Molecular Epidemiology and Evolution of Enterovirus 71 Strains Isolated from 1970 to 1998 (original) (raw)
Abstract
Enterovirus 71 (EV71) (genus Enterovirus, family Picornaviridae), a common cause of hand, foot, and mouth disease (HFMD), may also cause severe neurological diseases, such as encephalitis and poliomyelitis-like paralysis. To examine the genetic diversity and rate of evolution of EV71, we have determined and analyzed complete VP1 sequences (891 nucleotides) for 113 EV71 strains isolated in the United States and five other countries from 1970 to 1998. Nucleotide sequence comparisons demonstrated three distinct EV71 genotypes, designated A, B, and C. The genetic variation within genotypes (12% or fewer nucleotide differences) was less than the variation between genotypes (16.5 to 19.7%). Strains of all three genotypes were at least 94% identical to one another in deduced amino acid sequence. The EV71 prototype strain, BrCr-CA-70, isolated in California in 1970, is the sole member of genotype A. Strains isolated in the United States and Australia during the period from 1972 to 1988, a 1994 Colombian isolate, and isolates from a large HFMD outbreak in Malaysia in 1997 are all members of genotype B. Although strains of genotype B continue to circulate in other parts of the world, none have been isolated in the United States since 1988. Genotype C contains strains isolated in 1985 or later in the United States, Canada, Australia, and the Republic of China. The annual rate of evolution within both the B and C genotypes was estimated to be approximately 1.35 × 10−2 substitutions per nucleotide and is similar to the rate observed for poliovirus. The results indicate that EV71 is a genetically diverse, rapidly evolving virus. Its worldwide circulation and potential to cause severe disease underscore the need for additional surveillance and improved methods to identify EV71 in human disease.
Enterovirus 71 (EV71), the most recently described serotype of the genus Enterovirus (family Picornaviridae), causes a variety of neurological diseases, including aseptic meningitis, encephalitis, and poliomyelitis-like paralysis. This virus is also one of only a few enterovirus serotypes most often associated with large outbreaks of hand, foot, and mouth disease (HFMD) (14). EV71 has caused outbreaks of severe neurological disease in Australia (12, 15), Europe (2, 8, 22), Asia (25, 29), and the United States (1, 7, 9, 13, 26). Most recently, EV71 was associated with fatal cases of brain stem encephalitis during large HFMD outbreaks in Malaysia in 1997 (19, 31) and in Taiwan in 1998 (4, 6).
Like poliovirus, EV71 may display an affinity for anterior horn cells (8), and it is the most common nonpolio enterovirus associated with poliomyelitis-like paralysis (20). However, comparison of the complete genomic sequences of two EV71 strains to the polioviruses failed to reveal a genetic correlate for the neurovirulence associated with EV71 infection (3). EV71 has been associated with severe central nervous system disease, with a case fatality rate of 0 to 6% (17). During a large EV71 outbreak in Bulgaria in 1975 (705 reported cases), there were 149 cases of paralytic disease and 44 fatalities. Forty-five cases of EV71 infection were reported in the United States in 1987, including eight cases of paralysis and one fatality (1), and virus circulation was widespread, with isolates reported in at least 17 states. EV71 is most closely related genetically to coxsackievirus A16 (CA16), the other common agent of HFMD, but CA16 rarely causes paralysis or death.
Despite the wide variation in clinical presentation, little is known about the range of EV71 genetic diversity, either within an outbreak or among epidemiologically unrelated strains, and the rate of EV71 evolution is also unknown. To investigate genetic variability and its association with outbreaks, we have determined the complete sequences of the VP1 gene for 113 EV71 strains isolated in 23 states in the United States and in five other countries. Analysis of the sequences defined multiple EV71 genotypes and allowed estimation of the rate of EV71 evolution.
(This information was presented, in part, at the Annual Meeting of the American Society for Virology, 8 July 1995, in Austin, Tex. [3a].)
MATERIALS AND METHODS
Viruses.
The 113 EV71 strains examined in this study are listed in Table 1, with year and state or country of isolation and associated clinical symptoms, if known. The strains were isolated between 1970 and 1998 at the Centers for Disease Control and Prevention, Atlanta, Ga., in 25 different laboratories of state health departments in the United States, and in five national enterovirus laboratories in other countries. Viruses were isolated from original clinical specimens by using a variety of cell lines. Virus isolates sent to the Centers for Disease Control and Prevention were propagated in rhabdomyosarcoma cells prior to sequencing. Most isolates were typed by neutralization assay with monospecific rabbit anti-EV71 antiserum.
TABLE 1.
Isolates used in molecular analysis of EV71a
| Strainb | Yr of isolation | Origin | Outcomec | GenBank accession no. |
|---|---|---|---|---|
| BrCr-CA-70 | 1970 | California | Encephalitis | U22521 |
| 2228-NY-72 | 1972 | New York | NA | AF135867 |
| 2604-AUS-74 | 1974 | Australia | Meningitis | AF135883 |
| 2605-AUS-74 | 1974 | Australia | Meningitis | AF135884 |
| 2608-AUS-74 | 1974 | Australia | Meningitis | AF135885 |
| 2609-AUS-74 | 1974 | Australia | Meningitis | AF135886 |
| 2610-AUS-74 | 1974 | Australia | NA | AF135887 |
| 2229-NY-76 | 1976 | New York | NA | AF135868 |
| 2230-NY-76 | 1976 | New York | NA | AF135869 |
| 2231-NY-77 | 1977 | New York | NA | AF135870 |
| 2232-NY-77 | 1977 | New York | NA | AF135871 |
| 2234-NY-77 | 1977 | New York | NA | AF135872 |
| 2235-NY-77 | 1977 | New York | NA | AF135873 |
| 2236-NY-77 | 1977 | New York | NA | AF135874 |
| 2237-NY-77 | 1977 | New York | NA | AF135875 |
| 2238-NY-77 | 1977 | New York | NA | AF135876 |
| 2239-NY-77 | 1977 | New York | NA | AF135877 |
| 10181-NM-78 | 1978 | New Mexico | NA | AF138675 |
| 1011-ND-79 | 1979 | North Dakota | NA | AF135864 |
| 2241-NY-79 | 1979 | New York | NA | AF135878 |
| 2243-NY-79 | 1979 | New York | NA | AF135879 |
| 2258-CA-79 | 1979 | California | Tremors | AF135880 |
| 2114-TN-80 | 1980 | Tennessee | NA | AF135866 |
| 2952-SD-81 | 1981 | South Dakota | NA | AF135888 |
| 3663-MA-82 | 1982 | Massachusetts | NA | AF135889 |
| 3885-UT-82 | 1982 | Utah | NA | AF135891 |
| 3874-ND-82 | 1982 | North Dakota | NA | AF135890 |
| 3982-OH-82 | 1982 | Ohio | Rash | AF135892 |
| 3984-OH-82 | 1982 | Ohio | NA | AF009538 |
| 2259-CA-82 | 1982 | California | Diarrhea | AF135881 |
| 4224-MA-82 | 1982 | Massachusetts | Encephalitis | AF135893 |
| 4323-UT-83 | 1983 | Utah | NA | AF135894 |
| 4599-OR-83 | 1983 | Oregon | CNS disorder | AF135895 |
| 4644-AR-83 | 1983 | Arkansas | NA | AF135896 |
| 4826-CT-83 | 1983 | Connecticut | NA | AF135897 |
| 4827-CT-83 | 1983 | Connecticut | NA | AF009529 |
| 5115-TX-83 | 1983 | Texas | NA | AF135898 |
| 0667-CHN-85 | 1985 | Republic of China | HFMD | AF135934 |
| 2260-CA-86 | 1986 | California | Fever | AF135882 |
| 2623-AUS-86 | 1986 | Australia | HFMD | AF135945 |
| 6762-OK-86 | 1986 | Oklahoma | NA | AF135900 |
| 1410-CA-86 | 1986 | California | Paralysis | AF009525 |
| 0915-MA-87 | 1987 | California | Meningitis | AF009549 |
| 0916-MA-87 | 1987 | Massachusetts | NA | AF009550 |
| 1061-TN-87 | 1987 | Tennessee | NA | AF009528 |
| 1413-CA-87 | 1987 | California | Paralysis | AF009527 |
| 2219-IA-87 | 1987 | Iowa | Meningitis | AF009539 |
| 2246-NY-87 | 1987 | New York | Paralysis | AF009542 |
| 6910-OK-87 | 1987 | Oklahoma | Rash | AF135901 |
| 7234-AK-87 | 1987 | Alaska | Paralysis | AF009522 |
| 7235-AK-87 | 1987 | Alaska | Respiratory failure | AF135902 |
| 7237-AK-87 | 1987 | Alaska | Diarrhea | AF135951 |
| 7238-AK-87 | 1987 | Alaska | Rash | AF135952 |
| 7289-NC-87 | 1987 | North Carolina | NA | AF135903 |
| 7298-AK-87 | 1987 | Alaska | Fatality | AF135904 |
| 7423-MS-87 | 1987 | Mississippi | Paralysis | U22522 |
| 7628-PA-87 | 1987 | Pennsylvania | Paralysis | AF009530 |
| 7629-PA-87 | 1987 | Pennsylvania | Gastroenteritis | AF009531 |
| 7630-PA-87 | 1987 | Pennsylvania | Gastroenteritis | AF009532 |
| 7631-PA-87 | 1987 | Pennsylvania | Gastroenteritis | AF009533 |
| 7632-PA-87 | 1987 | Pennsylvania | Gastroenteritis | AF135905 |
| 7633-PA-87 | 1987 | Pennsylvania | Gastroenteritis | AF009534 |
| 7635-WA-87 | 1987 | Washington | Meningitis | AF135906 |
| 7673-CT-87 | 1987 | Connecticut | NA | AF009535 |
| 7962-PA-87 | 1987 | Pennsylvania | Paralysis | AF009523 |
| 7968-PA-87 | 1987 | Pennsylvania | NA | AF009524 |
| 8102-WA-87 | 1987 | Washington | Meningitis | AF009526 |
| 8209-MD-87 | 1987 | Maryland | NA | AF009536 |
| 8279-PA-87 | 1987 | Pennsylvania | NA | AF009537 |
| 2222-IA-88 | 1988 | Iowa | Fever | AF009540 |
| 8149-AL-88 | 1988 | Alabama | NA | AF135907 |
| 8495-VA-88 | 1988 | Virginia | NA | AF135953 |
| 9166-TX-89 | 1989 | Texas | NA | AF135954 |
| 9243-OK-89 | 1989 | Oklahoma | NA | AF135955 |
| 9323-TX-89 | 1989 | Texas | NA | AF135956 |
| 9541-TX-89 | 1989 | Texas | NA | AF009557 |
| 9718-TX-89 | 1989 | Texas | NA | AF135957 |
| 9837-WA-89 | 1989 | Washington | NA | AF135958 |
| 9873-NM-89 | 1989 | New Mexico | NA | AF135959 |
| 9978-TX-89 | 1989 | Texas | Rash | AF009558 |
| 0359-TX-90 | 1990 | Texas | NA | AF135931 |
| 0390-TX-90 | 1990 | Texas | Otitis media | AF135932 |
| 1411-CA-90 | 1990 | California | NA | AF009551 |
| 0443-TX-90 | 1990 | Texas | NA | AF135933 |
| 0925-OR-91 | 1991 | Oregon | Tremors | AF009547 |
| 0926-OR-91 | 1991 | Oregon | NA | AF009548 |
| 2261-CA-91 | 1991 | California | Meningitis | AF135938 |
| 2583-CAN-91 | 1991 | Quebec, Canada | NA | AF135944 |
| 2262-CA-92 | 1992 | California | Meningitis | AF135939 |
| 2251-NY-93 | 1993 | New York | NA | AF009543 |
| 1873-CT-94 | 1994 | Connecticut | Fatality | AF009559 |
| 1919-NM-94 | 1994 | New Mexico | Rash | AF009552 |
| 1924-AZ-94 | 1994 | Arizona | NA | AF009553 |
| 1997-NC-94 | 1994 | North Carolina | NA | AF135936 |
| 2006-CT-94 | 1994 | Connecticut | Rash | AF009554 |
| 2007-CT-94 | 1994 | Connecticut | NA | AF009555 |
| 2253-NY-94 | 1994 | New York | NA | AF009544 |
| 2254-NY-94 | 1994 | New York | NA | AF009545 |
| 2263-CA-94 | 1994 | California | Paralysis | AF135940 |
| 2264-CA-94 | 1994 | California | Meningitis | AF009546 |
| 6658-COL-94 | 1994 | Colombia | Paralysis | AF135899 |
| 2037-MD-95 | 1995 | Maryland | NA | AF009556 |
| 2132-VA-95 | 1995 | Virginia | NA | AF135937 |
| 2640-AUS-95 | 1995 | Australia | NA | AF135946 |
| 2641-AUS-95 | 1995 | Australia | HFMD | AF135947 |
| 2642-AUS-95 | 1995 | Australia | Encephalitis | AF135948 |
| 2644-AUS-95 | 1995 | Australia | NA | AF135949 |
| 0731-MAA-97 | 1997 | Malaysia (Sarawak) | Fatality | AF135911 |
| 0756-MAA-97 | 1997 | Peninsular Malaysia | NA | AF135935 |
| 2286-TX-97 | 1997 | Texas | NA | AF135941 |
| 2355-OK-97 | 1997 | Oklahoma | NA | AF135942 |
| 2381-MA-97 | 1997 | Massachusetts | Fatality | AF135943 |
| 2814-MO-98 | 1998 | Missouri | Meningitis | AF135950 |
RT-PCR.
Viral RNA was extracted from 200 μl of cell culture supernatant with UltraSpec III (Biotecx, Houston, Tex.) and resuspended in 20 μl of water or was extracted with the Qiamp viral RNA kit (Qiagen Inc., Valencia, Calif.). The primers used for reverse transcription-PCR (RT-PCR) and sequencing are listed in Table 2. The VP1 gene was amplified as a series of overlapping fragments in a one-tube RT-PCR mixture containing 2 μl of RNA, 20 pmol of each primer, a 100 μM concentration of each deoxynucleoside triphosphate, 2 mM MgCl2, 67 mM Tris-HCl (pH 8.8), 17 mM (NH4)2SO4, 1 mM β-mercaptoethanol, 0.2 mg of gelatin per ml, 10 U of placental RNase inhibitor (Boehringer Mannheim Biochemicals, Indianapolis, Ind.), 12 U of avian myeloblastosis virus reverse transcriptase (Boehringer Mannheim), and 5 U of Taq polymerase (Boehringer Mannheim) in a total volume of 50 μl. VP1-specific cDNA was synthesized by incubation of the reaction mixture for 30 min at 42°C and 3 min at 94°C, and it was amplified by 30 cycles of 94°C for 45 s, 42°C for 45 s, and 68°C for 1 min. DNA fragments used for sequencing were gel purified with the QIAquick gel extraction kit (Qiagen). Cycle sequencing was performed with the Prism Ready Reaction Dyedeoxy Terminator Cycle sequencing kit (Perkin-Elmer Corporation–Applied Biosystems, Foster City, Calif.). All sequences were determined on both strands.
TABLE 2.
Oligonucleotide primers used for RT-PCR and sequencing
| Primer | Sequenceb | Positiona | Use |
|---|---|---|---|
| 159 | ACYATGAAAYTGTGCAAGG | 2385–2403 | PCR |
| 197 | CTCTCGATAGTTTCTTCAGCAG | 2674–2695 | Sequencing |
| 172 | TTCAGTAGGGCAGGCTTGGTAGG | 2691–2714 | PCR |
| 204 | CTGCTGAAGAAACTATCGAGAG | 2698–2679 | Sequencing |
| 161 | CTGGGACATAGAYATAACWGG | 2766–2785 | PCR |
| 162 | CCRGTAGGKGTRCACGCRAC | 2869–2850 | PCR |
| 198 | CCGTCATAGAACCATTGATAAG | 3048–3037 | Sequencing |
| 163 | GAGCAYAARCAGGAGAAAGAYC | 3078–3100 | Sequencing |
| 169 | ATAYATGAGAATGAAGCAYGT | 3194–3215 | Sequencing |
| 174 | GCTGACCAAACTTTCCAAGGG | 3348–3328 | PCR |
| NP1A | GCICCICAYTGITGICCRAA | 3355–3336 | PCR |
Sequence analysis.
The assembled complete VP1 sequences were compared to one another with the GAP and PILEUP programs (11). Phylogenetic trees were constructed by the neighbor-joining method with PHYLIP, version 3.5 (10). Branch lengths were determined by the maximum-likelihood method implemented in Puzzle (28). The reliability of the neighbor-joining tree was estimated by bootstrap analysis with 1,000 pseudoreplicate data sets. Previously sequenced EV71 strains BrCr-CA-70 and 7423-MS-87 were also included in the analyses. The VP1 sequence of the CA16 prototype strain, G-10 (24), was included in the phylogenetic analysis as an outgroup.
Estimation of genetic distance and evolutionary rate.
Because of the lack of a true “founder” strain and the apparent presence of multiple lineages, sequences were selected based on their relationships, as depicted in Fig. 1, in order to estimate the evolutionary rate. Genetic distances were calculated by pairwise comparison according to the Kimura two-parameter method of the Distances program (11), using the oldest strain in each set as a reference. Two separate analyses were performed, one for all three positions (representative of both synonymous and nonsynonymous substitutions) and a second analysis for only synonymous substitutions. The evolutionary rate was calculated by linear regression of the genetic distance from the oldest isolate versus year of isolation. The synonymous substitution rate was calculated from the number of nucleotide substitutions per synonymous site by using the computer program Diverge (11) based on a method by Li et al. (18). The nonsynonymous rates, the numbers of nonsynonymous substitutions per nonsynonymous site, were less than 3 × 10−4 and were not included in the data.
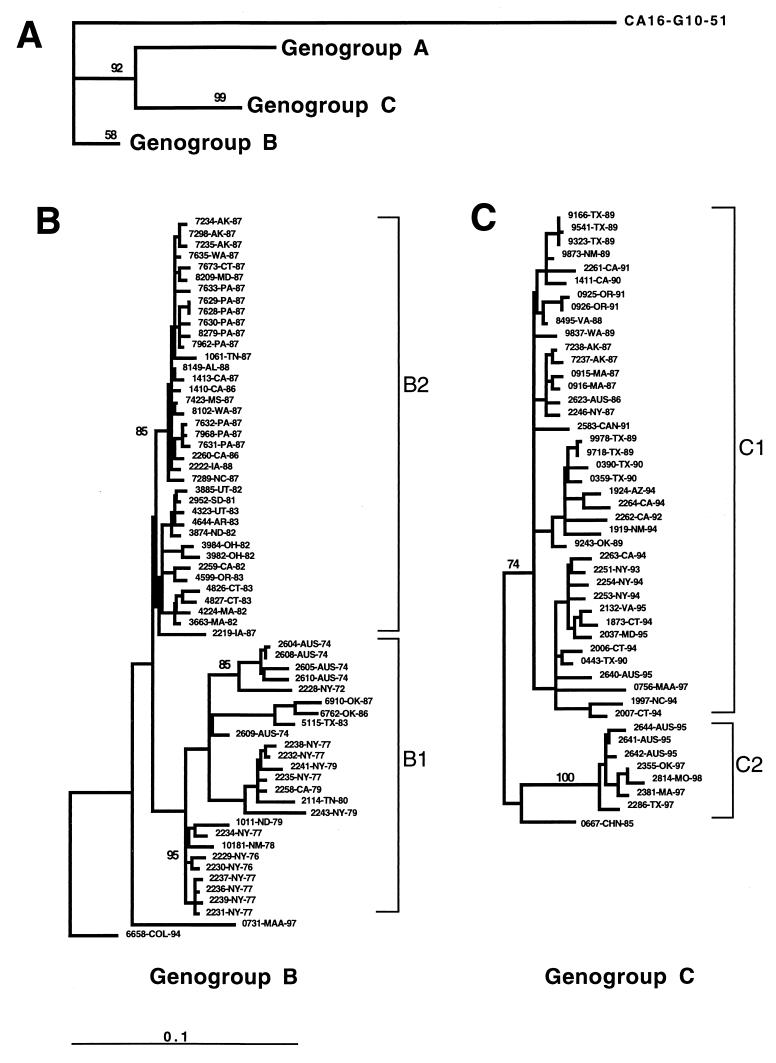
FIG. 1.
Dendrogram generated by the neighbor-joining method with the DNADIST distance measure program (PHYLIP, version 3.5). The phylogram was calculated based on the nucleotide divergence of the VP1 gene (position 2442 to 3332). The last four or five characters of each strain name indicate the state or country and year of isolation. Branch lengths are proportional to the number of nucleotide differences; the frequencies with which the branches for genotypes A, B, and C appeared in 1,000 bootstrap replications were 898, 543, and 999, respectively. Clades with bootstrap numbers are expressed in percentile. The marker denotes a measurement of the relative phylogenetic distance. (A) The branch length for the outgroup, CA16-G10-51, was reduced by 0.75 to save space.
Nucleotide sequence accession numbers.
The nucleotide sequence data reported in this paper have been deposited in the GenBank sequence database under accession no. AF009522 to AF009559, AF135867 to AF135949, AF135911, AF135935, and AF135941 to AF135950.
RESULTS
Nucleotide sequence comparisons.
The complete VP1 gene sequences (891 nucleotides) for 113 EV71 strains isolated in the United States, Australia, Colombia, the Republic of China, Canada, and Malaysia from 1970 to 1998 were determined. These EV71 strains are displayed in a phylogenetic tree constructed by the neighbor-joining method (Fig. 1). Comparisons with other enteroviruses indicate that EV71 strains are monophyletic with respect to other enterovirus serotypes (reference 23 and unpublished data). The strains are clustered in three distinct lineages (genotypes), designated A, B, and C. Genotype A contains a single member, BrCr-CA-70, the EV71 prototype, and differs from all other isolates by 16.5 to 19.7%. Genotype B is represented by 65 strains isolated from 1972 to 1997 in the United States, Australia, Colombia, and Malaysia (Sarawak, island of Borneo). Genotype C, represented by 47 strains isolated from 1986 to 1998, includes viruses from the United States, Australia, the Republic of China, Canada, and mainland Malaysia.
Genotypes B and C were further subdivided into clusters within each genotype, two for genotype B (Fig. 1B) and two for genotype C (Fig. 1C). Cluster B1 contains strains from the United States and Australia that were isolated during the 1970s, as well as a few U.S. isolates from the 1980s (2114-TN-80, 5115-TX-83, 6762-OK-86, and 6910-OK-87). Strains in cluster B1 were more diverse than the B2 strains, differing by up to 9.5% within the cluster and by 6.9 to 11.1% from other genotype B strains. Cluster B2 contains strains isolated in the United States from 1981 to 1987, including most isolates from the 1987 nationwide EV71 outbreak. Strain 6658-COL-94 is genetically distinct from all other genotype B strains (5.8 to 11.1% difference) but differs from strains of genotype C by 15.5 to 17.2%. Strain 0731-MAA-97, a typical representative of many Sarawak, Malaysia, strains, is also distinct from other genotype B strains, differing by 6.5 to 10.5%, and it differs from genotype C strains by 17.1 to 18.3%. The oldest genotype C strains in our collection were isolated in the Republic of China in 1985 and Australia in 1986 (Fig. 1C). Genotype C isolates differ from those of genotype B by 15.5 to 18.7%. Cluster C1 is composed of strains isolated in the United States and Australia from 1986 to 1995, as well as 1997 isolate from peninsular Malaysia. Cluster C2 is composed of U.S. and Australian strains isolated from 1995 to 1998. A 1985 isolate from the Republic of China appears to be intermediate to clusters C1 and C2. Viruses in cluster C1 differ from one another by 1.0 to 6.3% and from those in cluster C2 by 6.1 to 10.1%, while isolates in cluster C2 differ from one another by 0.7 to 1.1%.
Comparison of EV71 VP1 amino acid sequences.
Among all the EV71 isolates, 82% of the predicted VP1 amino acid residues are invariant (Fig. 2). In comparison, the VP1 amino acids of echovirus 30 isolates are at least 88% identical. The EV71 prototype strain, BrCr-CA-70 (genotype A), is 94.2 to 96.0% identical in VP1 amino acid sequence to all other EV71 isolates. VP1 amino acid sequences of genotype B isolates are at least 97.9% identical to one another, whereas those of the genotype C isolates are 98.9% identical to one another. Residues 58, 184, 240, and 289 vary among different genotype groups but are invariant within a genotype group. At four other sites (residues 43, 124, 249, and 292), the predominant amino acid differs between genotypes B and C (Fig. 2).
FIG. 2.
Alignment of genotype consensus VP1 amino acid sequences. The EV71 consensus sequence shows amino acid residues that are identical in at least 85% of all strains and those that are identical in at least 50% but less than 85% of all strains. Sites that are identical in all strains of all genotypes are double underlined; those that are identical in all strains of genotypes B and C, but different in BrCr-CA-70, are single underlined. The genotype consensus sequences indicate sites of at least 85% consensus among all strains of a given genotype (hyphens) and sites that are characteristic of one or more genotypes (uppercase letter, 85% consensus within genotype; lowercase letter, 50 to 85% consensus within genotype).
Estimation of the rate of EV71 evolution.
For the calculation of evolution rates, monophyletic clusters that spanned a period of at least 10 years were identified (Fig. 1 and Table 3). Within each cluster, one from genotype B and one from genotype C, the rate was calculated by plotting the number of nucleotide changes between each strain and the oldest strain in the lineage versus the year of isolation (data not shown). Synonymous and nonsynonymous changes were plotted separately for each of the two data sets. The slope of the linear regression line fitted to the data points is the calculated rate of evolution in substitutions per nucleotide per year. The overall evolutionary rates for all codon positions were 4.2 × 10−3 and 3.4 × 10−3 substitutions per nucleotide per year for the B and C genotypes, respectively. Approximately 93% of all substitutions in VP1 occurred in the third position, and 98% of all substitutions in the third position were synonymous, consistent with the very small number of amino acid changes observed among EV71 isolates. The synonymous substitution rates at the third position were 1.6 × 10−2 and 1.2 × 10−2 substitutions per nucleotide per year for the B and C genotypes, respectively (Table 3).
TABLE 3.
Estimation of the nucleotide substitution rate in the VP1 region of EV71
| Data set | Substitution rate (substitutions/nt/yr)a | |||
|---|---|---|---|---|
| Synonymous sites | (_R_2) | All sites | (_R_2) | |
| B1b | 1.6 × 10−2 | (0.74) | 4.2 × 10−3 | (0.68) |
| C1c | 1.2 × 10−2 | (0.85) | 3.4 × 10−3 | (0.73) |
| Avgd | 1.35 × 10−2 | 3.71 × 10−3 |
DISCUSSION
Based on limited virologic surveillance data, the isolation of EV71 is relatively uncommon in the United States, accounting for fewer than 2% of all enterovirus isolates in all years from 1970 to 1998 except 1994 (5.4%) and 1997 (2.9%) (5, 27). A seroepidemiological study conducted in New York in 1972 suggested that EV71 infection is relatively common, as 26% of the adults tested had antibody to the virus (9). Therefore, severe disease appears to be a rare consequence of a relatively common infection, a general property of most enteroviruses (21). There appears to be no correlation between the severity of disease and the genetic lineage of the virus isolated since viruses of all genotypes are capable of causing severe disease, as are viruses of multiple lineages within each genotype (Table 1 and Fig. 1). Malaysian isolates obtained from patients with uncomplicated HFMD and from fatal encephalitis cases in 1997 were virtually identical in the VP1 region. Preliminary studies indicate that EV71 strains isolated during a similar outbreak in Taiwan in 1998 (4, 6) were epidemiologically and genetically unrelated to those isolated in Malaysia in 1997. This observation also suggests that there is no obvious genetic correlation with clinical disease outbreaks or that viruses of many EV71 lineages may be capable of causing severe disease. However, since only the VP1 region was examined in this study, virulence determinants located elsewhere in the genome could be linked to many different VP1 genotypes via recombination. Further studies are needed, such as determinations of complete genome sequences of strains isolated from cases with a wide range of disease symptoms and severity, to determine whether other regions of the genome may correlate with severity of disease. We recognize that the isolates in this study may not be representative of all viruses in the population or in all countries. Furthermore, it is unknown whether the virus isolates in this study are representative of the virus quasispecies within an individual.
Phylogenetic analysis of complete VP1 sequences has identified three EV71 genotypes. The EV71 prototype strain, BrCr-CA-70, is the only example of genotype A that we identified, but members of genotypes B and C continue to circulate throughout the world. Strains of genotype B circulated widely in the United States from the early 1970s until the late 1980s, but none have been isolated in the United States since 1988. Strains of genotype C were first isolated in the United States in 1987, but the genotype was present in the Republic of China in 1985 and in Australia in 1986, suggesting that genotype C may have originated in the Far East. The limited data on strains from outside the United States suggest that type B strains continue to circulate over a wide geographic area: a B-type strain was isolated in Colombia in 1994 and in Malaysia in 1997. Both genotype B and C viruses were found in Malaysia in 1997, with B-type strains isolated in Sarawak on the island of Borneo and C-type viruses isolated on the mainland. Strains of both genotypes also cocirculated during the 1987 U.S. outbreak. For example, of five strains isolated in Alaska in 1987, three were of genotype B and two were of genotype C. Alaskan strains of the same genotype were closely related to one another, indicating an epidemiological link within genotype. Twenty-two of 27 isolates from the 1987 U.S. outbreak that were analyzed were genotype B strains. Twenty of these were closely related to one another and to two 1986 California isolates and a 1988 Iowa isolate. The 22 1987 outbreak isolates were also closely related to strains that circulated in the United States from 1981 to 1983. One 1987 B-type isolate (6910-OK-87) was related to strains found in the United States and Australia in the 1970s and early 1980s. The 1987 genotype B strains were from 11 states in widely separated regions of the United States (Alaska, California, Connecticut, Iowa, Maryland, Mississippi, North Carolina, Oklahoma, Pennsylvania, Tennessee, and Washington). The remaining five 1987 U.S. isolates were members of genotype C. They were from three states (Alaska, Massachusetts, and New York) and were closely related to one another and to a 1986 Australia isolate. The presence of three EV71 lineages of two genotypes in the United States, and two genotypes in one state, suggests that the 1987 outbreak was the result of coincident circulation of three genetically distinct viruses. Similarly, genotype B strains isolated in New York in 1977 fall into two distinct clusters, as do genotype C strains isolated in Texas in 1989, suggesting that the cocirculation of distinct strains is relatively common.
The apparent genetic separation of an isolate from Colombia (1994) and one from the Republic of China (1985) from their respective genotypes probably reflects the lack of additional strains from those countries and surrounding regions. Additional surveillance is required to ascertain whether strains similar to the Colombian isolate and the Chinese isolate continue to circulate and to further describe the genetic relationship of these viruses within their respective genotypes. Likewise, the lack of strict time ordering of the isolates and clusters shown in Fig. 1 could be the result of the absence of many truly genetically intermediate strains. Independent analysis of clusters B1 and C1 resulted in largely time-ordered lineages for the calculation of evolutionary rates. For example, U.S. and Australian strains isolated from 1995 to 1998 clustered closely together, indicating genetic and epidemiological linkages among those isolates, yet they were distinct from U.S. isolates from the period 1991 to 1994. The existence of two distinct clusters among the U.S. EV71 strains isolated since 1987 suggests the possibility that strains of genotype C have been introduced into the United States at least twice in the last 10 years.
The rate of EV71 evolution within a lineage was estimated to be 1.35 × 10−2 synonymous substitutions per nucleotide per year. This rate is similar to the rates calculated for other enteroviruses, such as poliovirus type 1 (3.36 × 10−2 substitutions per nucleotide per year) (16) and EV70 (2.2 × 10−2 substitutions per nucleotide per year) (30). The factors potentially influencing enterovirus evolution rates include replicase fidelity, rate of transmission (number of replication cycles per year), the number of progeny virions produced per infecting virion, and any effects of synonymous mutations on RNA structure and function. These factors are difficult to measure individually and are generally observed only in aggregate, making it difficult to determine which are the most important determinants influencing enterovirus evolution.
The association of severe neurological disease, including deaths, with recent large outbreaks of EV71 HFMD in Malaysia (31) and Taiwan (4, 6) underscores the need to understand the pathogenesis and epidemiology of EV71. The availability of sequence data for a large number of EV71 isolates from different parts of the world will make it possible to develop sensitive and specific molecular reagents for the rapid identification of EV71 during epidemics of HFMD or other enteroviral diseases. Increased surveillance, coupled with improved laboratory diagnostic tools, will enable public health authorities to rapidly recognize an outbreak of EV71 disease and to implement measures to limit further virus transmission.
ACKNOWLEDGMENTS
We acknowledge all of the laboratories that isolated the viruses necessary for this study. In particular, we thank Leo Grady (New York State Department of Health), Ron Cheshire (Arizona Department of Health), Norman Swack (University of Iowa Hygienic Laboratory), David Schnurr (California Department of Health Services), Nina Peláez (Instituto Nacional de Salud, Colombia), Lam Sai Kit (University of Malaysia), Mangalam Sinniah (Institute for Medical Research, Malaysia), and the Enterovirus-Respiratory staff at Fairfield Hospital, Melbourne, Australia. We thank W. S. Li for helpful discussions.
REFERENCES
- 1.Alexander J P, Baden L, Pallansch M A, Anderson L J. Enterovirus 71 infections and neurologic disease—United States, 1977–1991. J Infect Dis. 1994;169:905–908. doi: 10.1093/infdis/169.4.905. [DOI] [PubMed] [Google Scholar]
- 2.Blomberg J, Lycke E. New enterovirus type associated with epidemic of aseptic meningitis and/or disease. Lancet. 1974;ii:112–113. doi: 10.1016/s0140-6736(74)91684-5. [DOI] [PubMed] [Google Scholar]
- 3.Brown B A, Pallansch M A. Complete nucleotide sequence of enterovirus 71 is distinct from poliovirus. Virus Res. 1995;39:195–205. doi: 10.1016/0168-1702(95)00087-9. [DOI] [PubMed] [Google Scholar]
- 3a.Brown, B. A., M. S. Oberste, J. P. Alexander, Jr., M. L. Kennett, and M. A. Pallansch. 1995. Abstracts of the Annual Meeting of the American Society for Virology, abstr. T-50.
- 4.Centers for Disease Control and Prevention. Deaths among children during an outbreak of hand, foot and mouth disease—Taiwan, Republic of China, April–July 1998. Morbid Mortal Weekly Rep. 1998;47:629–632. [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. Nonpolio enterovirus surveillance—United States 1993–1996. Morbid Mortal Weekly Rep. 1997;46:748–750. [PubMed] [Google Scholar]
- 6.Chang L Y, Huang Y C, Lin T Y. Fulminant neurogenic pulmonary oedema with disease. Lancet. 1998;352:352–367. doi: 10.1016/S0140-6736(98)24031-1. [DOI] [PubMed] [Google Scholar]
- 7.Chonmaitree T, Menegus M A, Schervish-Swierkosz E M, Schwalenstocker E. Enterovirus 71 infection: report of an outbreak with two cases of paralysis and a review of the literature. Pediatrics. 1981;67:489–493. [PubMed] [Google Scholar]
- 8.Chumakov M, Voroshilova L, Shindarov I, Lavrova I, Gracheva L, Koroleva G, Vasilenko S, Brodvarova I, Nikolova M, Gyurova S, Gacheva M, Mitov G, Ninov N, Tsylka E, Robinson I, Frolova M, Bashkirtsev V, Martiyanova L, Rodin V. Enterovirus 71 isolated from cases of epidemic poliomyelitis-like disease in Bulgaria. Arch Virol. 1979;60:329–340. doi: 10.1007/BF01317504. [DOI] [PubMed] [Google Scholar]
- 9.Deibel R, Gross L L, Collins D N. Isolation of a new enterovirus (38506) Proc Soc Exp Biol Med. 1975;148:203–207. doi: 10.3181/00379727-148-38506. [DOI] [PubMed] [Google Scholar]
- 10.Felsenstein J. PHYLIP—phylogeny inference package (version 3.5) Cladistics. 1989;5:164–166. [Google Scholar]
- 11.Genetics Computer Group. Program manual for the GCG package, version 9.0 (1996). Madison, Wis: Genetics Computer Group; 1996. [Google Scholar]
- 12.Gilbert G L, Dickson K E, Waters M J, Kennett M L, Land S A, Sneddon M. Outbreak of enterovirus 71 infection in Victoria, Australia, with a high incidence of neurologic involvement. Pediatr Infect Dis J. 1988;7:484–488. doi: 10.1097/00006454-198807000-00007. [DOI] [PubMed] [Google Scholar]
- 13.Hayward J C, Gillespie S M, Kaplan K M, Packer R, Pallansch M A, Plotkin S, Schonberger L B. Outbreak of poliomyelitis-like paralysis associated with enterovirus 71. Pediatr Infect Dis J. 1989;8:611–616. doi: 10.1097/00006454-198909000-00009. [DOI] [PubMed] [Google Scholar]
- 14.Ishimaru Y, Kakano S, Yamaoka K, Takami S. Outbreaks of hand, foot, and mouth disease by enterovirus 71. High incidence of complication disorders of central nervous system. Arch Dis Child. 1980;55:583–588. doi: 10.1136/adc.55.8.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kennett M L, Birch C J, Lewis F A, Yung A P, Locarnini S A, Gust I D. Enterovirus type 71 infection in Melbourne. Bull W H O. 1974;51:609–615. [PMC free article] [PubMed] [Google Scholar]
- 16.Kew O M, Sutter R W, Nottay B K, McDonough M J, Prevots D R, Quick L, Pallansch M A. Prolonged replication of a type 1 vaccine-derived poliovirus in an immunodeficient patient. J Clin Microbiol. 1998;36:2893–2899. doi: 10.1128/jcm.36.10.2893-2899.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landry M L, Fonseca S N S, Cohen S, Bogue C W. Fatal enterovirus type 71 infection: rapid detection and diagnostic pitfalls. Pediatr Infect Dis J. 1995;14:1095–2000. doi: 10.1097/00006454-199512000-00013. [DOI] [PubMed] [Google Scholar]
- 18.Li W H, Wu C I, Luo C C. A new method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Mol Biol Evol. 1985;2:150–174. doi: 10.1093/oxfordjournals.molbev.a040343. [DOI] [PubMed] [Google Scholar]
- 19.Lum L C S, Wong K T, Lam S K, Chua K B, Goh A Y, Lim W L, Ong B B, Paul G, AbuBakar S, Lambert M. Fatal enterovirus 71 encephalomyelitis. J Pediatr. 1998;133:795–798. doi: 10.1016/s0022-3476(98)70155-6. [DOI] [PubMed] [Google Scholar]
- 20.Melnick J L. Enterovirus type 71 infections: a varied clinical pattern sometimes mimicking paralytic poliomyelitis. Rev Infect Dis. 1984;6:S387–S390. doi: 10.1093/clinids/6.supplement_2.s387. [DOI] [PubMed] [Google Scholar]
- 21.Melnick J L. Enteroviruses: polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses. In: Fields B N, Knipe D M, Howley P M, Channock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 655–712. [Google Scholar]
- 22.Nagy G, Takatsy S, Kukan E, Mihaly I, Domok I. Virological diagnosis of enterovirus type 71 infections: experiences gained during an epidemic of acute CNS diseases in Hungary in 1978. Arch Virol. 1982;71:217–227. doi: 10.1007/BF01314873. [DOI] [PubMed] [Google Scholar]
- 23.Oberste M S, Maher K, Kilpatrick D R, Pallansch M A. Molecular evolution of the human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification. J Virol. 1999;73:1941–1948. doi: 10.1128/jvi.73.3.1941-1948.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poyry T, Hyypia T, Horsnell C, Kinnunen L, Hovi T, Stanway G. Molecular analysis of coxsackievirus A16 reveals a new genetic group of enteroviruses. Virology. 1994;202:982–987. doi: 10.1006/viro.1994.1423. [DOI] [PubMed] [Google Scholar]
- 25.Samuda G M, Chang W, Yeung C, Tang P. Monoplegia caused by enterovirus 71: an outbreak in Hong Kong. Pediatr Infect Dis J. 1987;6:206–208. doi: 10.1097/00006454-198702000-00013. [DOI] [PubMed] [Google Scholar]
- 26.Schmidt N J, Lennette E H, Ho H H. An apparently new enterovirus isolated from patients with disease of the central nervous system. J Infect Dis. 1974;129:304–309. doi: 10.1093/infdis/129.3.304. [DOI] [PubMed] [Google Scholar]
- 27.Strikas R A, Anderson L J, Parker R A. Temporal and geographic pattern of nonpolio enterovirus in the United States. J Infect Dis. 1986;153:346–351. doi: 10.1093/infdis/153.2.346. [DOI] [PubMed] [Google Scholar]
- 28.Strimmer K, Haeseler A V. Quartet puzzling: a quartet maximum likelihood method for reconstructing tree topologies. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
- 29.Tagaya I, Moritsugu Y. Epidemic of disease in Japan. Jpn J Med Sci Biol. 1973;26:143–147. doi: 10.7883/yoken1952.26.143. [DOI] [PubMed] [Google Scholar]
- 30.Takeda N, Tanimura M, Miyamura K. Molecular evolution of the major capsid protein VP1 of enterovirus 70. J Virol. 1994;68:854–862. doi: 10.1128/jvi.68.2.854-862.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.World Health Organization. Outbreak of hand, foot and mouth disease in Sarawak. Cluster of deaths among infants and young children. Weekly Epidemiol Rec. 1997;72:211–212. [PubMed] [Google Scholar]