Replication fork blockage by RTS1 at an ectopic site promotes recombination in fission yeast (original) (raw)
Abstract
Homologous recombination is believed to play important roles in processing stalled/blocked replication forks in eukaryotes. In accordance with this, recombination is induced by replication fork barriers (RFBs) within the rDNA locus. However, the rDNA locus is a specialised region of the genome, and therefore the action of recombinases at its RFBs may be atypical. We show here for the first time that direct repeat recombination, dependent on Rad22 and Rhp51, is induced by replication fork blockage at a site-specific RFB (RTS1) within a ‘typical' genomic locus in fission yeast. Importantly, when the RFB is positioned between the direct repeat, conservative gene conversion events predominate over deletion events. This is consistent with recombination occurring without breakage of the blocked fork. In the absence of the RecQ family DNA helicase Rqh1, deletion events increase dramatically, which correlates with the detection of one-sided DNA double-strand breaks at or near RTS1. These data indicate that Rqh1 acts to prevent blocked replication forks from collapsing and thereby inducing deletion events.
Keywords: DNA replication, homologous recombination, replication fork barrier, Rqh1, RTS1
Introduction
The path of a replication fork is often littered with obstacles that prevent its easy passage. Such obstacles can be accidental, for example lesions in the DNA template or proteins bound to the DNA template, or can be programmed (Rothstein et al, 2000). The appropriate response to these obstacles is important for both efficient DNA synthesis and the maintenance of genome stability.
Proteins that promote homologous recombination play important roles in processing stalled and blocked replication forks (McGlynn and Lloyd, 2002; Michel et al, 2004). For example, they can promote fork reversal where the nascent strands are unwound from their templates and annealed to each other (McGlynn and Lloyd, 2002). Fork reversal may enable polymerases to switch templates and bypass DNA lesions (Higgins et al, 1976), as well as provide room for DNA repair enzymes to remove blocking lesions in the DNA template (Courcelle et al, 2003). Recombination proteins can also protect the replication fork from excessive nucleolytic attack (Courcelle et al, 2003), and help the restart of replication by generating DNA structures at which replication proteins can reload (Liu et al, 1999). However, the cell has to be circumspect about its use of recombination. Inappropriate recombination can result in genome instability, for example recombination between repetitive DNA elements can result in the expansion and contraction of tandem arrays, or even translocations between different chromosomes. Such events are hallmarks of a number of diseases including cancer.
Studies of replication fork stalling in Escherichia coli have established that the way in which a blocked replication fork is processed depends on the type of block (Michel et al, 2004). For example, an ultra violet light (UV)-induced lesion in the leading strand template may be side-stepped by a recombination-mediated transient switch of the polymerase onto the nascent lagging strand (Higgins et al, 1976), whereas a UV-induced lesion in the lagging strand template may be left unreplicated, the resultant daughter strand gap being repaired by recombination postreplication (Rupp and Howard-Flanders, 1968). At other types of stalled or blocked replication fork, it may be sufficient to simply stabilise the fork until the block is removed and DNA synthesis can be resumed. This appears to be true in yeast for forks that are stalled by dNTP depletion mediated by the drug hydroxyurea (HU), which inhibits ribonucleotide reductase. The maintenance of fork stability here depends on the intra-S checkpoint kinase Rad53 (Lopes et al, 2001; Sogo et al, 2002).
Members of the RecQ family of DNA helicases, which includes Sgs1 and Rqh1 in Saccharomyces cerevisiae and Schizosaccharomyces pombe, respectively, and BLM and WRN in humans, also play crucial roles in maintaining fork stability (Hickson, 2003). The importance of this is underlined by the fact that defects in BLM and WRN give rise to the cancer-prone diseases Blooms syndrome and Werners syndrome, respectively. In S. cerevisiae, Sgs1 prevents polymerase dissociation from replication forks stalled by HU treatment (Cobb et al, 2003). RecQ helicases may also reset reversed replication forks by branch migrating Holliday junctions (HJs) that are formed when nascent strands anneal to each other (Doe et al, 2000; Karow et al, 2000). It has been suggested that this activity might prevent replication fork breakage by the XPF-related endonuclease Mus81-Eme1 (Kaliraman et al, 2001; Doe et al, 2002). The ability to branch migrate HJs may also be used by RecQ helicases to abort inappropriate recombination at stalled forks, and, together with Topoisomerase III, to process double HJ intermediates as noncrossover recombinants (Ira et al, 2003; Wu and Hickson, 2003). These activities would reduce the risk of genome rearrangements caused by crossing over between repeated DNA sequences.
A growing number of programmed replication fork barriers (RFBs) or pause sites have been identified (Rothstein et al, 2000). Important roles for RFBs include ensuring the successful merging of opposing replication forks, and promoting the avoidance of collisions between replication forks and transcription complexes (Brewer and Fangman, 1988; Krabbe et al, 1997; Rothstein et al, 2000). Unlike most accidental blockades, the majority of programmed RFBs are unidirectional. Replication can therefore be completed when the blocked fork merges with the opposing fork. In theory, recombination at such blocked forks would be an unnecessary risk, and therefore should be suppressed. However, like accidental blockades, programmed RFBs can elicit different responses. In E. coli, replication forks blocked at Ter sites bound by the Tus protein remain stable, and only provoke recombination if a subsequent round of replication runs into the back of them to generate free double-strand ends (Bidnenko et al, 2002). In contrast, forks blocked at RFBs in the rDNA locus of S. cerevisiae appear to more directly stimulate recombination resulting in the expansion and contraction of the array, as well as the production of extrachromosomal rDNA circles (ERCs) (Kobayashi et al, 1998; Weitao et al, 2003b; Burkhalter and Sogo, 2004).
Here, we have analysed whether a third RFB, RTS1, provokes recombination in S. pombe. RTS1 ensures that replication of the mat1 locus in S. pombe occurs in the centromere-proximal direction (Dalgaard and Klar, 2001; Vengrova et al, 2002). This is necessary for laying down an imprint, which stimulates mating-type switching in the next generation (Dalgaard and Klar, 1999). Essential to RTS1 function are four repeated ∼55 bp motifs, whose function is enhanced by an ∼60 bp purine-rich region (Codlin and Dalgaard, 2003). The barrier function of these _cis_-acting elements depends on the Swi1–Swi3 complex and the _trans_-acting replication termination factors Rtf1 and Rtf2 (Dalgaard and Klar, 2000).
We show that replication fork blockage by RTS1 massively stimulates recombination between repeated DNA sequences with a bias toward conservative gene conversion events. This recombination, like that induced by stochastic events, depends on the S. pombe homologues of the recombinases Rad51 (called Rhp51) and Rad52 (called Rad22). These data provide the first demonstration that a site-specific replication fork blockage outside of the rDNA locus directly stimulates recombination. We also show that _RTS1_-induced recombination is constrained by Rqh1. In particular, _rqh1_− mutants exhibit a dramatic increase in deletion recombinants, which correlates with the detection of one-sided double-strand breaks (DSBs) in the vicinity of the RFB. This provides further evidence that RecQ DNA helicases protect stalled replication forks from collapse.
Results
Agents that stimulate direct repeat recombination
To measure inter/intra-chromosomal recombination in S. pombe, we have used a direct repeat of _ade6_− heteroalleles with an intervening his3+ gene positioned at the ade6 locus on chromosome 3. With this system, ade6+ recombinants can be divided into those that retain the intervening marker (conversion-types) and those that lose it (deletion-types) (Figure 1A). Under our normal assay conditions, approximately 3.0–4.5 Ade+ recombinants arise spontaneously in every 104 viable cells, and typically between 70 and 80% of these are deletion-types (Figure 1B and Table I). Various agents, which are known to impede replication forks, stimulate direct repeat recombination (Galli and Schiestl, 1999). However, a comparison of their affect on the ratio of deletion and conversion-types has not been made. UV, which generates lesions such as pyrimidine dimers that block the normal replicative polymerases, stimulates the formation of conversion-types more than it does deletion-types, altering the ratio of these recombinants from approximately 1:3 to 1:1 (Figure 1B) (Osman et al, 2000). In contrast camptothecin (CPT), which inhibits the religation step during the reaction cycle of topoisomerase I and consequently generates single-strand breaks in DNA at which replication forks collapse (Pommier et al, 2003), stimulates mostly deletion-type recombinants (Figure 1C). Finally, fork stalling induced by HU only slightly stimulates deletion-type recombinants (Figure 1C). These data confirm that the impedance of a replication fork can induce recombination, and that the nature of the impedance influences the type of recombinant that is generated. Intriguingly, the fact that recombinant formation is only weakly stimulated by acute HU exposure, suggests that replication fork stalling per se is insufficient to promote recombination.
Figure 1.
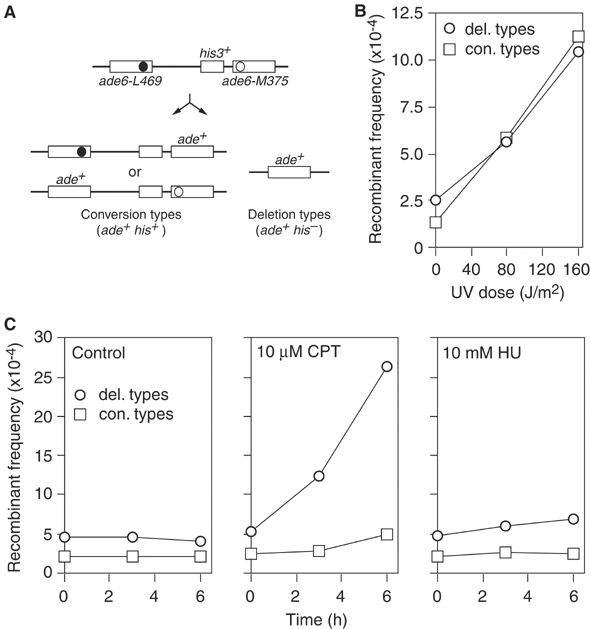
Induction of direct repeat recombination by different genotoxins. (A) Schematic of intrachromosomal recombination substrate and recombinant products. Solid and open circles represent the ade6-L469 and ade6-M375 mutations, respectively. (B) The effect of increasing doses of UV on the frequency and type of Ade+ recombinants in a wild-type strain (MCW39). (C) The effect of a timed exposure to CPT or HU on the frequency and type of Ade+ recombinants in a wild-type strain (MCW39). Note that the ∼2-fold increase in spontaneous Ade+ recombinants in these assays compared to that in ‘B' is due to the slightly different protocols that were used.
Table 1.
_RTS1_-induced mitotic recombination frequencies
| Strain | RTS1 | Mean frequency of Ade+ recombinants per 104 viable cells | % Conversion typesa | |||
|---|---|---|---|---|---|---|
| Site | Orientation | Total (His−+His+)a | Deletion type (His−)a | Conversion type (His+)a | ||
| Wild type (MCW39) | No RTS1 | 4.42 (±1.6) | 3.47 (±1.3) | 0.95 (±0.51) | 21.4 (±8.0) | |
| Wild type (MCW1262) | A | 1 | 3.26 (±1.0) | 2.18 (±0.76) | 1.08 (±0.49) | 33.6 (±11.0) |
| Wild type (MCW1433) | A | 2 | 189.2 (±114) | 86.2 (±60.1) | 103.0 (±61.9) | 55.9 (±10.9) |
| Wild type (MCW1256) | B | 1 | 6.57 (±4.63) | 5.75 (±4.36) | 0.82 (±0.51) | 13.4 (±7.2) |
| Wild type (MCW1260) | B | 2 | 36.1 (±14.1) | 24.2 (±11.2) | 11.9 (±4.5) | 35.1 (±11.6) |
| _swi1_Δ (MCW1362) | A | 1 | 33.7 (±18.7) | 25.1 (±14.8) | 8.6 (±6.8) | 25.2 (±12.0) |
| _swi1_Δ (MCW1358) | A | 2 | 47.9 (±27.4) | 36.2 (±24.2) | 11.7 (±7.5) | 27.0 (±11.0) |
| _swi1_Δ (MCW1395) | B | 1 | 37.6 (±21.7) | 30.8 (±19.0) | 6.77 (±4.15) | 19.1 (±9.4) |
| _swi1_Δ (MCW1377) | B | 2 | 20.9 (±13.1) | 16.8 (±10.9) | 4.1 (±3.3) | 19.3 (±9.2) |
| _rhp51_Δ (MCW1691) | A | 1 | 10.72 (±5.6) | 10.7 (±5.6) | 0.02 (±0.04) | 0.19 (±0.45) |
| _rhp51_Δ (MCW1692) | A | 2 | 34.2 (±17.3) | 32.6 (±16.7) | 1.55 (±1.06) | 4.71 (±3.4) |
| _rad22_Δ (MCW1687) | A | 1 | 1.02 (±0.49) | 1.0 (±0.48) | 0.016 (±0.037) | 1.2 (±2.7) |
| _rad22_Δ (MCW1688) | A | 2 | 3.39 (±1.6) | 3.27 (±1.59) | 0.12 (±0.17) | 3.57 (±4.52) |
| _rhp51_Δ _rad22_Δ (MCW1695) | A | 1 | 1.09 (±0.52) | 1.09 (±0.52) | 0.005 (±0.02) | 0.33 (±1.3) |
| _rhp51_Δ _rad22_Δ (MCW1696) | A | 2 | 2.52 (±1.33) | 2.49 (±1.36) | 0.028 (±0.057) | 1.75 (±3.36) |
| _rqh1_Δ (MCW1443) | A | 1 | 11.95 (±3.9) | 10.2 (±3.4) | 1.75 (±0.77) | 15.1 (±5.7) |
| _rqh1_Δ (MCW1447) | A | 2 | 2410 (±1579) | 2152 (±1530) | 258 (±179) | 13.1 (±12.1) |
| _rqh1_Δ (MCW1445) | B | 1 | 17.4 (±8.9) | 14.6 (±6.0) | 2.8 (±4.6) | 12.8 (±10.7) |
| _rqh1_Δ (MCW1438) | B | 2 | 748.7 (±266) | 678.8 (±243) | 69.9 (±43.4) | 9.2 (±4.7) |
| aThe values in parentheses are the standard deviations. |
The RTS1 RFB stimulates recombination when placed at the ade6 locus
To study the effect of a site-specific RFB on direct repeat recombination, we have engineered strains in which an 889 bp fragment containing the RTS1 element has been placed at two different positions within the _ade6_− heteroallelic repeat described above (Figure 2A). The two different positions for RTS1 are between the his3+ and ade6-M375 genes (site A), and within the ade6-L469 coding region (site B). As RTS1 is a polar RFB, strains have been constructed that contain RTS1 in both possible orientations at sites A and B. RTS1 in orientation 1 is active for blocking forks coming from the direction of the centromere (cen3), whereas orientation 2 blocks forks moving toward the centromere (Figure 2A). Although the direction of replication across the ade6 locus is unknown, the positions of putative origins of replication are such that there is a predicted bias in replication proceeding across the ade6 locus towards cen3 (Figure 2A) (Segurado et al, 2003). If true then only RTS1 in orientation 2 would result in replication forks being blocked within the _ade6_− direct repeat. To investigate this, we used a native two-dimensional (2-D) agarose gel electrophoresis technique for studying replication intermediates (Brewer and Fangman, 1987). Genomic DNA from logarithmically growing cells was digested with _Eco_NI, enriched for replication intermediates by fractionation on BND-cellulose, and separated in a 2-D gel. The gel was then analysed by Southern blotting using radiolabelled probe A, which is specific to the his3+ gene (Figure 2A). An uninterrupted arc of Y-shaped replication forks was detected in a strain without the RTS1 element and in one with RTS1 at site A in orientation 1 (Figure 2B). However, in the strain with RTS1 at site A in orientation 2 the replication forks accumulate as a comet-shaped spot at the top of the ascending part of the Y-arc (Figure 2B). Closer inspection of this comet-shaped signal reveals that it consists of at least two discrete spots (Figure 2B, inset). This is consistent with replication fork blockage by RTS1, which has previously been shown to consist of four repeated motifs that may each act as a barrier (Codlin and Dalgaard, 2003). These data are also consistent with the predicted bias in the direction of replication across the _ade6_− repeat.
Figure 2.
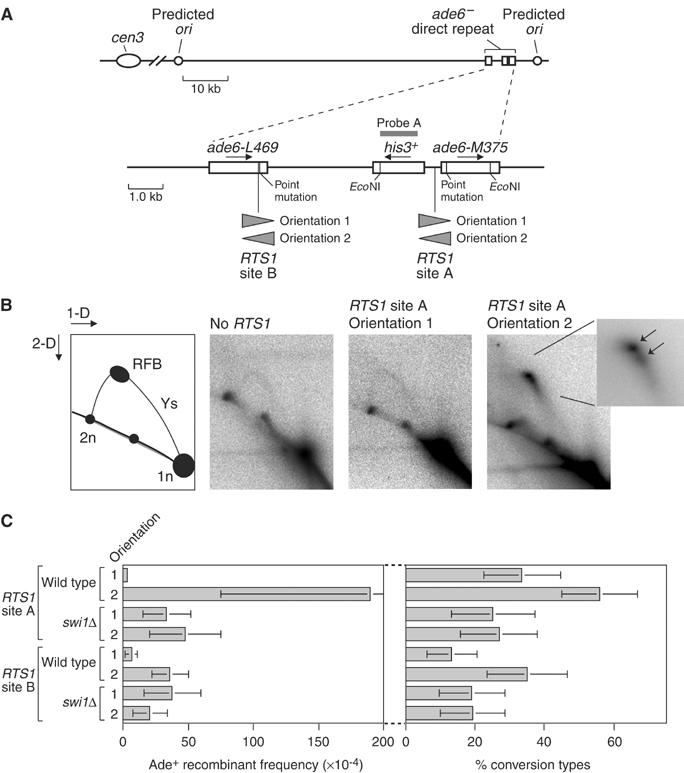
Effect of replication fork blockage by RTS1 on direct repeat recombination. (A) Schematic showing the position of the intrachromosomal recombination substrate on chromosome III. Sites of RTS1 insertion within the recombination substrate are also indicated as is probe (A) used to detect replication intermediates. (B) 2-D gel analysis of replication intermediates within the region delineated by the _Eco_NI restriction sites shown in ‘A'. The far left panel is a guide for interpreting 2-D gels. The panels to the right of this show the 2-D gel analysis of DNA from asynchronously grown cultures of the wild-type strains MCW39 (no RTS1), MCW1262 (RTS1 site A orientation 1), and MCW1433 (RTS1 site A orientation 2). The inset picture shows an enlargement of the RFB signal with arrows indicating that it consists of at least two discrete spots. (C) Bar charts showing the effect of RTS1 at different positions in the recombination substrate on the frequency and type of Ade+ recombinants in wild-type and _swi1_Δ mutant strains. In order of presentation the strains are MCW1262, MCW1433, MCW1362, MCW1358, MCW1256, MCW1260, MCW1395 and MCW1377. Error bars represent the standard deviations about the mean.
Having established that RTS1 acts as a strong RFB when placed within the _ade6_− direct repeat, we determined its effect on the frequency of recombination between the repeats (Figure 2C and Table I). In wild-type strains with RTS1 in orientation 1, either at site A or B, the frequency of Ade+ recombinants is not significantly different from that in a wild-type strain without the RTS1 element. However, strains with RTS1 at site B do produce fewer conversion-type recombinants. This is probably due to impedance of conversion-type recombination by the presence of a large chunk of heterologous sequence (the RTS1 element) within ade6-L469. In wild-type strains with RTS1 in orientation 2, the recombinant frequency increases by ∼50-fold at site A and ∼5-fold at site B compared to the equivalent strains with RTS1 in orientation 1 (Figure 2C and Table I). As replication fork blockage is specific to orientation 2 within the _ade6_− repeat, these data indicate that replication forks blocked by RTS1 strongly provoke recombination. Intriguingly, the proportion of these induced recombinants that are conversion-types is significantly greater than in the spontaneous recombinants (_P_=<8 × 10−9). In this respect, recombination induced by the RTS1 RFB resembles that induced by UV.
The site at which recombinases load normally acts as the recipient of genetic information during recombination. In our assay system, ade6-L469 to ade+ conversion-types can be distinguished from ade6-M375 to ade+ conversion-types by colony colour. The former appear as pale pink colonies, whereas the latter form white colonies (Osman et al, 1996). To see if we could get evidence that recombination was being initiated at the site of the blocked replication fork, we scored the conversion-types for the wild-type strains in Figure 2C for colony colour (data not shown). In all, >90% of conversion-types are ade6-M375 to ade+ irrespective of the position and orientation of RTS1. However, in the case of the strain containing RTS1 in orientation 2 at site A, 100% of conversion-types are ade6-M375 to ade+ compared to 92% for the equivalent strain with RTS1 in orientation 1. This difference is statistically significant (_P_=3.5 × 10−5). Similarly, the percentage of conversion-types that are ade6-L469 to ade+ is more with RTS1 at site B in orientation 2 (10%) than if it is in orientation 1 (4%). Again this difference is statistically significant (_P_=<0.05). These data are consistent with recombination being initiated at the site of the blocked replication fork.
Swi1 is required for the stimulation of recombination by RTS1
As mentioned in the Introduction, the RFB function of RTS1 is dependent on several _trans_-acting factors including Swi1. To provide further evidence that it is the RFB function of RTS1 that is responsible for stimulating recombination, we determined the effect of swi1 deletion on the frequency of recombination in our strains (Figure 2C and Table I). Swi1 is known to have a general role in maintaining the stability of stalled replication forks (Noguchi et al, 2003). In accord with this, the frequency of Ade+ recombinants in _swi1_Δ strains with RTS1 in either orientation 1 or 2 is significantly greater than in the equivalent wild-type strains with RTS1 in orientation 1. Importantly, however, there is little difference in the frequency of recombinants in _swi1_Δ strains containing RTS1 in different orientations. Furthermore, the recombination induced by RTS1 in orientation 2 at site A is abolished by _swi1_Δ. These data indicate that it is the normal RFB function of RTS1 that is responsible for stimulating recombination.
RTS1-induced recombination depends on Rhp51 and Rad22
The Rad22-dependent pathway is required for most of the direct repeat recombination in S. pombe, with a subpathway, dependent on Rhp51, generating conversion-types (Doe et al, 2004). Deletion-types form independently of Rhp51, and in fact _rhp51_Δ mutants exhibit a hyperrecombination phenotype for these recombinants (Doe et al, 2004). To determine whether the recombination induced by RTS1 is similarly dependent on these known pathways of recombination, we compared the frequency of Ade+ recombinants in wild-type, _rhp51_Δ, _rad22_Δ and _rhp51_Δ _rad22_Δ strains containing RTS1 at site A (Figure 3A and Table I). Virtually all of the induced recombinants are dependent on Rad22, with Rhp51 being additionally required for most of the induced conversion-types (Figure 3A and B). Surprisingly, Rhp51 is also required for more than half of the induced deletion-types (Table I). 2-D gel analysis of genomic DNA from these strains confirms that the RFB function of RTS1 is fully intact in _rhp51_Δ, _rad22_Δ and _rhp51_Δ _rad22_Δ strains (Figure 3C). Together, these data confirm that the recombination induced by RTS1 is mainly dependent on the known pathways of recombination in S. pombe. Furthermore, they indicate that strand invasion by Rhp51 is required for the majority of deletion events at blocked replication forks.
Figure 3.
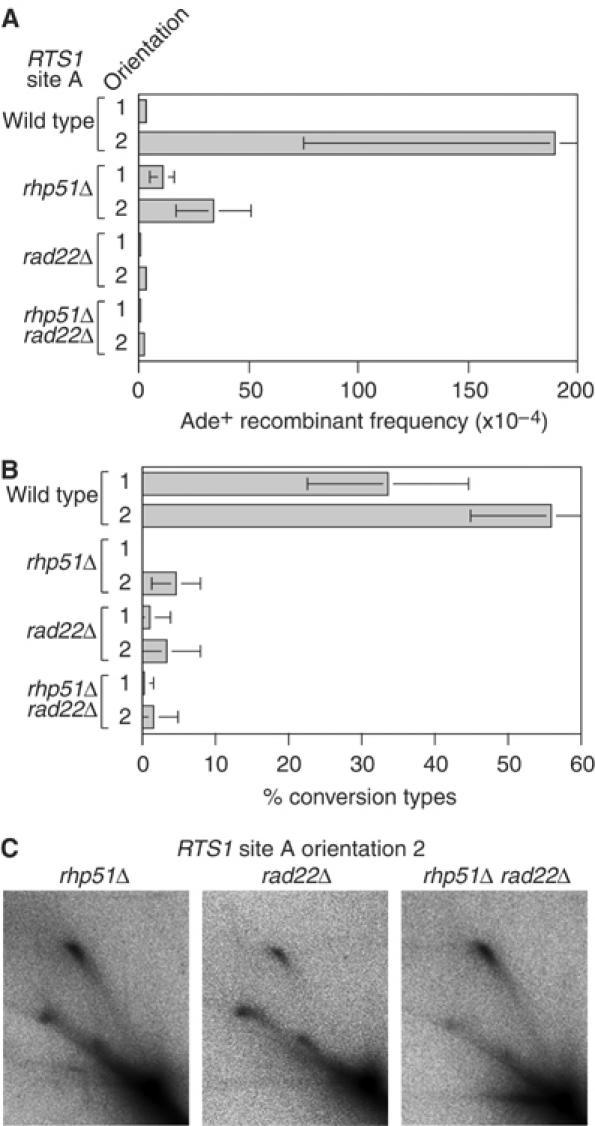
The dependency on Rhp51 and Rad22 for the recombination induced by replication fork blockage at RTS1. (A) Bar chart showing the frequency of Ade+ recombinants in wild-type, _rhp51_Δ, _rad22_Δ and _rhp51_Δ _rad22_Δ strains containing RTS1 at site A in either orientation 1 or 2 as indicated. In order of presentation, the strains are MCW1262, MCW1433, MCW1691, MCW1692, MCW1687, MCW1688, MCW1695 and MCW1696. (B) Bar chart showing the percentage of Ade+ recombinants in ‘A' that are conversion-types. (C) 2-D gel analysis of replication intermediates detected by probe A in the _Eco_NI DNA fragment shown in Figure 2A. The strains are MCW1692 (_rhp51_Δ), MCW1688 (_rad22_Δ) and MCW1696 (_rhp51_Δ _rad22_Δ).
Rqh1 constrains recombination induced by the RTS1 RFB
To see if Rqh1 limits recombination induced by the RTS1 RFB, we compared the frequency of Ade+ recombinants in wild-type and _rqh1_Δ strains containing RTS1 at sites A or B (Figure 4A and Table I). In _rqh1_Δ strains with RTS1 in orientation 1, at either site A or B, the frequency of recombinants is ∼3-fold higher than in the equivalent wild-type strain, and the percentage of these recombinants that are conversion-types is either the same or reduced compared to wild-type (Figure 4B and Table I). These differences are similar to those observed in strains without RTS1 (Doe et al, 2000). However, with RTS1 in orientation 2 the already high frequency of Ade+ recombinants is dramatically increased by _rqh1_Δ mutation. In the strain with RTS1 at site A, this increase is >12-fold over the equivalent wild-type strain, while the increase is >20-fold with RTS1 at site B. Again the percentage of these recombinants that are conversion-types is reduced significantly in a _rqh1_Δ mutant (Figure 4B and Table I). No difference in the growth/viability of _rqh1_− strains with or without RTS1 in either orientation 1 or 2 was detected (data not shown). The increased frequency of deletion-types is therefore not due to a detrimental effect on growth of RTS1 in orientation 2 in the absence of Rqh1. Instead, these data show that Rqh1 limits the recombination that is induced by the RTS1 RFB, and in particular it limits the formation of deletion-type recombinants.
Figure 4.
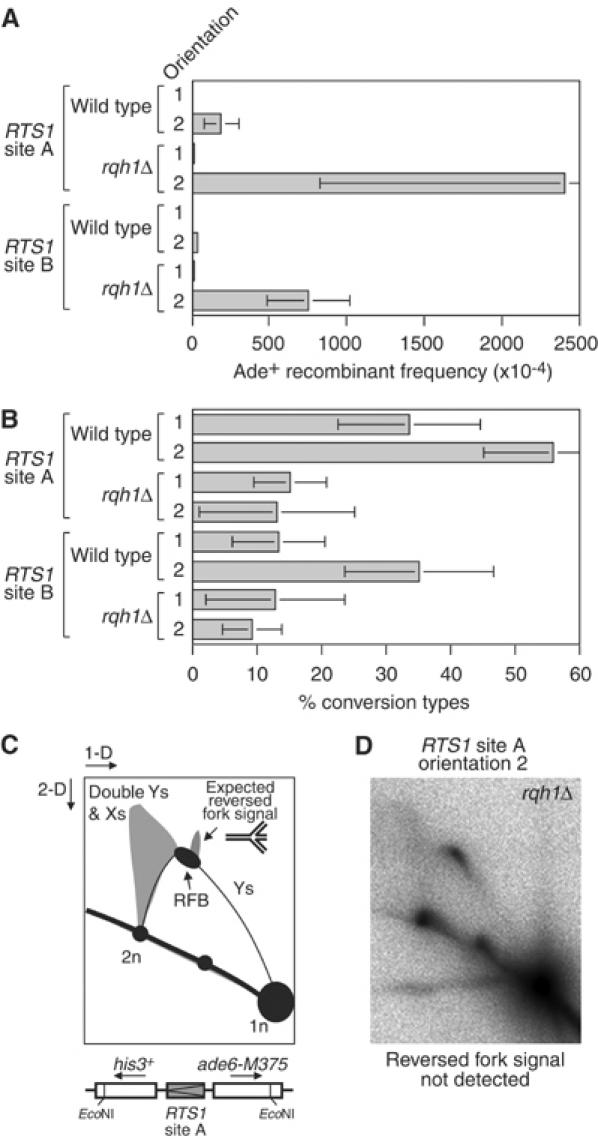
Effect of _rqh1_Δ mutation on the frequency and type of recombinants induced by replication fork blockage at RTS1. (A) Bar chart showing the frequency of Ade+ recombinants in wild-type and _rqh1_Δ strains containing RTS1 at site A or B in orientation 1 or 2 as indicated. In order of presentation, the strains are MCW1262, MCW1433, MCW1443, MCW1447, MCW1256, MCW1260, MCW1445 and MCW1438. (B) Bar chart showing the percentage of Ade+ recombinants in ‘A' that are conversion-types. (C) Schematic of a 2-D gel showing the expected position of the cone-shaped signal that would be indicative of fork reversal. (D) 2-D gel analysis of replication intermediates detected by probe A in the _Eco_NI DNA fragment from a _rqh1_Δ mutant containing RTS1 at site A in orientation 2 (MCW1447).
Stalled/blocked replication forks can reverse, either spontaneously or by an enzyme-driven reaction, to form a double-stranded tail of nascent DNA onto which recombinases can load and promote recombination (McGlynn and Lloyd, 2002). One way in which RecQ DNA helicases are thought to limit recombination at stalled/blocked replication forks is by resetting reversed forks (Doe et al, 2000; Karow et al, 2000). Reversed forks have recently been observed by 2-D gel analysis at the replication fork pause site within mat1 of S. pombe (Vengrova and Dalgaard, 2004). These reversed forks appear as a cone-shaped signal displaced up from the Y-arc at about the position of the replication pause site (Figure 4C). This signal was observed to increase in a _rqh1_Δ mutant consistent with the idea that Rqh1 resets reversed forks (Vengrova and Dalgaard, 2004). To see if forks blocked at RTS1 were undergoing reversal, we performed a 2-D gel analysis of genomic DNA from a _rqh1_Δ mutant that contained RTS1 in orientation 2 at site A (Figure 4D). Consistent with our other analyses of DNA from strains with RTS1 in this position, a strong RFB is detected. In addition, a triangular-shaped signal emanating from the descending portion of the Y-arc can be seen. Previous studies have shown that this signal represents a mixture of double-Y- and X-shaped molecules formed by converging replication forks and, in the case of the X-shaped molecules, by recombination. However, we are unable to detect a cone-shaped signal indicative of fork reversal. Therefore, if fork reversal occurs at the RTS1 RFB, it is below the detection limits of our assay. It should be noted that previous 2-D gel analyses of the RTS1 RFB have also not detected evidence of fork reversal in wild-type cells (Dalgaard and Klar, 2001; Codlin and Dalgaard, 2003).
Detection of one-sided DSBs at the RTS1 RFB in rqh1_Δ_ strains
It has been suggested that in the absence of Rqh1, stalled/blocked replication forks might be more susceptible to being cleaved by an endonuclease like Mus81-Eme1 (Kaliraman et al, 2001; Doe et al, 2002). This kind of endonucleolytic cleavage would detach one arm of the fork to produce a one-sided DSB. To see if this happens at the RTS1 RFB, genomic DNA from wild-type and _rqh1_Δ strains, which contain RTS1 at site A, was digested with _Pst_I, separated on a 1-D gel, and analysed by Southern blotting using radiolabelled probes A and B (Figure 5). Genomic DNA was prepared and restricted in agarose plugs to prevent it shearing. Probes A and B are specific to regions that flank RTS1 at site A (Figure 5A). Both probes detect the full-length _Pst_I fragment from wild-type and _rqh1_Δ DNA (Figure 5B and C). The full-length band is ∼9.2 kb but runs slightly slower (∼9.5 kb) than this when compared to the DNA ladder (lane e). This is probably because its migration is retarded by the agarose plug. An additional faster migrating band, which is indicative of a DSB, is detected with probe B in DNA from the _rqh1_Δ strain containing RTS1 in orientation 2 (i.e. the orientation in which replication fork blockage is detected) (Figure 5B, lane d). This band is ∼7.6 kb based on a comparison with the DNA ladder. However, as noted above, it is likely that the actual size of this band is a few hundred base pairs shorter. As this band is not detected by probe A (Figure 5C, lane d), the DSB site can be mapped approximately to the RTS1 element. If this was a two-sided DSB then a ∼2 kb band would be detected by probe A. However, only the full-length _Pst_I fragment is detected with this probe (Figure 5C, lane d). These data indicate that DSBs are formed at or near RTS1 in the replicated arms of the blocked replication fork in the absence of Rqh1. Quantification of the bands in Figure 5B lane d indicates that ∼1.9% of the genomic DNA is broken in this way.
Figure 5.
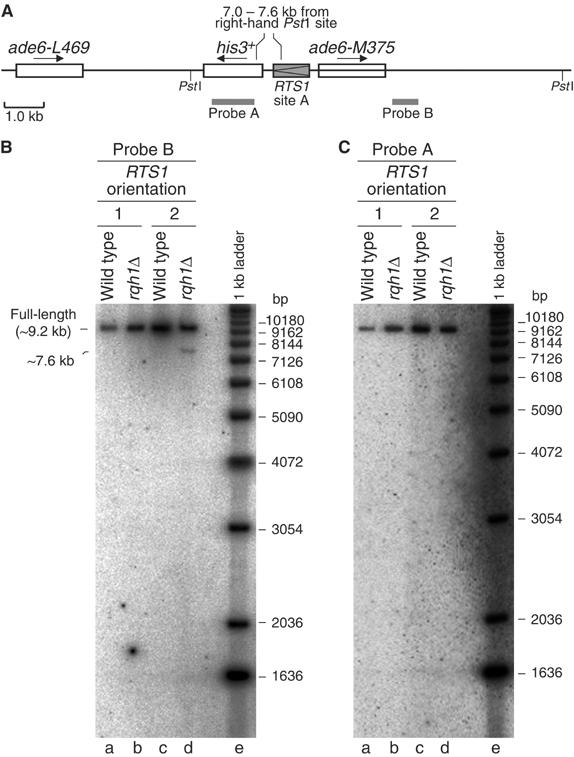
Detection of one-sided DSBs in a _rqh1_Δ mutant containing RTS1 at site A in orientation 2. (A) Schematic of the recombination substrate showing the position of _Pst_I sites and DNA probes. The region between 7 and 7.6 kb from the right-hand _Pst_I site, in which the DSB maps, is also marked. (B) Analysis of genomic DNA, prepared in agarose plugs and restricted with _Pst_I, on a 1-D gel using radiolabelled probe B. The strains are MCW1262 (lane a), MCW1443 (lane b), MCW1433 (lane c) and MCW1447 (lane d). (C) The same as ‘B' except that probe A was used instead of probe B.
To provide further evidence that one-sided DSBs are formed at the RTS1 RFB in _rqh1_Δ strains, we performed a similar analysis as above on DNA from strains containing RTS1 at site B (Figure 6A–C). In this case, the DNA was digested with _Bsu_36I and _Eco_RV, and analysed using probes C and D (Figure 6A). Like above, both probes detect the full-length _Bsu_36I–_Eco_RV genomic DNA fragment (∼8.2 kb) (Figure 6B and C, lanes b and c). Probe C also detects a band of ∼2.5 kb, which represents a portion of the normal his3 locus on chromosome 2 that has not been deleted in our strains (Figure 6B, lanes b and c; and D, lanes b–g). A faint nonspecific band of ∼2.5 kb is also detected by probe D (Figure 6C, lanes b and c; and E, lanes b–g). All of these bands are detected in both wild-type and _rqh1_Δ strains, irrespective of the orientation of the RTS1 element. However, probe C also detects a ∼5.1 kb band that is specific to DNA from _rqh1_Δ strains containing RTS1 in orientation 2 (Figure 6B, lane b). This band is indicative of a DSB, and, as it is not detected by probe D, it can be mapped to the RTS1 element. Furthermore, the failure to detect a band of ∼2.9 kb in this DNA sample with probe D indicates that the DSB is one-sided (note that the ∼11.3 kb band is formed by deletion-type recombination) (Figure 6C, lane b). These data are consistent with those for RTS1 at site A, and indicate that a significant portion of replication forks that are blocked at RTS1 are cleaved in the absence of Rqh1. Quantification of the blot in Figure 6B reveals that ∼1.3% of the genomic DNA contains a one-sided DSB at RTS1 at site B, which is similar to the amount of breakage detected at RTS1 at site A.
Figure 6.
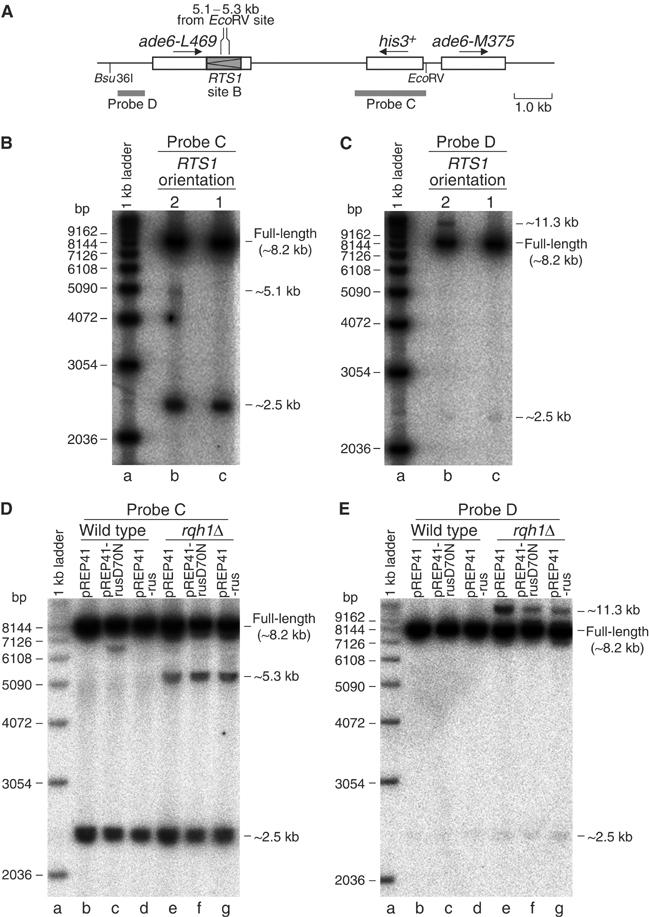
The detection of one-sided DSBs in a _rqh1_Δ mutant containing RTS1 at site B in orientation 2. (A) Schematic of the recombination substrate showing the position of _Bsu_36I and _Eco_RV sites, and DNA probes. The region between 5.1–5.3 kb from the _Eco_RV site, in which the DSB maps, is also marked. (B) Analysis of genomic DNA from the _rqh1_Δ mutant strains MCW1438 (lane b) and MCW1445 (lane c). Genomic DNA was prepared in agarose plugs and restricted with _Bsu_36I and _Eco_RV before analysis on a 1-D gel using radiolabelled probe C. (C) The same as ‘B' except probe D was used instead of probe C. (D) 1-D gel analysis of genomic DNA from wild-type (MCW1260) and _rqh1_Δ (MCW1438) strains carrying plasmids as indicated. Strains were grown in EMM without thiamine for ∼20 h to allow expression of RusA from the nmt1 promoter in pREP41. RusA and RusAD70N carry C-terminal green fluorescent protein tags so their expression in the wild-type and _rqh1_Δ strains was confirmed by fluorescent microscopy prior to harvesting the cells for the preparation of genomic DNA. The amounts of the ∼5.3 kb band in lanes e–g relative to the total probed DNA are 2.2% (lane e), 4.5% (lane f) and 3.6% (lane g). Repetitions of this experiment show that these differences in the amount of the ∼5.3 kb band are not significant. (E) The same as ‘D' except probe D was used instead of probe C.
An HJ resolvase does not increase fork breakage at the RTS1 RFB
One way in which a blocked replication fork can give rise to a one-sided break is if it reverses to form an HJ that is then cleaved by an HJ resolvase (Seigneur et al, 1998). Although our 2-D gel analysis had provided no evidence of fork reversal at the RTS1 RFB in an _rqh1_Δ mutant (Figure 4D), we looked to see whether the expression of an HJ resolvase would cause an increase in the amount of one-sided DSBs in these strains. For this, we used a recombinant form of the bacterial HJ resolvase RusA that has previously been shown to suppress both _rqh1_Δ and _mus81_Δ mutant phenotypes (Doe et al, 2000, 2002). Genomic DNA was prepared from wild-type and _rqh1_Δ strains containing RTS1 at site B in orientation 2, and the plasmid pREP41 or derivatives of pREP41 that express RusA (pREP41-rus) or a D70N mutant form of RusA that can bind but not cleave HJs (pREP41-rusD70N) (Doe et al, 2000). The genomic DNA was digested with _Bsu_36I and _Eco_RV and analysed using probes C and D as described in the previous section. No DSBs were detected in DNA from the wild-type strain containing either pREP41 or pREP41-rus (Figure 6D and E, lanes b and d). Curiously, a band of ∼6.5 kb was detected with probe C in DNA from the wild-type strain expressing RusAD70N (Figure 6D, lane c). We are currently uncertain of the significance of this band. Similar to the data in Figure 6B, a band of ∼5.3 kb was detected with probe C in each of the _rqh1_Δ DNA samples (Figure 6D, lanes e–g). This band is indicative of a one-sided DSB occurring within the RTS1 element. The slight discrepancy in size between it and the ∼5.1 kb band detected in Figure 6B, lane b is probably due to differences in the way the two gels have run. The amount of one-sided DSB product is approximately the same in each of the DNA samples (Figure 6D, lanes e–g). These data therefore provide no evidence for HJ formation by fork reversal at the RTS1 RFB in an _rqh1_Δ mutant.
Discussion
Direct repeats of mutant heteroalleles are widely used to monitor the frequency of inter/intra-chromatid recombination in mitotic cells (Klein, 1995). We have further developed this system here to monitor the effect of a site-specific RFB on recombination. Previous studies on the effects of RFBs on recombination in eukaryotes have concentrated on the RFB at the rDNA locus in S. cerevisiae. However, the rDNA is an atypical locus, making extrapolations to the wider genome uncertain. Furthermore, it contains other factors that stimulate recombination, namely the HOT1 site and collisions between replication forks and RNA polymerases (Voelkel-Meiman et al, 1987; Takeuchi et al, 2003), which complicate analysis of the contribution that the RFB makes to recombination. Our system avoids these complications and uncertainties by using the RTS1 RFB positioned at a ‘typical' genomic locus. With this we have confirmed that replication fork blockage by a programmed RFB can induce deletion recombination. It also strongly induces conservative gene conversion events, and we believe that this is the first time that this has been shown for a site-specific RFB. Recombination that is stimulated by replication fork blockage at RTS1 depends mainly on known pathways, which are catalysed by the Rhp51 and Rad22 recombinases (Doe et al, 2004). Furthermore, this recombination is controlled by the Rqh1 DNA helicase. In particular, Rqh1 greatly limits the formation of deletion recombinants seemingly by preventing replication fork breakage.
Recombination at blocked replication forks
Various agents, which are known to cause replication fork stalling or blockage, induce direct repeat recombination. Induction of recombination generally depends on passage through S-phase thereby linking replication fork stalling/blockage with recombinant formation (Galli and Schiestl, 1999). However, it is unclear to what extent recombination is initiated directly at the stalled/blocked fork as opposed to at DSBs that form as a consequence of it. In the case of HU, it seems that most recombinants are formed only after prolonged exposure when the stalled replication forks have broken (Saintigny et al, 2001). In accord with this, it takes ∼16 h for HU to induce Rad52 foci in S. cerevisiae as opposed to ⩽30 min for γ-irradiation, which induces DSBs directly (Lisby et al, 2004). This is also consistent with our data showing that very little direct repeat recombination is induced after a 6-h exposure to HU in S. pombe, even though all the cells are arrested in S-phase.
Similar to HU, the induction of recombination by both the Tus-Ter RFB in E. coli and the rDNA locus RFB in S. cerevisiae correlate with the detection of double-strand ends and DSBs, respectively (Bidnenko et al, 2002; Weitao et al, 2003b). In contrast, replication fork blockage by RTS1 only results in detectable DSBs in an _rqh1_Δ mutant. Furthermore, whereas a DSB at site A induces almost exclusively deletion-type recombinants (Osman et al, 1996), RTS1 at this site induces mostly conversion-types. Similarly, with a DSB at site B >90% of conversion-types are ade6-L469 to ade+ (Osman et al, 1996), compared to ∼10% with RTS1 at this site. Therefore, much of the recombination that is stimulated by replication fork blockage at RTS1 is unlikely to stem from DSB formation.
Intriguingly, UV, like RTS1, induces a high percentage of conversion-types. These may arise from recombination-mediated bypass of UV-induced lesions and/or the repair of daughter strand gaps postreplication (Rupp and Howard-Flanders, 1968; Higgins et al, 1976). Unlike UV, replication fork blockage at RTS1 is not expected to generate lesion-containing single-stranded gaps, which would necessitate recombination for their repair. However, gaps could be generated by replication fork convergence. The normal merging of replication forks is believed to generate fully replicated but intertwined DNA molecules, which are decatenated by topoisomerase II (Top2) (DiNardo et al, 1984; Holm et al, 1989). However, Top2 may be unable to decatenate converging replication forks that arrest prematurely. The concerted action of a RecQ helicase and topoisomerase III may be required here to untwine the DNA between the arrested forks and enable gap filling by a DNA polymerase (Wang, 1991). Possibly recombinases can load at these putative gaps before they are filled in. While this might be a tenable model for recombination induced by RTS1, we prefer the idea that recombinases load directly at the blocked fork and act before convergence. Is there enough time for this to happen, especially as there is no evidence that replication fork blockage at RTS1 induces a checkpoint response (unpublished data)? If we assume that the putative replication origins in Figure 2A fire at about the same time then, based on a fork rate of 3600 bp/min, the fork travelling toward cen3 would encounter RTS1 at site B, approximately 19 min before the opposing fork. Based on the dynamics of recombination proteins revealed by photobleaching studies in mammalian cells (Essers et al, 2002), and by ChIP analyses in S. cerevisiae (Sugawara et al, 2003), it is reasonable to think that this time interval is sufficient for recombination to occur.
The majority of recombinants that are induced by RTS1 depend on Rhp51. We suspect that Rhp51, aided by Rad22, loads onto regions of single-stranded parental or nascent DNA that become exposed at the blocked fork through the action of helicases and/or exonucleases. Strand invasion between different _ade6_− heteroalleles could then lead to gene conversion by mismatch repair and/or DNA synthesis. One way that this could happen is if Rhp51 assembles on the leading nascent strand so that strand invasion generates a 3′ end for priming DNA synthesis (Figure 7A, step 3). Exposure of this strand to Rhp51 might depend on the reversal of the replication fork (Figure 7A, steps 1–2). However, we have failed to detect evidence for this by 2-D gel analysis, so, if the leading strand is used, it might be exposed with little or no fork reversal (not shown). Following DNA synthesis or mismatch repair, the invading strand could be unwound by a DNA helicase (possibly Rqh1 or Srs2) thereby generating a conversion-type recombinant (Figure 7A, steps 3–5). Alternatively, the DNA junctions formed by fork reversal and/or strand invasion could be cleaved by an endonuclease, such as Mus81-Eme1, to generate a deletion-type recombinant (Figure 7A, steps 3a–5a).
Figure 7.
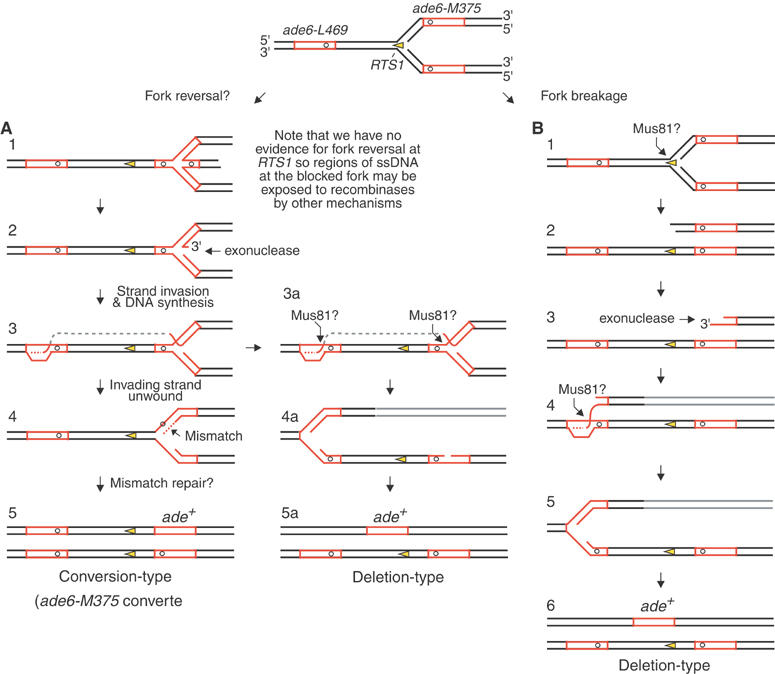
Hypothetical model for the formation of Ade+ recombinants following replication fork blockage at RTS1. Red boxes are the ade6 genes. The open circles indicate the position of point mutations within the ade6 genes. Further details are in the main text.
The role of Rqh1 in limiting recombination at the RTS1 RFB
An important finding of this study is the detection of one-sided DSBs at the RTS1 RFB in an _rqh1_Δ mutant. These are indicative of replication fork breakage and correlate with the increased levels of deletion-type recombinants in this mutant. Similar to _rqh1_Δ mutation, CPT also causes replication fork breakage and induces mainly deletion-type recombinants. These data indicate that one-sided DSBs generally give rise to deletion recombination, and suggest that a failure to prevent replication fork breakage is responsible for the hyper-recombination phenotype of an _rqh1_Δ mutant. The hyper-deletion-type recombination probably occurs by strand invasion from the broken DNA end into the ‘wrong' _ade6_− heteroallele (Figure 7B). However, there is also some stimulation of conversion-type recombination in an _rqh1_Δ mutant, which cannot be explained by the model in Figure 7B. This may be due to a defect in unwinding DNA junctions that are formed by strand invasion (Figure 7A, steps 3–4), which would make them longer-lived and more prone to give rise to conversion-type recombinants.
Similar to our study, increased levels of one-sided DSBs have been detected at the RFB in the rDNA locus of _S. cerevisiae sgs1_Δ mutants (Weitao et al, 2003a). Here, increased breakage is associated with increased pausing at the RFB (Weitao et al, 2003a). It is therefore unclear whether the extra breakage is due to a defect in maintaining fork stability, or simply that there are more paused forks available to be broken. In contrast to these data, we have not detected an increase in blocked forks at the RTS1 RFB in an _rqh1_Δ mutant. We can therefore make a more definite link between DNA breakage and a failure to maintain fork stability. However, we cannot exclude the possibility that the level of breakage in an _rqh1_Δ mutant is the same as in the wild type, and that the breaks are only detected in the mutant because their processing is impaired. This idea is plausible because in recBC mutants of E. coli, RecQ, together with the single-strand exonuclease RecJ, appear to perform DSB resection (Kuzminov, 1999). Nevertheless, the accumulating evidence that RecQ helicases act directly at stalled/blocked replication forks (Courcelle and Hanawalt, 1999; Cobb et al, 2003; Courcelle et al, 2003; Hishida et al, 2004; Vengrova and Dalgaard, 2004), together with the hyper-deletion-type recombination in an _rqh1_Δ mutant, encourages us to believe that the breaks we observe are a consequence of a failure to protect the blocked fork from nucleolytic attack.
Based on current knowledge of RecQ helicases, Rqh1 could help protect the fork in several different ways, including: promoting the stability of replisome components at the blocked fork (Cobb et al, 2003); exposing ssDNA at the blocked fork to enable recombinases to load, which in turn might protect the fork (Courcelle and Hanawalt, 1999; Hishida et al, 2004); and by unwinding DNA junctions that might otherwise be cleaved by an endonuclease (Doe et al, 2000, 2002; Karow et al, 2000). These DNA junctions could include HJs formed by fork reversal, which might be cleaved by an HJ resolvase in the absence of Rqh1 (Doe et al, 2000). However, although such reversed forks have been detected at the mat1 RFB (Vengrova and Dalgaard, 2004), and are observed in S. cerevisiae rad53 mutants following HU treatment (Sogo et al, 2002), we were unable to detect them at the RTS1 RFB. Furthermore, the expression of RusA in either a wild-type or _rqh1_Δ mutant failed to generate more one-sided DSBs. Possibly fork reversal is not that extensive at the RTS1 RFB and therefore tends not to form HJs. Instead, the replication fork may be blocked in such a manner to make it suitable for cleavage, with little or no reversal, by an endonuclease like Mus81-Eme1 (Whitby et al, 2003). Indeed, replication fork blockage at RTS1 may be similar to that at the RFB in the rDNA locus of S. cerevisiae. Here, the leading strand halts three bases behind the nascent lagging strand (Burkhalter and Sogo, 2004). In theory, Mus81 could cleave this blocked fork. Intriguingly, genetic studies indicate that Mus81-Eme1 shares an overlapping role with Rqh1 for processing DNA junctions (Boddy et al, 2000; Doe et al, 2002). This makes Mus81-Eme1 a prime candidate for making the breaks that we detect in an _rqh1_Δ mutant. Unfortunately, an _rqh1_Δ _mus81_Δ double mutant is unviable, and so prevents us from directly testing this idea.
Concluding remarks
As mentioned above, the normal function of RTS1 is to ensure the directionality of replication across mat1 (Dalgaard and Klar, 2001). One might have thought, therefore, that forks blocked at RTS1 would simply be stabilised to await fork convergence. Certainly, this is what appears to happen at the Tus-Ter RFB in E. coli, which only provokes recombination if a second round of DNA replication runs into the back of the blocked fork—a reaction that is unlikely to occur in S. pombe (Bidnenko et al, 2002). It is surprising, therefore, that recombination is stimulated at forks blocked by RTS1 positioned at an ectopic site. Possibly, RTS1 acts aberrantly when placed at an ectopic site, and at its normal genomic locus recombination is efficiently suppressed. Alternatively, the action of recombinases at RTS1 may be designed to help maintain fork stability by promoting sister chromatid interactions, and therefore could occur at both normal and ectopic RTS1 sites. This would only result in detectable genetic change when the block is within regions of repetitive DNA. However, we have not detected any difference in the growth or viability of recombination-deficient (e.g. _rad22_−) strains with or without RTS1 in orientation 1 or 2 (data not shown). It would seem, therefore, that recombinase action at forks blocked at RTS1 within the _ade6_− direct repeat is unnecessary for growth and survival. Nevertheless, regardless of its raison d'être, our data suggests that recombination may be a common feature at programmed RFBs in eukaryotes, especially knowing that RTS1 shares characteristics with RFBs in mammalian genomes (Codlin and Dalgaard, 2003). Further analysis of the events at RTS1 may also provide new clues to the actions of recombinases at accidental RFBs. These studies are in progress.
Materials and methods
Strains, plasmids and probes
Details of the strains, plasmids and probes are in Supplementary data.
Media and general genetic methods
Media for growing S. pombe and general genetic methods are as described (Gutz et al, 1974).
Recombination assays
Spontaneous, UV-induced and _RTS1_-induced recombinant frequencies were measured as described (Osman et al, 2000). The modifications to this procedure used to assess the effect of CPT and HU are described in Supplementary data. Recombinant frequencies are the mean from at least three independent assays where five independent colonies were tested in each assay. Two sample _t_-tests were used to determine the statistical significance of differences in recombination frequencies.
Preparations of S. pombe genomic DNA
DNA for 2-D gels was isolated by CTAB extraction (Lopes et al, 2003). Following restriction digestion, replicating DNA was enriched by fractionation on a benzoylated naphthoylated DEAE (BND)-cellulose (Sigma) column. The preparation of genomic DNA in agarose plugs was performed using a CHEF genomic DNA plug kit (Bio-Rad) according to the manufacturer's instructions. Further details are in Supplementary data.
2-D and 1-D gels
Neutral/neutral 2-D gels were run as described (Brewer and Fangman, 1987). 1-D gels were either 0.7 or 0.8% agarose and were run in 1 × TBE at 50–60 V for ∼16 h. Gels were Southern blotted and probed with the indicated 32P-labelled DNA fragments. Blots were analysed by phosphorImaging using a Fuji FLA3000 and Image Gauge software. Further details are in Supplementary data.
Supplementary Material
Supplementary Material
Acknowledgments
We thank Jacob Dalgaard for the gift of pBZ142 and the _swi1_Δ strain as well as for advice on 2-D gels, and Julie Dixon for technical support. This work was supported by a project grant (065278/Z/01/Z) and Senior Research Fellowship from The Wellcome Trust awarded to MCW.
References
- Bidnenko V, Ehrlich SD, Michel B (2002) Replication fork collapse at replication terminator sequences. EMBO J 21: 3898–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy MN, Lopez-Girona A, Shanahan P, Interthal H, Heyer WD, Russell P (2000) Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1. Mol Cell Biol 20: 8758–8766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer BJ, Fangman WL (1987) The localization of replication origins on ARS plasmids in S. cerevisiae. Cell 51: 463–471 [DOI] [PubMed] [Google Scholar]
- Brewer BJ, Fangman WL (1988) A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell 55: 637–643 [DOI] [PubMed] [Google Scholar]
- Burkhalter MD, Sogo JM (2004) rDNA enhancer affects replication initiation and mitotic recombination: Fob1 mediates nucleolytic processing independently of replication. Mol Cell 15: 409–421 [DOI] [PubMed] [Google Scholar]
- Cobb JA, Bjergbaek L, Shimada K, Frei C, Gasser SM (2003) DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J 22: 4325–4336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codlin S, Dalgaard JZ (2003) Complex mechanism of site-specific DNA replication termination in fission yeast. EMBO J 22: 3431–3440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle J, Donaldson JR, Chow KH, Courcelle CT (2003) DNA damage-induced replication fork regression and processing in Escherichia coli. Science 299: 1064–1067 [DOI] [PubMed] [Google Scholar]
- Courcelle J, Hanawalt PC (1999) RecQ and RecJ process blocked replication forks prior to the resumption of replication in UV-irradiated Escherichia coli. Mol Gen Genet 262: 543–551 [DOI] [PubMed] [Google Scholar]
- Dalgaard JZ, Klar AJ (1999) Orientation of DNA replication establishes mating-type switching pattern in S. pombe. Nature 400: 181–184 [DOI] [PubMed] [Google Scholar]
- Dalgaard JZ, Klar AJ (2000) swi1 and swi3 perform imprinting, pausing, and termination of DNA replication in S. pombe. Cell 102: 745–751 [DOI] [PubMed] [Google Scholar]
- Dalgaard JZ, Klar AJ (2001) A DNA replication-arrest site RTS1 regulates imprinting by determining the direction of replication at mat1 in S. pombe. Genes Dev 15: 2060–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNardo S, Voelkel K, Sternglanz R (1984) DNA topoisomerase II mutant of Saccharomyces cerevisiae: topoisomerase II is required for segregation of daughter molecules at the termination of DNA replication. Proc Natl Acad Sci USA 81: 2616–2620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe CL, Ahn JS, Dixon J, Whitby MC (2002) Mus81-Eme1 and Rqh1 involvement in processing stalled and collapsed replication forks. J Biol Chem 277: 32753–32759 [DOI] [PubMed] [Google Scholar]
- Doe CL, Dixon J, Osman F, Whitby MC (2000) Partial suppression of the fission yeast rqh1(−) phenotype by expression of a bacterial Holliday junction resolvase. EMBO J 19: 2751–2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe CL, Osman F, Dixon J, Whitby MC (2004) DNA repair by a Rad22-Mus81-dependent pathway that is independent of Rhp51. Nucleic Acids Res 32: 5570–5581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers J, Houtsmuller AB, van Veelen L, Paulusma C, Nigg AL, Pastink A, Vermeulen W, Hoeijmakers JH, Kanaar R (2002) Nuclear dynamics of RAD52 group homologous recombination proteins in response to DNA damage. EMBO J 21: 2030–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli A, Schiestl RH (1999) Cell division transforms mutagenic lesions into deletion-recombinagenic lesions in yeast cells. Mutat Res 429: 13–26 [DOI] [PubMed] [Google Scholar]
- Gutz H, Heslot H, Leopold U, Loprieno N (eds) (1974) Schizosaccharomyces pombe. New York: Plenum Press [Google Scholar]
- Hickson ID (2003) RecQ helicases: caretakers of the genome. Nat Rev Cancer 3: 169–178 [DOI] [PubMed] [Google Scholar]
- Higgins NP, Kato K, Strauss B (1976) A model for replication repair in mammalian cells. J Mol Biol 101: 417–425 [DOI] [PubMed] [Google Scholar]
- Hishida T, Han YW, Shibata T, Kubota Y, Ishino Y, Iwasaki H, Shinagawa H (2004) Role of the Escherichia coli RecQ DNA helicase in SOS signaling and genome stabilization at stalled replication forks. Genes Dev 18: 1886–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm C, Stearns T, Botstein D (1989) DNA topoisomerase II must act at mitosis to prevent nondisjunction and chromosome breakage. Mol Cell Biol 9: 159–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G, Malkova A, Liberi G, Foiani M, Haber JE (2003) Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell 115: 401–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaliraman V, Mullen JR, Fricke WM, Bastin-Shanower SA, Brill SJ (2001) Functional overlap between Sgs1-Top3 and the Mms4-Mus81 endonuclease. Genes Dev 15: 2730–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karow JK, Constantinou A, Li JL, West SC, Hickson ID (2000) The Bloom's syndrome gene product promotes branch migration of Holliday junctions. Proc Natl Acad Sci USA 97: 6504–6508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein HL (1995) Genetic control of intrachromosomal recombination. Bioessays 17: 147–159 [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Heck DJ, Nomura M, Horiuchi T (1998) Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev 12: 3821–3830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krabbe M, Zabielski J, Bernander R, Nordstrom K (1997) Inactivation of the replication–termination system affects the replication mode and causes unstable maintenance of plasmid R1. Mol Microbiol 24: 723–735 [DOI] [PubMed] [Google Scholar]
- Kuzminov A (1999) Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol Mol Biol Rev 63: 751–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Barlow JH, Burgess RC, Rothstein R (2004) Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118: 699–713 [DOI] [PubMed] [Google Scholar]
- Liu J, Xu L, Sandler SJ, Marians KJ (1999) Replication fork assembly at recombination intermediates is required for bacterial growth. Proc Natl Acad Sci USA 96: 3552–3555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M, Cotta-Ramusino C, Liberi G, Foiani M (2003) Branch migrating sister chromatid junctions form at replication origins through Rad51/Rad52-independent mechanisms. Mol Cell 12: 1499–1510 [DOI] [PubMed] [Google Scholar]
- Lopes M, Cotta-Ramusino C, Pellicioli A, Liberi G, Plevani P, Muzi-Falconi M, Newlon CS, Foiani M (2001) The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412: 557–561 [DOI] [PubMed] [Google Scholar]
- McGlynn P, Lloyd RG (2002) Recombinational repair and restart of damaged replication forks. Nat Rev Mol Cell Biol 3: 859–870 [DOI] [PubMed] [Google Scholar]
- Michel B, Grompone G, Flores MJ, Bidnenko V (2004) Multiple pathways process stalled replication forks. Proc Natl Acad Sci USA 101: 12783–12788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi E, Noguchi C, Du LL, Russell P (2003) Swi1 prevents replication fork collapse and controls checkpoint kinase Cds1. Mol Cell Biol 23: 7861–7874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman F, Adriance M, McCready S (2000) The genetic control of spontaneous and UV-induced mitotic intrachromosomal recombination in the fission yeast Schizosaccharomyces pombe. Curr Genet 38: 113–125 [DOI] [PubMed] [Google Scholar]
- Osman F, Fortunato EA, Subramani S (1996) Double-strand break-induced mitotic intrachromosomal recombination in the fission yeast Schizosaccharomyces pombe. Genetics 142: 341–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y, Redon C, Rao VA, Seiler JA, Sordet O, Takemura H, Antony S, Meng L, Liao Z, Kohlhagen G, Zhang H, Kohn KW (2003) Repair of and checkpoint response to topoisomerase I-mediated DNA damage. Mutat Res 532: 173–203 [DOI] [PubMed] [Google Scholar]
- Rothstein R, Michel B, Gangloff S (2000) Replication fork pausing and recombination or ‘gimme a break'. Genes Dev 14: 1–10 [PubMed] [Google Scholar]
- Rupp WD, Howard-Flanders P (1968) Discontinuities in the DNA synthesized in an excision-defective strain of Escherichia coli following ultraviolet irradiation. J Mol Biol 31: 291–304 [DOI] [PubMed] [Google Scholar]
- Saintigny Y, Delacote F, Vares G, Petitot F, Lambert S, Averbeck D, Lopez BS (2001) Characterization of homologous recombination induced by replication inhibition in mammalian cells. EMBO J 20: 3861–3870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segurado M, de Luis A, Antequera F (2003) Genome-wide distribution of DNA replication origins at A+T-rich islands in Schizosaccharomyces pombe. EMBO Rep 4: 1048–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seigneur M, Bidnenko V, Ehrlich SD, Michel B (1998) RuvAB acts at arrested replication forks. Cell 95: 419–430 [DOI] [PubMed] [Google Scholar]
- Sogo JM, Lopes M, Foiani M (2002) Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297: 599–602 [DOI] [PubMed] [Google Scholar]
- Sugawara N, Wang X, Haber JE (2003) In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol Cell 12: 209–219 [DOI] [PubMed] [Google Scholar]
- Takeuchi Y, Horiuchi T, Kobayashi T (2003) Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes Dev 17: 1497–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vengrova S, Codlin S, Dalgaard JZ (2002) _RTS1_—an eukaryotic terminator of replication. Int J Biochem Cell Biol 34: 1031–1034 [DOI] [PubMed] [Google Scholar]
- Vengrova S, Dalgaard JZ (2004) RNase-sensitive DNA modification(s) initiates S. pombe mating-type switching. Genes Dev 18: 794–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelkel-Meiman K, Keil RL, Roeder GS (1987) Recombination-stimulating sequences in yeast ribosomal DNA correspond to sequences regulating transcription by RNA polymerase I. Cell 48: 1071–1079 [DOI] [PubMed] [Google Scholar]
- Wang JC (1991) DNA topoisomerases: why so many? J Biol Chem 266: 6659–6662 [PubMed] [Google Scholar]
- Weitao T, Budd M, Campbell JL (2003a) Evidence that yeast SGS1, DNA2, SRS2, and FOB1 interact to maintain rDNA stability. Mutat Res 532: 157–172 [DOI] [PubMed] [Google Scholar]
- Weitao T, Budd M, Hoopes LL, Campbell JL (2003b) Dna2 helicase/nuclease causes replicative fork stalling and double-strand breaks in the ribosomal DNA of Saccharomyces cerevisiae. J Biol Chem 278: 22513–22522 [DOI] [PubMed] [Google Scholar]
- Whitby MC, Osman F, Dixon J (2003) Cleavage of model replication forks by fission yeast Mus81-Eme1 and budding yeast Mus81-Mms4. J Biol Chem 278: 6928–6935 [DOI] [PubMed] [Google Scholar]
- Wu L, Hickson ID (2003) The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature 426: 870–874 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material