Fast algorithms for large-scale genome alignment and comparison (original) (raw)
Abstract
We describe a suffix-tree algorithm that can align the entire genome sequences of eukaryotic and prokaryotic organisms with minimal use of computer time and memory. The new system, MUMmer 2, runs three times faster while using one-third as much memory as the original MUMmer system. It has been used successfully to align the entire human and mouse genomes to each other, and to align numerous smaller eukaryotic and prokaryotic genomes. A new module permits the alignment of multiple DNA sequence fragments, which has proven valuable in the comparison of incomplete genome sequences. We also describe a method to align more distantly related genomes by detecting protein sequence homology. This extension to MUMmer aligns two genomes after translating the sequence in all six reading frames, extracts all matching protein sequences and then clusters together matches. This method has been applied to both incomplete and complete genome sequences in order to detect regions of conserved synteny, in which multiple proteins from one organism are found in the same order and orientation in another. The system code is being made freely available by the authors.
INTRODUCTION
Genome sequence alignment research has developed highly efficient algorithms for alignment of protein sequences, which have been implemented in very widely used BLAST (1) and FASTA (2) systems. In 1999, as the number of complete genome sequences was rapidly increasing, we introduced a method for efficient alignment of large-scale DNA sequences, in the order of millions of nucleotides (3). This alignment system, called MUMmer, is capable of aligning complete bacterial genomes in <1 min on a standard desktop computer. The central algorithm of MUMmer takes two input sequences, either DNA or proteins, and finds all subsequences longer than a specified minimum length k that are identical between the two inputs. These matches are guaranteed to be maximal, in that they cannot be extended on either end without incurring a mismatch, and in the original system they are also unique, occurring exactly once in each input sequence. This last constraint has been relaxed in the new system, as explained below. This core algorithm is implemented using a suffix-tree data structure, which permits very fast and memory-efficient comparisons of the sequences.
The MUMmer system has been used to make a number of important discoveries about large-scale genome structure. Alignments of related bacterial species led to the discovery that chromosome-scale inversions are a common evolutionary phenomenon in these species, and that the inversions are nearly always symmetric about the origin of replication (4). These inversions show up as X-shaped alignments in the dot plot of all the DNA sequences conserved between two species. The X-alignments have been observed by running MUMmer to compare the following pairs of species: Escherichia coli and Vibrio cholerae, Mycobacterium tuberculosis and Mycobacterium leprae, Chlamydia trachomatis and Chlamydia pneumoniae, Staphylococcus aureus and Bacillus subtilis, Streptococcus pneumoniae and Streptococcus pyogenes, Pseudomonas aeruginosa and Pseudomonas putida, Helicobacter pylori strains 26695 and J99, and others.
MUMmer was used to construct alignments of the five chromosomes of the model plant Arabidopsis thaliana, which range in size from 17 to 29 million base pairs (Mb), against one another. These alignments revealed the striking discovery that the entire genome appears to have duplicated recently, and >60% of the genome, as it exists today, is part of large-scale duplications (5). An earlier MUMmer-aided analysis based on the first two completed chromosomes from that organism revealed a 5 Mb duplication between chromosomes 2 and 4 (6).
Despite these successful alignments, the original MUMmer system required relatively large amounts of memory, making it necessary to use large server computers to align genomes of more than a few million nucleotides. In addition, the original implementation could only handle DNA sequences. When the draft sequence of the human genome was nearing completion, we needed a method to align entire human chromosomes rapidly and accurately. We redesigned the algorithm, as described below, to require far less memory and in the process to run much faster. The new system was used successfully to align all the chromosomes of the human genome to each other and to the mouse genome, demonstrating that it can handle essentially all genome sequences, even those of mammals.
Another improvement in MUMmer 2 is the ability to align protein or DNA sequences. The need for this became necessary after our first attempts to align human chromosomes to each other (7), searching for duplications similar to those we had found in Arabidopsis. Our initial searches using DNA sequence alignments revealed no large-scale duplications in the human genome. We realized that if there had been more ancient duplications, these might yet be detectable in the protein sequences. Therefore, we enhanced MUMmer by giving it the ability to align large protein sequences, and applied it to human as follows. For each human chromosome, we concatenated all proteins in order (regardless of strand) to create 24 mini-proteomes. We then used MUMmer to align each chromosome to the entire genome at the protein level. This was much more successful, revealing hundreds of small-scale and many large-scale duplications all of them apparently quite ancient (7). For example, one of the most striking findings was that >70% of human chromosome 14 appears to be an ancient duplication of part of chromosome 2.
The complete system source code for MUMmer 2 is freely available from the TIGR website, at http://www.tigr.org/software/mummer/.
ALGORITHMIC IMPROVEMENTS
MUMmer uses suffix trees (8) to create an internal representation of a genome sequence, and based on this representation it can align two genomes in linear time and space. For example, MUMmer 1.0 aligns the 4.7 Mb genome of E.coli and the 3.0 Mb large chromosome of V.cholerae in 74 s on a 1 GHz desktop computer, requiring 293 megabytes (MB) of memory. The memory requirement of 38 bytes/bp, although it grows only linearly with the size of the input sequences, is still a limitation of the original system. This has been dramatically reduced in MUMmer 2. For the same two genomes, the new system computes the alignment in only 27 s and requires only 100 MB of memory. Both speed and memory usage has been improved by a factor of nearly three.
There are three significant technical improvements in the core algorithms of MUMmer 2. The first is a reduction in the amount of memory used to store suffix trees. By employing techniques described by Kurtz (9) the amount of memory used in the suffix tree was reduced to at most 20 bytes/bp (or amino acid, or other character). The maximum memory usage occurs in the case when each internal node in the suffix tree has only two children. In practice, however, many nodes have more than two children (particularly in the case of polypeptide sequences), which reduces the actual memory requirement.
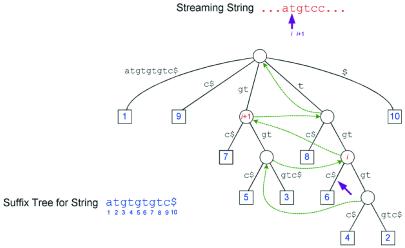
The second significant core improvement is an alternative algorithm to find initial exact matches. The original algorithm (3) built a suffix tree containing two input sequences, and then found all maximal unique matches (MUMs) between them (Fig. 1). A MUM is a subsequence that occurs in two exactly matching copies, once in each input sequence, and that cannot be extended in either direction. This algorithm is still available in the MUMmer 2 system, but the default algorithm is now a procedure that stores in the suffix tree only one sequence, which we call the reference sequence. The second sequence, which we call the query, is then ‘streamed’ against the suffix tree, exactly as if it were being added but without actually adding it. This technique was introduced by Chang and Lawler (10) and is fully described (8). Using this process we identify where the query sequence would branch off from the tree, thereby finding all matches to the reference sequence (Fig. 1). Wherever a branch occurs at a tree position with just a single leaf beneath it, the match is unique in the reference sequence. By checking the character immediately preceding the start of this match we can determine whether it is a maximal match.
Figure 1.
A sample suffix tree showing the streaming behavior for finding matches between a query and a reference.
Thus, in time proportional to the length of the query sequence, we can identify all maximal matches between it and a unique string in the reference sequence. Note that these matches are not necessarily unique in the query sequence. Because we stream through the query, outputting matches as we find them, we do not know what sequence will occur later in the query. The advantage of this method is that only one of the two sequences is stored in the suffix tree, reducing the memory requirement by at least half. Further, because of the streaming nature of the algorithm, once the suffix tree has been built for the reference sequence, arbitrarily long, multiple queries can be streamed against it. In fact, we have used these programs to compare two assemblies of the entire human genome (each approximately 2.7 billion characters), using each chromosome as a reference and then streaming the other entire genome past it (A.Halpern, personal communication).
Figure 1 illustrates how a string is streamed against a suffix tree to find its unique matches. Here the suffix tree represents the string atgtgtgtc$. Leaves are represented by squares labeled with the string position at which the suffix for that leaf starts. For example, leaf 6 represents the suffix tgtc$ that starts at position 6 in the string, which is formed by the sequence of edge labels from the root down to node 6. At the point shown in the figure, we have matched the input stream starting at position i, indicated by the arrow. The match extends to the corresponding arrow position in the tree. In this case we know that the match is unique because there is a single leaf below this position in the tree. The number label of the leaf gives the starting position of the match in the suffix-tree string.
To find the next match, we use the suffix links in the tree, indicated here by dotted arrows. These links are constructed for each internal node in the tree. A link points from node u to node v if the string label from the root to v is equal to the label from the root to u with the first character removed. For example, the string label of node i in Figure 1 is tgt and that of node i + 1 is gt. Note, that is exactly the tree position corresponding to the next position in the streaming string. From node i we can continue the match down the tree to determine how far the match can be extended. Because we match as far as possible in the tree, our matches are maximal on the right-hand side of the strings being compared. We check the maximality on the left-hand side by comparing the preceding characters in each string. In the above example, the match gtc starting at i + 1 in the streaming string and node 7 in the suffix-tree string is not maximal on the left because the preceding character in both strings is a t.
The one-sided uniqueness property of MUMmer 2 can be an advantage when comparing queries that represent only a partial genome assembly. In this case there may be overlapping contigs that were not joined. The overlapping regions would not be unique in the set of queries, but would have a unique match to the reference sequence. In general, because of the subsequent processing performed on matches, we have found that repetitiveness in the query sequence does not prevent the algorithm from finding alignments. In fact, because of the asymmetry of matches, we often process sequence pairs twice, swapping the reference and the query. MUMmer 2 also includes an additional program that finds all maximal exact matches (as were used in the analysis of E.coli strain O157; 11), with no uniqueness checking in either sequence.
The third technical improvement we have made is the addition of a new module to cluster matches. The original version of MUMmer presumed that two complete sequences were to be aligned, and that no major rearrangements would have occurred between them. Hence, it computed a single longest alignment between the sequences. To facilitate comparisons involving unfinished assemblies and genomes with significant rearrangements, we have added a module that first clusters matches together and then finds consistent paths within each cluster. As a result, the system outputs a series of separate, independent alignment regions. The clustering is performed by finding pairs of matches that are sufficiently close and on sufficiently similar diagonals in an alignment matrix (using thresholds set by the user), and then computing the connected components for those pairs. Within each component, a longest ascending subsequence computation is done to yield the most consistent sequence of matches in the cluster.
Additional modules are included in the system to compute alignments between, and extending out from the ends of, exact matches in a consistent chain. The entire package has been designed to be as modular as possible, with simple text input/output formats, to allow the user to mix and match components or add additional functionality.
ALIGNMENT OF INCOMPLETE GENOMES
Although genome sequencing has become much faster and cheaper in recent years, one of the major rate-limiting steps in a genome project is closing all the gaps, or ‘finishing’ the genome. In a typical whole-genome shotgun-sequencing (WGSS) project, the genome is broken up into millions of pieces, which under ideal conditions are a uniform random sample of the genome. These pieces, which may range in size from 2 to 150 kb, are then sequenced, usually from both ends (known as double-ended shotgun sequencing). Current sequencing technology generates sequence ‘reads’ of ~650 bp in length. According to the mathematical model of Lander and Waterman (12), if the reads are generated at random, then >99% of a genome will be covered by sequencing enough reads to cover the genome eight times. For a typical 2 Mb bacterial genome, this requirement for 8× coverage translates into approximately 25 000 reads. These reads are then assembled to reconstruct the genome. Because no one has yet been able to generate truly random fragments on real DNA, the result of assembly is usually a collection of large, unordered DNA sequences called contigs. The number of contigs for a WGSS project may range from a few dozen to many thousands for large genomes. Finishing is the process of determining the order and orientation of all the contigs, and then generating additional sequence to fill in all the gaps between them.
In order to speed the process of getting a picture of an organism, many sequencing projects now dispense with the finishing phase, or at least delay it indefinitely. Thus, the result of a genome sequencing effort will consist of hundreds of contigs that represent 3×, 5×, 8× or some other level of shotgun coverage. These projects are becoming especially popular when a reference genome—a sequence that is already completed—is available for a closely related organism. In order to extract the greatest value from such projects, we need a computational method for aligning multiple contigs to a genome, and for aligning one set of contigs to another.
There is also a need to compare different assemblies to one another. During the course of a WGSS project, the sequence reads may be assembled multiple times. In order to understand how the assemblies change as we go from 5× to 6× (for example), we need a method that can align the multiple contigs that result from assembling the genome at each level. The sequences will obviously be nearly identical, so a sensitive method like BLAST is unnecessary for this task. MUMmer alignments are the ideal solution, because their reliance on exact matches is well suited to the problem. A related need is for a system that allows us to compare the output of two sequence assembly programs with one another on the same input data. There are now at least three assemblers available: TIGR Assembler (13), phrap (www.phrap.org) and Cap3 (14), and several more in development. In order to evaluate the very complex output from these systems, we need to be able to align very large contigs quickly.
To solve both of these problems, we developed a multiple-contig alignment program that uses MUMmer 2 as its core alignment engine. This extension, called NUCmer (nucleotide MUMmer), takes as input two multi-fasta files representing partial or complete assemblies. The inputs may be different assemblies of the same genome, or of different genomes. The algorithm works as follows.
First, NUCmer creates a map of all contig positions within each of the multi-fasta files. It then concatenates the two files separately, and simply runs MUMmer to find all exact matches between the two genomes. These matches are then mapped back to the separate contigs. In its second step, NUCmer runs a clustering algorithm for all the MUMs along each contig. MUMs are clustered together if they are separated by no more than a user-specified distance. The system then runs a modified Smith–Waterman dynamic programming alignment algorithm (15) to align the sequences between the MUMs. In order to avoid excessive computation in this step, the algorithm permits only limited mismatches in these gaps between MUMs. The exact amount of mismatch is specified by the user.
The result of these steps is an alignment of every sequence contig in the first multi-fasta file to every sequence in the second. The percent identity is computed and included in the output. One of the outputs is a sorted list showing how every contig in the second file matches the first file. Therefore, if the first file contains a complete reference genome, the output of NUCmer allows one to simply read off the mapping of contigs in the second file to that genome. In a matter of minutes, one has complete order and orientation information about a partially sequenced genome with respect to a reference species.
The workings of NUCmer are best illustrated with an example. We took the sequence of Theileria parva (a parasite that causes East Coast Fever, a usually fatal disease affecting cattle in sub-Saharan Africa), a genome of ~8 Mb in four chromosomes. At the time of this analysis, T.parva was in the finishing stage, consistent of numerous contigs ranging from <1 kb to >1 Mb. We used NUCmer to compare the largest contig, 1.8 Mb, to the contigs that existed after 8× sequencing but before finishing. A portion of the comparison is shown in Table 1.
Table 1. A NUCmer alignment of a large contig from T.parva to multiple smaller contigs from an earlier assembly run.
| S1 | E1 | S2 | E2 | Len1 | Len2 | Percent identical | Contig |
|---|---|---|---|---|---|---|---|
| 1 | 19018 | 22244 | 3226 | 19018 | 19019 | 99.95 | 347625 |
| 17613 | 28621 | 1694 | 12702 | 11009 | 11009 | 99.22 | 347580 |
| 28243 | 46869 | 18627 | 1 | 18627 | 18627 | 99.97 | 347624 |
| 46855 | 112190 | 65337 | 1 | 65336 | 65337 | 100.00 | 347656 |
| 113996 | 117073 | 3079 | 1 | 3078 | 3079 | 99.97 | 347325 |
| 117842 | 131466 | 13625 | 1 | 13625 | 13625 | 99.99 | 347606 |
| 131485 | 183004 | 51519 | 1 | 51520 | 51519 | 100.00 | 347623 |
| 182996 | 254867 | 1 | 71872 | 71872 | 71872 | 100.00 | 347559 |
This large 1.8 Mb contig (approximately the same size as the entire genome of Haemophilus influenzae, the first genome ever completely sequenced; 16) was in 32 smaller pieces at the end of the shotgun phase. Table 1, which contains only the first eight contigs, shows how these contigs match the first 250 kb of the large contig. It is immediately clear from this data that some of the separate contigs overlap: for example, lines two to three show contigs 347580 and 347624, which overlap by nearly 400 bp in the large contig. The contigs in lines three to four overlap by only 15 bp, probably too little for an assembly program to join together. Lines four to five, in contrast, contain contigs separated by a gap of 1800 bp in the final assembly.
One important feature of the NUCmer output is that one can simply read off the order and orientation of one set of contigs with respect to another. In Table 1, the order of the contigs is given by reading down the last column, and the orientation of each one can be found by looking at the third and fourth columns, where numbers in ascending order indicate that the contigs are aligned the same way, while descending order indicates that the contigs are reversed with respect to the reference genome.
The output shown here demonstrates how an assembly has changed from the shotgun stage to a later, finished stage. If instead we use NUCmer to compare multiple contigs from an unfinished genome to a closely related species, then we can very quickly determine the order and orientation of those contigs. This mapping information would then have to be verified by PCR or other finishing steps, but such directed finishing is far more efficient than finishing a set of unmapped contigs.
USING MUMmer FOR COMPARATIVE GENOME ANNOTATION
Protein sequences remain conserved much longer, on an evolutionary time scale, than DNA sequences and, therefore, protein-based alignments can detect much older relationships than DNA alignments. The rate of large-scale genome rearrangements is slow enough that sets of homologous protein sequences can be found in related organisms even though the DNA sequence conservation is minimal. These conserved syntenic regions are extremely valuable as a source of insight into the functions of the proteins comprising them in both genomes: genes already characterized in one organism, for example, might be identified correctly in a second organism based on the synteny between the two. Syntenic regions are also valuable in focusing the search for conserved regulatory regions, which sometimes appear as short conserved stretches of DNA between the protein-coding regions.
Because the MUMmer 2 system can align protein sequences as well as DNA, it provides a good platform for a system to detect conserved synteny in protein sequences between two genomes that may or may not be completely sequenced. We have built a system extension called PROmer that can very rapidly compute the protein similarity between all possible protein-coding regions in two sets of DNA sequences.
Given two multi-fasta input files, PROmer will translate the DNA to amino acids and then compare each sequence in the first file to all of the sequences in the second file. Upon completion, the user is given a list of all significant matches, their respective sequence IDs, their coordinates, and their percent similarities. Each individual match usually represents a single exon or conserved protein domain; because the matching and clustering parameters are completely user defined, it is possible to expand the match sizes to include larger syntenic regions. The user can also view alignments of these matches to determine their significance, and in some cases to annotate the precise boundaries of highly conserved exons. The backbone of PROmer is the suffix-tree matching algorithm used by MUMmer, but turning the small exact matches identified by MUMmer into larger alignment regions takes more effort.
Initially, PROmer translates the DNA into amino acids for all input sequences in all six reading frames. An index is created that maps all protein sequences and lengths to the source DNA, which will be needed later to map the matches back. The amino acid translations are then filtered to remove sequences that have an excessive number of stop codons and thus not likely to be part of a protein. After these steps, each input genome, which began as a multi-fasta file containing an unrestricted number of DNA sequences, has been reduced to a single amino acid sequence that represents the concatenation of all potential proteins in the genome. These pseudo-proteomes are passed to MUMmer, which rapidly builds a suffix tree and finds all exact matches. The index is then used to translate these matches back onto the original DNA input.
After matches are identified, they are clustered according to their respective DNA coordinates. A series of consecutive matches that exceeds a user-specified minimum length and retains a consistent order is examined further. If the size of the interval between matches is less than a user-specified gap length, the matches are joined into a cluster. The resulting clusters may include amino acid sequences in competing (inconsistent) reading frames; thus the next step is to decide which frame, for each cluster, is the most likely. On the assumption that the correct reading frame will contain the greatest degree of protein sequence homology, the frame with the best matches between the two input genomes is preferred. At the end, each cluster has a series of amino acid hits in a single consistent reading frame.
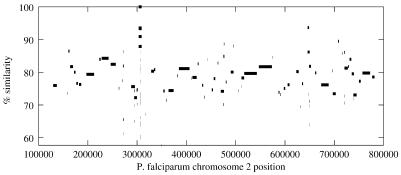
When the initial clustering step is complete, PROmer extends the clusters to enlarge the total coverage of the alignment region. For this step, the system uses a banded dynamic programming algorithm (15) that scores the protein matches using the BLOSUM62 matrix (17). The resulting alignment data is converted to a delta encoding, which represents the distance between inserts and deletions as a series of integers. Through this delta encoding, it is possible to reconstruct any of the alignments as needed. This makes it possible to tailor utility programs to parse the delta file and search for specific alignments, or to display the alignments graphically. The current system contains two such programs, the first of which provides a simple graphical display of the actual alignments. This display provides a quick overview of the overall similarity between the two input sequences; an example is shown in Figure 2.
Figure 2.
Alignment of multiple fragments from the partially sequenced P.yoelii genome to chromosome 2 of P.falciparum. Coordinates on chromosome 2 are displayed along the horizontal axis, and the percent similarity at the amino acid level is show on the vertical axis. Line segments represent regions detected as similar, not entire contigs; thus, a contig may contain multiple, distinct regions of similarity. Percent similarity refers only to the coding regions, not to entire length of each line segment. Contigs from P.yoelii are sorted according to their mapped position on P.falciparum.
Figure 2 shows all of the contigs from Plasmodium yoelii that mapped to Plasmodium falciparum chromosome 2 (17). We can see from the 5× assembly that many of the protein-coding regions in chromosome 2 are present in P.yoelii. (The telomeric regions are repeat-rich and contain very few genes; the sub-telomeric regions contain families of genes highly divergent in both species.) Another PROmer utility creates a table that captures all the alignment positions (5′ and 3′ ends), the identifiers of the respective contigs that contain the matching sequences, and their percent identity. Details from this mapping are illustrated in Table 2.
Table 2. Detail from the alignment of all contigs from the P.yoelii assembly at 5 × coverage to the completed chromosome 2 from P.falciparum.
| P.falciparum | P.yoelii contig | Contig ID | Length (bp) | Identity (%) | Similarity (%) | Frame | |||
|---|---|---|---|---|---|---|---|---|---|
| Start | End | Start | End | ||||||
| 587286 | 586992 | 384 | 81 | a1160 | 295 | 47.1 | 72.1 | –1 | –3 |
| 587626 | 587374 | 652 | 418 | a1160 | 253 | 55.8 | 76.7 | –3 | –2 |
| 588691 | 588271 | 1594 | 1180 | a1160 | 421 | 64.3 | 80.4 | –3 | –2 |
| 590531 | 591386 | 820 | 1639 | a1236 | 856 | 49.4 | 73.1 | 2 | 1 |
| 597754 | 598336 | 1496 | 2072 | a3475 | 583 | 57.6 | 80.3 | 1 | 2 |
| 598867 | 599386 | 2729 | 3242 | a3475 | 520 | 56.2 | 68.7 | 1 | 2 |
| 600503 | 600356 | 4990 | 4855 | a3475 | 148 | 62.0 | 80.0 | –2 | –1 |
| 605666 | 606053 | 3844 | 3481 | a2330 | 388 | 62.6 | 82.4 | 2 | –3 |
| 606674 | 607832 | 3190 | 1987 | a2330 | 1159 | 52.9 | 75.3 | 2 | –3 |
| 608543 | 609323 | 1234 | 454 | a2330 | 781 | 62.8 | 80.8 | 2 | –3 |
Table 2, which is extracted from a much longer list, makes it clear that P.yoelii contigs a1160, a1236, a3475 and a2330 align to positions 587k–609k of P.falciparum chromosome 2. Using this alignment as an anchor, one can examine the sequence more closely to find other genes that should align but that may be too distantly related to be detected. One can also find genes that have been lost from each genome within these syntenic regions.
The alignment between P.yoelii and P.falciparum highlights another valuable use of PROmer: creating a map that can be used for rapidly completing a genome. For genomes such as the Plasmodium species, where detailed map information is not available, direct alignment between the species provides a guide to finishing efforts. Although some rearrangements may have occurred since the species diverged, in most cases the bridge created by one sequence when aligned to the other will indicate the correct order and orientation of separate contigs. Directed PCR experiments can be conducted to bridge the gaps. By thoroughly pre- and post-processing the data from related species with MUMmer and PROmer, we can build accurate tiling paths and visual alignments in minutes. This approach gives results comparable with BLAST while consuming far less computing power. These methods are already in use to facilitate comparative annotation between the parasite genomes P.yoelii (5× coverage) and P.falciparum (coverage ranging from 8× to completely closed). For these two genomes, ~25 Mb each, PROmer analysis can be completed in <1 h, while equivalent Blast computations would take weeks of computing time. More specifically, a PROmer comparison between chromosome 2 of P.falciparum (1 Mb) and all the contigs from the 5× assembly of P.yoelii (~25 Mb) requires ~25 MB of memory and <12 min of runtime on a standard 500 MHz Pentium III Linux desktop.
The two Plasmodium species are sufficiently divergent that DNA sequence similarity is difficult to detect, even in syntenically conserved regions, but protein sequence similarity is usually very significant. For projects like these, where the genomes are just being sequenced and thousands of genomes are being discovered as part of the sequencing effort, the PROmer comparison makes it possible to find entire genes that were missed in annotation, and to adjust and correct the annotated coding regions for others.
At least two other systems are now available for comparing lengthy DNA sequences, and more are under development. The PIPmaker system (18) uses a hashing approach based on the BLAST algorithm, with improvements to handle large input sizes using only linear space (19). PIPmaker finds both approximate and exact alignments and also generates a very useful graphical display, showing which portions of the alignment match at different percent identities. The SSAHA system (20) uses a hash table to find matches to a query sequence, which is permitted to be as long as a whole chromosome, very quickly. With appropriate tuning of its parameters, it was able to perform the E.coli–V.cholerae alignment described above in <20 s, although it used 275 MB of memory. The memory usage of SSAHA is 4_k_ + 1 bytes for a minimum match size of k (e.g. 4 Gb for 15-bp matches), requiring one to use small values of k, which generates enormous amounts of output. However, the system includes filters that allow one to restrict the output so that it includes only longer exact matches, making it behave similarly to MUMmer.
CONCLUSION
The new MUMmer system is nearly three times faster, needs only one-third the memory needed by the original version, and now has algorithmic extensions that permit alignment and comparison of protein sequence and of multiple sequences from incomplete genomes. It has been used to compare sequences as long as entire human chromosomes, for which it was instrumental in finding large-scale ancient duplications (7), and it has also been used for whole-genome alignment of the DNA sequences of numerous bacterial species (4). As discussed here, it is proving highly valuable in the comparative analysis of related parasite species. The ability to align millions of nucleotides in a few minutes on a desktop computer provides the opportunity to conduct analyses that would be otherwise too computationally demanding for many researchers. The enhanced capabilities of the new system make it possible to detect large-scale relationships between more distantly related organisms, a feature that is becoming increasingly important as more and more genome sequences are completed.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported in part by grants IIS-9902923 to S.L.S. and IIS-9820497 to A.L.D. from the National Science Foundation, and by grant R01-LM06845 to S.L.S. from the National Institutes of Health. J.C. is supported by funds from NIAID R01-A142243. The TIGR P.yoelii whole genome shotgun project is funded by the Department of Army collaborative agreement DAMD17-98-2-8005.
REFERENCES
- 1.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 2.Pearson W.R. (2000) Flexible sequence similarity searching with the FASTA3 program package. Methods Mol. Biol., 132, 185–219. [DOI] [PubMed] [Google Scholar]
- 3.Delcher A.L., Kasif,S., Fleischmann,R.D., Peterson,J., White,O. and Salzberg,S.L. (1999) Alignment of whole genomes. Nucleic Acids Res., 27, 2369–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eisen J.A., Heidelberg,J.F., White,O. and Salzberg,S.L. (2000) Evidence for symmetric chromosomal inversions around the replication origin in bacteria. Genome Biol., 1, 1101–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature, 408,796–815. [DOI] [PubMed] [Google Scholar]
- 6.Lin X., Kaul,S., Rounsley,S., Shea,T.P., Benito,M.I., Town,C.D., Fujii,C.Y., Mason,T., Bowman,C.L., Barnstead,M. et al. (1999) Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature, 402, 761–768. [DOI] [PubMed] [Google Scholar]
- 7.Venter J.C., Adams,M.D., Myers,E.W., Li,P.W., Mural,R.J., Sutton,G.G., Smith,H.O., Yandell,M., Evans,C.A., Holt,R.A. et al. (2001) The sequence of the human genome. Science, 291, 1304–1351. [DOI] [PubMed] [Google Scholar]
- 8.Gusfield D. (1997) Algorithms on Strings, Trees and Sequences: Computer Science and Computational Biology. Cambridge University Press, New York.
- 9.Kurtz S. (1999) Reducing the space requirement of suffix trees. Software Pract. Experience, 29, 1149–1171. [Google Scholar]
- 10.Chang W.I. and Lawler,E.L. (1994) Sublinear expected time approximate string matching and biological applications. Algorithmica, 12, 327–344. [Google Scholar]
- 11.Perna N.T., Plunkett,G.,III, Burland,V., Mau,B., Glasner,J.D., Rose,D.J., Mayhew,G.F., Evans,P.S., Gregor,J., Kirkpatrick,H.A. et al. (2001) Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature, 409, 529–533. [DOI] [PubMed] [Google Scholar]
- 12.Lander E.S. and Waterman,M.S. (1988) Genomic mapping by fingerprinting random clones: a mathematical analysis. Genomics, 2, 231–239. [DOI] [PubMed] [Google Scholar]
- 13.Sutton G., White,O., Adams,M. and Kerlavage,A.R. (1995) TIGR Assembler: a new tool for assembling large shotgun sequencing projects. Genome Sci. Technol., 1, 9–19. [Google Scholar]
- 14.Huang X. and Madan,A. (1999) CAP3: a DNA sequence assembly program. Genome Res., 9, 868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waterman M.S. (1984) Efficient sequence alignment algorithms. J. Theor. Biol., 108, 333–337. [DOI] [PubMed] [Google Scholar]
- 16.Fleischmann R.D., Adams,M.D., White,O., Clayton,R.A., Kirkness,E.F., Kerlavage,A.R., Bult,C.J., Tomb,J.F., Dougherty,B.A., Merrick,J.M. et al. (1995) Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science, 269, 496–512. [DOI] [PubMed] [Google Scholar]
- 17.Henikoff J.G., Pietrokovski,S., McCallum,C.M. and Henikoff,S. (2000) Blocks-based methods for detecting protein homology. Electrophoresis, 21, 1700–1706. [DOI] [PubMed] [Google Scholar]
- 18.Schwartz S., Zhang,Z., Frazer,K.A., Smit,A., Riemer,C., Bouck,J., Gibbs,R., Hardison,R. and Miller,W. (2000) PipMaker—a web server for aligning two genomic DNA sequences. Genome Res., 10, 577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chao K.M., Zhang,J., Ostell,J. and Miller,W. (1995) A local alignment tool for very long DNA sequences. Comput. Appl. Biosci., 11, 147–153. [DOI] [PubMed] [Google Scholar]
- 20.Ning Z., Cox,A.J. and Mullikin,J.C. (2001) SSAHA: a fast search method for large DNA databases. Genome Res., 11, 1725–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]