The SWI/SNF Complex Creates Loop Domains in DNA and Polynucleosome Arrays and Can Disrupt DNA-Histone Contacts within These Domains (original) (raw)
Abstract
To understand the mechanisms by which the chromatin-remodeling SWI/SNF complex interacts with DNA and alters nucleosome organization, we have imaged the SWI/SNF complex with both naked DNA and nucleosomal arrays by using energy-filtered microscopy. By making ATP-independent contacts with DNA at multiple sites on its surface, SWI/SNF creates loops, bringing otherwise-distant sites into close proximity. In the presence of ATP, SWI/SNF action leads to the disruption of nucleosomes within domains that appear to be topologically constrained by the complex. The data indicate that the action of one SWI/SNF complex on an array of nucleosomes can lead to the formation of a region where multiple nucleosomes are disrupted. Importantly, nucleosome disruption by SWI/SNF results in a loss of DNA content from the nucleosomes. This indicates a mechanism by which SWI/SNF unwraps part of the nucleosomal DNA.
Regulatory elements of many genes show hypersensitivity to DNase I, an indication that the disruption of nucleosomes at these sites is required to relieve a repressive function of chromatin. Several studies indicate that the eukaryotic cell possesses large complexes which contribute to gene activation by disrupting nucleosomes positioned over regulatory regions of particular genes. Genetic studies have identified a 2-MDa multisubunit complex in Saccharomyces cerevisiae called SWI/SNF, which can bind to DNA, disrupt nucleosomes, and provide transcription factors with access to nucleosomal DNA (for a review, see reference 22). Genetic suppressors of mutant subunits of SWI/SNF in yeast have frequently been identified as components of chromatin, such as the core histones (11, 16, 23). Higher eukaryotes appear to have homologs of the yeast SWI/SNF complex, which are also capable of disrupting nucleosomes. For example, brahma (brm) is a Drosophila melanogaster homolog of SWI2 (26) and hbrm and BRG1 from human cells also appear to be functional homologs of yeast SWI2 (6, 15, 19). Other complexes may perform related functions, including RSC (a large complex like SWI/SNF), NURF, ACF, and CHRAC (a group of smaller related complexes) (4, 14, 27, 28, 30). Similarities among subunits of SWI/SNF, NURF, and other functional homologs indicate that the eukaryotic cell contains a variety of related but independent systems for activating genes by altering chromatin structure.
In biochemical studies the SWI/SNF complex has been shown to bind naked DNA and nucleosomes with nanomolar affinity (9, 24). The SWI/SNF complex uses the energy of ATP hydrolysis to remodel nucleosome structure, which increases the affinity of transcription factors for nucleosomal DNA (7, 9, 12, 13, 17, 21, 29, 33). The remodeled conformation of the nucleosome persists after depletion of ATP (13) and detachment of the SWI/SNF complex (9, 33). This remodeled nucleosome conformation will eventually revert to the original conformation on its own (9) or can be converted back by further action of SWI/SNF, indicating that SWI/SNF catalyzes the interconversion between these two nucleosome forms (33). A little is known about the remodeled nucleosome conformation. It retains all four core histones (33), although the affinity of the histone octamer for DNA is apparently diminished, as the octamer is more susceptible to displacement by the binding of multiple GAL4-AH dimers (21). A hallmark of the remodeled nucleosome conformation is an altered pattern of DNase I digestion, most apparent when SWI/SNF acts on nucleosomes in which the DNA is rotationally phased (7, 12, 17). On these nucleosomes, SWI/SNF disrupts the sequence-specific 10-bp periodic pattern of DNase cutting, which reflects the direction of DNA bending around the histone octamer. However, a sequence-independent 10-bp pattern of DNase cutting is retained over at least 70 bp of the nucleosome, indicating that, while SWI/SNF twists or alters the path of DNA bending, part of the DNA remains associated with the surface of the histone octamer (9).
When acting on arrays of nucleosomes, SWI/SNF functions catalytically (18), increasing restriction endonuclease sensitivity on multiple arrays. In addition, SWI/SNF can alter the translational phasing (i.e., location) of multiple nucleosomes within an array, although translational phasing on nucleosome positioning sequences is regained upon removal of the complex (21). To further investigate the interactions of SWI/SNF with nucleosome arrays and the consequences of its action on nucleosome structure within arrays, we have visualized SWI/SNF complexes on naked DNA and complexes of SWI/SNF with nucleosome arrays, using electron spectroscopic imaging (ESI) (1).
There are a number of advantages that ESI offers over conventional electron microscopy (EM) for imaging DNA-protein complexes (2, 3). First, because electron energy loss imaging provides high contrast, heavy-atom stains and shadows which can limit spatial resolution, are not required. Such agents also can exaggerate the presence of or fail to contrast particular biochemical entities. Second, images recorded from particular regions of the energy loss spectrum provide mass-sensitive information, so that molecular-mass estimates can be obtained. Finally, coupled with mass analysis, the detection of phosphorus and the production of phosphorus maps permits the calculation of stoichiometric relationships between the protein and the nucleic acid and is the basis for delineating the distribution of nucleic acid in a protein-DNA complex. The analysis presented here complements the biochemical data obtained previously and provides new information regarding the function of SWI/SNF. For example, multiple sites of DNA contact on its surface result in its ability to create loops in the DNA, bringing otherwise-distant sites into close proximity. Furthermore, the protein-DNA stoichiometric measurements provide insights into the nature of the conformation of SWI/SNF-remodeled nucleosomes. The results indicate an unwrapping of DNA from the histone octamer as the primary consequence of SWI/SNF action. Moreover, it is apparent that a single SWI/SNF complex can remodel a number of nucleosomes located in a region of chromatin constrained in an intervening loop created by the complex.
MATERIALS AND METHODS
Imaging of SWI/SNF-DNA complexes.
Electron spectroscopic images of SWI/SNF complexed to linear and relaxed closed-circular plasmid DNA were obtained as follows. The circular plasmid pJH28 was relaxed with Escherichia coli topoisomerase I as described previously (24). The SWI/SNF protein complex was purified as described previously (7). DNA or polynucleosome arrays (see Fig. 3 and 5) at a DNA concentration of 5 ng/μl were reacted with purified SWI/SNF at a final concentration of either 2 or 4 ng/μl in a binding buffer containing 20 mM HEPES (pH 7.5), 50 mM NaCl, 3 mM MgCl2, 1 μM ZnCl2, 2 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride, 5% glycerol, and 100 ng of insulin/μl. ATP or ATP-γS was added to a final concentration of 2 mM. The reactants were incubated for 30 min at 30°C, followed by a further fixation step with buffered EM-grade formaldehyde (1%) and glutaraldehyde (0.5%) for 15 min at 30°C. For ESI analysis, 1 μl of the reaction mixture was applied to a 4-μl drop of 3 mM MgCl2–10 mM HEPES (pH 8.0) on a 1,000-mesh EM specimen grid covered with a 3-nm-thick carbon film (3). After 1 min, the grid was washed with pure water (prepared for tissue culture by Gibco) and air dried.
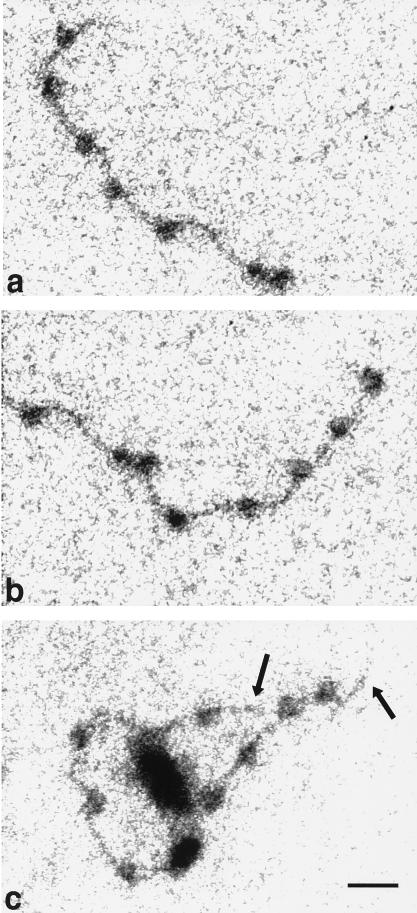
FIG. 3.
Electron spectroscopic images of a polynucleosome strand in the absence of SWI/SNF (a and b) and a polynucleosome strand with one SWI/SNF complex (c). The left and right halves of the same strand are shown in panels a and b, respectively. The polynucleosome arrays on a 12-mer repeat of the 208-bp sea urchin 5S rRNA gene nucleosome-positioning sequence (25) were formed by salt dialysis with histones purified from chicken erythrocytes (10). The final buffer contained 25 mM HEPES. All of the strands shown were subjected to incubation in buffer containing ATP. The images were recorded at an energy loss of 155 eV. Bar, 24 nm.
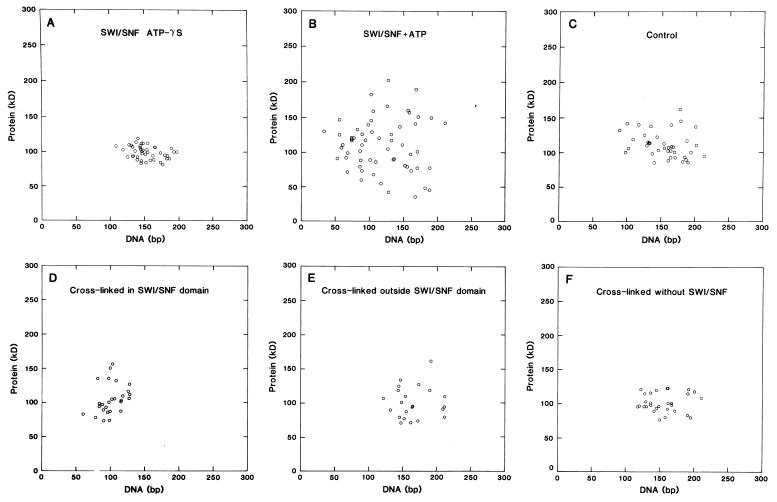
FIG. 5.
(A and B) Scatter plots of protein content versus DNA content of particles on polynucleosome strands bound by one SWI/SNF complex. Reactions were carried out in the presence of 2 mM ATP-γS (A) or 2 mM ATP (B). (C) Values for particles in polynucleosome strands exposed to ATP but not SWI/SNF in a mock reaction. The mean protein and DNA contents (± SD) are 100 ± 9 kDa and 151 ± 21 bp (n = 42) (A), 111 ± 45 kDa and 118 ± 43 bp (n = 59) (B), and 112 ± 22 kDa and 157 ± 34 bp (n = 40) (C). (D to F) Scatter plots of nucleosome arrays reconstituted with cross-linked histones. Nucleosomes within a loop created by a SWI/SNF complex (D), nucleosomes outside a loop or on strands not bound by SWI/SNF (E), and nucleosomes on arrays not exposed to SWI/SNF (mock-reacted with ATP) (F) were used to obtain the values for these plots. The average amounts of protein and DNA are 101 ± 26 kDa and 101 ± 17 bp (D), 101 ± 24 kDa and 166 ± 27 bp (E), and 102 ± 15 kDa and 155 ± 27 bp (F).
Electron spectroscopic images were recorded on a Zeiss EM902 transmission electron microscope equipped with a prism-mirror-prism electron-imaging spectrometer. A 90-μm-diameter objective and a 400-μm-diameter condenser aperture were used. The energy-selecting slit aperture corresponded to 20 eV. Phosphorus-enhanced images were recorded at 155 eV, and mass-sensitive reference images were recorded at 120 eV of energy loss as described previously (references 1 and 3 and references therein). Micrographs were recorded on electron image film (Kodak) and were processed in full-strength D-19 developer (Kodak) for 15 min.
Phosphorus and mass analyses of nucleosomes and DNA fibers were performed as described previously (3). Nucleosomes were delineated by choosing a boundary at 1 standard deviation (SD) above the mean of the background. The phosphorus content was calculated by arithmetically comparing a region in the 155-eV phosphorus-enhanced postedge image with the same region in the 120-eV preedge mass-sensitive image after the two images were aligned and normalized and background values in the region of the particles of interest had been subtracted. Mass was calculated in a similar way by using only the mass-sensitive 120-eV preedge image. Naked DNA and cospread tobacco mosaic virus were used as internal mass and phosphorus standards.
Several grids of each SWI/SNF-DNA or SWI/SNF-polynucleosome reaction were prepared to ensure that the structures were reproducible. In addition, reproducibility between reactions was established by reproducing every EM experiment at least three times.
Preparation of polynucleosomes and DNase I analysis of SWI/SNF-induced nucleosome disruption.
Polynucleosomes were reconstituted onto a 12-mer repeat of the 208-bp sea urchin 5S rRNA gene nucleosome-positioning sequence by high-salt dialysis with purified chicken erythrocyte nucleosomes (10). Reconstitution of mononucleosomes onto a GAL4 site DNA was accomplished by octamer transfer with H1-depleted HeLa oligonucleosomes purified and used as described previously (8). Histone cores were cross-linked with dimethyl suberimidate as described previously (31) during reconstitution in the DNase I nuclease sensitivity experiments. In these experiments, the nucleosomes (25 nM) were incubated in the presence or absence of 15 nM SWI/SNF and/or 100 nM GAL4-AH dimers and analyzed by DNase I digestion (7).
RESULTS
Interactions of SWI/SNF with linear and closed-circular DNA.
Images of SWI/SNF complexes with DNA were obtained by ESI. At a ratio of 2 ng of protein/μl to 5 ng of linear 1.4-kbp DNA/μl, we observed that 87 of 387 (22%) DNA molecules had a single SWI/SNF complex bound in the presence of ATP. The criteria for identifying a SWI/SNF complex were a molecular mass estimate of 1.5 to 2.0 MDa and the appearance of subunits within the complex. In the absence of ATP, the results were similar: 45 of 292 (15%) DNA molecules had interacted with a SWI/SNF complex. With a higher input ratio of protein to DNA, 4 ng of protein/μl to 5 ng of DNA/μl, 139 of 226 (62%) DNA molecules were complexed with a single SWI/SNF in the presence of ATP and 61 of 142 (43%) were complexed in the absence of ATP. Because SWI/SNF forms complexes with DNA in the absence of ATP and because ATP produces less than a 1.5-fold increase in the frequency of such complexes, we conclude that SWI/SNF binds DNA by an ATP-independent mechanism.
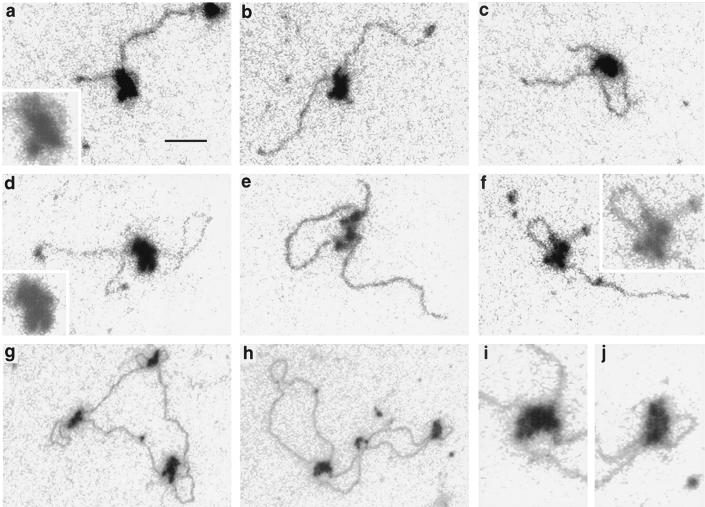
Images of unstained and unshadowed SWI/SNF-linear DNA complexes and SWI/SNF-relaxed closed-circular DNA complexes are shown in Fig. 1. There are a few features that should be noted. Though it is difficult to represent the full dynamic range of optical densities contributed by the low mass density of the DNA and the high mass density of the SWI/SNF complex in the same micrograph, the subunit nature of the SWI/SNF complex is apparent in most of the examples shown here: it is more obvious in Fig. 1e and f but less obvious in Fig. 1c. Lower-contrast and magnified images of some complexes are shown as insets. A second feature is that the SWI/SNF complex is able to create a loop in the DNA, the size of the loop being variable even on the same DNA fragment. The variable size of the loop indicates that there is not a stringent sequence dependence for the DNA contact sites or that the specific sequence of the contact site occurs frequently. At an input ratio of 4 ng of SWI/SNF/μl to 5 ng of DNA/μl in the presence of ATP, 90 of 177 SWI/SNF-DNA complexes, or 65%, had an obvious loop created by the SWI/SNF complex. In the absence of ATP, 109 of 225 SWI/SNF-DNA complexes, or 48%, displayed an obvious loop. Thus, ATP does not significantly enhance the formation of DNA loops. The fraction of complexes in which a loop is formed may be greater than indicated by these values. Though a loop is not always seen extending from the SWI/SNF complex, contour length measurements (data not shown) indicate that some complexes contain compacted DNA that could represent small loops superimposed on the complexes. We argue that the loops visualized are created by the SWI/SNF complex and are not the consequence of SWI/SNF binding to a loop created by the DNA spreading on the support film. The reason is that, in control experiments, a loop observed in a linear DNA fragment of 1 kbp occurs only in at most 2% of the molecules under the spreading conditions used in these experiments. With SWI/SNF, however, the frequency of loops is at least 25-fold higher. Furthermore, contour length measurements indicate that SWI/SNF causes a linear compaction of the DNA. Such a compaction would not be observed if SWI/SNF complexes were simply binding to intersections of the DNA strands on the support film. (In control experiments, the SDs of the contour lengths of 1-kbp fragments are between 1 and 2% [data not shown]).
FIG. 1.
Electron spectroscopic images of SWI/SNF complexes on linear plasmid DNA fragments (a to f) and relaxed closed circular DNA (g to j). (a to c and f) A 1.0-kb fragment from pBluescript; (d and e) 1.4-kb fragment containing promoter sequences of the human proenkephalin gene. The insets in panels a, d, and f are magnified twofold and reproduced at lower contrast to emphasize the subunit structure of SWI/SNF complexes. Panels i and j are magnified regions from panel h. Bar, 53 (a to f) and 160 (g and h) nm.
Because of the high mass density of the SWI/SNF complex relative to that of the DNA, it was generally not possible to form net phosphorus maps of the DNA-protein complexes, because the dynamic range of the film on which the images were collected was insufficient. In some cases, however, depending on (i) the exposure, (ii) the support film thickness, and (iii) the projection thickness of the SWI/SNF complex, we were able to obtain net phosphorus images. Some examples are shown in Fig. 2. Because of the high phosphorus content in DNA relative to that of the protein, the net phosphorus overlay reveals the DNA path in the complex if the phosphorus signal-to-noise ratio is sufficient. The noise in the phosphorus maps is sometimes a source of confusion in tracing the DNA path. Contour length measurements, however, by providing another parameter, can be used in conjunction with the phosphorus maps to delineate the most likely DNA path. The contours shown in the insets of Fig. 2 are the most likely, based on the phosphorus maps and the length measurements. The contours shown measure 1,044, 988, 1,002, and 1,009 bp, respectively. In Fig. 2c, for example, the noise along the left side of the complex is judged not to result from DNA because of the slightly lower phosphorus signal along that surface compared to the signal elsewhere along the DNA together with the fact that the length of the displayed contour is in agreement with the known length of the DNA fragment (1 kbp). More DNA on this left-hand surface would be incompatible with the contour length.
FIG. 2.
Electron spectroscopic images of SWI/SNF-DNA complexes. A mass-sensitive image is shown in grey levels, on which is superimposed the net phosphorus image in magenta. The short and long arrows indicate small and large loops, respectively, that have been created by SWI/SNF-DNA contacts. The insets reveal the most likely path of the DNA through or around the SWI/SNF-DNA complex, based on the phosphorus map, the contour length, and the topological consistency. Bar, 35 nm.
We observe that SWI/SNF has multiple sites which make contact with the DNA and which create small or large loops from the intervening DNA. From images of 23 complexes, we measured the contour length of DNA that appears to be in contact with the surface of a SWI/SNF complex. The average value was 133 ± 40 (SD) bp, with a range from 71 to 212 bp. The measurements were made from the point where DNA enters the complex to where it exits at a loop or at the final exit site. These contact length measurements are subject to underestimation because the two-dimensional projection length measured is equal to or less than the true length in three dimensions. The measurements, however, can also be overestimated because some of the DNA that overlaps with the SWI/SNF complex in projection may not actually be in contact with it in solution but deposited over it on the two-dimensional substrate used for EM imaging. Therefore, these values must be regarded as only estimates of DNA contact lengths. These limitations notwithstanding, the average length of DNA in contact with SWI/SNF is in agreement with the minimum DNA fragment length of 130 bp that is required for a stable SWI/SNF-DNA interaction (24).
The data presented above are a direct visualization of the SWI/SNF complex with DNA. Importantly, the large fraction of SWI/SNF-DNA complexes that generate DNA loops indicates that the complex has at least two high-affinity binding sites which can interact with different DNA sequences separated by long distances. The fact that the SWI/SNF-DNA interactions observed here are ATP independent is consistent with biochemical studies showing that the binding of SWI/SNF to DNA (24) and to nucleosomes (9) is ATP independent. The observed simultaneous interactions of SWI/SNF with more than one DNA site may provide new insights into how the complex can perturb multiple nucleosomes within an array (18, 21). To determine whether the SWI/SNF complex can also simultaneously interact with multiple nucleosomes, we examined the interactions of the SWI/SNF complex with nucleosome arrays.
Interactions of SWI/SNF with polynucleosomal DNA.
The SWI/SNF complex has been shown to utilize the energy of ATP hydrolysis to alter nucleosome structure, leading to an increased affinity of transcription factors (see the introduction). Very little, however, is known about the molecular mechanisms of this function. To obtain structural information on chromatin bound by SWI/SNF, we imaged nucleosomes assembled onto a 12-mer repeat of the sea urchin 5S rRNA gene sequence, a sequence with a well-characterized ability to position a nucleosome (25). A histone-DNA ratio was chosen so that the strands would be nearly saturated with nucleosomes. An example of a polynucleosomal strand after reconstitution (and incubation in ATP) is shown in Fig. 3a and b. Generally, the spacing of the nucleosomes was uniform, though on this strand, the sixth and seventh nucleosomes from the right end are unevenly spaced. Either the seventh nucleosome has “slid” to contact the sixth, or it has bound in a minor positioning frame. Initially, we did not observe differences between the numbers of nucleosomes in the arrays incubated in the presence of ATP and those in the arrays incubated in the presence of ATP-γS (a nonhydrolyzable form of ATP, not usable by SWI/SNF in other assays). In a control experiment, the number of nucleosomes per strand was 8.7 ± 2.6 (n = 129) in the presence of ATP and 9.2 ± 2.0 (n = 107) in the presence of ATP-γS. From these numbers we concluded that ATP itself does not affect the structure or stability of the nucleosomes on this 12-mer repeat DNA sequence. The binding of the SWI/SNF complex to the polynucleosomal array is illustrated in Fig. 3c. Reminiscent of its interaction with naked DNA templates (Fig. 3a and b), the SWI/SNF complex is clearly able to make contacts with distant sites on the polynucleosome strand, creating an intervening loop of nucleosomal DNA. The binding of the SWI/SNF complex to polynucleosome arrays did not appear to be significantly affected by the presence of ATP relative to its binding in the presence of ATP-γS. Strands with evenly distributed nucleosome-like particles and with one SWI/SNF complex attached could be found in the presence of either ATP or ATP-γS.
ESI analysis did reveal effects that SWI/SNF has on the polynucleosome strands, effects which are dependent on a hydrolyzable form of ATP. The effect of hydrolyzable ATP became apparent when ESI was used to measure the mass and the phosphorus content of the nucleosomes on the strands (1, 3). Although a superficial examination indicated that a SWI/SNF complex had bound to an array of normal nucleosomes, the phosphorus and mass analyses of these nucleosomes indicated otherwise (Fig. 4). With the measurements of total mass and total phosphorus content, the protein and DNA stoichiometry can be determined. The average protein and DNA contents of particles on strands with one SWI/SNF complex in the presence of ATP were 111 ± 45 kDa and 118 ± 43 bp, respectively (n = 59). Whereas the average protein content is consistent with that of canonical nucleosomes, the DNA content was significantly less than that expected for nucleosomes. Scatter plots of protein versus DNA (Fig. 5B) show a large degree of heterogeneity in protein and DNA stoichiometry. The average DNA content of 118 bp is a value well below that expected for a canonical nucleosome. In contrast, the average protein and DNA contents of nucleosomes on strands complexed with SWI/SNF in the presence of ATP-γS were 100 ± 9 kDa and 151 ± 21 bp, respectively (n = 42). The scatter plots show that there is far less heterogeneity in protein and DNA stoichiometry when ATP-γS is used in place of hydrolyzable ATP (Fig. 5A). In a mock reaction (minus SWI/SNF) in the presence of ATP, polynucleosomes also have normal stoichiometry, containing on average 112 ± 22 kDa of protein and 157 ± 34 bp of DNA (n = 40) (Fig. 5C). From this stoichiometric analysis, we conclude that SWI/SNF has the ability to disrupt nucleosome structure in an ATP-dependent manner, resulting in a reduction in and increased heterogeneity of the extent of DNA interacting with each histone octamer.
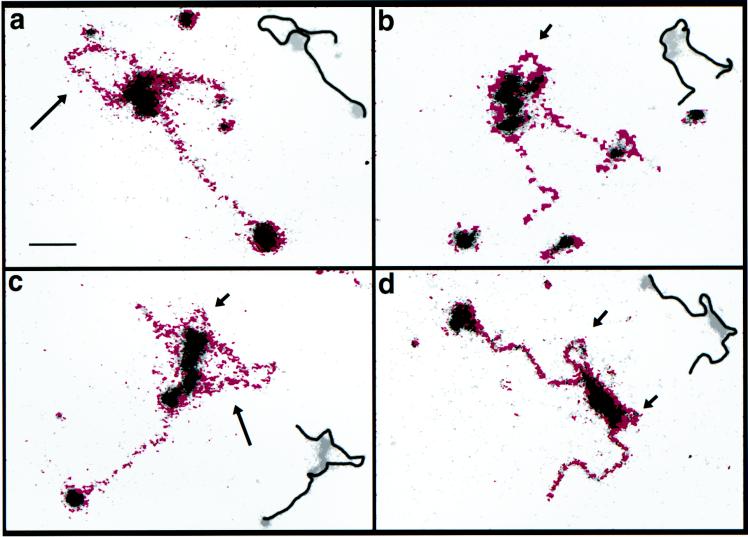
FIG. 4.
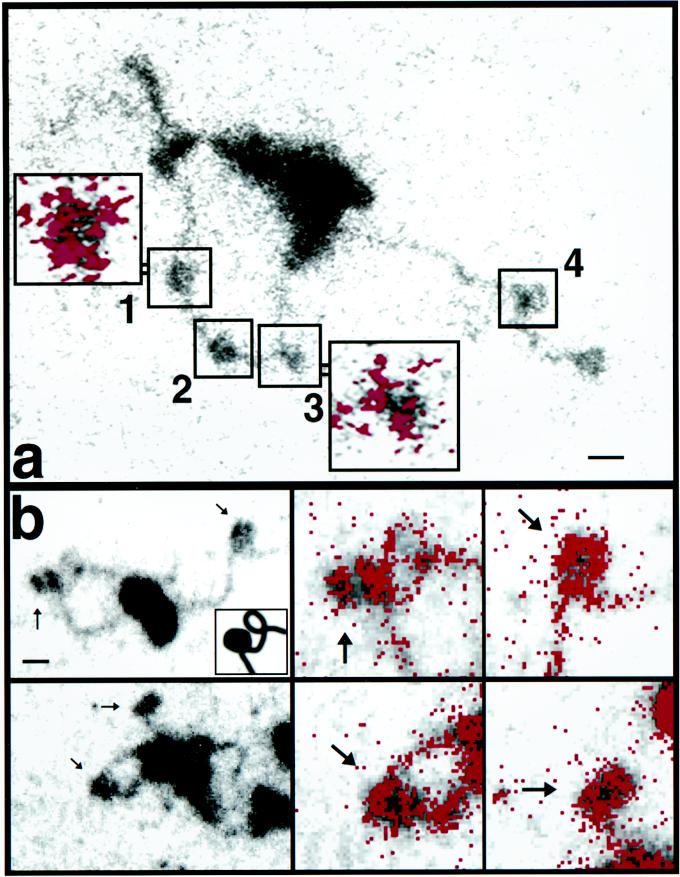
Electron spectroscopic images of SWI/SNF-polynucleosome strands with histones not chemically cross-linked (a) and with cross-linked histone octamers (b). The mass and phosphorus contents of SWI/SNF-DNA and nucleosome complexes were determined as described previously (3). (a) Protein and DNA contents of the nucleosomes in the numbered boxes are presented in the text. The colored insets show the net phosphorus distributions of the indicated nucleosomes. Bar, 24 nm (insets, 12 nm). (b) Nucleosomes indicated by arrows in the low-magnification views on the left are shown on the right at higher magnification, with the phosphorus represented as red. The inset in the upper-left panel is included to clarify that a protein-free DNA writhe lies just to the right of the nucleosome at the left of the field. Bar, 11 (left) and 5.5 (right) nm.
The protein-DNA stoichiometric disruption was often reflected in a disruption in morphology, which was particularly apparent in the phosphorus distribution maps. In Fig. 4a, for example, particles 1 (80 kDa; 152 bp) and 2 (120 kDa; 138 bp) have near-normal levels of protein and DNA. Particle 1 has the normal en face morphology commonly seen in ESI images of nucleosomes, where the phosphorus (inset) is concentrated on the particle’s periphery. In contrast, particle 3 (104 kDa; 85 bp) has a higher-than-average protein/DNA ratio, probably reflecting the loss of a turn of DNA. The inset for particle 3, showing the phosphorus (or DNA) distribution, also indicates that this nucleosome has a disrupted morphology. The protein content of particle 4 (80 kDa; 170 bp), located outside the loop created by the SWI/SNF complex, is close to the canonical value, and there has been no loss of DNA in the particle. A value greater than 146 bp of DNA may indicate that linker DNA is superimposed on the particle in the projection.
The altered morphology of SWI/SNF-remodeled nucleosomes is further illustrated in Fig. 4b. The DNA distribution, based on phosphorus imaging of the nucleosome in the SWI/SNF-induced loop (upper-left panel), does not have the characteristic “doughnut” profile typical of ESI images of nucleosomes, such as the nucleosome outside of the loop (see the high-magnification phosphorus maps of both nucleosomes in the right-hand panels). (Because of the low contrast of the DNA in the image reproduction, the inset was used for clarification, showing that a protein-free writhe in the DNA is present just to the upper right of the nucleosome.) This nucleosome in the SWI/SNF loop has a protein mass of 107 kDa and a DNA content of 107 bp, with a protein/DNA mass ratio of 1.53. The nucleosome in the SWI/SNF-induced loop in the lower-left panel appears less disrupted than its counterpart in the upper-left panel. It does, however, show a larger amount of phosphorus on its left than on its right side. Its protein content is normal (114 kDa), but it has a reduced amount of DNA (100 bp) compared to that of a canonical nucleosome. The nucleosome outside the loop is located at the end of the polynucleosome strand. Its phosphorus profile is similar to that of its counterpart in the panel above, typical of en face views of intact nucleosomes in arrays visualized by ESI (3, 20).
The analytical microscope emphasizes important features of nucleosome disruption by SWI/SNF. First, structures within loops created by SWI/SNF may appear to be nucleosomes, based on a superficial examination. Comparisons of mass- and phosphorus-sensitive images indicate, however, that only some structures have the characteristics of canonical nucleosomes while other structures have abnormally low amounts of phosphorus compared to those expected for canonical nucleosomes. Earlier in this paper we reported that polynucleosome arrays complexed with one SWI/SNF were observed with both ATP-γS and ATP. Subsequent comparisons of mass and phosphorus contents, however, indicate that not all of the structures initially scored as nucleosomes were actually canonical nucleosomes. SWI/SNF together with ATP, but not ATP-γS, does indeed affect the number of normal or intact nucleosomes on an array. Second, only some nucleosomes within loops are disrupted (e.g., Fig. 4a, particle 3) while some appear intact (Fig. 4a, particles 1 and 2). Based on the mean values of protein and DNA content (Fig. 5B), we conclude that SWI/SNF is able to remove DNA from the histone octamer. The heterogeneity in the protein content, however, indicates that protein loss can also occur, perhaps as a consequence of reduced DNA binding. An H2A-H2B dimer, for example, may be lost along with the disruption of DNA from the histone core on some particles. Particles with higher-than-average protein content may arise from atypical associations with additional histones liberated from other disrupted nucleosomes.
SWI/SNF disruption involves loss of histone-DNA contacts but does not require loss of histones.
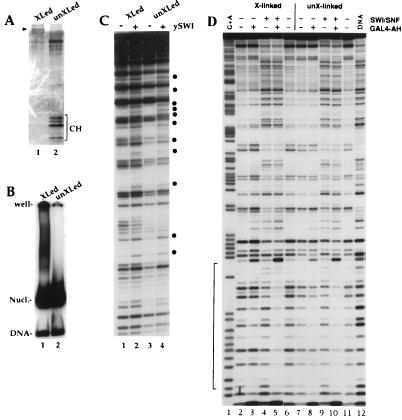
The images and stoichiometric analysis by ESI (Fig. 4 and 5) indicate that ATP-dependent disruption of multiple nucleosomes in an array by SWI/SNF favors a loss of DNA over a loss of histones from the particles. Thus, disruption of nucleosomes by SWI/SNF might only require alteration of histone-DNA contacts, resulting in loss of the DNA, perhaps by unwinding, without the loss of histones from the nucleosome core. To test the possibility that SWI/SNF disrupts nucleosomes without requiring the loss of histones from the octamer core, we analyzed SWI/SNF disruption of nucleosomes biochemically, using nucleosome cores which contained covalently cross-linked histone octamers. Histone-histone cross-linking was performed with dimethyl suberimidate (31). This converts the histone octamer into a single 100-kDa complex (Fig. 6A), which retains the ability to reconstitute DNA into nucleosome cores (Fig. 6B) (reference 31 and references therein). Note that, although the same amount of protein was loaded, the cross-linked octamer is poorly contrasted because it takes up less silver than the control histones. The individual core histones are resolved in the control octamer lane but are not present in the cross-linked octamer lane. Though the silver staining suggests that the core histones are not present in equal stoichiometric amounts, the Coomassie stain (not shown) confirmed that they are present at equal levels.
FIG. 6.
Histone octamer cross-linking does not affect nucleosomal DNA perturbation and enhanced GAL4-AH binding by SWI/SNF. Histone cross-linking conditions with dimethyl suberimidate and nucleosome reconstitution by transfer were described previously (31). (A) Sodium dodecyl sulfate-18% polyacrylamide gel showing that cross-linking conditions with dimethyl suberimidate create a protein band at around 100 kDa (arrowhead shows migration of 97.5-kDa marker) with no detectable free histone or partially cross-linked product (50 ng of histone loaded; 172-bp 5S DNA probe [21] used in the reconstitution). Note that the cross-linked histones take up less stain than control histones. The band that migrates more slowly than the core histones corresponds to bovine serum albumin, used to maintain stability. (B) Mobility shift gel showing that the cross-linked octamers do not significantly change the migration of the 5S mononucleosome. (C) Nucleosomes (25 nM) from the preparation used for panel B were incubated with 1 mM ATP in the presence (+) or absence (−) of purified SWI/SNF (2.5 nM) and analyzed by DNase I digestion. The bullets show where the intensity of a band is modified by SWI/SNF action. (D) A single-GAL4-site DNA probe (7) was reconstituted in nucleosome cores by using cross-linked (lanes 2 to 6) or non-cross-linked (lanes 7 to 11) histone octamers. The nucleosomes (25 nM) were incubated in the presence (+) or absence (−) of 15 nM SWI/SNF and/or 100 nM GAL4-AH dimers as indicated and were analyzed by DNase I digestion. Mg-ATP (1 mM) is also present in all of the lanes. The bracket indicates the GAL4-AH binding site. XLed and X-linked, cross-linked; unXLed and unX-linked, not cross-linked.
Treatment of the cross-linked nucleosomes with the SWI/SNF complex in the presence of ATP led to a disruption of the nucleosomal DNase I digestion pattern that is indistinguishable from the disruption observed with the control nucleosomes, which were not cross-linked (Fig. 6C, compare lane 1 to lane 3, and Fig. 6D, compare lanes 2 and 4 to lanes 7 and 9). The effect of SWI/SNF on the mononucleosome is less in Fig. 6C than in 6D, because the molar ratio of SWI/SNF to mononucleosomes was 10-fold higher in the reaction represented in 6D. Furthermore, we observe that the SWI/SNF complex is also able to stimulate the binding of GAL4-AH to cross-linked nucleosomes with the same efficiency as to control nucleosomes (Fig. 6D, compare lanes 3 and 5, and 8 and 10). Thus, the consequences of SWI/SNF action on the nucleosomes composed of cross-linked histones and on the control nucleosome cores were indistinguishable by the DNase I protection assay. This assay of mononucleosomes fully supports the conclusions from the ESI data on nucleosomal arrays that showed that SWI/SNF disrupts nucleosomes by targeting histone-DNA contacts without requiring the eviction of histones from the core particle.
To provide structural information to help interpret the DNase digestion studies of cross-linked histone nucleosomes, we used ESI to examine arrays of nucleosomes reconstituted with cross-linked octamers. Nucleosomes reconstituted into arrays with cross-linked octamers were morphologically indistinguishable from control nucleosomes (images not shown; compare Fig. 5F). Quantitative analysis revealed that nucleosomes within loops created by SWI/SNF had a protein content equivalent to that of canonical nucleosomes (101 ± 26 [SD] kDa) but had lost a significant amount of DNA (101 ± 17 bp); the protein/DNA mass ratio was 1.61 ± 0.36 (Fig. 5D). Cross-linked nucleosomes outside a SWI/SNF-induced loop or on strands not occupied by a SWI/SNF complex had near-normal levels of protein and DNA (101 ± 24 kDa of protein and 166 ± 27 bp of DNA; protein/DNA mass ratio, 0.95 ± 0.26) (Fig. 5E). Similarly, nucleosomes on arrays not exposed to SWI/SNF but mock-reacted in the presence of ATP were canonical on the basis of their protein and DNA contents (102 ± 15 kDa of protein and 155 ± 27 bp of DNA; protein/DNA mass ratio, 1.02 ± 0.24) (Fig. 5F). In contrast to nucleosomes that were not cross-linked, the cross-linked nucleosomes were more homogeneous in protein content in the presence of SWI/SNF and ATP (compare Fig. 5B and D). Thus, the cross-linked nucleosomes underwent only loss of DNA without substantial protein redistribution, yet they displayed the biochemical hallmarks of the SWI/SNF-remodeled nucleosomes (i.e., altered DNase digestion patterns and increased factor binding [Fig. 6]). This indicates that protein redistribution is a secondary consequence of nucleosome disruption which is not required for enhancement of transcription factor binding. Protein redistribution, however, may be a requirement in other aspects of transcriptional activation or elongation.
DISCUSSION
The structural studies presented here extend the previous biochemical analysis in two important respects. (i) While the SWI/SNF complex has been shown to perturb the DNase I digestion pattern of nucleosomal DNA (7, 12, 17), this effect could easily result from either the loss of histone proteins or the unwinding of nucleosomal DNA. Our stoichiometric measurements and DNase I protection analysis of cross-linked histone octamers both demonstrate that it is DNA that is generally lost from the nucleosome, supporting a mechanism where SWI/SNF peels DNA off the histone octamer surface. (ii) While previous biochemical studies have shown that the SWI/SNF complex can perturb multiple nucleosomes within an array (18, 21), it was unclear whether this would occur sequentially, perhaps requiring multiple SWI/SNF complexes per polynucleosome array. The structural analysis presented here illustrates clearly that one SWI/SNF complex can lead to the disruption of multiple nucleosomes in a polynucleosome array. This multinucleosome disruption is also in agreement with our previous report, in which we showed that nucleosome arrays can be disrupted efficiently at very low SWI/SNF ratios (18).
These data argue that the action of the SWI/SNF complex on an array of nucleosomes can lead to a region of nucleosome disruption. We think that the domains in which nucleosomes appear disrupted result from an interaction with only the SWI/SNF complex that is visualized and not with the visualized complex and another complex that had previously interacted and then moved on. One reason for this interpretation is that the input ratio of SWI/SNF to DNA is quite low, so that only about one-third of the nucleosome strands have one SWI/SNF complex associated and strands with more than one SWI/SNF complex are extremely rare (≤1%). Secondly, cross-linked nucleosomes outside of SWI/SNF loops and on strands that are not occupied by SWI/SNF are not, on average, disrupted, based on mass and phosphorus analyses. Nevertheless, though it is unlikely, we cannot definitively rule out the possibility that disruption of nucleosomes around a bound SWI/SNF could have involved another SWI/SNF molecule in a “hit-and-run” mechanism. However, we favor a mechanism whereby a loop domain is created by SWI/SNF, followed by a sequential disruption of nucleosomes within that domain, probably by direct contacts with SWI/SNF by linear diffusion, while the domain is maintained in a topologically constrained state. Evidence for a topologically constrained region is frequently observed in SWI/SNF-nucleosome complexes formed in the presence of ATP, where the DNA in the loops is interwound with itself. Such interwound DNA is rarely seen when ATP-γS is used instead (data not shown). Indeed, the fact that the SWI/SNF complex can interact with multiple DNA sites, generating a loop between them, raises the intriguing possibility that the SWI/SNF complex might be able to simultaneously disrupt nucleosomes at distal regulatory elements, i.e., enhancers and promoters.
ACKNOWLEDGMENTS
This work was supported by research grants from the Natural Sciences and Engineering Research Council of Canada to D.P.B.-J., from the Medical Research Council of Canada to J.C., and from the NIH-NIGMS to C.L.P. and J.L.W. J.C. was a Centennial Fellow of the Medical Research Council of Canada, and C.L.P. is a Scholar of the Leukemia Society of America. J.L.W. is an Associate Investigator of the Howard Hughes Medical Institute.
We thank Michael J. Hendzel for comments on the manuscript and Manfred Herfort for excellent technical assistance.
REFERENCES
- 1.Bazett-Jones D P. Empirical basis for phosphorus mapping and structure determination of DNA: protein complexes by electron spectroscopic imaging. Microbeam Anal. 1993;2:69–79. [Google Scholar]
- 2.Bazett-Jones D P, Leblanc B, Herfort M, Moss T. Short-range DNA looping by the Xenopus HMG-box transcription factor, xUBF. Science. 1994;264:1134–1137. doi: 10.1126/science.8178172. [DOI] [PubMed] [Google Scholar]
- 3.Bazett-Jones D P, Mendez E, Czarnota G J, Ottensmeyer F P, Allfrey V G. Visualization and analysis of unfolded nucleosomes associated with transcribing chromatin. Nucleic Acids Res. 1996;24:321–329. doi: 10.1093/nar/24.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cairns B R, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg R D. RSC, an essential, abundant chromatin re-modeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 5.Carlson M, Laurent B C. The SWI/SNF family of global transcriptional activators. Curr Opin Cell Biol. 1994;6:396–402. doi: 10.1016/0955-0674(94)90032-9. [DOI] [PubMed] [Google Scholar]
- 6.Chiba H, Muramatsu M, Nomoto A, Kato H. Two human homologs of Saccharomyces cerevisiae SWI2/SNF2 and Drosophila brahma are transcriptional coactivators cooperating with the estrogen receptor and the retinoic acid receptor. Nucleic Acids Res. 1994;22:1815–1820. doi: 10.1093/nar/22.10.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Côté J, Quinn J, Workman J L, Peterson C L. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 8.Côté J, Utley R T, Workman J L. Methods Mol. Genet. 1995;6:108–127. [Google Scholar]
- 9.Côté J, Peterson C L, Workman J L. Perturbation of nucleosome core structure by the SWI/SNF complex after its detachment, enhancing subsequent transcription factor binding. Proc Natl Acad Sci USA. 1998;95:4947–4952. doi: 10.1073/pnas.95.9.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hansen J C, Lohr D J. Assembly and structural properties of subsaturated chromatin arrays. J Biol Chem. 1993;268:5840–5848. [PubMed] [Google Scholar]
- 11.Hirschhorn J N, Bortvin A L, Ricupero-Hovasse S L, Winston F. A new class of histone H2A mutations in Saccharomyces cerevisiae causes specific transcriptional defects in vivo. Mol Cell Biol. 1995;15:1999–2009. doi: 10.1128/mcb.15.4.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imbalzano A N, Kwon H, Green M R, Kingston R E. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature. 1994;370:481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- 13.Imbalzano A N, Schnitzler G R, Kingston R E. Nucleosome disruption by human SWI/SNF is maintained in the absence of continued ATP hydrolysis. J Biol Chem. 1996;271:20726–20733. doi: 10.1074/jbc.271.34.20726. [DOI] [PubMed] [Google Scholar]
- 14.Ito T, Bulger M, Pazin M J, Kobayashi R, Kadonaga J T. ACD, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- 15.Khavari P A, Peterson C L, Tamkun J W, Mendel D B, Crabtree G R. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- 16.Kruger W, Peterson C L, Sil A, Coburn G, Arents G, Moudrianakis E N, Herskowitz I. Amino acid substitutions in the structured domains of histones H3 and H4 partially relieve the requirement of the yeast SWI/SNF complex for transcription. Genes Dev. 1995;9:2770–2779. doi: 10.1101/gad.9.22.2770. [DOI] [PubMed] [Google Scholar]
- 17.Kwon H, Imbalzano A N, Khavarl P A, Kingston R E, Green M R. Nucleosome disruption and enhancement of activator binding by a human SWI/SNF complex. Nature. 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 18.Logie C, Peterson C L. Catalytic activity of the yeast SWI/SNF complex on reconstituted arrays. EMBO J. 1997;16:6772–6782. doi: 10.1093/emboj/16.22.6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muchardt C, Yaniv M. A human homolog of Saccharomyces brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 1993;12:4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oliva R, Bazett-Jones D P, Dixon G H. Histone hyperacetylation can induce unfolding of the nucleosome core particle. Nucleic Acids Res. 1990;18:2739–2747. doi: 10.1093/nar/18.9.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Owen-Hughes T, Utley R T, Côté J, Peterson C L, Workman J L. Persistent site-specific remodeling of a nucleosome array by transient action of the SWI/SNF complex. Science. 1996;273:513–516. doi: 10.1126/science.273.5274.513. [DOI] [PubMed] [Google Scholar]
- 22.Peterson C L, Tamkun J W. The SWI/SNF complex: a chromatin remodeling machine? Trends Biochem Sci. 1995;20:143–146. doi: 10.1016/s0968-0004(00)88990-2. [DOI] [PubMed] [Google Scholar]
- 23.Prelich G, Winston F. Mutations that suppress the deletion of an upstream activating sequence in yeast: involvement of a protein kinase and histone H3 in repressing transcription in vivo. Genetics. 1993;135:665–676. doi: 10.1093/genetics/135.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quinn J, Fyrberg A M, Ganster R W, Schmidt M C, Peterson C L. DNA-binding properties of the yeast SWI/SNF complex. Nature. 1996;379:844–847. doi: 10.1038/379844a0. [DOI] [PubMed] [Google Scholar]
- 25.Simpson R T, Thoma F, Brubaker J M. Chromatin reconstituted from tandemly repeated cloned DNA fragments and core histones: a model system for study of higher order structure. Cell. 1985;42:799–808. doi: 10.1016/0092-8674(85)90276-4. [DOI] [PubMed] [Google Scholar]
- 26.Tamkun J W, Deuring R, Scott M P, Kissinger M, Pattatucci A M, Kaufman T C, Kennison J A. brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- 27.Tsukiyama T, Wu C. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell. 1995;83:1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- 28.Tsukiyama T, Daniel C, Tamkun T, Wu C. ISWI, a member of the SWI2/SNF2 ATPase family, encodes the 140 kDa subunit of the nucleosome remodeling factor. Cell. 1995;83:1021–1026. doi: 10.1016/0092-8674(95)90217-1. [DOI] [PubMed] [Google Scholar]
- 29.Utley R T, Côté J, Owen-Hughes T, Workman J L. SWI/SNF stimulates the formation of disparate activator-nucleosome complexes but is partially redundant with cooperative binding. J Biol Chem. 1997;272:12642–12649. doi: 10.1074/jbc.272.19.12642. [DOI] [PubMed] [Google Scholar]
- 30.Varga-Weisz P D, Wilm M, Bonte E, Dumas K, Mann M, Becker P B. Chromatin-remodeling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- 31.Walter P P, Owen-Hughes T A, Côté J, Workman J L. Stimulation of transcription factor binding and histone displacement by nucleosome assembly protein 1 and nucleoplasmin requires disruption of the histone octamer. Mol Cell Biol. 1995;15:6178–6187. doi: 10.1128/mcb.15.11.6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang W, Côté J, Xue Y, Zhou S, Khavari P A, Biggar S R, Muchardt C, Kalpana G V, Goff S P, Yaniv M, Workman J L, Crabtree G R. Purification and biochemical heterogeneity of the mammalian SWI/SNF complex. EMBO J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 33.Schnitzler G, Sif S, Kingston R E. Human SWI/SNF interconverts a nucleosome between its base state and a stable remodeled state. Cell. 1998;94:17–27. doi: 10.1016/s0092-8674(00)81217-9. [DOI] [PubMed] [Google Scholar]