Probabilistic prediction of Saccharomyces cerevisiae mRNA 3′-processing sites (original) (raw)
Abstract
We present a tool for the prediction of mRNA 3′-processing (cleavage and polyadenylation) sites in the yeast Saccharomyces cerevisiae, based on a discrete state-space model or hidden Markov model. Comparison of predicted sites with experimentally verified 3′-processing sites indicates good agreement. All predicted or known yeast genes were analyzed to find probable 3′-processing sites. Known alternative 3′-processing sites, both within the 3′-untranslated region and within the protein coding sequence were successfully identified, leading to the possibility of prediction of previously unknown alternative sites. The lack of an apparent 3′-processing site calls into question the validity of some predicted genes. This is specifically investigated for predicted genes with overlapping coding sequences.
INTRODUCTION
The availability of complete genome sequences has made possible large-scale sequence analysis to identify and model complex biological phenomena such as regulatory control (or signal) sequences (1–7). We are studying the control sequences that determine the position of 3′-processing (cleavage and polyadenylation) of pre-mRNA, in order to develop predictive tools that will locate unknown 3′-processing sites in a genomic sequence.
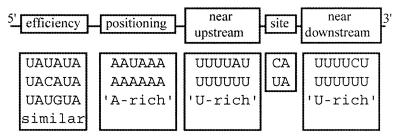
The control sequences involved in 3′-processing in the yeast Saccharomyces cerevisiae have been extensively studied (8–10), yet the creation of a predictive model has remained elusive. Experimental mutagenesis studies have identified many different (though often related) sequences that act as elements in selection and processing of the 3′-ends of yeast mRNA transcripts. Recent computational studies (11,12) of large sets (thousands of sequences) of putative 3′-processing sites have improved our understanding. A general pattern (Fig. 1) of the control sequences required for 3′-processing in yeast has emerged as follows. (i) An ‘efficiency element’, typically found between 35 and 60 nt upstream from the 3′-processing site. Mutagenesis studies and computational analysis have identified the best sequence (word) for this element as UAUAUA. (ii) A ‘positioning element’, typically found between 10 and 30 nt upstream of the 3′-processing site. The best word for this element is AAUAAA (as in mammalian sequences), however, it is commonly described only as ‘A-rich’, since many functional sequences are characterized only by their adenosine content. (iii) A ‘near-upstream element’, typically occurring within 10 nt upstream of the 3′-processing site, and best characterized as ‘U-rich’. (iv) A ‘near-downstream element’, typically 10 nt or less downstream of the 3′-processing site and also characterized as ‘U-rich’. (v) The 3′-processing site itself, which has been experimentally shown to consist most commonly of a pyrimidine followed by one or more A residues (13). Significantly, the functionality of the U-rich control elements near the 3′-processing site was first postulated based on computational sequence analysis (11,12), and subsequently verified through laboratory experiments (14). Recent work (15–17) has called the naming conventions of the control elements into question; therefore, in the work reported here, we refer simply to elements 1–4, plus the 3′-processing site.
Figure 1.
A simplified representation of the arrangement of control elements (with example sequences) that identify the 3′-processing site in yeast mRNA.
Analysis of verified 3′-processing sites often reveals that one or more of the elements may bear little or no resemblance to its optimal form, yet the complete 3′-processing site is functional. Furthermore, it has been found through mutagenesis studies that the deletion of sequence elements with known function fails to disable 3′-processing of the transcript, but instead reduces its efficiency. It has been postulated (9,18,19) that the complete control sequence acts cooperatively, allowing comparatively ‘strong’ words of some elements to compensate for sub-optimal or missing words in the remaining elements. Alternatively, several ‘weak’ words with acceptable positioning can have an additive effect, giving a net efficiency similar to a single ‘strong’ word.
The analysis and prediction of 3′-processing sites in yeast is further complicated by the multiplicity of alternative functional sites. Two forms of alternative polyadenylation of yeast transcripts have been demonstrated: (i) regulatory alternatives (20,21), which are typically separated by hundreds of nucleotides or more; and (ii) apparently non-regulatory alternatives, in which alternative sites are separated by tens of nucleotides or fewer. In one extreme case, 13 separate polyadenylation sites were identified for the yeast HIS3 gene (22). Alternate polyadenylation in animal cells has been reported only as the widely spaced, regulatory form, whereas studies of plant cells have potentially indicated both forms. From our previous studies of yeast 3′-processing sites, we believe that all control elements for a given site are within 100 nt of the 3′-processing site; therefore, the control sequence of regulatory alternative sites are not likely to overlap, and as such will probably not complicate the development of predictive tools. However, the non-regulatory alternatives will most probably have overlapping control elements, and therefore will complicate prediction. The need to accurately model potentially overlapping control sequences specifically led to the use of the forward or filtering algorithm, as described below.
A predictive model for 3′-processing sites has many possible uses. A principal use would be in the enhancement of gene prediction (23–26). Most current gene prediction software uses naïve models for the prediction of 3′-processing sites. The simple models typically are based primarily on a search for the canonical AAUAAA positioning element or its most common variant AUUAAA (27,28). Previous attempts at prediction of 3′-processing sites have focused on mammalian sequences (27–31). While the focus on the positioning element is reasonable for modeling 3′-processing sites in mammalian sequences, it is completely inadequate in describing the control sequences that are observed in either yeast or plants (9,19,32).
Improvement of gene prediction algorithms goes hand-in-hand with improvement in the annotation of newly generated genomic sequence. Currently, such annotation is typically limited to identification of functional RNA (tRNA and rRNA), coding sequences, and other large-scale phenomena. Improvements in transcript prediction (as opposed to simply coding sequence) will also help in troubleshooting and designing large-scale genetic expression measurements. The probes necessary for such hybridization are primarily generated from the coding sequence. Expression measurements are made on complete transcripts; therefore, knowledge of the genomic limits of the transcript will improve the ability to avoid systematic problems such as cross-hybridization of probes due to similar untranslated region (UTR) sequences.
As we have shown previously (11,19), in a large set of aligned 3′-processing sequences, the control elements appear with a distinctly non-random positioning with respect to the 3′-processing site. By plotting the relative frequency of occurrence of all words as a function of position with respect to the 3′-processing site, we can cluster words based on similar distributions. We assume that the different words in each of these clusters perform the same biological function.
We present a probabilistic method for prediction of S.cerevisiae 3′-processing sites, based on a discrete state-space model (DSM), which is represented mathematically as a hidden Markov model (HMM), but with designed rather than trained parameters.
A web-server interface for our S.cerevisiae 3′-processing site predictions is available at http://bmerc-www.bu.edu/polyA/.
MATERIALS AND METHODS
Putative S.cerevisiae 3′-processing sequences were identified through genomic/EST alignment and analysis as described previously (11). This set totals 1352 3′-processing sites, associated with 861 different genes. Our predictive models are based on the assumption that the functional elements of the 3′-processing control sequence can be identified by their positioning with respect to the 3′-processing site. Based on our previous work, as well as published experimental evidence (9), we have built a probabilistic model to include information from a relative position of –110 to +40 with respect to the 3′-processing site (see Fig. 3). All protein-coding bases in the training set were replaced with Ns so that the resulting models would emphasize the functional elements of the 3′-processing control sequence, rather than the end of the protein coding sequence (CDS).
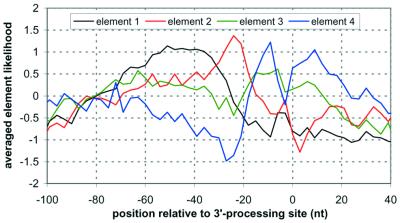
Figure 3.
The characteristic logarithmic distributions for positioning the four functional elements used in our model of the 3′-processing control sequence, as determined by a _k_-means clustering of words with similar positioning with respect to the processing site. The elements are normalized to a uniform distribution.
The mathematical structure of the DSM is that of a HMM (33). A HMM models sequence generation as a traversal of a set of discrete states. Each state is defined by emission probabilities, the probability for each of the four nucleotides to be observed in that state, and transitions from any specific state to any other are governed by transition probabilities. We used the filtering, or forward, algorithm (33–36), which computes the probability that a query sequence will be generated by a given model. The smoothing algorithm (34–36) [also referred to as the forward–backward algorithm or posterior decoding (37)] calculates the probability for each model state at each position of the query sequence. Note that the smoothing algorithm will give an answer (in our case, the most likely positions where 3′-processing will occur) regardless of the probability that the model would have produced the query sequence. As shown below, the output of both algorithms is necessary to find 3′-processing sites.
The combination of the filtering and smoothing algorithms is a natural choice to compensate for many of the complications described in the Introduction, including non-regulatory alternative 3′-processing sites and multiple, additive elements for a single 3′-processing site. The filtering and smoothing algorithms assess a query sequence by computing all paths through the model that could generate the sequence, in contrast with optimal path methods such as the Viterbi algorithm (37), which use dynamic programming to model only the most likely path through the model. By evaluating all paths, the filtering–smoothing combination allows for and incorporates multiple occurrences of any modeled element, whether it is one of the control elements or the 3′-processing site itself. The presence of multiple 3′-processing sites in a query sequence would be signaled by a high filtering probability and multiple maxima in the conditional probability of the model state associated with the 3′-processing site (Fig. 2, element c).
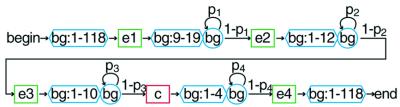
Figure 2.
The structure of the HMM used to model 3′-processing control sequences for yeast. All state-to-state transitions not explicitly labeled have a probability of 1.0. The hexagonal elements are background elements that can take on any length in the given range with equal probability. The functional elements e1–e4 are hexamers, with individual nucleotide probabilities determined through analysis with the Gibbs sampler (38). Probabilities p1–p4 were optimized empirically in analysis of known processing sites. The position of the cleavage and polyadenylation is the center of the c element. Nucleotide probabilities for the c element were obtained from the 1352 training sequences. Complete details, including nucleotide emission probabilities, are available as Supplementary Material.
The structure of the model developed for 3′-processing prediction in S.cerevisiae is shown in Figure 2. As shown, the model consists of four hexamer elements (e1–e4) and the 3′-processing site (c), separated by variable length background sequences. Nucleotide emission frequencies for each position in the four control elements (e1, e2, e3, e4) were obtained with the Gibbs sampler (38). In order to optimize the Gibbs Sampler’s ability to locate the elements in question, separate analyses were made for each element, with the input sequences restricted to the region where the element is most likely to occur (based on the results shown in Fig. 3). While the Gibbs sampler was used in this work, any methodology used to assign the nucleotide emission probabilities to the control elements would work, for example MEME (39) or AlignACE (2), or even simple statistical averaging if the training set is large enough. The 3′-processing site was modeled as a pentamer, centered on the cleavage site. Nucleotide emission frequencies for the 3′-processing site, (c), were obtained through direct tabulation of the frequencies of each base in the 5 nt centered on the polyadenylation site in our 1352 training sequences. The nucleotide emission frequencies for each of the elements are available as Supplementary Material or at our website (http://bmerc-www.bu.edu/polyA/).
In order to determine the positioning of the elements, we extended our previous analysis of S.cerevisiae 3′-processing sequences (11), grouping (presumably functionally) similar words with a _k_-means clustering algorithm (40) based on a Pearson correlation coefficient (41) comparison of their positional distributions. Based on our previous work, we chose to create four clusters representing the four elements. The characteristic distribution of each element was determined by averaging all words clustered into that element. Figure 3 shows the characteristic distributions for the four clusters used in the generation of the yeast 3′-processing predictor.
The spaces between the functional elements were modeled as states emitting nucleotides at background sequence frequencies (using statistics for typical yeast mRNA transcripts). As shown in Figure 2, the length distributions for these spacer states between functional elements were given a uniform probability over a specified range, and an exponentially decaying upper limit. The minimum distance between the functional elements is fixed as shown, whereas the maximum distance is mediated by the four non-unity transition probabilities p1–p4, which were determined primarily through examination of the data summarized in Figure 3, followed by optimization through testing of known 3′-processing sites. The final values were p1 = 0.8, p2 = 0.65, p3 = 0.5 and p4 = 0.65. Additional knowledge from previous studies was also used, specifically the previously reported minimum distance (10 nt) that can separate functional copies of the first two elements (8,42). The HMM is described in complete detail in the Supplementary Material or at our website.
As stated in the Introduction, a principal goal of our work is to aid in the annotation of new genomes and, as such, we need to be able to analyze large sequences (tens or hundreds of thousands of nucleotides). In contrast, the control sequences for which we are searching are limited to ∼150 nt. In order to reconcile this, we have chosen to test the model on a sliding 150 nt-wide window, stepping along the entire sequence, combining the results using the following equation for the score at nucleotide x:
S(x) = log10 {[Σ_w_ PF w ¥ PS w,_x_]/[(PF w ¥ PS w,x)bg]}
where the summation is over all windows (w) that include nucleotide x. PF w is the filtering algorithm’s probability that our model generates the sequence in the window w. PS w,x is the smoothing algorithm’s probability that nucleotide x is a 3′-processing site in window w. The denominator includes a normalization that makes the average score in a random sequence set (described below) equal to 0. In principle, the model should be tested and combined in all possible windows, however, to reduce computation time, we tested various step sizes between consecutive windows and found that up to 20 nt could be safely used without changing the predictions (data not shown). Specific positions within query sequences are classified as likely 3′-processing sites on the basis of whether or not S(x) exceeds a user-specified threshold value. Statistical assessment of the threshold value for S(x) is discussed below.
RESULTS
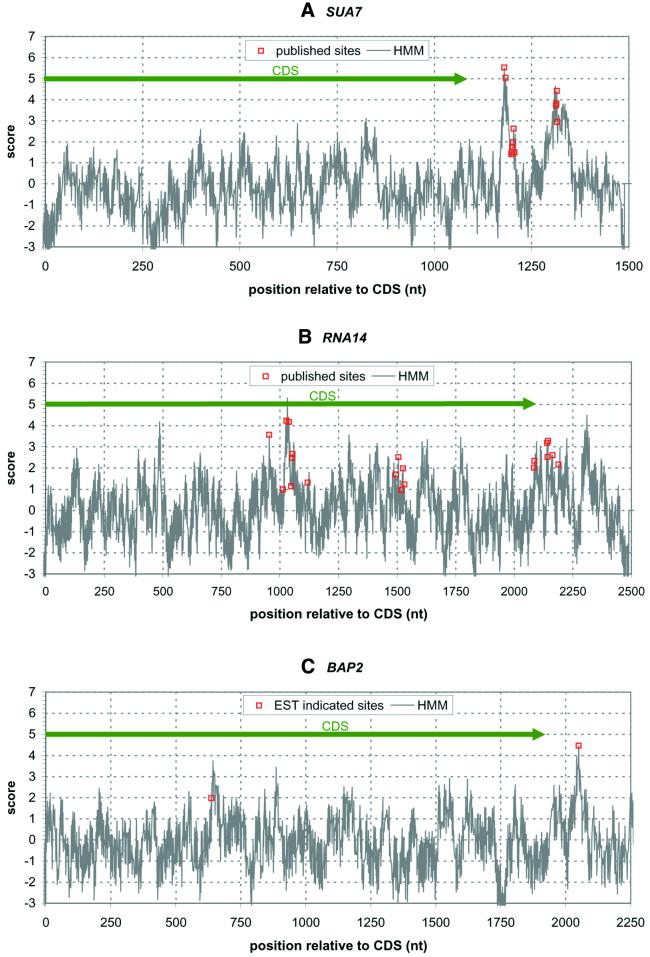
Figure 4 shows examples of the output of our analysis (see Materials and Methods) for three different mRNA sequences, SUA7, RNA14 and BAP2. The plot shown for SUA7 (Fig. 4A) is typical in most ways (see the discussion on regulatory alternative 3′-processing below). The CDS (delineated by the large green arrow) is characterized by either small or negative scores and a few isolated, low value (<3) peaks in the HMM plot. The plot shows clusters of peaks in the 3′-UTR. Several previously published 3′-processing sites (21) for SUA7 are also shown in Figure 4A. The published sites are well aligned with our prediction; however, our algorithms indicate further potential sites not previously reported.
Figure 4.
Examples of 3′-processing predictions for yeast genes (A) SUA7, (B) RNA14 and (C) BAP2. The large green arrow marks the CDS, and the red markers mark previously reported 3′-processing sites for (A) SUA7 (21), (B) RNA14 (20) or EST-identified 3′-processing sites for (C) BAP2.
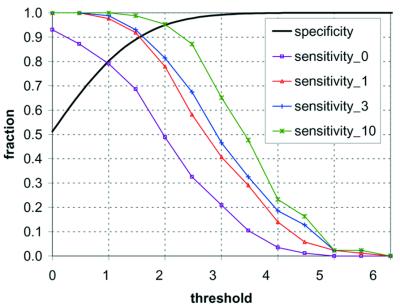
In order to test the statistical significance of our predictions, we calculated the sensitivity and specificity. The sensitivity is the fraction of verified sites correctly predicted by our model. For positive controls, we obtained a set of 86 polyadenylation sites previously reported in experimental studies of 3′-processing in yeast. We restricted this set to sites that were identified through RACE–PCR or clone sequencing, as RNase protection-based determination has been shown to be less reliable in exact positioning (43). This analysis was also restricted to sites that are positioned after the stop codon (in the 3′-UTR). Figure 5 shows the results of our analysis. For the sensitivity analysis, we counted as true positives any predicted site that matched an experimental site exactly, or within 1, 3 or 10 nt (sensitivity_0, sensitivity_1, sensitivity_3 and sensitivity_10, respectively, in Fig. 5). When the threshold value of S(x) was set to 2.5, the resulting sensitivities are 0.33, 0.58, 0.67 and 0.87, respectively. It is possible that this measure of the sensitivity is overly conservative, as we are treating our set of 86 sites as equivalent, whereas it is possible that the strength (or efficiency) of these sites is variable.
Figure 5.
Sensitivity and specificity determined for the HMM. The sensitivity was tested on 86 reported 3′-processing sites for the yeast genes GCN4 (56), HIS3 (22), FBP1 (57), SUA7 (21), ARO4 (58), TRP4 (59), SIR1, AEP2, CBP1 and RNA14 (20), and ACT1, CYC1, ADH1 and YPT1 (60). The four measured curves for sensitivity represent exact position agreement between predicted and reported site (sensitivity_0), or mismatch by up to 1, 3 or 10 nt (sensitivity_1, sensitivity_3 and sensitivity_10, respectively). The specificity was estimated from analysis of 200 000 nt of randomly generated sequence that preserved second-order (trinucleotide) statistics of putative yeast transcripts.
Specificity can be expressed as one minus the false positive fraction of the predicted 3′-processing sites. The widespread occurrence of alternative 3′-processing sites in yeast, most of which have never been identified, makes the definitive classification of any site as non-processing problematic. It is unlikely that all possible 3′-processing sites have been identified for most yeast genes, with the possible exception of CYC1 or a few other thoroughly investigated genes. (Our EST analysis supported the existence of many previously undetected 3′-processing sites.) Therefore, we chose to estimate the fraction of false positives by analyzing randomly generated sequences that preserve the trinucleotide (second-order) probabilities of putative yeast mRNA transcripts (coding sequence plus 5′- and 3′-UTR). 200 000 nt of such random sequence were generated and subjected to the HMM analysis. The specificity was then estimated as one minus the fraction of positions that would be classified as 3′-processing sites, based on S(x) above a given threshold value. The variation of the specificity with threshold value is shown in Figure 5.
Several genes in yeast have been shown to have alternative 3′-processing activity that varies with growth conditions. We have analyzed the four genes that were reported by Sparks et al. (20), RNA14, CBP1, SIR1 and AEP2, as well as SUA7 (21). These genes have been shown to utilize alternative 3′-processing sites differentially as experimental conditions are changed. Figure 4A and B show our results for SUA7 and RNA14, respectively. (Results for CBP1, SIR1 and AEP2 can be found at our website.) Most of the reported 3′-processing sites, including the regulatory sites internal to the CDS, correlate well with our predictions. In Figure 4B, there are predicted sites near position 1770 and near position 500 that do not correspond to previously measured 3′-processing sites. These sites are either false positives of our method or alternative 3′-processing sites that have not yet been detected. In addition, the known sites around position 1500 are not predicted terribly well by our method.
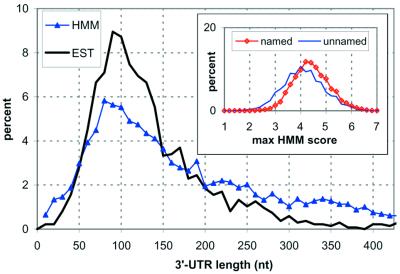
The 6281 predicted and known genes from yeast, as defined by the Saccharomyces Genome Database (SGD) at Stanford University (44,45), were analyzed for predicted 3′-processing sites. The putative transcript for each gene included 50 nt of the 5′-UTR sequence and 500 nt of the 3′-UTR sequence. The limit of 500 nt downstream was chosen based on our previous finding that <3% of the genes are likely to have 3′-UTRs that extend further (11). The following parameters were measured for each gene: (i) the maximum value of the HMM score [S(x)max] after the stop codon; (ii) the distance from the stop codon to the position of S(x)max; and (iii) the number of peaks higher than a given threshold that occur inside the CDS. Assuming that the position of S(x)max after the stop codon is the most likely position of 3′-processing, we can compute a projected length of the 3′-UTR. The distribution of lengths is plotted in Figure 6, along with the distribution determined through analysis of the ESTs. The distribution of S(x)max for all genes is plotted in the inset to Figure 6. In the inset, the 6281 predicted genes from SGD have been grouped based on whether or not the gene has an alternative name listed. Genes with an assigned name are assumed to have been identified experimentally or have shown significant sequence similarity to other known genes. A large percentage of the gene sequences that fail to reach a maximum S(x)max of at least 3.0 come from the ‘unnamed’ group. This is a possible indication of a spurious open reading frame (ORF) not encoding a functional protein.
Figure 6.
A comparison of the distribution 3′-UTR lengths for yeast genes, determined by analysis of 1352 yeast EST sequences (11) (black line), and from analysis of all 6281 predicted yeast genes by HMM (blue line). The inset shows the distribution of maximum HMM scores for the 6281 yeast genes, with the genes separated by the presence (3651 genes, shown in red) or absence (2630 genes, shown in blue) of an alternative name in the gene description, beyond the systematic yeast gene name.
To further test this, we analyzed a set of 213 pairs of predicted genes in S.cerevisiae, previously identified as problematic (46) because of overlap in predicted CDS, a rare phenomenon in eukaryotes. Our analysis included only pairs where one of the genes was named and where the predicted genes occurred on opposite strands. We tested our ability to discriminate the named gene from the unnamed gene based on putative 3′-processing signals, under the assumption that only the named gene in each pair was valid. Initial classification was made based on the difference in the maximum scores [r = S(x)max,named – S(x) max,unnamed], setting r > 0.3 correct, 0.3 > r > –0.3 indeterminate and r < –0.3 incorrect. This classification resulted in 150 (70.4%) correct, 30 (14.1%) incorrect and 33 (15.5%) indeterminate calls. On closer examination of the 63 pairs initially classified as either incorrect or indeterminate, we found many cases where the presence of a 3′-processing signal for the unnamed gene could be explained by the presence of a third nearby gene. Re-classification following elimination of such signals resulted in 183 (85.9%) correct, 13 (6.1%) incorrect, and 17 (8.0%) indeterminate calls. In an intriguing case of incorrect classification, the overlapping gene pair YHR022C and ECM12 (YHR021W-A) had r = –2.2, indicating an extreme preference for the unnamed gene YHR022C. A search for homologs of the respective protein sequences revealed that YHR022C is probably a GTPase protein, while ECM12 has no known homologs. We believe that, in this case, the ‘incorrect’ classification based on 3′-processing sequences is a sign of incorrect annotation of the genes. The complete listing of all compared pairs of overlapping genes is available at our website.
Predicted 3′-processing sites internal to the CDS can indicate either regulatory alternative 3′-processing of the transcript or false positive predictions of our model. Given the analysis of genes with known internal sites (such as RNA14, Fig. 4B), it is likely that some of these predicted sites are genuine. The number of counted internal peaks is, of course, strongly dependent on the value set for the threshold. Table 1 summarizes the results of this analysis. With a threshold S(x)threshold = 3, we found that 1969 (31.3%) of the ORFs investigated had no internal peaks. If the threshold is raised to 3.8 (the value at which the estimated false positive rate is ∼1 in 1000), there are 4870 (77.5%) genes with no internal sites. The expected total number of internal peaks was computed by multiplying our estimated false positive rate by the total number of coding bases in yeast (8.9 × 106). Note that for all threshold values there are approximately three times fewer internal peaks above threshold counted than would be expected based on a second order Markov chain, possibly indicating a suppression of 3′-processing control sequences within the CDS.
Table 1. Summary of potential internal 3′-processing sites in S.cerevisiae pre-mRNA.
| S(x) threshold value | Genes with no internal sites | Total number of internal sites | Expected number of internal sites |
|---|---|---|---|
| 2 | 235 | 141 469 | 448 392 |
| 2.5 | 744 | 69 090 | 190 009 |
| 3 | 1969 | 23 476 | 69 886 |
| 3.5 | 3630 | 7784 | 21 695 |
| 3.8 | 4870 | 3047 | 8935 |
| 4 | 5369 | 1786 | 5468 |
In our initial genomic analysis of the yeast ESTs, we identified several ESTs that indicated poly(A) tails internal to the CDS that could not be explained through any of the systematic problems with EST creation (11). Of particular interest was the BAP2 gene, which has a 1939 nt CDS. Three ESTs aligned to the genome identically, in a position that would indicate a 3′-processing site at position 635 in the CDS. Figure 4C shows our analysis for BAP2, along with the 3′-processing sites indicated by EST matches. (There was one EST indicating a full-length coding sequence.) As can be seen, the prediction has a peak very close to the internal EST locations. Based on the combination of the EST evidence and the predictions of our methods, we believe that this is a regulatory alternative 3′-processing site, and not a false positive prediction.
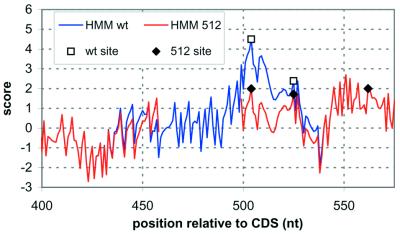
As a final test of our ability to identify 3′-processing sites, we analyzed the effects of mutation on 3′-processing likelihood. Many of the earliest reported mutagenesis experiments were on the CYC1 test system (13,42,47–51). Figure 7 shows a comparison of 3′-processing likelihoods for wild-type CYC1 and the CYC1-512 mutant, which had a 38 nt deletion that drastically reduced 3′-processing efficiency. The reduction in S(x) for CYC1-512 represents the loss of efficiency rather well. The CYC1-512 mutant, while greatly reduced in efficiency, did still produce a few processed transcripts at positions as shown in Figure 7. Interestingly, the three sites still correspond to local maxima, though with greatly reduced S(x). Similar tests have been made on other reported mutations, with varying results. These are available for viewing at our website.
Figure 7.
A comparison of the 3′-processing likelihood for wild-type yeast CYC1 (blue line), and the mutated form CYC1-512 (red line). CYC1-512 has a 38 nt deletion (from position 461 to 499) that greatly reduced 3′-processing efficiency. A gap is inserted into the plot for the CYC1-512 likelihood plot to keep the downstream sequences, and thus the processing sites, correctly aligned. Squares, experimentally determined 3′-processing sites in CYC1; diamonds, the experimentally measured positions in CYC1-512.
DISCUSSION
We have developed a DSM/HMM method for identification of putative yeast 3′-processing sites, based upon the idea that the 3′-processing control elements have predictable positioning and sequence content. The exact structure of the HMM was created manually from a merging of multiple sources of information, including statistical studies of putative element positioning and previously published reports. It is assumed that these elements act in a cooperative fashion, implying no absolute requirement for the sequence of any single element. While not required, it is assumed that the different 3′-processing elements interact with distinct components of the 3′-processing complex (see figure 3 in ref..52).
As shown in Figure 5, the choice of threshold for assessing the predictions is a trade-off between sensitivity and specificity. For threshold value 3 and higher, the false positive rate approaches zero, however, the sensitivity, even allowing up to a 10 base error in positioning, also drops. Any sequence with S(x) > 4.0 clearly is extremely unlikely to arise randomly, and is therefore a strong prediction. At lower threshold values, the prediction becomes increasingly tenuous, as the possibility of random occurrences rises.
As stated in the Introduction, the ability to identify likely 3′-processing sites has the potential to improve gene predictions. In the inset to Figure 6 some of the predicted genes for S.cerevisiae seemingly lack a 3′-processing control sequence, possibly indicating a non-functional ORF. Our analysis of the pairs of overlapping ORFs demonstrated this more clearly. Using the comparison of strength of putative 3′-processing signals for these pairs of genes, we were able to clearly identify the named gene in 86% of the test cases. In addition, our analysis of the putative 3′-processing sites has called into question the relative merits of the pair of overlapping genes ECM12 and YHR022C.
Our analysis can point to alternative 3′-processing sites. Of particular interest is RNA14 (Fig. 4B), which is the yeast homolog of the 77 kDa subunit of cleavage stimulation factor (CstF-77k) and the Drosophila melanogaster protein suppressor of forked Su(f), a known component of the 3′-processing machinery. Studies of the fruit fly gene (53) have indicated a negative self-regulatory effect, in which excessive amounts of Su(f) protein activate an alternative 3′-processing site within the coding sequence that results in a truncated, non-functional transcript. In yeast, the alternative sites are differentially activated with changes in growth conditions (20). The alternative site in the fruit fly sequence is intronic, whereas in yeast, the protein is encoded by a single exon. The existence of alternative 3′-processing sites inside the coding sequence for the same gene in different organisms brings up the intriguing possibility of a conserved regulatory mechanism. Searches for similar phenomena in other organisms revealed ESTs that indicated a truncated version of the human CstF-77k mRNA. We also analyzed the sequences of the other known components (9) of the 3′-processing machinery, and found evidence for potential truncated transcripts in CLP1, YSH1, YHH1 and PCF11. In addition, RNA15 and PTA1 have evidence of regulatory alternative 3′-processing sites in the 3′-UTR. The analyses of all 3′-processing genes are available at our website.
In our analysis of the complete set of known and predicted yeast genes, the distribution of the predicted maxima (corresponding to the most likely 3′-processing site) implies more genes with long UTRs than were found through EST analysis (Fig. 6). However, we believe that there are reasons that mRNA 3′-processing sites would display a bias towards shorter UTRs. In the presence of multiple possible sites (a relatively common occurrence in yeast), sites that occur closer to the stop codon have a competitive advantage, since they are available for interaction with the 3′-processing factors sooner. ‘Weaker’ sites (as defined by our analysis) can win out over ‘stronger’ sites by arising first.
Our model appears to have a number of false positive predictions (dependent on the threshold value) due to inhibitory sequences occurring between control elements. Mutagenesis experiments previously demonstrated that the local sequence environment could significantly inhibit sequences that were elsewhere known to be functional (13,49). The model currently has no ability to penalize such inhibitory sequences. We are investigating more sophisticated background models that may improve this performance.
Our model is also limited by the fact that we are explicitly modeling sequence elements that define all sites in our training set. There are clearly differences in individual sites, as demonstrated by previously reported instances (20,21,54,55) of regulatory alternative 3′-processing sites. It is a distinct possibility that the poor predictions at position 1500 of RNA14 (a known alternative 3′-processing site) can be explained by regulatory differences in the sequences. A larger training set would give us the ability to generate separate models for regulatory alternative sites. This is not available for yeast, but as we expand our investigations to other organisms with larger amounts of training data, such an approach will become possible.
The ever-increasing availability of complete genomic sequences will provide both new data and new systems to study. The tools that we have developed and presented here can be applied to any regulatory sequences that satisfy the requirements defined above: a relatively large data set for training and a regular, predictable positioning of the control elements relative to the processing site.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
[Supplementary Data]
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank the scientists of CAB and BMERC at Boston University, especially Charles Cantor, for continued support and guidance. Jim White, Scott Mohr and Martin Frith provided critical reviews of the manuscript. J.H.G. is partially funded by Sequenom, Inc., and by NSF/KDI grant MCB-9980088. T.F.S. and J.H.G. are supported by NHLBI grant U01 HL66678.
REFERENCES
- 1.Dandekar T., Beyer,K., Bork,P., Kenealy,M.R., Pantopoulos,K., Hentze,M., Sonntag-Buck,V., Flouriot,G., Gannon,F. and Schreiber,S. (1998) Systematic genomic screening and analysis of mRNA in untranslated regions and mRNA precursors: combining experimental and computational approaches. Bioinformatics, 14, 271–278. [DOI] [PubMed] [Google Scholar]
- 2.Hughes J.D., Estep,P.W., Tavazoie,S. and Church,G.M. (2000) Computational identification of _cis_-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J. Mol. Biol., 296, 1205–1214. [DOI] [PubMed] [Google Scholar]
- 3.McGuire A.M., Hughes,J.D. and Church,G.M. (2000) Conservation of DNA regulatory motifs and discovery of new motifs in microbial genomes. Genome Res., 10, 744–757. [DOI] [PubMed] [Google Scholar]
- 4.Wasserman W.W., Palumbo,M., Thompson,W., Fickett,J.W. and Lawrence,C.E. (2000) Human–mouse genome comparisons to locate regulatory sites. Nature Genet., 26, 225–228. [DOI] [PubMed] [Google Scholar]
- 5.McCue L., Thompson,W., Carmack,C., Ryan,M.P., Liu,J.S., Derbyshire,V. and Lawrence,C.E. (2001) Phylogenetic footprinting of transcription factor binding sites in proteobacterial genomes. Nucleic Acids Res., 29, 774–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wasserman W.W. and Fickett,J.W. (1998) Identification of regulatory regions which confer muscle-specific gene expression. J. Mol. Biol., 278, 167–181. [DOI] [PubMed] [Google Scholar]
- 7.GuhaThakurta D. and Stormo,G.D. (2001) Identifying target sites for cooperatively binding factors. Bioinformatics, 17, 608–621. [DOI] [PubMed] [Google Scholar]
- 8.Guo Z. and Sherman,F. (1996) 3′-end-forming signals of yeast mRNA. Trends Biochem. Sci., 21, 477–481. [DOI] [PubMed] [Google Scholar]
- 9.Zhao J., Hyman,L. and Moore,C. (1999) Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev., 63, 405–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keller W. and Minvielle-Sebastia,L. (1997) A comparison of mammalian and yeast pre-mRNA 3′-end processing. Curr. Opin. Cell Biol., 9, 329–336. [DOI] [PubMed] [Google Scholar]
- 11.Graber J.H., Cantor,C.R., Mohr,S.C. and Smith,T.F. (1999) Genomic detection of new yeast pre-mRNA 3′-end-processing signals. Nucleic Acids Res., 27, 888–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Helden J., del Olmo,M. and Perez-Ortin,J.E. (2000) Statistical analysis of yeast genomic downstream sequences reveals putative polyadenylation signals. Nucleic Acids Res., 28, 1000–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Russo P., Li,W.Z., Guo,Z. and Sherman,F. (1993) Signals that produce 3′ termini in CYC1 mRNA of the yeast Saccharomyces cerevisiae. Mol. Cell. Biol., 13, 7836–7849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barabino S.M., Ohnacker,M. and Keller,W. (2000) Distinct roles of two Yth1p domains in 3′-end cleavage and polyadenylation of yeast pre-mRNAs. EMBO J., 19, 3778–3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gross S. and Moore,C.L. (2001) Rna15 interaction with the a-rich yeast polyadenylation signal is an essential step in mRNA 3′-end formation. Mol. Cell. Biol., 21, 8045–8055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helmling S., Zhelkovsky,A. and Moore,C.L. (2001) Fip1 regulates the activity of Poly(A) polymerase through multiple interactions. Mol. Cell. Biol., 21, 2026–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dichtl B. and Keller,W. (2001) Recognition of polyadenylation sites in yeast pre-mRNAs by cleavage and polyadenylation factor. EMBO J., 20, 3197–3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beyer K., Dandekar,T. and Keller,W. (1997) RNA ligands selected by cleavage stimulation factor contain distinct sequence motifs that function as downstream elements in 3′-end processing of pre-mRNA. J. Biol. Chem., 272, 26769–26779. [DOI] [PubMed] [Google Scholar]
- 19.Graber J.H., Cantor,C.R., Mohr,S.C. and Smith,T.F. (1999) In silico detection of control signals: mRNA 3′-end-processing sequences in diverse species. Proc. Natl Acad. Sci. USA, 96, 14055–14060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sparks K.A. and Dieckmann,C.L. (1998) Regulation of poly(A) site choice of several yeast mRNAs. Nucleic Acids Res., 26, 4676–4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoopes B.C., Bowers,G.D. and DiVisconte,M.J. (2000) The two Saccharomyces cerevisiae SUA7 (TFIIB) transcripts differ at the 3′-end and respond differently to stress. Nucleic Acids Res., 28, 4435–4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahadevan S., Raghunand,T.R., Panicker,S. and Struhl,K. (1997) Characterisation of 3′ end formation of the yeast HIS3 mRNA. Gene, 190, 69–76. [DOI] [PubMed] [Google Scholar]
- 23.Burge C.B. and Karlin,S. (1998) Finding the genes in genomic DNA. Curr. Opin. Struct. Biol., 8, 346–354. [DOI] [PubMed] [Google Scholar]
- 24.Burset M. and Guigo,R. (1996) Evaluation of gene structure prediction programs. Genomics, 34, 353–367. [DOI] [PubMed] [Google Scholar]
- 25.Guigo R., Agarwal,P., Abril,J.F., Burset,M. and Fickett,J.W. (2000) An assessment of gene prediction accuracy in large DNA sequences. Genome Res., 10, 1631–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guigo R. (1997) Computational gene identification: an open problem. Comput. Chem., 21, 215–222. [DOI] [PubMed] [Google Scholar]
- 27.Tabaska J.E. and Zhang,M.Q. (1999) Detection of polyadenylation signals in human DNA sequences. Gene, 231, 77–86. [DOI] [PubMed] [Google Scholar]
- 28.Tabaska J.E., Davuluri,R.V. and Zhang,M.Q. (2001) Identifying the 3′-terminal exon in human DNA. Bioinformatics, 17, 602–607. [DOI] [PubMed] [Google Scholar]
- 29.Kondrakhin Yu V., Shamin,V.V. and Kolchanov,N.A. (1994) Construction of a generalized consensus matrix for recognition of vertebrate pre-mRNA 3′-terminal processing sites. Comput. Appl. Biosci., 10, 597–603. [DOI] [PubMed] [Google Scholar]
- 30.Matis S., Xu,Y., Shah,M., Guan,X., Einstein,J.R., Mural,R. and Uberbacher,E. (1996) Detection of RNA polymerase II promoters and polyadenylation sites in human DNA sequence. Comput. Chem., 20, 135–140. [DOI] [PubMed] [Google Scholar]
- 31.Salamov A.A. and Solovyev,V.V. (1997) Recognition of 3′-processing sites of human mRNA precursors. Comput. Appl. Biosci., 13, 23–28. [DOI] [PubMed] [Google Scholar]
- 32.Rothnie H.M. (1996) Plant mRNA 3′-end formation. Plant Mol. Biol., 32, 43–61. [DOI] [PubMed] [Google Scholar]
- 33.Rabiner L.R. (1989) A tutorial on hidden Markov models and select applications in speech recognition. Proc. Inst. Electrical Electronics Eng., 77, 257–286. [Google Scholar]
- 34.White J.V. (1988) Chapter 10: Modeling and filtering for discretely valued time series. In Spall,J.C. (ed.), Bayesian Analysis of Time Series and Dynamic Models. Marcel Dekker, New York, pp. 255–283.
- 35.White J.V., Stultz,C.M. and Smith,T.F. (1994) Protein classification by stochastic modeling and optimal filtering of amino-acid sequences. Math. Biosci., 119, 35–75. [DOI] [PubMed] [Google Scholar]
- 36.Stultz C.M., White,J.V. and Smith,T.F. (1993) Structural analysis based on state-space modeling. Protein Sci., 2, 305–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Durbin R., Eddy,S., Krogh,A. and Mitchison,G. (1998) Biological Sequence Analysis: Probabilistic Models of Proteins and Nucleic Acids. Cambridge University Press, Cambridge.
- 38.Lawrence C.E., Altschul,S.F., Boguski,M.S., Liu,J.S., Neuwald,A.F. and Wootton,J.C. (1993) Detecting subtle sequence signals: a Gibbs sampling strategy for multiple alignment. Science, 262, 208–214. [DOI] [PubMed] [Google Scholar]
- 39.Grundy W.N., Bailey,T.L., Elkan,C.P. and Baker,M.E. (1997) Meta-MEME: motif-based hidden Markov models of protein families. Comput. Appl. Biosci., 13, 397–406. [DOI] [PubMed] [Google Scholar]
- 40.Hartigan J.A. (1975) Clustering Algorithms. John Wiley & Sons, New York.
- 41.Pearson K. (1896) Regression, heredity and panmixia. Phil. Trans. R. Soc. Lon., Ser. A, 187, 253–318. [Google Scholar]
- 42.Guo Z. and Sherman,F. (1996) Signals sufficient for 3′-end formation of yeast mRNA. Mol. Cell. Biol., 16, 2772–2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hyman L.E. and Moore,C.L. (1993) Termination and pausing of RNA polymerase II downstream of yeast polyadenylation sites. Mol. Cell. Biol., 13, 5159–5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chervitz S.A., Hester,E.T., Ball,C.A., Dolinski,K., Dwight,S.S., Harris,M.A., Juvik,G., Malekian,A., Roberts,S., Roe,T. et al. (1999) Using the Saccharomyces Genome Database (SGD) for analysis of protein similarities and structure. Nucleic Acids Res., 27, 74–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cherry J.M., Adler,C., Ball,C., Chervitz,S.A., Dwight,S.S., Hester,E.T., Jia,Y., Juvik,G., Roe,T., Schroeder,M. et al. (1998) SGD: Saccharomyces Genome Database. Nucleic Acids Res., 26, 73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith T.F. and Zhang,X. (1997) The challenges of genome sequence annotation or ‘the devil is in the details’. Nat. Biotechnol., 15, 1222–1223. [DOI] [PubMed] [Google Scholar]
- 47.Guo Z. and Sherman,F. (1995) 3′-end-forming signals of yeast mRNA. Mol. Cell. Biol., 15, 5983–5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo Z., Russo,P., Yun,D.F., Butler,J.S. and Sherman,F. (1995) Redundant 3′ end-forming signals for the yeast CYC1 mRNA. Proc. Natl Acad. Sci. USA, 92, 4211–4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Russo P., Li,W.Z., Hampsey,D.M., Zaret,K.S. and Sherman,F. (1991) Distinct _cis_-acting signals enhance 3′ endpoint formation of CYC1 mRNA in the yeast Saccharomyces cerevisiae. EMBO J., 10, 563–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Henikoff S., Kelly,J.D. and Cohen,E.H. (1983) Transcription terminates in yeast distal to a control sequence. Cell, 33, 607–614. [DOI] [PubMed] [Google Scholar]
- 51.Henikoff S. and Cohen,E.H. (1984) Sequences responsible for transcription termination on a gene segment in Saccharomyces cerevisiae. Mol. Cell. Biol., 4, 1515–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gavin A.C., Bosche,M., Krause,R., Grandi,P., Marzioch,M., Bauer,A., Schultz,J., Rick,J.M., Michon,A.M., Cruciat,C.M. et al. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature, 415, 141–147. [DOI] [PubMed] [Google Scholar]
- 53.Audibert A. and Simonelig,M. (1998) Autoregulation at the level of mRNA 3′ end formation of the suppressor of forked gene of Drosophila melanogaster is conserved in Drosophila virilis. Proc. Natl Acad. Sci. USA, 95, 14302–14307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mandart E. and Parker,R. (1995) Effects of mutations in the Saccharomyces cerevisiae RNA14, RNA15 and PAP1 genes on polyadenylation in vivo. Mol. Cell. Biol., 15, 6979–6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mandart E. (1998) Effects of mutations in the Saccharomyces cerevisiae RNA14 gene on the abundance and polyadenylation of its transcripts. Mol. Gen. Genet., 258, 16–25. [DOI] [PubMed] [Google Scholar]
- 56.Duvel K. and Braus,G.H. (1999) Different positioning elements select poly(A) sites at the 3′-end of GCN4 mRNA in the yeast Saccharomyces cerevisiae. Nucleic Acids Res., 27, 4751–4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aranda A., Perez-Ortin,J.E., Moore,C. and del Olmo,M. (1998) The yeast FBP1 poly(A) signal functions in both orientations and overlaps with a gene promoter. Nucleic Acids Res., 26, 4588–4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Springer C., Valerius,O., Strittmatter,A. and Braus,G.H. (1997) The adjacent yeast genes ARO4 and HIS7 carry no intergenic region. J. Biol. Chem., 272, 26318–26324. [DOI] [PubMed] [Google Scholar]
- 59.Duvel K., Egli,C.M. and Braus,G.H. (1999) A single point mutation in the yeast TRP4 gene affects efficiency of mRNA 3′ end processing and alters selection of the poly(A) site. Nucleic Acids Res., 27, 1289–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heidmann S., Obermaier,B., Vogel,K. and Domdey,H. (1992) Identification of pre-mRNA polyadenylation sites in Saccharomyces cerevisiae. Mol. Cell. Biol., 12, 4215–4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplementary Data]