The Breakpoint Region of the Most Common Isochromosome, i(17q), in Human Neoplasia Is Characterized by a Complex Genomic Architecture with Large, Palindromic, Low-Copy Repeats (original) (raw)
Abstract
Although a great deal of information has accumulated regarding the mechanisms underlying constitutional DNA rearrangements associated with inherited disorders, virtually nothing is known about the molecular processes involved in acquired neoplasia-associated chromosomal rearrangements. Isochromosome 17q, or “i(17q),” is one of the most common structural abnormalities observed in human neoplasms. We previously identified a breakpoint cluster region for i(17q) formation in 17p11.2 and hypothesized that genome architectural features could be responsible for this clustering. To address this hypothesis, we precisely mapped the i(17q) breakpoints in 11 patients with different hematologic malignancies and determined the genomic structure of the involved region. Our results reveal a complex genomic architecture in the i(17q) breakpoint cluster region, characterized by large (∼38–49-kb), palindromic, low-copy repeats, strongly suggesting that somatic rearrangements are not random events but rather reflect susceptibilities due to the genomic structure.
Introduction
Isochromosome 17q, or “i(17q),” is the most common isochromosome characterizing human neoplasia and has been described as both a primary and a secondary abnormality, indicating an important pathogenetic role in both tumor development and progression (Mertens et al. 1994). The average frequency of i(17q) as a single or additional chromosomal abnormality in carcinomas and hematologic malignancies is ∼3% (Mitelman Database of Chromosome Aberrations in Cancer Web site). Hematologic malignancies with i(17q) are characterized by adverse prognosis, and i(17q) is one of the most frequent changes observed during disease progression of chronic myeloid leukemia (CML), occurring in ∼21% of the cases with chromosomal abnormalities in addition to the pathognomonic reciprocal translocation t(9;22)(q34;q11) (McClure et al. 1999; Johansson et al. 2002). The i(17q) rearrangement has also been described as the most common chromosomal aberration in primitive neuroectodermal tumors and medulloblastomas (Biegel et al. 1995).
The high frequency of i(17q) formation in leukemia and solid tumors compared with other isochromosomes may potentially be explained by the presence of unique DNA sequence characteristics in the breakpoint region of i(17q) that favor genetic rearrangement, in combination with a selective advantage conferred by dysregulated genes in the vicinity of the breakpoint—or, perhaps more likely, conferred by gene dosage imbalances resulting from the loss of 17p and gain of 17q material (Fioretos et al. 1999). To date, however, limited knowledge is available regarding either the mechanism by which this chromosomal abnormality is formed or the resulting molecular genetic consequences. Loss of the tumor-suppressor gene TP53, located in 17p13, has been suggested to be a critical outcome resulting from i(17q) formation, but mutation analysis of the remaining TP53 allele has produced conflicting data, with a low overall incidence of mutations (Nakai and Misawa 1995; Fioretos et al. 1999).
In a previous study, we characterized i(17q) in hematologic malignancies through use of FISH with several YAC clones (Fioretos et al. 1999) and showed that this aberration, in most cases, is not a monocentric but a dicentric isochromosome (isodicentric), as evidenced by the detection of two centromeres through use of molecular cytogenetics methods. Hence, it should be more precisely referred to as “idic(17)(p11.2).” However, for historical reasons and because it is both more general in its description and a less cumbersome designation, the term “i(17q)” remains ensconced in the literature. The i(17q) breakpoints were shown to cluster within 17p11.2, either in the very pericentromeric region or within a 900-kb YAC clone (828b9) located in the Smith-Magenis syndrome (SMS) common deletion region (Chen et al. 1997; Fioretos et al. 1999). A similar clustering of genomic breaks in 17p for i(17q) has also been reported in a set of 18 leukemia and medulloblastoma cases (Scheurlen et al. 1999).
The SMS region is a genetically unstable region that contains multiple low-copy repeats (LCRs) or segmental duplications. Deletions of this region are characteristic features of the congenital disorder SMS (Chen et al. 1997; Stankiewicz et al. 2003). The SMS common-deletion interval is ∼4 Mb and is flanked by large (200 kb), highly homologous, LCR gene clusters termed “SMS-REPs” (Bi et al. 2002; Park et al. 2002). The proximal and distal SMS-REPs likely act as substrates for nonallelic homologous recombination (NAHR), resulting in both common deletions and reciprocal duplications of the same chromosome segment (Chen et al. 1997; Potocki et al. 2000; Shaw et al. 2002; Bi et al. 2002, 2003).
During recent years, LCRs have been increasingly implicated as mediators of NAHR, resulting in a number of congenital genomic disorders, including, for example, Charcot-Marie Tooth disease type 1A, DiGeorge syndrome, Williams-Beuren syndrome, and the Angelman and Prader-Willi syndromes (Stankiewicz and Lupski 2002_a_).
In the present study, we have used FISH analysis with a large set of BAC and PAC clones to further delineate the breakpoints of i(17q) in hematologic malignancies. Sequencing of individual BAC, PAC, and fosmid clones covering the breakpoint region and subsequent sequence comparison of individual clones enabled us to generate a physical map of an ∼240-kb genomic interval including the i(17q) breakpoint cluster region. The delineated region contains two unique LCRs, organized in a complex manner, with the capability to form a long DNA palindromic structure. Mapping of all breakpoints to within this unique genome architecture suggests its important role in the genesis of i(17q).
Material and Methods
Patients and Cytogenetic Analysis
Eleven patients with hematologic malignancies—eight patients with CML in blast crisis (BC), one patient with myelodysplastic syndrome (MDS), and two patients with acute myeloid leukemia—carrying i(17q), either as a sole anomaly or as an additional abnormality, were investigated (table 1). The study was considered and approved by the local ethical committee at Lund University. The cases were selected on the basis of the availability of stored cell pellets in fixative, obtained after routine cytogenetic investigation at the Department of Clinical Genetics, Lund University Hospital (Sweden), or at the Department of Human Genetics, University of Leuven (Belgium).
Table 1.
Clinical Data, G-Banding Karyotypes, and FISH Mapping Results of the 11 Patients with Hematologic Malignancies with i(17)(q10)
| PatientNumbera | Sex/Agein Years | HematologicMalignancyb | Karyotype | FISH Breakpoints |
|---|---|---|---|---|
| 1 | F/50 | CML BC | 47,XX,+8,t(9;22)(q34;q11),i(17)(q10)[22] | RP11-1113L8 (FISH signal absent); RP11-2354J3 (FISH signal present) |
| 2 | F/45 | CML BC | 46,XX,t(9;22)(q34;q11),i(17)(q10)[23]/47,idem,+mar[2] | Within RP11-160E2/RP11-744A16 |
| 3 | M/53 | AML | 46,XY,i(17)(q10)[9]/46,XY[6] | Within RP11-160E2/RP11-744A16 |
| 4 | M/74 | CML BC | 47,XY,add(4)(p12),+8,t(9;22)(q34;q11),i(17)(q10)[7]/48,idem,+der(22)t(9;22)[2] | Within RP11-160E2/RP11-744A16 |
| 5 | M/44 | CML BC | 46,XY,t(9;22)(q34;q11)[5]/47,idem,i(17)(q10),+19[4]/48,idem,+8,i(17)(q10),+19[6] | Within RP11-160E2/RP11-744A16 |
| 6 | M/72 | MDS | 46,XY,i(17)(q10)[24] | Within RP11-160E2/RP11-744A16 |
| 7 | M/35 | CML BC | 47,XY,+8,t(9;22)(q34;q11),i(17)(q10)[5] | Within RP11-160E2/RP11-744A16 |
| 8 | F/50 | CML BC | 46,X,i(X)(p10),t(9;22)(q34;q11),i(17)(q10)[9]/50,idem,+1,+8,+13,+19[2] | Within RP11-160E2/RP11-744A16 |
| 9 | M/72 | MDS-AML | 46,XY,i(17)(q10)[19] | Within RP11-160E2/RP11-744A16 |
| 10 | M/16 | CML BC | 46,XY,t(9;22)(q34;q11)[3]/49,idem,+8,+14,i(17)(q10), +19[5] | Within RP11-160E2/RP11-744A16 |
| 11 | M/69 | CML BC | 46,XY,t(9;22)(q34;q11)[2]/46,idem,?dup(13)(q22q32),i(17)(q10)[12] | Within RP11-160E2/RP11-744A16 |
FISH Analysis
FISH analysis using BAC or PAC clones and whole chromosome painting probes (ALTechnologies) were performed as described elsewhere (Barbouti et al. 2002). The BAC/PAC contig, located between the middle and proximal SMS-REP, included the following clones (GenBank accession numbers are listed in parentheses): CTC-457L16 (AC003957), RP11-160E2 (AC007952), RP11-744A16 (AC106017), RP11-970O14 (AC036110), CTB-187M2 (AC004448), RP11-1113L8 (AC025627), CTD-2354J3 (AC015935), RP11-311F12 (AC005722), RP11-78O7 (AC015726), RP11-209D14 (AC005730), and CTD-2010G8 (AC007963) (Bi et al. 2002). To verify the genome architecture of REPAs and REPBs identified by electronic calculations, we performed fiber-FISH experiments with REPA- and REPB-specific fosmids L29248 and L29227 as probes (Heiskanen et al. 1994).
Bioinformatics and Sequence Analysis
The Blast 2 browser (Blast 2 Sequences Web site) and Sequencher software (Gene Codes) were used for sequence comparisons between the BACs or PACs CTC-457L16, RP1-149D14 (GenBank accession number AJ009617), RP11-744A16, CTD-2525F10 (AC109313), and RP11-135L13 (AC124066); fosmids L29232 (AC139083), L29246 (AC139093), L29227 (AC139315), L29233 (AC140106), L29228 (AC144507), L29248 (AC139138), L29225 (AC144506), and L29280 (AC139077); and/or their pieces with the overlapping BAC clone RP11-160E2. The Mfold program (Mfold Server Web site) was used for the prediction of the secondary structure of the palindromic sequence identified in sequenced piece 1 of BAC RP11-744A16 (nts 1–5824) and in the BAC RP11-160E2 (nts 38912–47643). Repeat sequence analysis was performed with RepeatMasker 2 (RepeatMasker Web Server), and the restriction maps of the sequenced pieces of individual BACs or fosmid clones were based on Webcutter 2.0 (Webcutter 2.0 Web site) and Sequencher analysis.
Results
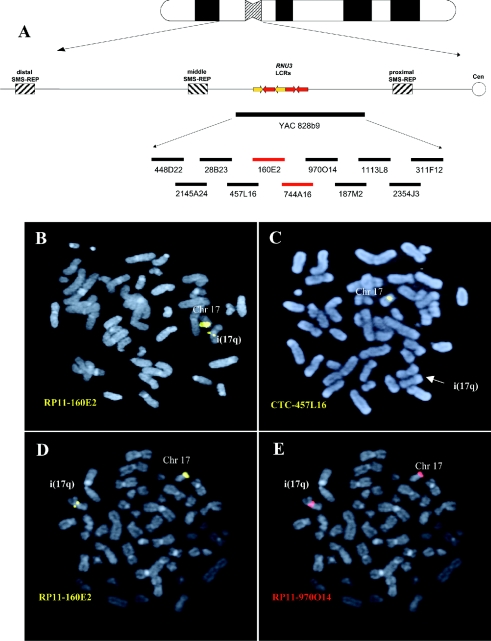
To refine the 17p11 breakpoints, 10 overlapping BAC and PAC clones, from within the 900-kb genomic region contained within the YAC 828b9 that spanned the breakpoint cluster region for i(17q), were used to analyze 11 patients with hematologic malignancies (table 1). Metaphase FISH analyses revealed that the breakpoints occurred within BAC clone RP11-160E2 (chr17:21,075,577–21,080,896; University of California Santa Cruz Genome Browser, July 2003 assembly) or the overlapping clone RP11-744A16 in 10/11 patients with i(17q) (patients 2–11 [fig. 1; table 1]). One patient (patient 1 [table 1]) harbored a more proximal breakpoint, within the two overlapping BAC clones RP11-1113L8 and RP11-2354J3.
Figure 1.
i(17q) breakpoint mapping. A, Ideogram of chromosome 17 with a schematic representation of the 17p11.2 genomic region. Thick, horizontal lines at the bottom of the panel represent large insert YAC and BAC clones. The BACs harboring the breakpoints in 10/11 cases are indicated in red. B and C, FISH analysis of patient 2 with the BAC probes RP11-160E2 and CTC-457L16. B, One RP11-160E2 signal is present on the normal chromosome 17 and one RP11-160E2 (yellow) signal of lower intensity is located on the i(17q). C, CTC-457L16 signal (yellow) is present on the normal chromosome 17 but absent on the i(17q). D and E, Dual color hybridization in patient 3, with the adjacent BACs RP11-160E2 (yellow) and RP11-970O14 (red) on the same metaphase. D, One RP11-160E2 signal is located on the normal chromosome 17 and one signal of lower intensity is present on the i(17q). E, Two RP11-970O14 (red) signals, one present on the normal chromosome 17 and one of slightly higher intensity present on the i(17q).
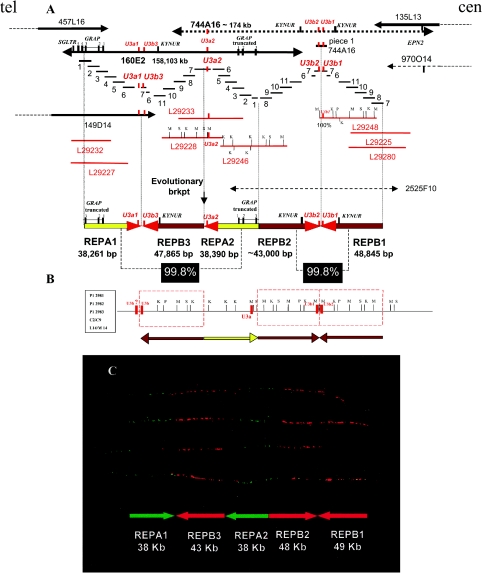
The fact that the DNA sequences of BAC RP11-160E2 and the overlapping clones were present in a draft format and were reported only as several unordered sequenced pieces initially hampered a detailed refinement of the region. To characterize further the genomic architecture within and around these BAC clones, we assembled the DNA sequences of this region by performing extensive sequence comparisons between overlapping, partially sequenced BAC, PAC, and fosmid clones. An electronic analysis of the restriction map of the aligned sequences from the overlapping clones, as well as subsequent comparisons with a previous restriction map of this region (Gao et al. 1997), facilitated the generation of a physical map covering an ∼240-kb genomic interval (fig. 2). The delineated region between BAC clones CTC-457L16 and RP11-135L13 contains two types of distinct LCRs, which we designated “REPA” and “REPB,” organized in a complex manner, with the capability to form a long DNA palindromic structure. The two copies of REPA—REPA1 and REPA2—are both ∼38,000 bp, display 99.8% identity, are arrayed inversely with respect to each other, and are separated by another LCR, the 47,865-bp REPB3 (fig. 2).
Figure 2.
High-resolution physical map of the i(17q) breakpoint cluster region, revealing a complex genome architecture. A, Schematic contig reconstruction of the genomic region based on DNA sequence alignment among completely sequenced clones: BACs CTC-457L16, RP11-160E2, and RP11-135L13; PAC RP1-149D14; fosmids L29232, L29246, L29227, L29233, L29228, L29248, L29225, and L29280; and pieces of unfinished BAC clones, RP11-744A16, CTD-2525F10, and RP11-970O14. The individually sequenced pieces (_short blue horizontal bars labeled “1–11_”) of RP11-160E2 were used together with sequence information of available fosmid clones (red horizontal lines) to arrive at the final assembly of RP11-160E2. Similarly, sequenced pieces of RP11-744A16 and fosmid sequences were used to reconstruct the genomic region between RP11-160E2 and RP11-135L13. The structure of the sequence assembly of RP11-160E2 is well supported by read pairs from plasmid templates of two sizes spanning the region. We have similar support for RP11-744A16 and all the sequenced fosmids. Each clone/fragment is represented as a horizontal bar and is labeled with a number or clone name. The dashed vertical lines mark the border between different LCR copies. The high DNA sequence identity (>99.8%) and, thus, small differences (1 in 500–1,000 nts) among analyzed copies were identified by distinguishing among _cis_-morphisms (differences among LCR copies), polymorphisms (differences among libraries), and DNA sequencing errors, each of which is represented at a similar frequency. Yellow and brown arrows refer to REPA and REPB copies, respectively. LCRs are arranged according to their orientation and structure. Note that REPB2 is truncated when compared with REPB1 and REPB3 copies. Both REPA copies contain exons 1–3 of the GRAP gene, and the remaining exons map telomeric to the REPA1. This indicates that REPA2 originated from REPA1; the position of the putative evolutionary breakpoint (“brkpt”) is indicated with a vertical black arrow. Three U3b and two U3a genes map to the arrowhead portion of REPB and REPA, respectively. The red arrowheads of REPAs and REPBs represent a nearly identical ∼4-kb sequence shared among both REPAs and REPBs. B, Restriction enzyme map of the region. The red dashed rectangles represent the REPB copies. Enzyme abbreviations are as follows: K = _Kpn_I; M = _Mlu_I; S = _Sal_I; P = _Pac_I. An alternative order (horizontally flipped) of REPB2, REPA2, and REPB3 has been deduced from a previously reported restriction map of the analyzed region (Gao et al. 1997). It is possible that the different order may represent population polymorphism. KYNUR = kynureninase related; SGLTR = Na/glucose-transporter related. C, Dual color fiber-FISH using fosmid clones L292248 (red) and L292227 (green) as probes on stretched control genomic DNA supported the genome architecture proposed by in silico analysis.
REPA1 harbors the first three exons of the GRAP gene, encoding a GRB2-related adaptor protein, whereas the telomeric and adjacent region contains the remaining two exons of this gene. The first three exons of GRAP and the 5′ region of this gene are also present in REPA2, whereas exons 4 and 5 are absent; thus, REPA2 contains a truncated version of the GRAP gene.
The organization of the three REPB copies (REPB1, REPB2, and REPB3) is more complex. REPB1 (∼48,845 bp) and the truncated REPB2 (∼43,000 bp) share 99.8% sequence identity and are inverted with respect to one another (inverted repeats [IRs]). They are separated by an ∼400-bp spacer region and, hence, represent a palindrome. REPB3 (∼47,865 bp) is oriented inversely with respect to REPB2 (99.8% identity) (fig. 2A and 2_B_). In the center of this palindromic structure, two U3b genes (U3b2 and U3b1) are located in an inverted orientation. A similar DNA structure of 8,731 bp is present in the junction region between REPA1 and REPB3 (RP11-160E2; nts 38912–47643). The U3a or U3b genes are located at the head portion of each REPA and REPB. They display >98.5% identity and belong to the U3 (RNU3) evolutionarily conserved gene family of small nuclear RNAs required for the processing of pre-18S rRNA (Gao et al. 1997; Dragon et al. 2002). In each REPB, we also identified a gene encoding a protein with 75% identity to kynureninase, which is involved in tryptophan metabolism. The very high degree of similarities among the three REPBs and independently among two REPAs indicates that the duplication events that generated these LCRs are of recent origin, as suggested by evolutionary studies of other highly homologous LCRs (Bailey et al. 2002; Inoue et al. 2002; Stankiewicz and Lupski 2002_b_).
Fiber-FISH studies supported the genome architecture proposed on the basis of computational analyses, confirming both the size and order of the REPA and REPB repeats (fig 2C). We found no evidence for REPA and REPB duplications in the syntenic regions of mouse (RP23-278F12 [GenBank accession number AC084044]) and rat (CH230-200D5 [AC105503]; CH230-255K6 [AC134746]). Unfortunately, electronic comparisons among the primate genome regions syntenic to the i(17q) breakpoint were not available.
Another remarkable feature of this region is the high frequency of Alu repeats. Repetitive sequence analyses of BAC clones RP11-160E2, CTD-2525F10, and RP11-744A16 revealed that they were composed of 31%, 33%, and 29% Alu sequences, respectively. The mean Alu frequency in the draft human genome is 10%, whereas the Alu content in the genomic regions surrounding the REPA/REPB cluster consists of 18% in the distally flanking BAC clone CTC-457L16, 17% in the proximally adjacent BAC clone RP11-135L13, ∼11% in the SMS-REPs, and 21% in the ∼1.1-Mb SMS critical region (Lander et al. 2001; Venter et al. 2001; Bi et al. 2002; Park et al. 2002). It is interesting that >80% of the 4 kb flanking the center of the palindrome are repetitive sequences, with 50% consisting of Alu sequences. Furthermore, each of the five REPs have been found to be flanked by Alu sequences. The high Alu repeat frequencies potentially could be important in the genesis of the large segmental duplications present in this region (Bailey et al. 2003).
Discussion
We propose that the identified LCRs—REPA and REPB—present in two and three copies, respectively, are important mediators of recombination events leading to the formation of i(17q). Inverted repeats or palindromic sequences are well-established hotspots of genomic instability in different model organisms (Mizuuchi et al. 1982; Nasar et al. 2000) and have been implicated as causative in congenital human genomic disorders, as well as in constitutional balanced translocations. The breakpoints of the most frequent constitutional balanced translocation, t(11;22)(q23;q11), were recently found to reside within 166 bp and ⩾852 bp of AT-rich palindromes on chromosomes 11 and 22, respectively (Edelmann et al. 2001; Kurahashi and Emanuel 2001). Moreover, the constitutional reciprocal translocation t(17;22)(q11;q11) has been identified in two patients with neurofibromatosis type 1 (NF1), and, in both cases, AT-rich inverted repeat sequences were detected at the breakpoints (Kehrer-Sawatzki et al. 1997; Kurahashi et al. 2003). Furthermore, the 4.5-Mb _AZF_c deleted region associated with male infertility was found to coincide with 3-Mb-long highly homologous (99.97%) palindromes (Kuroda-Kawaguchi et al. 2001), and palindromic structures have been suggested to be important features in the mechanism of class switch recombination and somatic hypermutation (Honjo et al. 2002).
The large, ∼86-kb-long palindrome generated by REPB1 and REPB2 is a genomic architectural feature that potentially could trigger a recombination event. IRs or palindromes are known to generate hairpins or cruciform structures, facilitated by intrastrand pairing of complementary single-strand DNA sequences assembled in the lagging strand during DNA replication (Mizuuchi et al. 1982; Nasar et al. 2000). A double-strand break (DSB) introduced in these hairpins can result in deletions or in either inter- or intrachromosomal recombination events, mediated by the homologous recombination machinery (Akgun et al. 1997). Critical factors implicated in the rate of such rearrangements are the extent of sequence identity between the inverted repeats, their size, and the length of the spacer (sequences present between the palindromic sequences). The rate of genomic rearrangements rises as the length of the repeat increases and spacer sequence length decreases (Lobachev et al. 1998). The palindrome identified in the i(17q) breakpoint cluster region is characterized by two inverted repeats, ∼48 kb and 43 kb in length, separated by a 400-bp spacer. This is within the range associated with recombinogenic events in somatic cells.
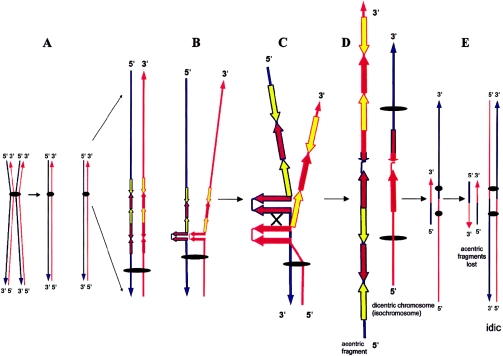
If broken, DNA ends generated in the hairpin/cruciform structures of the i(17q) breakpoint can trigger the DSB-repair machinery. A subsequent NAHR event between the repeats with opposite orientations located in the two sister chromatids (i.e., sister chromatid exchange) could result in the formation of an isodicentric chromosome 17 and of an acentric fragment (fig. 3).
Figure 3.
Molecular mechanism for i(17q) formation. A, Division of a metaphase chromosome. The double-strand DNA of each chromatid is depicted in red and blue, as is its 3′ and 5′ orientation. B, The formation of an REPB1 and REPB2 palindrome with subsequent breakage (C) and reunion between palindromes on sister chromatids (D) results in the origin of both dicentric and acentric structures. E, Dicentric and acentric structures after replication. The acentric material is lost in dividing cells because of the lack of a centromere. The (iso-)dicentric structure (isochromosome) is retained and will not become disrupted during anaphase because one of the two centromeres is inactivated as a result of their close proximity. Yellow and brown arrows refer to REPA and REPB copies, respectively, and the “×” depicts interchromatid mispairing of direct repeats. According to this model and our present and previously reported data (Fioretos et al. 1999), i(17q) should be formally designated “idic(17)(p11.2).”
Similar to i(17q), breakpoint analyses of constitutional isochromosomes of the long arm of the X chromosome, i(Xq), have revealed that the vast majority of i(Xq) rearranged chromosomes are, in fact, dicentrics with breakpoints mapping within several duplicated loci in Xp11.21. A U-type breakage/reunion mechanism between sister chromatids or homologous X chromosomes has been proposed for the origin of the constitutional i(Xq) rearrangement (Wolff et al. 1996). The same mechanism between sister chromatids or homologous chromosomes has been suggested for the origin of the most common constitutional isodicentric chromosomes—that is, inv dup(15) and inv dup(22). However, recent characterization of the LCRs in the pericentromeric region of chromosomes 15, 17, 22, and X indicates that these isodicentric abnormalities more likely result from NAHR between inverted LCRs (Stankiewicz and Lupski 2002_a_).
At the molecular level, i(17q) formation leads to a loss of genetic material located distally to the breakpoints described above, whereas material located proximally becomes duplicated. It is presently unknown, however, what the consequences of such relatively large genomic alterations are for the genes in the region. We favor the idea that dosage imbalances of several genes encoded by these regions are pathogenetically important. Nevertheless, we cannot rule out the possibility that gene(s) located in the breakpoint region could become deleted or altered in their expression or structure as a result of either chromosomal disruption or a position effect.
LCRs are now well-established mediators of erroneous meiotic recombination events generating constitutional chromosomal rearrangements associated with congenital human genomic disorders (Lupski 1998, 2003; Stankiewicz and Lupski 2002_a_). Recently, similar structures in the form of two large “duplicons” were identified in the proximity of the breakpoints in the t(9;22)(q34;q11) associated with chronic myeloid leukemia (Saglio et al. 2002). The present study strongly implicates a role for LCRs in i(17q) formation and provides a mechanism for generating this chromosome rearrangement, further strengthening the assumption that genome architecture may be important in the generation of other somatic chromosomal rearrangements associated with human neoplasia.
Acknowledgments
We thank Kurt Labutti and Holly Cordum for their assistance in independently assembling the sequence of RP11-160E2. This work was supported by the Swedish Cancer Society, the Swedish Children’s Cancer Foundation, the Medical Faculty of Lund University, and United States National Institute of Child Health and Human Development grant PO1 HD39420.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Blast 2 Sequences, http://www.ncbi.nlm.nih.gov/blast/bl2seq/bl2.html (for the Blast 2 browser)
- GenBank, http://www.ncbi.nih.gov/Genbank/ (for CTC-457L16 [accession number AC003957], RP11-160E2 [accession number AC007952], RP11-744A16 [accession number AC106017], RP11-970O14 [accession number AC036110], CTB-187M2 [accession number AC004448], RP11-1113L8 [accession number AC025627], CTD-2354J3 [accession number AC015935], RP11-311F12 [accession number AC005722], RP11-78O7 [accession number AC015726], RP11-209D14 [accession number AC005730], CTD-2010G8 [accession number AC007963], RP1-149D14 [accession number AJ009617], CTD-2525F10 [accession number AC109313], RP11-135L13 [accession number AC124066], L29232 [accession number AC139083], L29246 [accession number AC139093], L29227 [accession number AC139315], L29233 [accession number AC140106], L29228 [accession number AC144507], L29248 [accession number AC139138], L29225 [accession number AC144506], L29280 [accession number AC139077], mouse RP23-278F12 [accession number AC084044], rat CH230-200D5 [accession number AC105503], and rat CH230-255K6 [accession number AC134746])
- Mfold Server, http://www.bioinfo.rpi.edu/applications/mfold/old/rna/form1.cgi
- Mitelman Database of Chromosome Aberrations in Cancer, http://cgap.nci.nih.gov/Chromosomes/Mitelman
- RepeatMasker Web Server, http://repeatmasker.genome.washington.edu/cgi-bin/RepeatMasker
- University of California Santa Cruz Genome Browser, http://genome.ucsc.edu/cgi-bin/hgGateway
- Webcutter 2.0, http://www.firstmarket.com/cutter/cut2.html
References
- Akgun E, Zahn J, Baumes S, Brown G, Liang F, Romanienko PJ, Lewis S, Jasin M (1997) Palindrome resolution and recombination in the mammalian germ line. Mol Cell Biol 17:5559–5570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, Schwartz S, Adams MD, Myers EW, Li PW, Eichler EE (2002) Recent segmental duplications in the human genome. Science 297:1003–1007 10.1126/science.1072047 [DOI] [PubMed] [Google Scholar]
- Bailey JA, Liu G, Eichler EE (2003) An Alu transposition model for the origin and expansion of human segmental duplications. Am J Hum Genet 73:823–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbouti A, Johansson B, Höglund M, Mauritzson N, Strömbeck B, Nilsson PG, Tanke HJ, Hagemeijer A, Mitelman F, Fioretos T (2002) Multicolor COBRA-FISH analysis of chronic myeloid leukemia reveals novel cryptic balanced translocations during disease progression. Genes Chromosomes Cancer 35:127–137 10.1002/gcc.10099 [DOI] [PubMed] [Google Scholar]
- Bi W, Park S-S, Shaw CJ, Withers MA, Patel PI, Lupski JR (2003) Reciprocal crossovers and a positional preference for strand exchange in recombination events resulting in deletion or duplication of chromosome 17p11.2. Am J Hum Genet 73:1302–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi W, Yan J, Stankiewicz P, Park S-S, Walz K, Boerkoel CF, Potocki L, Shaffer LG, Devriendt K, Nowaczyk MJ, Inoue K, Lupski JR (2002) Genes in a refined Smith-Magenis syndrome critical deletion interval on chromosome 17p11.2 and the syntenic region of the mouse. Genome Res 12:713–728 10.1101/gr.73702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biegel JA, Rorke LB, Janss AJ, Sutton LN, Parmiter AH (1995) Isochromosome 17q demonstrated by interphase fluorescence in situ hybridization in primitive neuroectodermal tumors of the central nervous system. Genes Chromosomes Cancer 14:85–96 [DOI] [PubMed] [Google Scholar]
- Chen KS, Manian P, Koeuth T, Potocki L, Zhao Q, Chinault AC, Lee CC, Lupski JR (1997) Homologous recombination of a flanking repeat gene cluster is a mechanism for a common contiguous gene deletion syndrome. Nat Genet 17:154–163 [DOI] [PubMed] [Google Scholar]
- Dragon F, Gallagher JE, Compagnone-Post PA, Mitchell BM, Porwancher KA, Wehner KA, Wormsley S, Settlage RE, Shabanowitz J, Osheim Y, Beyer AL, Hunt DF, Baserga SJ (2002) A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417:967–970 10.1038/nature00769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann L, Spiteri E, Koren K, Pulijaal V, Bialer MG, Shanske A, Goldberg R, Morrow BE (2001) AT-rich palindromes mediate the constitutional t(11;22) translocation. Am J Hum Genet 68:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fioretos T, Strömbeck B, Sandberg T, Johansson B, Billstrom R, Borg A, Nilsson PG, Van Den Berghe H, Hagemeijer A, Mitelman F, Hoglund M (1999) Isochromosome 17q in blast crisis of chronic myeloid leukemia and in other hematologic malignancies is the result of clustered breakpoints in 17p11 and is not associated with coding TP53 mutations. Blood 94:225–232 [PubMed] [Google Scholar]
- Gao L, Frey MR, Matera AG (1997) Human genes encoding U3 snRNA associate with coiled bodies in interphase cells and are clustered on chromosome 17p11.2 in a complex inverted repeat structure. Nucleic Acids Res 25:4740–4747 10.1093/nar/25.23.4740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiskanen M, Karhu R, Hellsten E, Peltonen L, Kallioniemi PP, Palotie A (1994) High resolution mapping using fluorescence in situ hybridization to extended DNA fibers prepared from agarose-embedded cells. Biotechniques 17:928–934 [PubMed] [Google Scholar]
- Honjo T, Kinoshita K, Muramatsu M (2002) Molecular mechanism of class switch recombination: linkage with somatic hypermutation. Annu Rev Immunol 20:165–196 10.1146/annurev.immunol.20.090501.112049 [DOI] [PubMed] [Google Scholar]
- Inoue K, Osaka H, Thurston VC, Clarke JTR, Yoneyama A, Rosenbarker L, Bird TD, Hodes ME, Shaffer LG, Lupski JR (2002) Genomic rearrangements resulting in PLP1 deletion occur by nonhomologous end joining and cause different dysmyelinating phenotypes in males and females. Am J Hum Genet 71:838–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson B, Fioretos T, Mitelman F (2002) Cytogenetic and molecular genetic evolution of chronic myeloid leukemia. Acta Haematol 107:76–94 10.1159/000046636 [DOI] [PubMed] [Google Scholar]
- Kehrer-Sawatzki H, Haussler J, Krone W, Bode H, Jenne DE, Mehnert KU, Tummers U, Assum G (1997) The second case of a t(17;22) in a family with neurofibromatosis type 1: sequence analysis of the breakpoint regions. Hum Genet 99:237–247 10.1007/s004390050346 [DOI] [PubMed] [Google Scholar]
- Kurahashi H, Emanuel BS (2001) Long AT-rich palindromes and the constitutional t(11;22) breakpoint. Hum Mol Genet 10:2605–2617 10.1093/hmg/10.23.2605 [DOI] [PubMed] [Google Scholar]
- Kurahashi H, Shaikh T, Takata M, Toda T, Emanuel BS (2003) The constitutional t(17;22): another translocation mediated by palindromic AT-rich repeats. Am J Hum Genet 72:733–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda-Kawaguchi T, Skaletsky H, Brown LG, Minx PJ, Cordum HS, Waterston RH, Wilson RK, Silber S, Oates R, Rozen S, Page DC (2001) The _AZF_c region of the Y chromosome features massive palindromes and uniform recurrent deletions in infertile men. Nat Genet 29:279–286 10.1038/ng757 [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, et al (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- Lobachev KS, Shor BM, Tran HT, Taylor W, Keen JD, Resnick MA, Gordenin DA (1998) Factors affecting inverted repeat stimulation of recombination and deletion in Saccharomyces cerevisiae. Genetics 148:1507–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR (1998) Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet 14:417–422 10.1016/S0168-9525(98)01555-8 [DOI] [PubMed] [Google Scholar]
- ——— (2003) Genomic disorders: recombination-based disease resulting from genome architecture. Am J Hum Genet 72:246–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure RF, Dewald GW, Hoyer JD, Hanson CA (1999) Isolated isochromosome 17q: a distinct type of mixed myeloproliferative disorder/myelodysplastic syndrome with an aggressive clinical course. Br J Haematol 106:445–454 10.1046/j.1365-2141.1999.01537.x [DOI] [PubMed] [Google Scholar]
- Mertens F, Johansson B, Mitelman F (1994) Isochromosomes in neoplasia. Genes Chromosomes Cancer 10:221–230 [DOI] [PubMed] [Google Scholar]
- Mizuuchi K, Mizuuchi M, Gellert M (1982) Cruciform structures in palindromic DNA are favored by DNA supercoiling. J Mol Biol 156:229–243 [DOI] [PubMed] [Google Scholar]
- Nakai H, Misawa S (1995) Chromosome 17 abnormalities and inactivation of the p53 gene in chronic myeloid leukemia and their prognostic significance. Leuk Lymphoma 19:213–221 [DOI] [PubMed] [Google Scholar]
- Nasar F, Jankowski C, Nag DK (2000) Long palindromic sequences induce double-strand breaks during meiosis in yeast. Mol Cell Biol 20:3449–3458 10.1128/MCB.20.10.3449-3458.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SS, Stankiewicz P, Bi W, Shaw C, Lehoczky J, Dewar K, Birren B, Lupski JR (2002) Structure and evolution of the Smith-Magenis syndrome repeat gene clusters, SMS-REPs. Genome Res 12:729–738 10.1101/gr.82802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potocki L, Chen KS, Park SS, Osterholm DE, Withers MA, Kimonis V, Summers AM, Meschino WS, Anyane-Yeboa K, Kashork CD, Shaffer LG, Lupski JR (2000) Molecular mechanism for duplication 17p11.2: the homologous recombination reciprocal of the Smith-Magenis microdeletion. Nat Genet 24:84–87 10.1038/71743 [DOI] [PubMed] [Google Scholar]
- Saglio G, Storlazzi CT, Giugliano E, Surace C, Anelli L, Rege-Cambrin G, Zagaria A, Jimenez Velasco A, Heiniger A, Scaravaglio P, Torres Gomez A, Roman Gomez J, Archidiacono N, Banfi S, Rocchi M (2002) A 76-kb duplicon maps close to the BCR gene on chromosome 22 and the ABL gene on chromosome 9: possible involvement in the genesis of the Philadelphia chromosome translocation. Proc Natl Acad Sci USA 99:9882–9887 10.1073/pnas.152171299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheurlen WG, Schwabe GC, Seranski P, Joos S, Harbott J, Metzke S, Dohner H, Poustka A, Wilgenbus K, Haas OA (1999) Mapping of the breakpoints on the short arm of chromosome 17 in neoplasms with an i(17q). Genes Chromosomes Cancer 25:230–240 [DOI] [PubMed] [Google Scholar]
- Shaw CJ, Bi W, Lupski JR (2002) Genetic proof of unequal meiotic crossovers in reciprocal deletion and duplication of 17p11.2. Am J Hum Genet 71:1072–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR (2002_a_) Genome architecture, rearrangements and genomic disorders. Trends Genet 18:74–82 10.1016/S0168-9525(02)02592-1 [DOI] [PubMed] [Google Scholar]
- ——— (2002_b_) Molecular-evolutionary mechanisms for genomic disorders. Curr Opin Genet Dev 12:312–319 10.1016/S0959-437X(02)00304-0 [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Shaw CJ, Dapper JD, Wakui K, Shaffer LG, Withers M, Elizondo L, Park SS, Lupski JR (2003) Genome architecture catalyzes nonrecurrent chromosomal rearrangements. Am J Hum Genet 72:1101–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, et al (2001) The sequence of the human genome. Science 291:1304–1351 10.1126/science.1058040 [DOI] [PubMed] [Google Scholar]
- Wolff DJ, Miller AP, Van Dyke DL, Schwartz S, Willard HF (1996) Molecular definition of breakpoints associated with human Xq isochromosomes: implications for mechanisms of formation. Am J Hum Genet 58:154–160 [PMC free article] [PubMed] [Google Scholar]