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okada S, Akahane Y, Suzuki H, Okamoto H, Mishiro S. The degree of variability in the amino terminal region of the E2/NS1 protein of hepatitis C virus correlates with responsiveness to interferon therapy in viremic patients. Hepatology. 1992;16:619–624. doi: 10.1002/hep.1840160302. [DOI] [PubMed] [Google Scholar]
- 17.Okamoto H, Nishizawa T, Kato N, Ukita M, Ikeda H, Iizuka H, Miyakawa Y, Mayumi M. Molecular cloning and characterization of a novel DNA virus (TTV) associated with posttransfusion hepatitis of unknown etiology. Hepatol Res. 1998;10:1–16. [Google Scholar]
- 18.Okamoto H, Akahane Y, Ukita M, Fukuda M, Tsuda F, Miyakawa Y, Mayumi M. Fecal excretion of a nonenveloped DNA virus (TTV) associated with posttransfusion non-A-G hepatitis. J Med Virol. 1998;56:128–132. [PubMed] [Google Scholar]
- 19.Okamoto H, Takahashi M, Nishizawa T, Ukita M, Fukuda M, Miyakawa Y, Mayumi M. Marked genomic heterogeneity and frequent mixed infection of TT virus demonstrated by PCR with primers from coding and noncoding regions. Virology. 1999;259:428–436. doi: 10.1006/viro.1999.9770. [DOI] [PubMed] [Google Scholar]
- 20.Okamoto H, Nishizawa T, Ukita M, Takahashi M, Fukuda M, Iizuka H, Miyakawa Y, Mayumi M. The entire nucleotide sequence of a TT virus isolate from the United States (TUS01): comparison with reported isolates and phylogenetic analysis. Virology. 1999;259:437–448. doi: 10.1006/viro.1999.9769. [DOI] [PubMed] [Google Scholar]
- 21.Renshaw R W, Soine C, Weinkle T, O’Connell P H, Ohashi K, Watson S, Lucio B, Harrington S, Schat K A. A hypervariable region in VP1 of chicken infectious anemia virus mediates rate of spread and cell tropism in tissue culture. J Virol. 1996;70:8872–8878. doi: 10.1128/jvi.70.12.8872-8878.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simmonds P, Davidson F, Lycett C, Prescott L E, MacDonald D M, Ellender J, Yap P L, Ludlam C A, Haydon G H, Gillon J, Jarvis L M. Detection of a novel DNA virus (TTV) in blood donors and blood products. Lancet. 1998;352:191–195. doi: 10.1016/s0140-6736(98)03056-6. [DOI] [PubMed] [Google Scholar]
- 23.Summers J, Mason W S. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell. 1982;29:403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- 24.Takahashi K, Ohta Y, Mishiro S. Partial -2.4-kb sequences of TT virus (TTV) genome from eight Japanese isolates: diagnostic and phylogenetic implications. Hepatol Res. 1998;10:110–120. [Google Scholar]
- 25.Tanaka H, Okamoto H, Luengrojanakul P, Chainuvati T, Tsuda F, Tanaka T, Miyakawa Y, Mayumi M. Infection with an unenveloped DNA virus (TTV) associated with posttransfusion non-A to G hepatitis in hepatitis patients and healthy blood donors in Thailand. J Med Virol. 1998;56:234–238. doi: 10.1002/(sici)1096-9071(199811)56:3<234::aid-jmv10>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 26.Tanaka Y, Mizokami M, Orito E, Nakano T, Kato T, Ding X, Ohno T, Ueda R, Sonoda S, Tajima K, Miura T, Hayami M. A new genotype of TT virus (TTV) infection among Colombian native Indians. J Med Virol. 1999;57:264–268. doi: 10.1002/(sici)1096-9071(199903)57:3<264::aid-jmv9>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 27.Tanaka Y, Mizokami M, Orito E, Ohno T, Nakano T, Kato T, Kato H, Mukaide M, Park Y M, Kim B S, Ueda R. New genotypes of TT virus (TTV) and a genotyping assay based on restriction fragment length polymorphism. FEBS Lett. 1998;437:201–206. doi: 10.1016/s0014-5793(98)01231-9. [DOI] [PubMed] [Google Scholar]
- 28.Tsuda F, Okamoto H, Ukita M, Tanaka T, Akahane Y, Konishi K, Yoshizawa H, Miyakawa Y, Mayumi M. Determination of antibodies to TT virus (TTV) and application to blood donors and patients with post-transfusion non-A to G hepatitis in Japan. J Virol Methods. 1999;77:199–206. doi: 10.1016/s0166-0934(98)00154-2. [DOI] [PubMed] [Google Scholar]
- 29.Weiner A J, Brauer M J, Rosenblatt J, Richman K H, Tung J, Crawford K, Bonino F, Saracco G, Choo Q L, Houghton M, Han J H. Variable and hypervariable domains are found in the regions of HCV corresponding to the flavivirus envelope and NS1 proteins and the pestivirus envelope glycoproteins. Virology. 1991;180:842–848. doi: 10.1016/0042-6822(91)90104-j. [DOI] [PubMed] [Google Scholar]