Phylogenetic Analysis of Enterovirus 71 Strains Isolated during Linked Epidemics in Malaysia, Singapore, and Western Australia (original) (raw)
Abstract
Enterovirus 71 (EV71) is a frequent cause of hand, foot, and mouth disease (HFMD) epidemics associated with severe neurological sequelae in a small proportion of cases. There has been a significant increase in EV71 epidemic activity throughout the Asia-Pacific region since 1997. Recent HFMD epidemics in this region have been associated with a severe form of brainstem encephalitis associated with pulmonary edema and high case fatality rates. In this study, we show that four genetic lineages of EV71 have been prevalent in the Asia-Pacific region since 1997, including two previously undescribed genogroups (B3 and B4). Furthermore, we show that viruses belonging to genogroups B3 and B4 have circulated endemically in Southeast Asia during this period and have been the primary cause of several large HFMD or encephalitis epidemics in Malaysia, Singapore, and Western Australia.
Since the initial description of enterovirus 71 (EV71) in the medical literature in 1974 (27), outbreaks of infection with this virus have occurred periodically throughout the world (1, 9, 12, 13, 15, 16, 26). EV71 infection manifests most frequently as a mild childhood exanthem known as hand, foot, and mouth disease (HFMD) and is clinically indistinguishable from HFMD caused by the closely related virus coxsackievirus A16 (CA16). In addition, a small proportion of acute EV71 infections are associated with severe neurological disease (1, 9, 12).
EV71 and CA16 are two distinct serotypes belonging to the human enterovirus A species (18). A recent study of EV71 evolution (2) has demonstrated the development of three independent genetic lineages (A, B, and C) over a 30-year period. Genogroup A includes a single virus, the prototype strain BrCr-CA-70. All other EV71 strains so far identified belong to either genogroup B or C, which are divided into two sublineages, B1 and B2 and C1 and C2, respectively (2).
Since 1997, several large epidemics of EV71 infection have occurred in the Asia-Pacific region, the first being reported in Sarawak (Malaysian Borneo) in 1997 (5, 6), followed by smaller outbreaks in peninsular Malaysia in 1997 (20) and Singapore in 1998 (28). As with previous EV71 epidemics, numerous cases of HFMD were reported, with neurological complications arising in a small proportion of cases. In addition, many cases of brainstem encephalitis with pulmonary edema and a high case fatality rate (7, 21) were described during these outbreaks. Twenty-nine fatal cases of this disease were reported in Sarawak (5, 6) and twelve in peninsular Malaysia (20). During 1998, a large HFMD epidemic due to EV71 occurred in Taiwan in which 405 cases of severe neurological disease and 78 fatal cases of brainstem encephalitis and neurogenic pulmonary edema were reported (14, 19). In 1999, a large epidemic of HFMD due to EV71 occurred in Perth, Western Australia (WA) (23), and included 14 cases of severe neurological disease, although no fatal cases were identified. EV71 epidemic activity has continued in the region during 2000–2001, with EV71 isolation from cases of HFMD and encephalitis in Sarawak, peninsular Malaysia, Singapore, and WA.
In this study, we report on the molecular epidemiology of EV71 outbreaks in Malaysia, Singapore, and WA between 1997 and 2001. The 66 EV71 strains examined in this study, together with the year of isolation, source of virus isolate, associated clinical illnesses, and GenBank accession numbers are listed in Table 1. Virus isolation was undertaken in cell culture, and all isolates were formally identified by neutralization using the EV71-specific polyclonal antiserum 385JS (16).
TABLE 1.
Clinical isolates used in the phylogenetic analysis of EV71a
Viral RNA was extracted from cell culture supernatant using either the QIAamp viral RNA extraction kit (Qiagen) or the High Pure viral nucleic acid extraction kit (Roche) and the VP1 gene amplified by reverse transcriptase-PCR (RT-PCR). RT-PCR was undertaken in a one-tube reaction (50 μl) containing 5μl of viral RNA, 25 pmol of each primer, 50 mM Tris-HCl (pH 8.3), 2 mM MgCl2, 0.2 mM deoxynucleoside triphosphate mix, 16 U of RNase inhibitor, 12 U of avian myeloblastosis virus reverse transcriptase, and 2 U of Taq DNA polymerase. First-strand cDNA synthesis was for 30 min at 42°C. Samples were then subjected to 35 cycles of denaturation at 94°C for 30 s, annealing at 47°C for 45 s, and extension at 68°C for 1 min. The VP1 gene was amplified in two overlapping fragments using primer pairs 159/162 and 161/NP1A (2). The two enterovirus VP1 gene cDNA fragments were gel purified, and both strands were sequenced.
Alignment of the complete VP1 gene sequences was undertaken using the ClustalW program (11, 29). Phylogenetic trees were constructed by neighbor joining using the Kimura two-parameter distance method (17) with Phylip, version 3.5 (10), and trees were drawn with the program TreeView (24). The reliability of the neighbor-joining tree was estimated by bootstrap analysis using 1,000 pseudoreplicate data sets generated by the program Seqboot (11). The previously published VP1 sequences of BrCr-CA-70, 2609-AUS-74, 2610-AUS-74, 2623-AUS-86, 2640-AUS-95, 2641-AUS-95, 2642-AUS-95, 2644-AUS-95, 2258-CA-79, 7628-PA-87, 7673-CT-87, 7238-AK-87, 2222-IA-88, 0926-OR-91, and 2286-TX-97 (2, 3) were used to reconstruct the three EV71 genogroups (A, B1/2, and C1/2) identified by Brown et al. (2). In addition, the VP1 sequences of five Taiwanese EV71 strains from the 1998 epidemic, TW-1465-98 (AF116814), TW-1567-98 (AF116810), TW-1743-98 (AF116816), TW-2086-98 (AF119796), and NCKU9822 (AF136379) (30), two VP1 sequences from EV71 strains isolated in peninsular Malaysia during 1997, 2289-MAA-97 (AF135914) and 0756-MAA-97 (AF135935) (2), and a VP1 sequence from the 1997 Singaporean EV71 isolate 18-SIN-97 (AF251359) (28) were obtained from GenBank. The VP1 sequence of the prototype CA16 strain G-10 (U05876) (25) was included in the phylogenetic analysis as an outgroup.
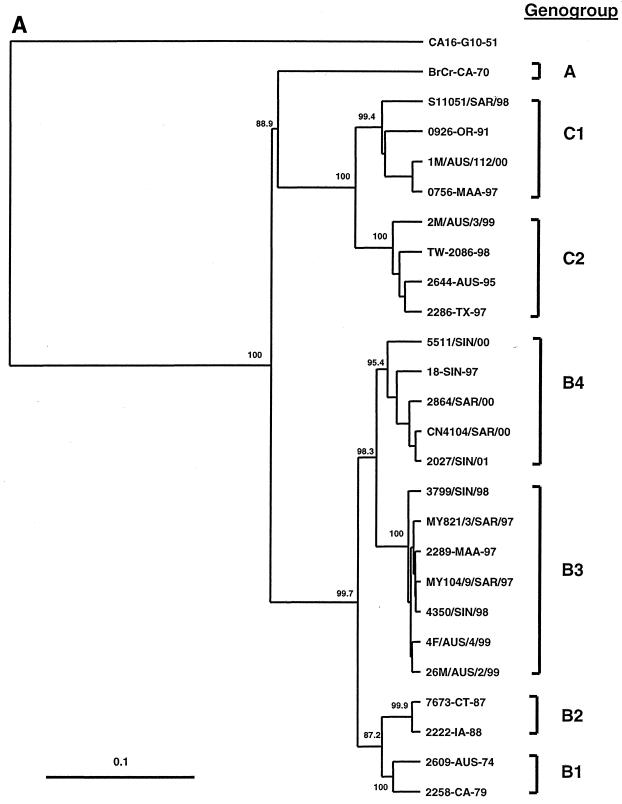
Phylogenetic analysis of the 66 EV71 isolates was based on the alignment of complete (891 nucleotide) VP1 gene sequences and construction of phylogenetic trees using the neighbor-joining method (Fig. 1). The majority of the EV71 strains isolated in Sarawak, Singapore, and WA since 1997 belong to two new genetic lineages within genogroup B (Fig. 1A and 1B); we have called these new lineages B3 (Malaysia 1997, Singapore 1998, and WA 1999) and B4 (Malaysia and Singapore 2000–2001) to distinguish them from the previously described B1 and B2 lineages (2). The separation of genogroups B3 and B4 from one another and from genogroups B1 and B2 is strongly supported by bootstrap analysis. Our interpretation of the phylogenetic data contrasts with that of Singh et al. (28), who suggested that recent EV71 isolates from Singapore and Malaysia belong to a completely new genogroup, which they labeled genogroup D. In the Singh et al. study (28), phylogenetic analysis was based on a smaller number of nucleotides within VP1 (341 nucleotides) than in our study, in which the complete VP1 gene (891 nucleotides) was analyzed. As shown in Fig. 1A, our analysis has accurately reproduced the A, B1/2, and C1/2 genogroups identified by Brown et al. (2). Using this approach, it is clear that the recent Southeast Asian EV71 strains are more closely related to EV71 strains within genogroup B, with which they show 89 to 91% nucleotide sequence identity, than to viruses from genogroups A and C, with which they show 79 to 83% nucleotide sequence identity.
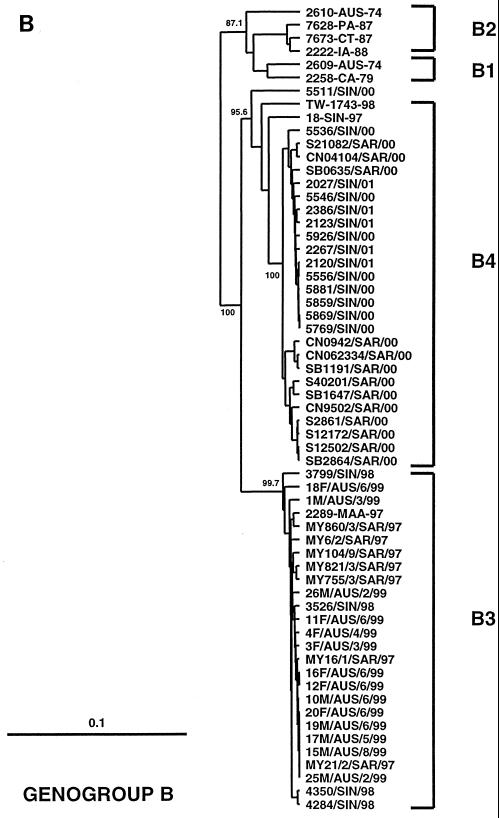
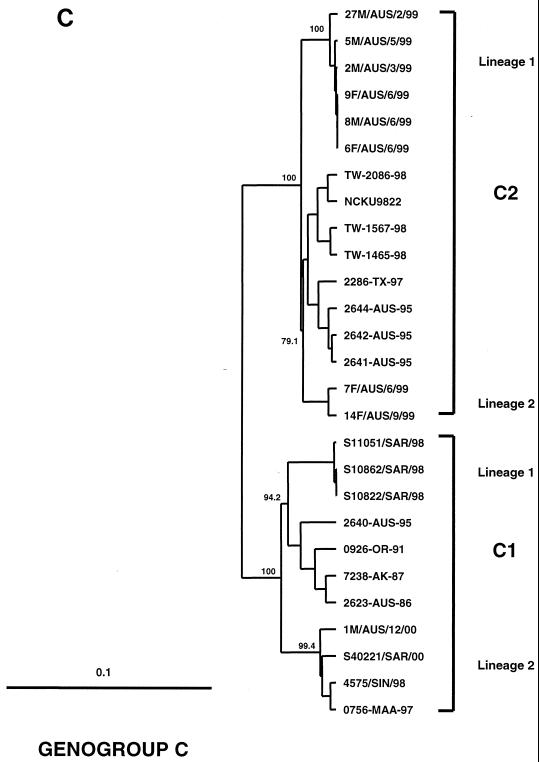
FIG. 1.
Phylogenetic trees showing genetic relationships among 66 EV71 field isolates based on alignment of the complete VP1 gene sequence (nucleotide positions 2442 to 3332). Branch lengths are proportional to the number of nucleotide differences. Most strain names indicate a unique number/country or U.S. state of isolation/year of isolation. The trees were constructed by neighbor joining using the Kimura two-parameter distance method (17). The bootstrap values in 1,000 pseudoreplicates for major lineages within the tree are shown as percentages. The marker denotes a measurement of relative phylogenetic distance. In panels B and C, the branches for genogroup A (BrCr-CA-70) and the outgroup (CA16-G10-51) have been removed from the dendrograms to save space. (A) Simplified dendrogram showing genogroups A, B, and C identified by Brown et al. (2). (B) Detailed dendrogram of strains belonging to genogroup B. (C) Detailed dendrogram of strains belonging to genogroup C.
Genogroup B3 (Fig. 1B) comprises viruses isolated in peninsular Malaysia, Sarawak, Singapore, and WA between 1997 and 1999; the nucleotide sequence identity of virus strains within genogroup B3 is >97%. Genogroup B4 is composed primarily of viruses isolated in Sarawak and Singapore during 2000–2001; the nucleotide sequence identity of virus strains within genogroup B4 is >98%. Genogroups B3 and B4 show 90 to 92% nucleotide sequence identity with one another and 89 to 91% nucleotide sequence identity with viruses from genogroups B1 and B2. Although viruses belonging to genogroups B3 and B4 appear to be derived from an unidentified common ancestor, their phylogenetic relationship to one another remains unclear. Genogroup B3 viruses circulated widely in Southeast Asia and WA between 1997 and 1999 but have not been isolated since that time. Although genogroup B4 viruses have been predominant in the region during 2000–2001, it is of interest that viruses belonging to this genogroup were identified in Singapore as early as 1997 (18-SIN-97) and in Taiwan during the 1998 epidemic (TW-1743-98). These data clearly indicate that genogroups B3 and B4 have cocirculated in the Asia-Pacific region since 1997 and that genogroup B4 has not evolved recently from genogroup B3.
It is also apparent that several sublineages of genogroups B3 and B4 have circulated in Malaysia and Singapore (Fig. 1B), suggesting that these genetic lineages circulated endemically in Southeast Asia for a considerable time prior to 1997. By contrast, genogroup B3 viruses isolated during the WA HFMD epidemic appear to belong to a single genetic lineage and are closely related to viruses isolated in Sarawak during 1997 (>99% nucleotide sequence identity). This, together with the failure to identify EV71 by routine surveillance until 1 month prior to the epidemic (P. C. McMinn, unpublished data), suggests that EV71 was introduced into WA immediately prior to the onset of the epidemic.
The phylogenetic relationships between recent Asian and Australian EV71 isolates belonging to genogroup C are presented in Fig. 1C. Several viruses belonging to genogroup C1 were isolated from cases of uncomplicated HFMD in Singapore and Sarawak during 1998 and in Sarawak and WA during 2000. Two lineages of genogroup C1 have circulated in the Asia-Pacific region between 1997 and the present. Lineage 1 (>98% nucleotide sequence identity) appears to have been confined to Sarawak during 1998 (S10822/SAR/98, S10862/SAR/98, and S11051/SAR/98). Lineage 2 (>98% nucleotide sequence identity) circulated more widely within the region during 1997 (0756-MAA-97), 1998 (4575/SIN/98), and 2000 (S40221/SAR/00 and 1M/AUS/12/00). These two lineages show 94 to 95% nucleotide sequence identity with one another and 91 to 93% identity with other members of genogroup C1.
A group of viruses isolated during the 1999 HFMD epidemic in WA belong to genogroup C2. These viruses show 98 to 99% nucleotide sequence identity to one another and 95 to 97% identity to other members of genogroup C2. The WA genogroup C2 viruses are most closely related to EV71 strains isolated in Victoria (eastern Australia) in 1995 (96 to 97% nucleotide sequence identity) and to genogroup C2 viruses isolated in Taiwan in 1998 (95 to 96% nucleotide sequence identity). The WA genogroup C2 viruses have formed two independent genetic lineages (Fig. 1C), both of which are closely related to viruses isolated in Victoria in 1995, suggesting that these viruses have descended from strains previously circulating in Australia. From our analysis, it appears that genogroup C2 was the major source of virus for the Taiwanese HFMD epidemic in 1998 (Fig. 1C), although an isolate belonging to genogroup B4 (TW-1743-98) was also identified (Fig. 1B). In contrast to our data, Chu et al. (8) have suggested that a new genetic lineage within genogroup C, which they have called genogroup C3, was primarily responsible for the 1998 Taiwanese HFMD epidemic. Unfortunately, direct comparison with our data is not possible, as the VP4 region rather than VP1 was chosen for the phylogenetic analysis in this study.
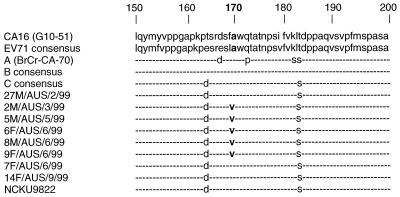
The genetic determinants of EV71 virulence remain unknown. During the EV71 epidemic in WA, viruses belonging to lineage 1 of genogroup C2 were isolated from all of the identified cases of severe neurological disease and from only one case of uncomplicated HFMD (Table 1, Fig. 1C). By contrast, genogroup B3 viruses were isolated mainly from children with uncomplicated HFMD and aseptic meningitis and from cases of postinfectious neurological disease (Table 1, Fig. 1B). The VP1 gene has been shown to be a source of virulence determinants in animal models of both coxsackievirus B4 (4) and poliovirus chimeras (22). In order to determine if mutations in the VP1 gene were associated with the observed differences in neurovirulence of the WA viruses, we compared the VP1 deduced amino acid sequences of genogroup C2 (lineage 1) viruses from WA with VP1 consensus amino acid sequences for EV71 (2), genogroups A, B, and C (2), and CA16 (25). A partial VP1 deduced amino acid sequence alignment is presented in Fig. 2. Amino acid position 170 is part of a highly conserved region of the enterovirus VP1 protein and has an alanine residue in CA16 and in all of the EV71 consensus sequences. Five genogroup C2 (lineage 1) isolates (2M/AUS/3/99, 5M/AUS/5/99, 6F/AUS/6/99, 8M/AUS/6/99, and 9F/AUS/6/99), obtained from children with severe neurological disease during the WA epidemic (23), have an identical amino acid sequence in VP1, including an alanine to valine (A→V) substitution at position 170. The earliest virus isolate belonging to this lineage (27M/AUS/2/99), obtained from a case of uncomplicated HFMD in February 1999, has alanine (wild type) at position 170 in VP1. This is the only amino acid difference in VP1 between 27M/AUS/2/99 and the five other members of the lineage. All other EV71 isolates examined in this study, including the two WA genogroup C2 (lineage 2) isolates 7F/AUS/6/99 and 14F/AUS/9/99, have alanine at VP1 position 170. These data indicate that the VP1 170(A→V) substitution may be associated with increased neurovirulence of EV71. However, as we have not examined the complete nucleotide sequences of the six WA genogroup C2 (lineage 1) viruses, we cannot rule out the possibility that observed differences in the clinical outcome of infection are due to other factors, such as attenuating mutations within other regions of the viral genome, or to different host susceptibility factors.
FIG. 2.
Partial alignment of deduced VP1 amino acid sequences (residues 150 to 200) of the EV71 isolates from WA belonging to genogroup C2. The deduced amino acid sequence in the same region of VP1 is also shown for CA16-G10-51 (25), the EV71 consensus sequence (2), genogroup A (BrCr-CA-70), consensus sequences for genogroups B and C (2), and the 1998 Taiwanese isolate NCKU9822 (30). Amino acid residues that are identical to those in the EV71 consensus sequence are denoted with hyphens.
In contrast to the WA genogroup C2 viruses, there does not appear to be a single “neurovirulent” strain of EV71 associated with fatal cases in the Asia-Pacific region. Genogroup B3 and B4 viruses have been isolated from fatal cases in Sarawak, peninsular Malaysia, and Singapore, and a genogroup C2 virus was isolated from a fatal case in Taiwan (30). Furthermore, fatal pulmonary edema cases were not seen in WA despite the isolation of genogroup B3 strains with a VP1 nucleotide sequence almost identical to that of viruses isolated from fatal cases in Sarawak and Singapore (Fig. 1B). Examination of the deduced VP1 amino acid sequences of genogroup B3 and B4 viruses did not provide evidence for specific amino acid residues linked to severe or fatal cases (data not shown). Alternative explanations for the pathogenesis of brainstem encephalitis and pulmonary edema may be that virulence determinants for this syndrome are located within other regions of the EV71 genome or that the disease results from an interaction between EV71 and other concurrent viral infections, as has been suggested for adenoviruses (5). In this study, viruses from genogroup C1 were cultured only from isolated cases of uncomplicated HFMD, suggesting that this genogroup may have lower epidemic and neurovirulence potential than viruses belonging to genogroups B3, B4, and C2. Other recent genogroup C1 isolates from this region were also obtained from cases of uncomplicated HFMD, including 2640-AUS-95 (M. Kennett, personal communication) and 0756-MAA-97 (2).
In conclusion, recent EV71 epidemic activity in the Asia-Pacific region (1997 to 2001) has been caused by viruses belonging to at least three genogroups, B3 (Southeast Asia and WA), B4 (Southeast Asia and Taiwan), and C2 (Taiwan and WA). Viruses belonging to genogroup C1 have shown low-level endemic activity in the region over the same time period. Genogroup C2 (lineage 1) viruses isolated during the WA epidemic were strongly linked to severe neurological disease. The increased neurovirulence of these viruses is associated with an A→V mutation at amino acid position 170 in VP1. The VP1 170(A→V) mutation appears to be a marker for a neurovirulent lineage, and it is possible that this mutation is a virulence determinant. Further studies on the role of this residue in the neurovirulence of EV71 will be undertaken by site-directed mutagenesis of an infectious cDNA clone of EV71 (R. J. Hurrelbrink and P. C. McMinn, unpublished data).
Acknowledgments
This study was supported by grants from the Princess Margaret Hospital for Children Research Fund and the Royal College of Pathologists of Australasia and a special grant from the state government of Sarawak, Malaysia.
We thank Margery Kennett, Victorian Infectious Diseases Reference Laboratory, Melbourne, Australia, for provision of the rabbit polyclonal EV71 antiserum 385JS. We also thank Margaret Laasonen for expert assistance with cell cultures and Seng Eng Hong for invaluable assistance with neutralization assays.
REFERENCES
- 1.Alexander J P, Baden L, Pallansch M A, et al. Enterovirus infections and neurologic disease — United States, 1977–91. J Infect Dis. 1994;169:905–908. doi: 10.1093/infdis/169.4.905. [DOI] [PubMed] [Google Scholar]
- 2.Brown B A, Oberste M S, Alexander J P, et al. Molecular epidemiology and evolution of enterovirus 71 strains isolated from 1970 to 1998. J Virol. 1999;73:9969–9975. doi: 10.1128/jvi.73.12.9969-9975.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown B A, Pallansch M A. Complete nucleotide sequence of enterovirus 71 is distinct from poliovirus. Virus Res. 1995;39:195–205. doi: 10.1016/0168-1702(95)00087-9. [DOI] [PubMed] [Google Scholar]
- 4.Caggana M, Chan P, Ramsingh A. Identification of a single amino acid residue in the capsid protein VP1 of Coxsackievirus B4 that determines the virulence phenotype. J Virol. 1993;67:4797–4803. doi: 10.1128/jvi.67.8.4797-4803.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cardosa M J, Krishnan S, Tio P H, et al. Isolation of subgenus B adenovirus during a fatal outbreak of enterovirus 71-associated hand, foot and mouth disease in Sibu, Sarawak. Lancet. 1999;354:987–991. doi: 10.1016/S0140-6736(98)11032-2. [DOI] [PubMed] [Google Scholar]
- 6.Chan L G, Parashar U D, Lye M S, et al. Deaths of children during an outbreak of hand, foot and mouth disease in Sarawak, Malaysia: clinical and pathological characteristics of the disease. Clin Infect Dis. 2000;31:678–683. doi: 10.1086/314032. [DOI] [PubMed] [Google Scholar]
- 7.Chang L-Y, Huang Y-C, Lin T Y. Fulminant neurogenic pulmonary oedema with hand, foot and mouth disease. Lancet. 1998;352:367–368. doi: 10.1016/S0140-6736(98)24031-1. [DOI] [PubMed] [Google Scholar]
- 8.Chu P Y, Lin K H, Hwang K P, Chou L C, Wang C F, Shih S R, Wang J R, Shimada Y, Ishiko H. Molecular epidemiology of enterovirus 71 in Taiwan. Arch Virol. 2001;146:589–600. doi: 10.1007/s007050170164. [DOI] [PubMed] [Google Scholar]
- 9.Chumakov M, Voroshilova M, Shindarov I, et al. Enterovirus 71 isolated from cases of epidemic poliomyelitis-like disease in Bulgaria. Arch Virol. 1979;60:329–360. doi: 10.1007/BF01317504. [DOI] [PubMed] [Google Scholar]
- 10.Felsenstein J. PHYLIP—phylogeny inference package (version 3.5) Cladistics. 1989;5:164–166. [Google Scholar]
- 11.Genetics Computer Group. Program manual for the Wisconsin GCG package, version 8.0 1994. Madison, Wis: Genetics Computer Group; 1994. [Google Scholar]
- 12.Gilbert G L, Dickson K E, Waters M, et al. Outbreak of enterovirus 71 in Victoria, Australia, with a high incidence of neurologic involvement. Pediatr Infect Dis J. 1988;7:484–488. doi: 10.1097/00006454-198807000-00007. [DOI] [PubMed] [Google Scholar]
- 13.Hayward J C, Gillespie S M, Kaplan K M, et al. Outbreak of poliomyelitis-like paralysis associated with enterovirus 71. Pediatr Infect Dis J. 1989;8:611–616. doi: 10.1097/00006454-198909000-00009. [DOI] [PubMed] [Google Scholar]
- 14.Ho M, Chen E-R, Hsu K-H, et al. An epidemic of enterovirus 71 infection in Taiwan. N Engl J Med. 1999;341:929–935. doi: 10.1056/NEJM199909233411301. [DOI] [PubMed] [Google Scholar]
- 15.Ishimaru Y, Nakano S, Yamaoka K, et al. Outbreaks of hand, foot and mouth disease by enterovirus 71. Arch Dis Childhood. 1980;55:583–588. doi: 10.1136/adc.55.8.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kennett M L, Birch C J, Lewis F A, et al. Enterovirus type 71 infections in Melbourne. WHO Bull. 1974;51:609–615. [PMC free article] [PubMed] [Google Scholar]
- 17.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 18.King A M Q, Brown F, Christian P, Hovi T, Hyypiä T, et al. Picornaviridae. In: Van Regenmortel M H V, Fauquet C M, Bishop D H L, Calisher C H, et al., editors. Virus taxonomy: seventh report of the International Committee for the Taxonomy of Viruses. New York, N.Y: Academic Press; 2000. pp. 657–673. [Google Scholar]
- 19.Liu C-C, Tseng H-W, Wang S-M, et al. An outbreak of enterovirus 71 infection in Taiwan, 1998: epidemiologic and clinical manifestations. J Clin Virol. 2000;7:23–30. doi: 10.1016/s1386-6532(00)00068-8. [DOI] [PubMed] [Google Scholar]
- 20.Lum L C, Wong K T, Lam S K, et al. Fatal enterovirus 71 encephalomyelitis. J Pediatr. 1998;133:795–798. doi: 10.1016/s0022-3476(98)70155-6. [DOI] [PubMed] [Google Scholar]
- 21.Lum L C S, Wong K T, Lam S K, Chua K B, Goh A Y T. Neurogenic pulmonary oedema and enterovirus 71 encephalomyelitis. Lancet. 1998;352:1391. doi: 10.1016/s0140-6736(05)60789-1. [DOI] [PubMed] [Google Scholar]
- 22.Martin A, Wychowski C, Couderc T, Crainic R, Hogle J, Girard M. Engineering a poliovirus type 2 antigenic site on a type 1 capsid results in a chimaeric virus that is neurovirulent in mice. EMBO J. 1988;7:2839–2847. doi: 10.1002/j.1460-2075.1988.tb03140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McMinn P C, Stratov I, Nagarajan L, Davis S. Neurological manifestations of enterovirus 71 infection in children during a hand, foot and mouth disease outbreak in Western Australia. Clin Infect Dis. 2001;32:236–242. doi: 10.1086/318454. [DOI] [PubMed] [Google Scholar]
- 24.Page R D. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 25.Poyry T, Hyypiä C, Horsnell L, et al. Molecular analysis of Coxsackievirus A16 reveals a new genetic group of enteroviruses. Virology. 1994;202:982–987. doi: 10.1006/viro.1994.1423. [DOI] [PubMed] [Google Scholar]
- 26.Samuda G M, Chang W-K, Yeung C-Y, et al. Monoplegia caused by enterovirus 71: an outbreak in Hong Kong. Pediatr Infect Dis J. 1987;6:206–208. doi: 10.1097/00006454-198702000-00013. [DOI] [PubMed] [Google Scholar]
- 27.Schmidt N J, Lennette E H, Ho H H. An apparently new enterovirus isolated from patients with disease of the central nervous system. J Infect Dis. 1974;129:304–309. doi: 10.1093/infdis/129.3.304. [DOI] [PubMed] [Google Scholar]
- 28.Singh S, Chow V T K, Chan K P, Ling A E, Poh C L. RT-PCR, nucleotide, amino acid and phylogenetic analyses of enterovirus type 71 in Asia. J Virol Methods. 2000;88:193–204. doi: 10.1016/s0166-0934(00)00185-3. [DOI] [PubMed] [Google Scholar]
- 29.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan J-J, Wang J-R, Liu C-C, et al. An outbreak of enterovirus 71 infection in Taiwan 1998: a comprehensive pathological, virological and molecular study on a case of fulminant encephalitis. J Clin Virol. 2000;17:13–22. doi: 10.1016/s1386-6532(00)00067-6. [DOI] [PubMed] [Google Scholar]