Evidence of Two Lyssavirus Phylogroups with Distinct Pathogenicity and Immunogenicity (original) (raw)
Abstract
The genetic diversity of representative members of the Lyssavirus genus (rabies and rabies-related viruses) was evaluated using the gene encoding the transmembrane glycoprotein involved in the virus-host interaction, immunogenicity, and pathogenicity. Phylogenetic analysis distinguished seven genotypes, which could be divided into two major phylogroups having the highest bootstrap values. Phylogroup I comprises the worldwide genotype 1 (classic Rabies virus), the European bat lyssavirus (EBL) genotypes 5 (EBL1) and 6 (EBL2), the African genotype 4 (Duvenhage virus), and the Australian bat lyssavirus genotype 7. Phylogroup II comprises the divergent African genotypes 2 (Lagos bat virus) and 3 (Mokola virus). We studied immunogenic and pathogenic properties to investigate the biological significance of this phylogenetic grouping. Viruses from phylogroup I (Rabies virus and EBL1) were found to be pathogenic for mice when injected by the intracerebral or the intramuscular route, whereas viruses from phylogroup II (Mokola and Lagos bat viruses) were only pathogenic by the intracerebral route. We showed that the glycoprotein R333 residue essential for virulence was naturally replaced by a D333 in the phylogroup II viruses, likely resulting in their attenuated pathogenicity. Moreover, cross-neutralization distinguished the same phylogroups. Within each phylogroup, the amino acid sequence of the glycoprotein ectodomain was at least 74% identical, and antiglycoprotein virus-neutralizing antibodies displayed cross-neutralization. Between phylogroups, the identity was less than 64.5% and the cross-neutralization was absent, explaining why the classical rabies vaccines (phylogroup I) cannot protect against lyssaviruses from phylogroup II. Our tree-axial analysis divided lyssaviruses into two phylogroups that more closely reflect their biological characteristics than previous serotypes and genotypes.
The etiologic agent of rabies encephalitis was believed to be unique until 1956, when the first rabies-related viruses were isolated in Africa and Europe (for reviews, see references 1, 26, and 43). To account for this increasing diversity, the cross-reactivity of internal antigens (the ribonucleoprotein complex) was used to identify the Lyssavirus genus within the Rhabdoviridae family (44). Virus-neutralizing antibodies (VNAbs), which recognize the membrane glycoprotein (G), subdivided the genus into three serotypes (44), and monoclonal antibody studies further refined the classification into four serotypes (10). Comparison of the viral nucleoprotein gene (N) delineated six genotypes: four matched the previously described serotypes (1, Rabies virus; 2, Lagos bat virus; 3, Mokola virus; and 4, Duvenhage virus), and two additional genotypes were created for European bat lyssavirus (EBL) type 1 (5, EBL1) and type 2 (6, EBL2) (6). Finally, an Australian bat lyssavirus (ABL) responsible for human cases (23, 24) was proposed to inaugurate a seventh new genotype, which is closely related to genotype 1 (22).
The worldwide Rabies virus (genotype 1) is found in various domestic and wild mammals, mainly carnivores, but also in American bats (33, 47). Rabies-related viruses have so far been isolated in limited geographic regions. Lagos Bat, Mokola, and Duvenhage viruses have been isolated in subequatorial and southern African countries, mostly from frugivorous megachiropterans (Eidolon and Epomophorus spp.), micromammals, and insectivorous microchiropterans (Miniopterus and Nycteris spp.), respectively (26). EBL1 and EBL2 are widely distributed in Europe, from Russia to Spain, mainly in coastal regions (43). They preferentially infect insectivorous microchiropterans of Eptesicus and Myotis spp., respectively (1, 5). ABL was isolated along the Australian East Coast, mainly from frugivorous megachiropterans (Pteropus spp.) (24), but also from insectivorous microchiropterans (23).
Virus strains of commercially available vaccines belong to genotype 1. Their spectrum of protection against the rabies-related viruses is variable (25, 31). Pasteur virus (PV) elicits VNAbs against genotypes 1, 4, 5, and 6 but fails to protect against genotypes 2 and 3 (3, 16, 59). Differences also exist in the pathogenicity of virus strains; genotypes 1 and 5 are pathogenic for mice by the peripheral route, while genotype 3 is not (37). However, all genotypes except genotype 2 have caused human and/or animal deaths in nature.
The rabies virus transmembrane glycoprotein is involved in tropism and pathogenicity. It is the main protecting antigen, inducing a complete immune response with the production of VNAbs (30, 58). The mature glycoprotein without its cleaved signal peptide (SP) forms a trimer (19). It is composed of an endodomain (ENDO), which interacts with internal proteins (9, 35, 57); a transmembrane (TM) region, and an ectodomain (ECTO), protruding from the viral membrane. The ectodomain carries B- and T-cell antigenic sites (4, 28) and the regions responsible for receptor recognition (32, 51, 54, 55) and membrane fusion (13). Several amino acid residues important for virulence were identified in the glycoprotein (8, 12, 38, 39, 45).
Because of these attributes, we compared the glycoprotein sequence in representative lyssaviruses from the seven genotypes and identified two phylogroups. We evaluated the biological significance of this phylogenetic grouping by investigating immunological and pathological properties in lyssaviruses. This is the first global approach to studying the diversity in lyssaviruses that combines genetic, pathogenicity, and immunogenicity studies.
MATERIALS AND METHODS
Viruses.
Sixteen lyssaviruses representing the seven genotypes (minimum of two per genotype except genotype 7) were included in this study (Table 1). Fifteen of them were wild isolates, and one was a vaccine strain (genotype 1). Of these isolates, 11 were previously described (5, 6, 22, 34, 41), and 5 were received from collaborative laboratories. Bob Swanepoel (National Institute for Virology, Johannesburg, South Africa), Donald Lodmell (Rocky Mountain Laboratory, Hamilton, Mont.), and Hervé Bourhy (Pasteur Institute, Paris, France) generously provided isolates from South Africa (LagSAF1, LagSAF2, and MokSAF), Montana (USA7-BT), and the Central African Republic (LagCAR), respectively. Isolates consisted of either the original infected brain or suckling mouse brain after limited passages.
TABLE 1.
Isolates studied
| Virus | Genotype | Origin | Isolation | Yr of isolation | Strain | Sourceb | Accession no.a |
|---|---|---|---|---|---|---|---|
| PV | 1 | France | Vaccine strain | 1882 | PV | IPP | A14671* |
| USA7-BT | 1 | USA | Bat (Myotis sp.) | 1982 | bat#01 | RML | AF298141 |
| ABL | 7 | Australia | Bat (Pteropus alecto) | 1997 | ABL | CSIRO | AF006497* |
| EBL1POL | 5 | Poland | Bat (Eptesicus serotinus) | 1985 | 8615POL | IPP | AF298142 |
| EBL1FRA | 5 | France | Bat (Eptesicus serotinus) | 1989 | 8918FRA | IPP | AF298143 |
| EBL2FIN | 6 | Finland | Human bitten by a bat | 1986 | 9007FIN | IPP | AF298144 |
| EBL2HOL | 6 | Holland | Bat (Myotis dasycneme) | 1986 | 9018HOL | IPP | AF298145 |
| DuvSAF1 | 4 | South Africa | Human bitten by a bat | 1970 | 86132SA | IPP | AF298146 |
| DuvSAF2 | 4 | South Africa | Bat (Miniopterus schreibersii) | 1981 | 9020SA | IPP | AF298147 |
| LagNGA | 2 | Nigeria | Bat (Eidolon helvum) | 1956 | 8619NGA | IPP | AF298148 |
| LagCAR | 2 | Central African Republic | Bat (Micropterus pusillus) | 1974 | 8620CAR | IPP | AF298149 |
| LagSAF1 | 2 | South Africa | Bat (Epomophorus wahlbergi) | 1980 | OP640/80 | OVI | AF298225 |
| LagSAF2 | 2 | South Africa | Bat (Epomophorus wahlbergi) | 1980 | OP1248/80 | OVI | AF298226 |
| MokSAF | 3 | South Africa | Cat | 1970 | OP700/70 | OVI | AF298227 |
| MokZIM | 3 | Zimbabwe | Cat | 1981 | ? | IPP | S59447* |
| MokETH | 3 | Ethiopia | Cat | 1990 | Eth-16 | ? | U17064* |
RNA extraction to sequence analysis.
Total RNA extraction, cDNA synthesis, PCR amplification, and sequencing were done as described by Sacramento et al. (42) with minor modifications. Several amplicons were sequenced automatically using the cycling and dye terminator technology in an ABI 346 analyzer. Only positive-stranded primers were used for cDNA synthesis from the negative-stranded viral genome, and the consensus sequence of the G genes was determined without subsequent cloning, eliminating the influence of individual variabilities on the observed genetic polymorphism. For sequence determination, we used 46 oligodeoxynucleotide primers (sequences available upon request) located between nucleotide positions 2901 (upstream matrix gene M2) and 5543 (downstream polymerase gene L) with reference to the PV genome (52, 53). Their structure was deduced either from conserved coding regions between lyssaviruses or from genotype-specific noncoding regions.
Sequence analysis and phylogenetic studies were performed using various packages: GCG version 9.1 (21), ClustalW (50), Phylip version 3.5 (17), and PAUP version 3.1 (49).
Pathogenicity and immunogenicity studies.
BALB/c, C3H, and Swiss mice were purchased from the Centre d'élevage et de Recherche (Janvier, Legenest St. Isle, France). The intracerebral (i.c.) lethal dose 50% (LD50 i.c.) was determined by injecting 30 μl of virus into Swiss male mice (20 g) by the i.c. route. BALB/c and C3H female mice, 6 to 8 weeks old, were injected (100 μl) by the intramuscular (i.m.) route in the thigh with 2 × 105 (PV), 6 × 105 (EBL1FRA), 3 × 107 (MokZIM), or 3 × 105 (LagNGA) LD50 i.c. PV, EBL1FRA, and MokZIM were used as purified viruses, and LagNGA was used as a concentrated infected BHK-21 cell supernatant as previously described (37).
Cross-neutralization was determined as described by Bahloul et al. (3). Briefly, mice were immunized i.m. with 50 μg of plasmid DNA encoding the glycoprotein of either PV rabies virus or Mokola virus. The capacity of mouse sera to neutralize different lyssaviruses was evaluated 39 days postimmunization by the rapid fluorescent focus inhibition test (48). A VNAb titer higher than 0.5 IU/ml reflects positive seroconversion. Average VNAb titers from three mice were compared to the percent G ectodomain amino acid identity (% ECTO aa identity).
RESULTS
Diversity of Lyssavirus glycoproteins.
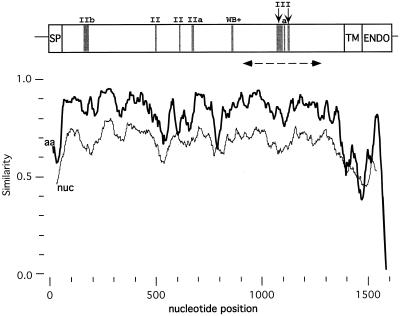
To account for the genetic variability within and between Lyssavirus genotypes, at least two isolates per genotype (except genotype 7) were studied. These isolates were obtained over a 40-year period. Four G gene sequences (indicated in Table 1) were retrieved from the GenBank database. Nine new G gene sequences were determined from the G to the L mRNA transcription start signals (≅2,100 bases). The sequence covers the entire G coding sequence (≅1,575 bases), including the noncoding Ψ region (≅450 bases). Using ClustalW, the glycoprotein coding region was easily aligned among all lyssaviruses. However, the 3′ noncoding region (Ψ) exhibited significant alignments within but not between genotypes (data not shown). Figure 1 shows the nucleotide and amino acid similarity profiles resulting from alignment of the G gene coding region of lyssaviruses of the seven genotypes. Figure 2 shows the alignment of the deduced amino acid sequences of 13 full-length glycoproteins from the seven genotypes and three partial glycoproteins from Lagos bat (LagSAF1 and LagSAF2) and Mokola (MokSAF) viruses. Both figures clearly show that the G ectodomain is more conserved (75% average aa identity) than the SP (35%), the TM (50%), and ENDO (40%) regions, where the hydrophobic and hydrophilic nature, not a specific sequence, should be conserved. The same profile was observed when only genotype 1 isolates were compared (unpublished results), which confirms the functional importance of the G ectodomain for lyssaviruses. In this ectodomain, the nucleotide conservation is globally lower than the amino acid conservation (Fig. 1), and the ratio between synonymous and nonsynonymous substitutions (ds/dn) is clearly greater than 1 (data not shown). This result suggests that positive selection is not acting on this region.
FIG. 1.
Lyssavirus similarity profile along the G gene coding region. Nucleotide (nuc, thin line) and amino acid (aa, bold line) sequence similarity profiles among 13 complete Lyssavirus glycoproteins are shown (similarityplot program of GCG; window, 100 nucleotides or 50 aa, step 1). SP, signal peptide; TM, transmembrane domain; ENDO, endodomain. Gray boxes indicate two major antigenic sites (II and III), the minor antigenic site (a), and the neutralizing linear epitope VI (WB+, Western blot positive). The discontinuous line with arrowheads indicates the region sequenced in LagSAF1, LagSAF2, and MokSAF (Fig. 2).
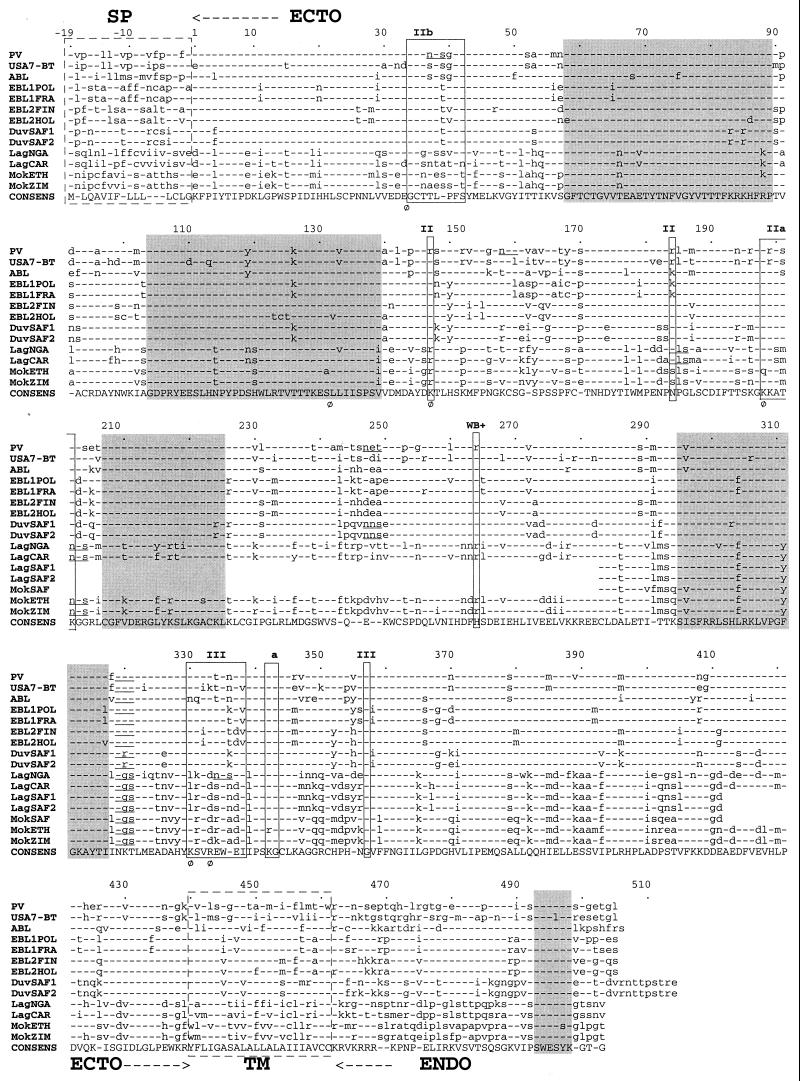
FIG. 2.
Multiple alignment of Lyssavirus glycoproteins. Alignment of complete or partial deduced amino acid sequences of the glycoprotein from genotypes 1 (PV and USA7-BT), 2 (LagNGA, LagCAR, LagSAF1, and LagSAF2), 3 (MokSAF, MokETH, and MokZIM), 4 (DuvSAF1 and DuvSAF2), 5 (EBL1POL and EBL1FRA), 6 (EBL2FIN and EBL2HOL), and 7 (ABL). Dashes indicate amino acids agreeing with the consensus sequence (CONSENS). Boxes with discontinuous lines show the hydrophobic signal peptide (SP) and transmembrane domain (TM). ECTO, ectodomain; ENDO, endodomain. Boxes with continuous lines outline the main antigenic sites and epitopes. Underlined NX(S/T) motifs in the ectodomain are potential N-glycosylation sites. Phi (Φ) indicates residues involved in pathogenicity. Gray boxes show the five most conserved blocks.
The glycoprotein of lyssaviruses consists of between 503 and 514 aa. This size heterogeneity exclusively affects the ENDO (42 to 53 aa) region, but not the SP (19 aa), the ECTO (439 aa), or the TM (22 aa) region. Size and sequence diversity of ENDO could influence the interaction of the glycoprotein with internal viral and/or cellular proteins (35). It is noteworthy in this context that a small stretch in the ENDO region (positions 493 to 498) is strongly conserved (Fig. 2).
Cysteine residues are strongly conserved, indicating constraints on the ectodomain structure. Four conserved blocks are distinguishable within the ectodomain and corresponded to segments 58 to 89, 104 to 139, 207 to 225, and 295 to 317 (Fig. 2). These blocks are not part of the antigenic sites or epitopes (34 to 42, 198 to 202, 264, and 330 to 338) (4, 28), which are among the most variable regions. Likewise, these blocks are not within the neurotoxin-like loop domain (175 to 203), which binds to the nicotinic acetylcholine receptor (32). However, one block (104 to 139) is within the domain (102 to 179) proposed to promote the fusion of the viral and the endosomal membranes, thus releasing the RNP into the cytoplasm (13). The potential N-glycosylation site at position 319 is the only one conserved in all genotypes and is located in the most conserved region with the vesicular stomatitis virus glycoprotein (40). This is the only site present in the G ectodomain of the European (EBL1 and EBL2), Australian (ABL), and American (USA7-BT) bat lyssaviruses. Therefore, it may constitute the minimal glycosylation site sufficient to ensure adequate maturation and routing of the glycoprotein through the endoplasmic reticulum and Golgi apparatus (18, 46). Additional potential glycosylation sites have been identified at positions 202 (genotype 3), 247 (genotype 4), 184, 202, and 334 (genotype 2), and 37 (genotype 1) (unpublished results). It is not known whether these potential sites are used in vivo. However, glycoproteins from rabies PV and Mokola viruses having, respectively, four (37, 157, 247, and 319) and two (202 and 319) potential sites were glycosylated to the same degree (personal observation), suggesting that some of these sites are not used. The effect of glycosylation on the antigenicity has been reported previously (11), and one might expect it to interfere in virus-host interactions (2). For instance, some potential glycosylation sites may be located in the major antigenic site II (202, genotypes 2 and 3) or III (334, LagNGA isolate). The glycoprotein palmitoylation site (C461) (20) located at the TM-ENDO junction is conserved in all viruses except the PV strain, which has C460.
Phylogenetic relationships between lyssaviruses.
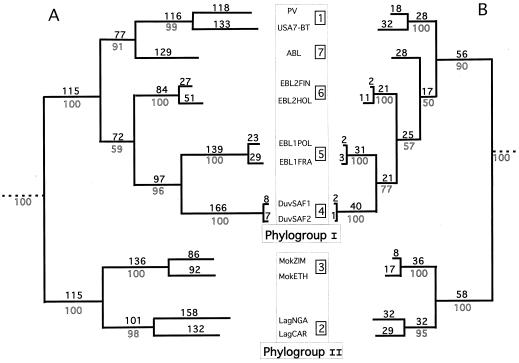
Different phylogeny methods (neighbor joining, maximum likelihood, and parsimony) were tested on different regions of the G gene and produced phylogenetic trees with similar topologies. Figure 3A shows the estimated phylogenetic tree by comparing the ectodomain nucleotide sequences. Seven distinct lineages with significant bootstrap values (>90%) were predicted. Six of them matched the six genotypes already defined by the N gene sequence comparison (6). The seventh lineage (ABL), which branches with genotype 1, corresponds to the recently proposed seventh genotype (22). However, comparison of the ectodomain amino acid sequences (Fig. 3B) revealed that ABL branched with genotypes 4, 5, and 6 (low bootstrap value of 50). This suggests that genotype 7 has an intermediate phylogenetic position between genotype 1 and genotypes 4, 5, and 6.
FIG. 3.
Lyssavirus phylogeny. Estimated rooted phylogenetic trees using nucleotide (A) or amino acid (B) sequences of the G ectodomain. The PAUP program (parsimony) was used with the following options: branch and bound search, bootstrap with 50% majority rule consensus, and collapsed zero-length branches. Outline numbers are bootstrap values of 100 replicates, testing the robustness of their corresponding internal branches. Bold numbers are steps occurring on each branch (branch lengths). Numbers inside squares indicate the seven genotypes. Chandipura and Piry viruses from the Vesiculovirus genus were used as an outgroup.
The main outcome of this phylogenetic analysis is the division of the Lyssavirus genus into two clearly distinct phylogroups supported with the strongest bootstrap values (Fig. 3). Phylogroup I comprised genotypes 1 and 4 to 7, and phylogroup II included genotypes 2 and 3. This grouping was also supported by the ectodomain sequence identity (Table 2). The ectodomain amino acid identity was at least 74% within each phylogroup and less than 64% between phylogroups. The _t_-test showed that pairwise ECTO aa identities were significantly higher within than between phylogroups (P = 0.0004, 99% confidence). Genotypes 4, 5, and 6 are more homogeneous (97 to 98.5% ECTO aa identity) than genotypes 2 and 1, which had the highest intragenotype heterogeneities (86 and 88.5%, respectively). Between genotypes, conservation varies from 61 to 83.5% ECTO aa identity.
TABLE 2.
Nucleotide and amino acid identities from glycoprotein ectodomain pairwise comparisonsa
| Virus (genotype) | % Nucleotide/aa identity | ||||||
|---|---|---|---|---|---|---|---|
| Phylogroup I | Phylogroup II | ||||||
| Rabies virus | ABL | EBL2 | EBL1 | Duvenhage virus | Lagos bat virus | Mokola virus | |
| Rabies virus (1) | 81.5/88.5 | 80.5 | 77.5 | 76.0 | 74.0 | 62.0 | 62.0 |
| ABL (7) | 73.5 | NA/NA | 83.5 | 80.5 | 78.0 | 64.0 | 62.0 |
| EBL2 (6) | 72.0 | 74.5 | 94.0/97.0 | 83.5 | 82.0 | 61.5 | 61.0 |
| EBL1 (5) | 72.0 | 72.0 | 76.0 | 96.0/98.5 | 82.5 | 63.0 | 62.5 |
| Duvenhage virus (4) | 68.5 | 71.0 | 74.0 | 74.5 | 98.0/98.5 | 61.5 | 61.0 |
| Lagos bat virus (2) | 62.0 | 63.5 | 64.0 | 63.5 | 62.5 | 79.0/86.0 | 79.0 |
| Mokola virus (3) | 62.0 | 63.0 | 62.0 | 64.0 | 64.5 | 72.0 | 86.0/94.0 |
Pathogenicity and immunogenicity investigations.
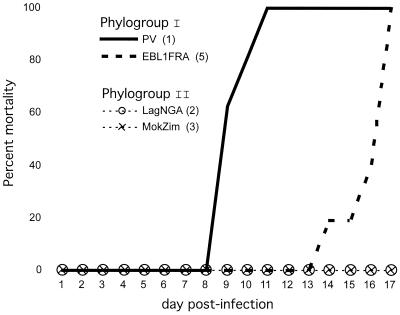
Particular residues of the ectodomain considerably influence viral pathogenicity (Fig. 2). It was shown that the presence of an R333 (or K333) in the glycoprotein is essential for the virulence of the ERA and CVS strains of rabies virus (56). Phylogroup I viruses possess R333, whereas phylogroup II members have an R333D replacement (Fig. 2). Thus, we tested one representative of each of the two genotypes of phylogroup II (MokZIM and LagNGA) to determine whether they were pathogenic to adult BALB/c and C3H mice. PV and EBL1FRA were used as controls for phylogroup I (Fig. 4). The four viruses were fully pathogenic when injected by the i.c. route. However, when 105 to 107 LD50 i.c. were injected by the i.m. route, phylogroup I viruses were pathogenic but phylogroup II viruses were not. Thus as predicted, the natural absence of R333 has a negative effect on the pathogenicity of lyssaviruses.
FIG. 4.
Pathogenicity of lyssaviruses by the i.m. route. Eight BALB/c mice were injected by the i.m. route in the thigh with 105 to 107 LD50 i.c. of PV, EBL1FRA, LagNGA, or MokZIM. For each virus, the genotype number is indicated in parentheses. Results are expressed as the percentage of dead animals monitored up to 17 days postinfection.
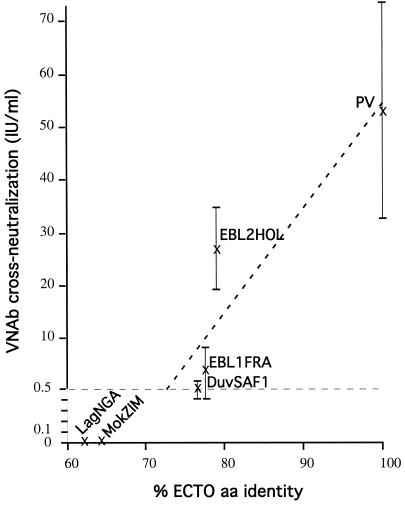
The segregation of lyssaviruses into two phylogroups was also consistent with a previous analysis of serological cross-neutralization in mice after DNA immunization with a plasmid carrying the PV (phylogroup I) or MokZIM (phylogroup II) G gene (3). Another study demonstrated cross-neutralization between genotypes 1 and 7 (22). Lyssavirus genotypes appeared to be immunologically separated in the same two phylogroups. Table 3 shows the variation in mouse serum cross-neutralization levels against representatives of six genotypes (estimated 39 days after i.m. injection of 50 μg of plasmid DNA encoding the G protein) according to the percent ECTO aa identity. Immunization with the PV G gene produced a high titer of VNAbs against viruses from genotypes 1 (53 IU/ml, 100% ECTO aa identity) and 6 (27 IU/ml, 79%) and a significant titer against viruses from genotypes 5 (4.2 IU/ml, 77.5%) and 4 (0.6 IU/ml, 76.5%). However, there was no cross-neutralization against viruses from genotypes 3 and 2 (0 IU/ml, ≤64.5%). Conversely, immunization with the MokZim G gene produced a high titer of VNAbs against genotype 3 virus (48 IU/ml, 100%), a significant titer against genotype 2 virus (2.5 IU/ml, 78.5%), but very weak or no neutralization against viruses from genotypes 1, 4, 5, and 6 (≤0.2 IU/ml, ≤64.5%). Figure 5 shows that in phylogroup I there is a very good correlation (coefficient = 0.92) between the genetic distance from PV (% ECTO aa identity) and the neutralizing capacity of the anti-G PV sera. Anti-Duvenhage virus VNAb titers (0.6 IU/ml) were slightly above the accepted limit for seroconversion (0.5 IU/ml). However, the value increased to 8 IU/ml 160 days after immunization, whereas titers against viruses from phylogroup II remained nonsignificant (≤0.4 IU/ml) (3). Figure 5 suggests that two lyssaviruses will cross-neutralize each other if they have more than 72% ECTO aa identity. All these data indicate that cross-neutralization exists within but not between Lyssavirus phylogroups.
TABLE 3.
Glycoprotein ectodomain amino acid identity versus cross-neutralization.
| Virus (genotype) | Mean VNAb titer (IU/ml) 39 days postimmunization ± SD (% ECTO aa identity) | |||||
|---|---|---|---|---|---|---|
| Phylogroup I | Phylogroup II | |||||
| PV (1) | EBL2 (6) | EBL1 (5) | Duvenhage virus (4) | Lagos bat virus (2) | Mokola virus (3) | |
| PV (1) | 53 ± 21 (100) | 27 ± 7 (79) | 4.2 ± 3.8 (77.5) | 0.6 ± 0.2 (76.5) | 0 (62.5) | 0 (64.5) |
| Mok2ZIM (3) | 0 (64.5) | 0 (60) | 0 (63) | 0.2 ± 0.2 (61) | 2.5 ± 2.5 (78.5) | 48 ± 17 (100) |
FIG. 5.
Curve of ectodomain amino acid identity versus cross-neutralization. We correlated the cross-neutralization (mean ± standard deviation VNAb titers at day 39 post-DNA immunization) and the G ectodomain percent identity (% ECTO aa identity) between PV and four phylogroup I members (PV, EBL1FRA, EBL2HOL, and DuvSAF1) (see Table 3). No significant titers were found against two phylogroup II members (MokZIM and LagNGA). A linear correlation between VNAb cross-neutralization and % ECTO aa identity was observed (dashed oblique line) with a high coefficient (r = 0.92). The horizontal dashed line at 0.5 IU/ml corresponds to the accepted minimal titer for seroconversion. The vertical axis was enlarged between 0 and 0.5 for convenience.
DISCUSSION
Since the first isolation of a rabies-related virus in Africa in 1956, the number of viral variants implicated in rabies encephalitis etiology has increased considerably (1, 26, 43). This led to the creation of the Lyssavirus genus in the Rhabdoviridae family. Serotyping and genotyping are two methods widely used to differentiate between lyssaviruses. Serotyping showed a limitation when EBLs were isolated (5), and genotyping lacks biological significance. The specific objective of this work was to combine phylogenetic analysis with the study of biological (immunopathological) characteristics (Fig. 6). Indeed, we approached the diversity of lyssaviruses by looking at genetic, immunogenic, and pathogenic properties. This three-axis analysis distinguished two genetically and immunogenetically distant phylogroups; one is worldwide and pathogenic for mice by the i.m. route of inoculation, and the other is African and apathogenic by the i.m. route.
FIG. 6.
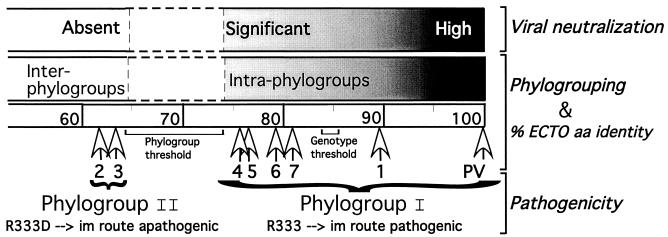
Schematic representation of the three-axis analysis. % ECTO aa identity is a scale of distances where the seven white arrowheads show the positions of the seven genotypes (only their numbers are given) from PV. Phylogrouping is represented by a horizontal box with a directional color gradient from black to gray (intraphylogroup comparisons, ≥74% ECTO aa identity) to white (interphylogroup comparisons, ≤64% ECTO aa identity). The phylogroup and genotype thresholds are shown within ranges from 64 to 74% and 83.5 to 86% ECTO aa identity, respectively. VNAb titer is represented by a horizontal box with a directional color gradient from black (high titer) to gray (significant titer) to white (no protection, about 74% ECTO aa identity). For pathogenicity, the presence of R333 in the glycoprotein of phylogroup I members (PV and genotypes 1, 4, 5, 6, and 7) is an indication of their pathogenicity for mice by the i.m. route, whereas the R333D replacement in the glycoprotein of phylogroup II members (genotypes 2 and 3) concords with their apathogenicity for mice by the i.m. route.
The genetic diversity of lyssaviruses was studied on the glycoprotein, which is crucial for the virus-host interaction, immunogenicity, and pathogenicity. The complete nucleotide sequence of the G gene of representative lyssaviruses was determined (≅2,100 nucleotides). Phylogenetic analysis using different methods and different regions of the G gene gave trees with similar topologies, indicating that evolution has acted uniformly throughout the glycoprotein. Lyssaviruses were segregated into seven main clusters corresponding to the seven genotypes previously noted by comparing the N gene (6, 22). However, they can be divided into two distinct phylogroups, predicted with the strongest bootstrap values. This dichotomy was also supported by the genetic distance; pairwise ECTO aa identities within phylogroups were significantly higher (≥74%) than those between phylogroups (≤64%). The worldwide phylogroup I included genotypes 1, 4, 5, 6, and 7. Phylogroup II comprised genotypes 2 and 3, which are limited to subequatorial and southern African countries. The available data suggested that the genotype and phylogroup thresholds were within the ranges from 86 to 83.5% and 79 to 64% ECTO aa identity, respectively. These values may be refined in the future by studying additional isolates.
We investigated the biological significance of the phylogrouping in relation to the pathogenicity and immunogenicity of the lyssaviruses. When inoculated in adult mice, genotype 1 and 6 viruses (phylogroup I) were pathogenic by both the i.c. and i.m. routes. However, genotype 2 and 3 viruses (phylogroup II) were only pathogenic by the i.c. route. Two positively charged residues within the antigenic site III, K330 and R333, have been shown to considerably influence viral pathogenicity (8, 12, 45). Selected antigenic mutants of rabies virus laboratory strains in which R333 was replaced with another residue (except lysine) were totally apathogenic for adult mice (i.c. and i.m. routes) (56). This was possibly due to a restriction in the type of infected neurons and in the transmission at interneurons (7, 27, 29). An antigenic double mutant (K330N and R333M) was unable to penetrate either motor or sensory neurons (8). Phylogroup II members were found in nature to carry similar replacements. In fact, viruses from genotype 3 resemble a single (R333D) mutant, and viruses from genotype 2 resemble a double mutant (K330L and R333D). In a genotype 1 ectodomain background, both types of mutants would be completely apathogenic for adult mice (i.c. and i.m. routes). Thus, it appears that mutations in positions 330 and 333 have different pathological consequences; they are less deleterious in a phylogroup II (i.c. pathogenic) than in a genotype 1 (both i.c. and i.m. apathogenic) ectodomain background. This modulation may be due to the local amino acid context. It has recently been proposed that region 319 to 340 (including both the K330 and R333 residues) is involved in the recognition of “high-affinity” neuron-specific receptors (8). It seems that the conservation of at least one positively charged residue is necessary and sufficient for receptor recognition and penetration into sensory and motor neurons. In summary, although the presence of an arginine (or lysine) at position 333 is crucial, genotype 2 and 3 viruses still remain pathogenic by the i.c. route because they possess compensatory positively charged residues at position 331 or 334, not present in the glycoprotein of phylogroup I viruses.
Cross-neutralization between lyssaviruses was measured after DNA immunization of mice with the G gene of a representative of each of the two phylogroups, PV strain (genotype 1) from phylogroup I and Mokola virus (genotype 3) from phylogroup II. Once again, lyssaviruses were segregated into two antigenic groups, which corresponded to the two phylogroups. In addition, a very good correlation was observed between the genetic distance (% ECTO aa identity) from PV to a Lyssavirus and the capacity of the anti-G PV serum to neutralize this Lyssavirus. It is generally recognized that seroconversion is obtained when the VNAb titer is over 0.5 IU/ml; thus, the range of identity where cross-neutralization disappears is between 74% (genotype 1 versus 4, still present) and 64.5% (genotype 1 versus 2 or 3, already absent). Indeed, the correlation curve predicts a cross-neutralization threshold at about 72% ECTO aa identity (Fig. 5). Thirty informative positions throughout the G ectodomain have nonconservative substitutions distinguishing lyssaviruses in the two defined phylogroups. It is of interest that six of them (20%) are located in the 26 residues (6% of the ECTO) that form the two main conformational antigenic sites. Some or all these six nonconservative substitutions may be implicated in the lack of cross-neutralization between the two phylogroups. Only site-specific mutagenesis can decipher their role in immunogenicity.
Due to their African distribution, their reduced pathogenicity in mice, and the small number of human cases and animal epizootics reported so far (14, 15), lyssaviruses of phylogroup II (genotypes 2 and 3) could appear to be less dangerous for public and veterinary health. However, Mokola virus repeatedly emerged in South Africa between 1995 and 1998 (36), and its reservoir is still unknown. In addition, the great genetic diversity in phylogroup II despite a very small number of isolates should be stressed and suggests an even greater diversity in nature. For example, four lyssaviruses of genotype 2 isolated in neighboring countries (Nigeria and the Central African Republic) displayed only 86% ECTO aa identity. In contrast, about 300 lyssaviruses of genotype 1 isolated worldwide still have 88.5% identity (unpublished results) and about 50 EBLs of genotypes 5 and 6 exhibited 97 and 99% identity, respectively (1). The greater genetic heterogeneity in phylogroup II may provide molecular flexibility. On the one hand, it could explain why several animals vaccinated with animal vaccines (genotype 1) were protected from challenge with Lagos bat virus (genotype 2) (16). On the other hand, it could favor the emergence of natural D333R or D333K mutants with increased pathogenicity. It was shown that virulent revertants can be easily generated in vivo from avirulent strains following a single base change at position 333 (56). One might predict that replacing D333 of phylogroup II viruses with K (or R) could have a dramatic positive effect on their pathogenicity, while they may remain uncontrolled by classic vaccines (genotype 1). Such substitutions may even be positively selected during oral vaccination campaigns of wild vectors with classic vaccines. These campaigns are currently limited to Europe and North America but hopefully will be extended to Africa in the future. DNA-mediated immunization, combining glycoprotein segments from various genotypes, has demonstrated its potential to protect against the whole Lyssavirus genus (3, 25). The use of such a wide-spectrum vaccine could prevent the emergence of escaping virulent lyssaviruses.
ACKNOWLEDGMENTS
We thank Corinne Jallet for technical assistance. We thank Hervé Bourhy (Centre National de Référence, Institut Pasteur, Paris, France), Courtney Meredith (Rabies Reference Centre, OIE Regional Collaborative Centre for Africa, Onderstepoort, South Africa), Bob Swanepoel (National Institute for Virology, Johannesburg, South Africa), and Donald Lodmell (Rocky Mountain Laboratory, Hamilton, Mont.), who provided us with isolates of rabies or rabies-related viruses. A special thank you to A. R. Gould (Commonwealth Scientific and Industrial Research Organization), who shared the ABL G protein sequence before publication. The help of F. Tekaia in handling computer facilities was much appreciated. We thank C. Roth for critical reading of the manuscript. We are grateful to two anonymous reviewers whose constructive criticism and suggestions improved the manuscript.
H.B. was a recipient of fellowships from the Moroccan Government and from the French-Moroccan cooperation. C.B. was a recipient of a Tunisian Government fellowship.
REFERENCES
- 1.Amengual B, Whitby J E, King A, Cobo S, Bourhy H. Evolution of European bat lyssaviruses. J Gen Virol. 1997;78:2319–2328. doi: 10.1099/0022-1317-78-9-2319. [DOI] [PubMed] [Google Scholar]
- 2.Atanasiu P, Tsiang H, Perrin P, Favre S. Demonstration of sialic acid in the rabies virus. Consequences of its removal on infectious and hemagglutinating properties. C R Acad Sc Hebd Seances Acad Sci D. 1976;283:111–114. [PubMed] [Google Scholar]
- 3.Bahloul C, Jacob Y, Tordo N, Perrin P. DNA-based immunisation for exploring the enlargement of immunological cross-reactivity against the lyssaviruses. Vaccine. 1998;16:417–425. doi: 10.1016/s0264-410x(97)00204-1. [DOI] [PubMed] [Google Scholar]
- 4.Benmansour A, Leblois H, Coulon P, Tuffereau C, Gaudin Y, Flamand A, Lafay F. Antigenicity of rabies virus glycoprotein. J Virol. 1991;65:4198–4203. doi: 10.1128/jvi.65.8.4198-4203.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourhy H, Kissi B, Lafon M, Sacramento D, Tordo N. Antigenic and molecular characterization of bat rabies virus in Europe. J Clin Microbiol. 1992;30:2419–2426. doi: 10.1128/jcm.30.9.2419-2426.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bourhy H, Kissi B, Tordo N. Molecular diversity of the Lyssavirus genus. Virology. 1993;194:70–81. doi: 10.1006/viro.1993.1236. [DOI] [PubMed] [Google Scholar]
- 7.Coulon P, Derbin C, Kucera P, Lafay F, Prehaud C, Flamand A. Invasion of the peripheral nervous systems of adult mice by the CVS strain of rabies virus and its avirulent derivate AvO1. J Virol. 1989;63:3550–3554. doi: 10.1128/jvi.63.8.3550-3554.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coulon P, Ternaux J P, Flamand A, Tuffereau C. An avirulent mutant of rabies virus is unable to infect motoneurons in vivo and in vitro. J Virol. 1998;72:273–278. doi: 10.1128/jvi.72.1.273-278.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delagneau J F, Perrin P, Atanasiu P. Structure of rabies virus: spacial relationships of the proteins G, M1, M2 and N. Ann Virol (Inst Pasteur) 1981;132E:473–493. [Google Scholar]
- 10.Dietzschold B, Rupprecht C E, Tollis M, Lafon M, Mattei J, Wiktor T J, Koprowski H. Antigenic diversity of the glycoprotein and nucleocapsid proteins of rabies and rabies-related viruses: implications for epidemiology and control of rabies. Rev Infect Dis. 1988;10:S785–S798. doi: 10.1093/clinids/10.supplement_4.s785. [DOI] [PubMed] [Google Scholar]
- 11.Dietzschold B, Wiktor T J, MacFarlan R, Varrichio A. Antigenic structure of rabies virus glycoprotein: ordering and immunological characterization of the large CNBr cleavage fragments. J Virol. 1982;44:595–602. doi: 10.1128/jvi.44.2.595-602.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dietzschold B, Wunner W H, Wiktor T J, Lopes A D, Lafon M, Smith C L, Koprowski H. Characterization of an antigenic determinant of the glycoprotein that correlates with pathogenicity of rabies virus. Proc Natl Acad Sci USA. 1983;80:70–74. doi: 10.1073/pnas.80.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Durrer P, Gaudin Y, Ruigrok R W H, Graf R, Brunner J. Photolabeling identifies a putative fusion domain in the envelope glycoprotein of rabies and vesicular stomatitis virus. J Biol Chem. 1995;270:17575–17581. doi: 10.1074/jbc.270.29.17575. [DOI] [PubMed] [Google Scholar]
- 14.Familusi J B, Moore D L. Isolation of a rabies related virus from the cerebrospinal fluid of a child with ‘aseptic meningitis.’. Afr J Med Sci. 1972;3:93–96. [PubMed] [Google Scholar]
- 15.Familusi J B, Osunkoya B O, Moore D L, Kemp G E, Fabiyi A. A fatal human infection with Mokola virus. Am J Trop Med Hyg. 1972;21:959–963. doi: 10.4269/ajtmh.1972.21.959. [DOI] [PubMed] [Google Scholar]
- 16.Fekadu M, Shaddock J H, Sanderlin D W, Smith J S. Efficacy of rabies vaccines against Duvenhage virus isolated from European house bats (Eptesicus serotinus), classic rabies and rabies-related viruses. Vaccine. 1988;6:533–539. doi: 10.1016/0264-410x(88)90107-7. [DOI] [PubMed] [Google Scholar]
- 17.Felsenstein J. PHYLIP: phylogeny inference package, version 3.52c. Seattle, Wash: University of Washington; 1993. [Google Scholar]
- 18.Gaudin Y. Folding of rabies virus glycoprotein, epitope acquisition, and interaction with endoplasmic reticulum chaperones. J Virol. 1997;71:3742–3750. doi: 10.1128/jvi.71.5.3742-3750.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaudin Y, Ruigrok R W H, Tuffereau C, Knossow M, Flamand A. Rabies virus glycoprotein is a trimer. Virology. 1992;187:627–632. doi: 10.1016/0042-6822(92)90465-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaudin Y, Tuffereau C, Benmansour A, Flamand A. Fatty acylation of rabies virus proteins. Virology. 1991;184:441–444. doi: 10.1016/0042-6822(91)90866-a. [DOI] [PubMed] [Google Scholar]
- 21.Genetics Computer Group. Wisconsin package, version 9.1. Madison, Wis: Genetics Computer Group; 1997. [Google Scholar]
- 22.Gould A R, Hyatt A D, Lunt R, Kattenbelt J A, Hengstberger S, Blacksell S D. Characterisation of a novel lyssavirus isolated from pteropid bats in Australia. Virus Res. 1998;54:165–187. doi: 10.1016/s0168-1702(98)00025-2. [DOI] [PubMed] [Google Scholar]
- 23.Hanna J N, Carney I K, Smith G A, Tannenberg A E, Deverill J E, Botha J A, Serafin I L, Harrower B J, Fitzpatrick P F, Searle J W. Australian bat lyssavirus infection: a second human case, with a long incubation period. Med J Aust. 2000;172:597–599. doi: 10.5694/j.1326-5377.2000.tb124126.x. [DOI] [PubMed] [Google Scholar]
- 24.Hooper P T, Lunt R A, Gould A R, Samaratunga H, Hyatt A D, Gleeson L J, Rodwell B J, Rupprecht C E, Smith J S, Murray P K. A new lyssavirus—the first endemic rabies-related virus recognized in Australia. Bull Inst Pasteur. 1997;95:209–218. [Google Scholar]
- 25.Jallet C, Jacob Y, Bahloul C, Drings A, Desmézières E, Tordo N, Perrin P. Chimeric lyssavirus glycoproteins with increased immunological potential. J Virol. 1999;73:225–233. doi: 10.1128/jvi.73.1.225-233.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.King A A, Meredith C D, Thompson G R. The biology of Southern African lyssavirus variants. In: Rupprecht C E, Dietzschold B, Koprowski H, editors. Lyssaviruses. Berlin, Germany: Spinger-Verlag; 1994. pp. 267–295. [DOI] [PubMed] [Google Scholar]
- 27.Kucera P, Dolivo P, Coulon P, Flamand A. Pathways of the early propagation of virulent and avirulent rabies strains from the eye to the brain. J Virol. 1985;55:158–162. doi: 10.1128/jvi.55.1.158-162.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lafay F, Benmansour A, Chebli K, Flamand A. Immunodominant epitopes defined by a yeast-expressed library of random fragments of the rabies virus glycoprotein map outside major antigenic sites. J Gen Virol. 1996;77:339–346. doi: 10.1099/0022-1317-77-2-339. [DOI] [PubMed] [Google Scholar]
- 29.Lafay F, Coulon P, Astic L, Saucier D, Riche D, Holley A, Flamand A. Spread of the CVS strain of rabies virus and of the avirulent mutant AvO1 along the olfactory pathways of the mouse after intra-nasal inoculation. Virology. 1991;183:320–330. doi: 10.1016/0042-6822(91)90145-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lafon M. Immunobiology of lyssaviruses: the basis for immunoprotection. In: Rupprecht C E, Dietzschold B, Koprowski H, editors. Lyssaviruses. Berlin, Germany: Springer-Verlag; 1994. pp. 145–160. [DOI] [PubMed] [Google Scholar]
- 31.Lafon M, Bourhy H, Sureau P. Immunity against the European bat rabies (Duvenhage) virus induced by rabies vaccines: an experimental study in mice. Vaccine. 1988;6:362–368. doi: 10.1016/0264-410x(88)90184-3. [DOI] [PubMed] [Google Scholar]
- 32.Lentz T L, Wilson P T, Hawrot E, Speicher D W. Amino acid sequence similarity between rabies virus glycoprotein and snake venom curaremimetic neurotoxins. Science. 1984;226:847–848. doi: 10.1126/science.6494916. [DOI] [PubMed] [Google Scholar]
- 33.McColl K A, Tordo N, Aguilar Setien A. Bat lyssavirus infections. Rev Sci Tech Off Int Epizoot. 2000;19:177–196. doi: 10.20506/rst.19.1.1221. [DOI] [PubMed] [Google Scholar]
- 34.Mebatsion T, Cox J H, Frost J W. Isolation and characterization of 115 street rabies virus isolates from Ethiopia by using monoclonal antibodies: identification of 2 isolates as Mokola and Lagos bat viruses. J Infect Dis. 1992;166:972–977. doi: 10.1093/infdis/166.5.972. [DOI] [PubMed] [Google Scholar]
- 35.Mebatsion T, Weiland F, Conzelmann K K. Matrix protein of rabies virus is responsible for the assembly and budding of bullet-shaped particles and interacts with the transmembrane spike glycoprotein G. J Virol. 1999;73:242–50. doi: 10.1128/jvi.73.1.242-250.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nel L, Jacobs J, Jaftha J, von Teichman B, Bingham J. New cases of Mokola virus infection in South Africa: A genotypic comparison of southern African virus isolates. Virus Genes. 2000;20:103–106. doi: 10.1023/a:1008120511752. [DOI] [PubMed] [Google Scholar]
- 37.Perrin P, Tino De Franco M, Jallet C, Fouque F, Morgeaux S, Tordo N, Colle J-H. The antigen-specific cell-mediated immune response in mice is suppressed by infection with pathogenic lyssaviruses. Res Virol. 1996;147:289–299. doi: 10.1016/0923-2516(96)82287-4. [DOI] [PubMed] [Google Scholar]
- 38.Préhaud C, Coulon P, Diallo A, Martinet-Edelist C, Flamand A. Characterization of a new temperature-sensitive and avirulent mutant of the rabies virus. J Gen Virol. 1989;70:133–143. doi: 10.1099/0022-1317-70-1-133. [DOI] [PubMed] [Google Scholar]
- 39.Préhaud C, Coulon P, Lafay F, Thiers C, Flamand A. Antigenic site II of the rabies virus glycoprotein: structure and role in viral virulence. J Virol. 1988;62:1–7. doi: 10.1128/jvi.62.1.1-7.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rose T K, Doolittle R F, Anilionis A, Curtis P J, Wunner W H. Homology between the glycoproteins of vesicular stomatitis virus and rabies virus. J Virol. 1982;43:361–364. doi: 10.1128/jvi.43.1.361-364.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sacramento D, Badrane H, Bourhy H, Tordo N. Molecular epidemiology of rabies in France: comparison with vaccinal strains. J Gen Virol. 1992;73:1149–1158. doi: 10.1099/0022-1317-73-5-1149. [DOI] [PubMed] [Google Scholar]
- 42.Sacramento D, Bourhy H, Tordo N. PCR technique as an alternative method for diagnosis and molecular epidemiology of rabies virus. Mol Cell Probes. 1991;6:229–240. doi: 10.1016/0890-8508(91)90045-l. [DOI] [PubMed] [Google Scholar]
- 43.Schneider L G, Cox J H. Bat lyssaviruses in Europe. In: Rupprecht C E, Dietzschold B, Koprowski H, editors. Lyssaviruses. Berlin, Germany: Spinger-Verlag; 1994. pp. 207–218. [Google Scholar]
- 44.Schneider L G, Dietzschold B, Dierks R E, Matthaeus W, Enzmann P J, Strohmaier K. Rabies group-specific ribonucleoprotein antigen and a test system for grouping and typing of rhabdoviruses. J Virol. 1973;11:748–755. doi: 10.1128/jvi.11.5.748-755.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seif I, Coulon P, Rollin P E, Flamand A. Rabies virulence: effect on pathogenicity and sequence characterization of rabies virus mutations affecting antigenic site III of the glycoprotein. J Virol. 1985;53:926–934. doi: 10.1128/jvi.53.3.926-934.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shakin-Eshleman S H, Remaley A T, Eshleman J R, Wunner W H, Spitalnik S L. N-linked glycosylation of rabies virus glycoprotein. Individual sequons differ in their glycosylation efficiencies and influence on cell surface expression. J Biol Chem. 1992;267:10690–10698. [PubMed] [Google Scholar]
- 47.Smith J S, Orciari L A, Yager P A. Molecular epidemiology of rabies in the United States. Semin Virol. 1995;6:387–400. [Google Scholar]
- 48.Smith J S, Yager P A, Baer G M. A rapid fluorescent focus inhibition test (RFFIT) for determining rabies virus-neutralizing antibody. In: Meslin F-X, Kaplan M M, Koprowski H, editors. Laboratory techniques in rabies. Geneva, Switzerland: 4th World Health Organization; 1996. pp. 181–192. [Google Scholar]
- 49.Swofford D L. Phylogenetic analysis using parsimony. Champaign, Ill: Illinois Natural History Survey; 1991. [Google Scholar]
- 50.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thoulouze M I, Lafage M, Schachner M, Hartmann U, Cremer H, Lafon M. The neural cell adhesion molecule is a receptor for rabies virus. J Virol. 1998;72:7181–7190. doi: 10.1128/jvi.72.9.7181-7190.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tordo N, Poch O, Ermine A, Keith G, Rougeon F. Completion of the rabies virus genome sequence determination: highly conserved domains along the L (polymerase) proteins of unsegmented negative-strand RNA viruses. Virology. 1988;165:565–576. doi: 10.1016/0042-6822(88)90600-9. [DOI] [PubMed] [Google Scholar]
- 53.Tordo N, Poch O, Ermine A, Keith G, Rougeon F. Walking along the rabies genome: is the large G-L intergenic region a remnant gene? Proc Natl Acad Sci USA. 1986;83:3914–3918. doi: 10.1073/pnas.83.11.3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tuffereau C, Bénéjean J, Blondel D, Kieffer B, Flamand A. Low-affinity nerve-growth factor receptor (P75NTR) can serve as a receptor for rabies virus. EMBO J. 1998;17:7250–7259. doi: 10.1093/emboj/17.24.7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tuffereau C, Benejean J, Roque Alfonso A-M, Flamand A, Fishman M C. Neuronal cell surface molecules mediate specific binding to rabies virus glycoprotein expressed by a recombinant baculovirus on the surfaces of lepidoptera cells. J Virol. 1998;72:1085–1091. doi: 10.1128/jvi.72.2.1085-1091.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tuffereau C, Leblois H, Bénéjean J, Coulon P, Lafay F, Flamand A. Arginine or lysine in position 333 of ERA and CVS glycoprotein is necessary for rabies virulence in adult mice. Virology. 1989;172:206–212. doi: 10.1016/0042-6822(89)90122-0. [DOI] [PubMed] [Google Scholar]
- 57.Whitt M A, Buonocore L, Prehaud C, Rose J K. Membrane fusion activity, oligomerization, and assembly of the rabies virus glycoprotein. Virology. 1991;185:681–688. doi: 10.1016/0042-6822(91)90539-n. [DOI] [PubMed] [Google Scholar]
- 58.Wiktor T J, Gyorgy E, Schlumberger H D, Sokol F, Koprowski H. Antigenic properties of rabies virus components. J Immunol. 1973;110:269–276. [PubMed] [Google Scholar]
- 59.Wiktor T J, Macfarlan R I, Foggin C M, Koprowski H. Antigenic analysis of rabies and Mokola virus from Zimbabwe using monoclonal antibodies. Dev Biol Stand. 1984;57:199–211. [PubMed] [Google Scholar]