Phylogeography of Y-Chromosome Haplogroup I Reveals Distinct Domains of Prehistoric Gene Flow in Europe (original) (raw)
Abstract
To investigate which aspects of contemporary human Y-chromosome variation in Europe are characteristic of primary colonization, late-glacial expansions from refuge areas, Neolithic dispersals, or more recent events of gene flow, we have analyzed, in detail, haplogroup I (Hg I), the only major clade of the Y phylogeny that is widespread over Europe but virtually absent elsewhere. The analysis of 1,104 Hg I Y chromosomes, which were identified in the survey of 7,574 males from 60 population samples, revealed several subclades with distinct geographic distributions. Subclade I1a accounts for most of Hg I in Scandinavia, with a rapidly decreasing frequency toward both the East European Plain and the Atlantic fringe, but microsatellite diversity reveals that France could be the source region of the early spread of both I1a and the less common I1c. Also, I1b*, which extends from the eastern Adriatic to eastern Europe and declines noticeably toward the southern Balkans and abruptly toward the periphery of northern Italy, probably diffused after the Last Glacial Maximum from a homeland in eastern Europe or the Balkans. In contrast, I1b2 most likely arose in southern France/Iberia. Similarly to the other subclades, it underwent a postglacial expansion and marked the human colonization of Sardinia ∼9,000 years ago.
Haplogroup (Hg) I-M170 is a component of the European Y-chromosome gene pool, accounting, on average, for 18% of the total paternal lineages. Its virtual absence elsewhere, including the Near East, suggests that it arose in Europe, likely before the Last Glacial Maximum (LGM) (Semino et al. 2000).
Previous studies revealed that Hg I reached a frequency of ∼40%–50% in two distinct regions—in Nordic populations of Scandinavia and, in southern Europe, around the Dinaric Alps—each showing different background STR modal haplotypes (Semino et al. 2000; Passarino et al. 2002; Barać et al. 2003). In addition, subclade I-M26 (Underhill et al. 2000) reaches a very high frequency (∼40%) (Semino et al. 2000; Passarino et al. 2001; Francalacci et al. 2003) in Sardinia, particularly in the “archaic area” (Cappello et al. 1996; Zei et al. 2003), and is associated with the peculiar YCAIIb-11 allele (Ciminelli et al. 1995; Caglia et al. 1997; Quintana-Murci et al. 1999; Malaspina et al. 2000; Scozzari et al. 2001). Overall, these observations suggest that Hg I could have played a central role in the process of human recolonization of Europe from isolated refuge areas after the LGM and suggest the likelihood that a comprehensive phylogeographic study should be able to localize the in situ origin and spread of principal male founders.
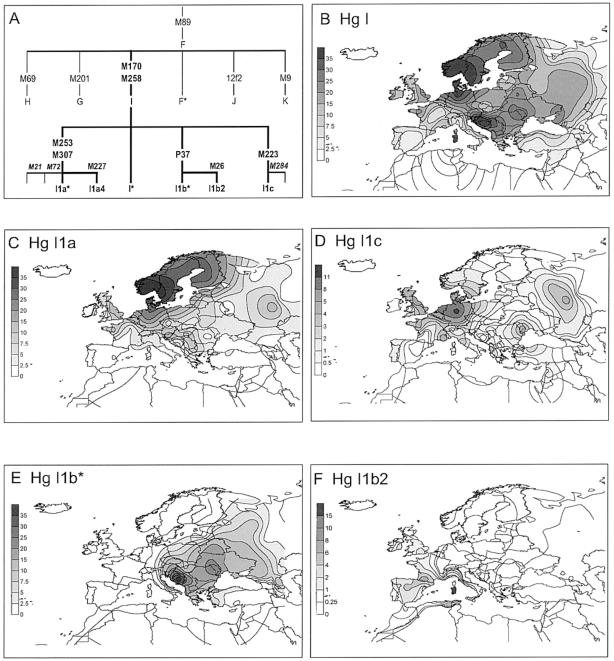
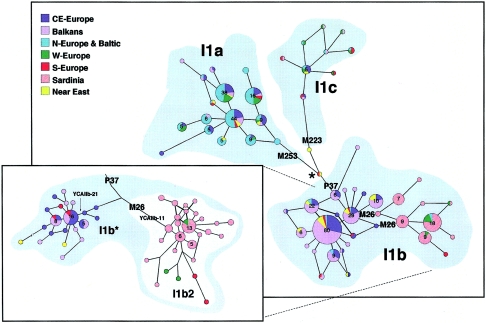
In the present study, the M170 A→C transversion, which defines Hg I, was assessed in a total of 7,574 subjects, including 6,095 Y chromosomes from 48 European populations and 1,479 individuals from 12 populations of surrounding regions (the Near East, Macaronesia, Central Asia, and the Caucasus). The results are reported in table 1, together with 407 additional members of Hg I out of 3,859 Y chromosomes extracted from the literature. Of the 1,104 Y chromosomes from the present study (1,060 from European subjects and 44 from the adjacent regions) that showed the derived M170 C-allele, 236, representative of the entire collection, were first examined for all the Hg I mutations known to date—namely, M21, M26, P37, M72, M223, M227, M253, M258, M284, and M307, whose phylogenetic relationships are illustrated in figure 1_A_. Genotyping was performed in a hierarchical way, and methods are provided in the legend to figure 1. The M258 and M307 mutations were observed in all of the Hg I and I1a Y chromosomes, respectively, whereas the M21, M72, and M284 mutations were not found. Thus, all of these were subsequently omitted in the remainder of the survey. To evaluate the differentiation of the I subclades (fig. 2), 533 Hg I Y chromosomes from 34 population samples were examined for the microsatellites DYS19, DYS388, DYS390, DYS391, DYS392, and DYS393 (Roewer et al. 1992, 1996; Thomas et al. 1998, 1999); 183 of the same chromosomes (from 20 populations) were also genotyped for the DYS389 (Roewer et al. 1996), YCAIIa, and YCAIIb (Mathias et al. 1994) loci; and 128 were also genotyped for the 49a,f system (for a review, see Poloni et al. [1997]). The results obtained were used to construct the network illustrated in figure 2. The STR and 49a,f data are available upon request.
Table 1.
Frequencies of Haplogroup I and its Subhaplogroups
| Hg I | Frequency of I Subhaplogroupa(%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Region and Population | SampleSize | N | % | I* M170 | I1a* M253 | I1a4 M227 | I1b*****P37 | I1b2 M26 | I1c M223 | hb |
| Western Europe: | ||||||||||
| Portuguesec | 303 | 16 | 5.3 | 1.3 | 1.3 | .7 | .3 | 1.6 | .808 | |
| Andalusianc,d | 103 | 4 | 3.9 | 2.9 | 1.0 | |||||
| Catalanc,d | 32 | 1 | 3.1 | 3.1 | ||||||
| Basque (Spanish, French)c,d | 100 | 6 | 6.0 | 6.0 | .000 | |||||
| Bearnaisc | 26 | 2 | 7.7 | 7.7 | ||||||
| French (Southern France)c | 38 | 6 | 15.8 | 5.3 | 5.3 | 5.3 | .800 | |||
| French (Low Normandy)c | 42 | 10 | 23.8 | 4.8 | 11.9 | 2.4 | 4.8 | .733 | ||
| French (Lyon, Poitier)c | 99 | 4 | 4.0 | 2.0 | 1.0 | 1.0 | ||||
| Swissc | 144 | 11 | 7.6 | .7 | 5.6 | 1.4 | .473 | |||
| Irish (Rush)e | 76 | 8 | 10.5 | NT | NT | NT | NT | 2.6 | NT | |
| Welshe | 196 | 16 | 8.1 | NT | NT | NT | NT | .5 | NT | |
| Englishe | 945 | 174 | 18.4 | NT | NT | NT | NT | .7 | NT | |
| Scottishe | 178 | 20 | 11.2 | NT | NT | NT | NT | NT | ||
| Scottish (Scottish Isles)e | 272 | 45 | 16.5 | NT | NT | NT | NT | .4 | NT | |
| Germanc,d | 16 | 6 | 37.5 | 25.0 | 12.5 | .733 | ||||
| Dutchc,d | 30 | 8 | 26.7 | 16.7 | 10.0 | .535 | ||||
| Danishf | 194 | 75 | 38.7 | NT | NT | NT | NT | NT | NT | |
| Macaronesia: | ||||||||||
| Madeiran (Portugal)c | 132 | 6 | 4.5 | 1.5 | .7 | .7 | 1.5 | .866 | ||
| Azorean (Portugal)c | 121 | 7 | 5.8 | 1.6 | 2.5 | 1.6 | .762 | |||
| Cape Verde Islandersc | 201 | 1 | .5 | .5 | ||||||
| Southern Europe: | ||||||||||
| Italian (northern Italy)c | 194 | 9 | 4.6 | 2.6 | 1.0 | 1.0 | .688 | |||
| Italian (central Italy)c | 196 | 14 | 7.1 | 1.0 | 2.0 | 1.0 | 3.0 | .747 | ||
| Italian (Calabria)c,d | 148 | 8 | 5.4 | 2.0 | .7 | .7 | .7 | 1.4 | .857 | |
| Italian (Albanese origin)c | 78 | 5 | 6.4 | 1.3 | 1.3 | 3.8 | ||||
| Italian (Apulia)c | 78 | 2 | 2.6 | 1.3 | 1.3 | |||||
| Italian (Sicily)g | 51 | 4 | 8.8 | NT | NT | NT | NT | NT | ||
| Italian (Sardinia)c,d | 142 | 60 | 42.3 | 40.9 | 1.4 | .098 | ||||
| Corsicang | 34 | 1 | 3.9 | NT | NT | NT | NT | NT | ||
| Balkans: | ||||||||||
| Slovenianc | 55 | 21 | 38.2 | 3.6 | 10.9 | 1.8 | 20.0 | 1.8 | .663 | |
| Croat (mainland)c,d,h | 189 | 72 | 38.1 | .5 | 5.3 | .5 | 31.2 | .5 | .312 | |
| Bosnianc | 100 | 42 | 42.0 | 2.0 | 40.0 | .092 | ||||
| Albanianc | 106 | 25 | 23.6 | 2.8 | 17.0 | 3.8 | .581 | |||
| Macedonian (northern Greece)c | 50 | 15 | 30.0 | 2.0 | 8.0 | 2.0 | 18.0 | .600 | ||
| Romanianc | 361 | 80 | 22.2 | .8 | 1.7 | 17.7 | 1.9 | .356 | ||
| Moldavianc | 60 | 17 | 28.3 | 3.3 | 21.7 | 3.3 | .411 | |||
| Gagauz (Moldova)c | 79 | 25 | 31.6 | 2.5 | 24.1 | 5.1 | .406 | |||
| Greekc,d | 261 | 36 | 13.8 | 1.5 | 2.3 | 8.4 | 1.5 | .590 | ||
| Northern Europe: | ||||||||||
| Swedish (South Sweden)c | 168 | 68 | 40.5 | .6 | 35.7 | .6 | 3.6 | .216 | ||
| Swedish (North Sweden)c | 57 | 15 | 26.3 | 26.3 | .000 | |||||
| Norwegianc,i | 72 | 29 | 40.3 | 38.9 | 1.4 | .058 | ||||
| Saamic,j | 35 | 11 | 31.4 | 2.9 | 28.6 | .182 | ||||
| Baltic: | ||||||||||
| Estonianc,j | 210 | 39 | 18.6 | 14.8 | .5 | 2.9 | .5 | .310 | ||
| Latvianc,j | 86 | 6 | 7.0 | 4.7 | 1.2 | 1.2 | .600 | |||
| Central-eastern Europe: | ||||||||||
| Polishc,d | 191 | 34 | 17.8 | 5.8 | 1.0 | 9.9 | 1.0 | .593 | ||
| Czech and Slovakc,d | 198 | 27 | 13.6 | .5 | 4.5 | .5 | 7.1 | 1.0 | .635 | |
| Hungarianc,d | 162 | 37 | 22.8 | .6 | 9.9 | 11.1 | 1.2 | .588 | ||
| Byelorussianc | 147 | 28 | 19.0 | .7 | 2.7 | 15.0 | .7 | .373 | ||
| Ukrainianc,d | 585 | 128 | 21.9 | .2 | 4.8 | .3 | 16.1 | .5 | .415 | |
| Russian (northern, Pinega)c | 127 | 6 | 4.7 | .8 | 3.9 | .333 | ||||
| Russian (Kostroma region)c | 53 | 10 | 18.9 | 6.0 | 9.4 | 3.7 | .688 | |||
| Russian (Smolensk region)c | 120 | 13 | 10.8 | 1.7 | 9.1 | .283 | ||||
| Russian (Belgorod region)c | 144 | 24 | 16.7 | 3.5 | 12.5 | .7 | .634 | |||
| Russian (Cossacks)c | 97 | 22 | 22.7 | 1.0 | 4.1 | 15.5 | 2.0 | .515 | ||
| Russian (Adygea)c | 78 | 19 | 24.4 | 1.3 | 5.1 | 16.7 | 1.3 | .508 | ||
| Russian (Bashkortostan)c | 50 | 3 | 6.0 | 4.0 | 2.0 | |||||
| Udmurtc,d | 132 | 3 | 2.3 | NT | NT | NT | NT | NT | NT | |
| Mordvinc | 83 | 16 | 19.3 | 12.0 | 2.4 | 4.8 | .566 | |||
| Komic | 110 | 5 | 4.5 | 3.6 | .9 | |||||
| Chuvashesc | 80 | 9 | 11.3 | 7.5 | 1.3 | 2.5 | .555 | |||
| Tatarc | 123 | 6 | 4.9 | 1.6 | .8 | 2.4 | .733 | |||
| Near East: | ||||||||||
| Turkishc,d,k | 741 | 38 | 5.1 | 1.1 | .9 | 2.3 | .7 | .723 | ||
| Lebanesec,d | 66 | 3 | 4.5 | 1.5 | 1.5 | 1.5 | ||||
| Jewishc,d | 150 | 2 | 1.3 | .7 | .7 | |||||
| Iraqic,l | 176 | 1 | .6 | .6 | ||||||
| Iranianc | 83 | 0 | ||||||||
| Caucasus, Central Asia: | ||||||||||
| Nogaysc | 61 | 3 | 4.9 | 4.9 | ||||||
| Adygeisc | 138 | 6 | 4.3 | 1.4 | 2.9 | .533 | ||||
| Karachaisc | 70 | 5 | 7.1 | 7.1 | ||||||
| Northern Caucasianm | 114 | 7 | 6.1 | NT | NT | NT | NT | NT | NT | |
| Southern Caucasianm | 249 | 10 | 4.0 | NT | NT | NT | NT | NT | NT | |
| Georgianc,d | 63 | 0 | ||||||||
| Central Asiann | 984 | 15 | 1.5 | NT | NT | NT | NT | NT | NT |
Figure 1.
A, Phylogram of Hg I and its subclades within the context of the superhaplogroup F. Mutation labeling follows the Y Chromosome Consortium nomenclature (Y Chromosome Consortium 2002; Jobling and Tyler-Smith 2003). Markers M21 and M72 (Underhill et al. 2001) and the three new markers—M258 (a T→C transition at position 123), M284 (ACAAdel at position 105), and M307 (a G→A transversion at position 282)—were examined in a subset of 236 Y chromosomes, representative of the entire collection, by using the DHPLC method. The primers used for the new markers were as follows: F, 5′-tatatagcatatgttaaatgtttaggt-3′ and R, 5′-gacttttgaataatttgcatctttc-3′ for M258; F, 5′-ggcagttttcatttaagcaga-3′and R, 5′-agcgaaactttcagcacttc-3′ for M284; and F, 5′-ttattggcatttcaggaagtg-3′ and R, 5′-gggtgaggcaggaaaatagc-3′ for M307. When the DHPLC method was not used, M170 was detected as described by Ye et al. (2002); M253 was detected by using published primers (Cinnioğlu et al. 2004) and restriction analysis with _Hin_cII; P37 was assayed by _Taa_I digestion using the primers given by YCC (2002); and M223, M26, and the novel M227 (a C→G transversion at position 157) were studied by sequencing using published primers (Underhill et al. 2001) and the primers F, 5′-gagtgccaagctgaggatg-3′ and R, 5′-tccttgcagccgctgaggag-3′, respectively. A minority (_n_=67) of widely geographically distributed Hg I Y chromosomes (table 1) not tested for M258 and not harboring derived alleles at the sites M253, P37, and M223 were aggregated into paragroup I*. B–F, Frequency distribution of haplogroup I (B) and its subclades: I1a (C), I1c (D), I1b* (E), and I1b2 (F). Maps were obtained by applying the frequencies from table 1 in Surfer (version 7) software (Golden Software).
Figure 2.
Network of haplogroup I. The network was obtained by using the biallelic markers and six STR loci (DYS19, DYS388, DYS390, DYS391, DYS392, and DYS393) in 533 Hg I chromosomes from 34 populations (Andalusian, Basques [French and Spanish], Bearnais, French [southern France, Low Normandy, Lyon, and Poitier], Swiss, Dutch, Italian [northern Italy, central Italy, Calabria1, Calabria2 of Albanese origin, Apulia, and Sardinia], Croat, Bosnian, Albanian, Macedonian, Moldavian, Gagauz, Greek, Swedish, Norwegian, Saami, Estonian, Polish, Czech, Slovak, Hungarian, Ukrainian, Turkish, and Jewish). The phylogenetic relationships between the 58 microsatellite haplotypes (out of the 156 observed) with frequency >1 were determined by using the program NETWORK 4.0b (Fluxus Engineering Web site). Networks were calculated by the median-joining method (ɛ=0) (Bandelt et al. 1995), weighting the STR loci according to the average of their relative variability in the haplogroup I subclades and after having processed the data with the reduced-median method. Circles represent microsatellite haplotypes. Unless otherwise indicated by a number on the pie, the area of the circles and the area of the sectors are proportional to the haplotype frequency in the haplogroup (the smallest circle corresponds to two individuals) and in the geographic area indicated by the color. The inset reveals, in more detail, the relationship between I1b2 and I1b* in a subsample of 103 Y chromosomes from 20 populations. The network was determined as described above but, in this case, in addition to the above mentioned six STR loci, the YCAIIa, YCAIIb, and DYS389 microsatellites and the 49a,f system were also considered. Here, the smallest circle of the network corresponds to a single Y chromosome. Very stable YCAII motifs characterize both I1b* (YCAIIa-21/YCAIIb-21) and I1b2 (YCAIIa-21/YCAIIb-11), supporting the hypothesis that single-banded patterns and very short alleles are due to deletion events rather than stepwise mutations.
Hg I accounts for more than one-third of paternal lineages in two distinct regions of Europe: among Scandinavian populations and in the northwestern Balkans (table 1; fig. 1_B_). Relatively high frequencies are also characteristic of some French regions, like Low Normandy and southern France. Interestingly, a lower frequency of haplogroup I distinguished the Baltic-speaking Latvians (7.0%; 90% CI 3.8%–13.2%) from their northern neighbors, the Finnic-speaking Estonians (18.6%; 90% CI 14.6%–23.4%). Similar cases of even more significant frequency change over a short geographic distance occur between the southern Slavic-speaking populations and their adjacent neighbors: namely, the Slovenians versus the northern Italians (38.2%; 99.9% CI 19.5%–60.2% vs. 4.6%; 99.9% CI 1.4%–11.7%), and Macedonians versus Greeks (30.0%; 95% CI 19.1%–43.8% vs. 13.8%; 95% CI 10.1%–18.5%).
As reported in table 1 and illustrated in figure 1_A_, three subhaplogroups, defined by the M253 (I1a), P37 (I1b), and M223 (I1c) markers, account for 95% of the Hg I Y chromosomes, and the remaining subhaplogroups belong to paragroup I*.
Subhaplogroup I1a is mostly found in northern Europe, with its highest frequencies in Scandinavian populations, where it accounts for 88%–100% of Norwegian, Swedish, and Saami M170 lineages. I1a has a decreasing gradient from its peak frequency in Scandinavia toward both the Urals and the Atlantic periphery (fig. 1_C_). Its I1a4 subclade has been found mainly as single observations scattered in eastern and southeastern Europe. Since the Scandinavian Peninsula was completely depopulated during the LGM, two main European refugia, the Iberian Peninsula/southern France and the Ukraine/Central Russian Plain (Dolukhanov 2000), can be considered as possible source regions of Scandinavian I1a chromosomes. Although Hg I occurs in the Ukraine at a higher incidence (22%) than in western Europe (11% in France), the virtual absence in Scandinavia of the most represented eastern European I1b* subclade, together with the higher I1a microsatellite diversity background, point to western Europe as the source of the Scandinavian I1a chromosomes, since the STR diversity of I1a decreases from western to eastern Europe, showing a significant negative correlation (_r_=-0.63; significance level 0.99) with longitude. In France, I1a is the leading subclade, representing 45% of the Hg I lineages, which, however, occur in a focal rather than clinal pattern. Hg I is more frequent in Low Normandy (I = 23.8%; I1a = 11.9%) and southern France (I = 15.8%; I1a = 5.3%), whereas it has a much lower occurrence in the Poitier and Lyon interior regions (I = 4.0%; I1a = 2.0%). Interestingly, subclade I1a shows a distribution similar to the second PC of the synthetic maps based on classical genetic markers (Cavalli-Sforza et al. 1994) and reveals a significantly positive correlation with mtDNA haplogroups V and U5b (r _V_=0.47; r _U_5_b_=0.60; significance level 0.999), which have been suggested to mark a postglacial population expansion from Iberia (Torroni et al. 1998, 2001; Tambets et al. 2004).
A different scenario has to be envisioned for subhaplogroup I1b*, which is the most frequent clade in eastern Europe and the Balkans. It reaches its highest incidences in Croatia (31%) and Bosnia (40%), encompassing almost 80%–90% of I (table 1). In western Europe, its subclade I1b2 (M26) (fig. 1_F_) (Semino et al. 2000; Bosch et al. 2001; Capelli et al. 2003; Maca-Meyer et al. 2003) is found at a very low frequency (<5%), except in Sardinia (41%), Castile (19%) (Flores et al., in press), Bearnais (8%), and in the Basques (6%). Although subclade I1b2 and the paragroup I1b* (the latter present at marginal frequencies) co-occur west of the Italian Apennines, only I1b* is present east of the Adriatic. I1b* Y chromosomes rapidly dissipate west of the Balkans—they are virtually absent among Italians, Germans, French, and Swiss (table 1; fig. 1_E_)—but extend eastward at notable frequencies among Slavic-speaking populations. This finding suggests that, similar to I1a, I1b* also may have expanded from a glacial refuge area. However, this area was most likely located in eastern Europe or the Balkans.
In central and eastern Europe, subhaplogroups I1a and I1b show overlapping frequency gradients, although with opposite post-LGM spreading. The divergent distributions of I1b2 and I1b* suggest that their separation occurred before the LGM and that the M26 mutation arose in a I1b Y chromosome from western Europe, most likely in a population in Iberia/southern France. The exceptionally high incidence of I1b2 in the archaic zone of Sardinia (Cappello et al. 1996; Zei et al. 2003) can be explained by the presence of I1b2 chromosomes among the first humans who colonized the island, ∼9,000 years ago, followed by isolation and genetic drift. The extremely low frequency of I1b2 in the Scandinavian Peninsula, where the “western European” I1a Y chromosomes account for the large majority of Hg I, suggests, in addition, that the ancestral western European population(s), characterized by the M26 mutation, probably played a minor role in the colonization of that region. A geographic and genetic subdivision within the broad western refuge area, together with differences in initial sample size, genetic drift, and expansions, could also explain the quite different distribution of Hg I subhaplogroups with respect to the west-east decreasing gradient displayed by R1b, the most frequent subhaplogroup in western Europe.
The high STR diversity of the I1b* lineages in Bosnia supports the view that the P37 SNP might have been present in the Balkan area before the LGM, as previously proposed by Semino et al. (2000). Diversity h values based on STR haplotypes for I1a are highest near Iberia but vary substantially in different populations (table 2). For I1b*, conversely, the highest h values are in the Balkan populations—among Bosnians (0.93) and Croats (0.85)—coinciding with the area of its frequency peak, but equally high values were also observed for Czechs and Slovaks (0.90). The lowest h values of I1b* were detected among Turks (0.76) and in our Moldavian sample (0.41).
Table 2.
Haplotype Diversity of I1a and I1b*
| Diversity ofa | ||
|---|---|---|
| Population | I1a | I1b* |
| French | .972 | … |
| Italian | .933 | … |
| Swiss | .750 | … |
| Norwegian | .809 | … |
| Saami | .806 | … |
| Swedish | .926 | … |
| Estonian | .895 | .867 |
| Hungarian | .884 | .746 |
| Ukrainian | .782 | .802 |
| Czech and Slovak | .857 | .904 |
| Polish | … | .818 |
| Croat | .830 | .845 |
| Bosnian | … | .929 |
| Gagauz (Moldova) | … | .900 |
| Moldavian | … | .410 |
| Turkish | .800 | .755 |
Subhaplogroup I1c (fig. 1_D_) covers a wide range in Europe, with the highest frequencies (∼5%–12%) in northwestern Europe and lower frequencies elsewhere. Its geographic and linguistic correlations across the continent were insignificant. However, the 49a,f system and the microsatellites YCAIIa-YCAIIb reveal that I1a and I1c harbor an identical compound haplotype (49a,f Ht12/YCAIIa-21/YCAIIb-19), which is different from those of I1b* (49a,f Ht10/YCAIIa-21/YCAIIb-21) and I1b2 (49a,f Ht12/YCAIIa-21/YCAIIb-11). These results indicate that subhaplogroups I1a and I1c may be part of a single monophyletic clade whose deep biallelic mutations are still undefined and that they probably share a common history of expansion. This scenario is also supported by the high positive correlation between the geographic distributions of I1a and I1c (_r_=0.75 when Fennoscandia is excluded; significance level 0.999).
Anatolia is at the easternmost fringe of the spread of haplogroup I, where it is found at higher frequencies in the regions that are geographically closer to Europe (Cinnioğlu et al. 2004). This observation, combined with a low haplotype diversity in Turkey plus exact haplotype matches with Europe, suggests that haplogroup I Y chromosomes in Turkey are due to migrations from Europe, as has been argued for a fraction of the Turkish mtDNAs (Richards et al. 2000).
A temporal interpretation of the phylogeography based on the results of the STR length variation in the individual subhaplogroups of I (Zhivotovsky et al. 2004) is reported in table 3. The age of STR variation for I* was estimated as 24,000±7,100 years, a value that is very close to the population divergence time (23,000±7,700 years). This finding supports the earlier suggestion that haplogroup I originated from a pool of European pre-LGM, middle Upper Paleolithic Y chromosomes (Semino et al. 2000). Our time estimates hint that its initial spread in Europe may be linked to the diffusion of the largely pan-European Gravettian technology ∼28,000–23,000 years ago (Djindjian 2000; Perles 2000). On the other hand, these values represent the lower limit of the age of M170 mutation. The precedent mutation (M89) (fig. 1_A_) defines the overarching superhaplogroup F, whose representatives span the entire non-African gene pool, likely predating the peopling of Europe (some 40,000–50,000 years ago). Potentially more informative are the estimates of subclade divergence times. Thus, it appears that I1a, I1b, and I1c all diverged from I* in the Late Upper Paleolithic/Mesolithic period (table 3), possibly during the recolonization of Europe after the LGM. However, the expansion phase of I1a and I1b, displaying contrasting phylogeographies, seems to have occurred later, around the early Holocene. Only the less frequent subclade I1c, spread thinly over much of Europe, from Mordvin in the Volga region to southern France (table 1; fig. 1_D_), shows a somewhat earlier age for its STR variation (table 3), suggesting that the corresponding mutation arose earlier.
Table 3.
Age Estimates and Divergence Times of Haplogroup I Subclades
| Haplogroup I Subclade | |||||
|---|---|---|---|---|---|
| Age Estimate orDivergence Time | I* | I1a | I1b* | I1b2 | I1c |
| Time since subclade divergencea | … | 15.9 ± 5.2b | 10.7 ± 4.8b | 9.3 ± 7.6c | 14.6 ± 3.8b |
| Age of STR variationd | 24.0 ± 7.1 | 8.8 ± 3.2 | 7.6 ± 2.7 | 8.0 ± 4.0 | 13.2 ± 2.7 |
| Time since population divergencee | 23.0 ± 7.7f | 6.8 ± 1.9g | 7.1 ± 2.5 | 7.9 ± 3.6h | 11.2 ± 2.3i |
In conclusion, although haplogroup I represents only a single piece in the puzzle of European genetic variation, its essential continental specificity and the clearly defined phylogeographic patterns of its subclades contribute uniquely to understanding the human settlement of Europe. Haplogroup I provides an exceptional record of European-specific paternal heritage, including pre-LGM differentiation followed by contraction, isolation, and subsequent post-LGM expansions and spread. Still, the wide CIs in the time estimates dictate caution in definitively linking the phylogeography of this haplogroup with known prehistoric and historic scenarios. Nonetheless, the I1a data in Scandinavia are consistent with a post-LGM recolonization of northwestern Europe from Franco-Cantabria, whereas the expansion of I1b* in the east Adriatic–North Pontic continuum probably reflects demographic processes that began in a refuge area located in that region.
Acknowledgments
We are grateful to all the donors for providing blood samples and to the people who contributed to their collection. We thank Ille Hilpus and Jaan Lind for technical assistance. We wish to thank the reviewers, whose comments and suggestions helped us to improve the quality of the manuscript. This research was supported by Estonian basic research grant 514 and European Commission Directorate General Research grant ICA1CT20070006 (to R.V.); Estonian basic research grant 5574 (to T.K.); Russian Foundation for Basic Research project numbers 01-04-48487a (to E.K.), 04-06-80260a (to E.B.), and 04-04-49664a (to O.B.); Ministry of Sciences and Technology of Russia (to E.K.); Progetto Finalizzato Consiglio Nazionale della Ricerche (CNR) “Beni Culturali” (to A.S.S.-B.); Progetto MIUR-CNR Genomica Funzionale-Legge 449/97 (to A.T.); Fondo d’Ateneo per la Ricerca dell’Università di Pavia (to A.S.S.-B. and A.T.); the Italian Ministry of the University: Progetti Ricerca Interesse Nazionale 2002 and 2003 (to A.T.); National Institutes of Health grant GM28428 (to the Stanford researchers); and Ministry of Science and Technology of the Republic of Croatia project number 0196005 (to P.R.).
Electronic-Database Information
The URL for data presented herein is as follows:
- Fluxus Engineering, http://www.fluxus-engineering.com/ (for NETWORK 4.0b)
References
- Al-Zahery N, Semino O, Benuzzi G, Magri C, Passarino G, Torroni A, Santachiara-Benerecetti AS (2003) Y-chromosome and mtDNA polymorphisms in Iraq, a crossroad of the early human dispersal and of post-Neolithic migrations. Mol Phylogenet Evol 28:458–472 10.1016/S1055-7903(03)00039-3 [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Sykes BC, Richards MB (1995) Mitochondrial portraits of human populations using median networks. Genetics 141:743–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barać L, Pericic M, Klaric IM, Rootsi S, Janicijevic B, Kivisild T, Parik J, Rudan I, Villems R, Rudan P (2003) Y chromosomal heritage of Croatian population and its island isolates. Eur J Hum Genet 11:535–542 10.1038/sj.ejhg.5200992 [DOI] [PubMed] [Google Scholar]
- Bosch E, Calafell F, Comas D, Oefner PJ, Underhill PA, Bertranpetit J (2001) High-resolution analysis of human Y chromosome variation shows a sharp discontinuity and limited gene flow between Northwestern Africa and Iberian Peninsula. Am J Hum Genet 68:1019–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caglia A, Novelletto A, Dobosz M, Malaspina P, Ciminelli BM, Pascali VL (1997) Y-chromosome STR loci in Sardinia and continental Italy reveal islander-specific haplotypes. Eur J Hum Genet 5:288–292 [PubMed] [Google Scholar]
- Capelli C, Redhead N, Abernethy JK, Gratrix F, Wilson JF, Moen T, Hervig T, Richards M, Stumpf MP, Underhill PA, Bradshaw P, Shaha A, Thomas MG, Bradman N, Goldstein DB (2003) A Y chromosome census of the British Isles. Curr Biol 13:979–984 10.1016/S0960-9822(03)00373-7 [DOI] [PubMed] [Google Scholar]
- Cappello N, Rendine S, Griffo R, Mameli GE, Succa V, Vona G, Piazza A (1996) Genetic analysis of Sardinia: I. data of 12 polymorphisms in 21 linguistic domains. Ann Hum Genet 60:125–141 [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Piazza A, Menozzi P (1994) The history and geography of human genes. Princeton University Press, Princeton, New Jersey [Google Scholar]
- Ciminelli BM, Pompei F, Malaspina P, Hammer M, Persichetti F, Pignatti PF, Palena A, Anagnou N, Guanti G, Jodice C, Terrenato L, Novelletto A (1995) Recurrent simple tandem repeat mutations during human Y-chromosome radiation in Caucasian subpopulations. J Mol Evol 41:966–973 [DOI] [PubMed] [Google Scholar]
- Cinnioğlu C, King R, Kivisild T, Kalfoglu E, Atasoy S, Cavalleri GL, Lillie AS, Roseman CC, Lin AA, Prince K, Oefner PJ, Shen P, Semino O, Cavalli-Sforza LL, Underhill PA (2004). Excavating Y-chromosome haplotype strata in Anatolia. Hum Genet 114:127–148 10.1007/s00439-003-1031-4 [DOI] [PubMed] [Google Scholar]
- Djindjian F (2000) The Mid Upper Palaeolithic (30,000 to 20,000 bp) in France. In: Roebroeks W, Mussi M, Svoboda J, Fennema K (eds) Hunters of the golden age: the Mid Upper Palaeolithic of Eurasia 30,000–20,000 BP: Analecta Praehistorica Leidensia. University of Leiden, Leiden, pp 313–324 [Google Scholar]
- Dolukhanov PM (2000) “Prehistoric revolutions” and languages in Europe. In: Künnap A (ed) The roots of peoples and languages of Northern Eurasia II and III. University of Tartu, Tartu, pp 71–78 [Google Scholar]
- Flores C, Maca-Meyer N, González AM, Oefner PJ, Shen P, Pérez JA, Rojas A, Larruga JM, Underhill PA. Genetic structure of Iberian Peninsula revealed by Y chromosome analysis. Eur J Hum Genet (in press) [DOI] [PubMed] [Google Scholar]
- Francalacci P, Morelli L, Underhill PA, Lillie AS, Passarino G, Useli A, Madeddu R, Paoli G, Tofanelli S, Calo CM, Ghiani ME, Varesi L, Memmi M, Vona G, Lin AA, Oefner P, Cavalli-Sforza LL (2003) Peopling of three Mediterranean islands (Corsica, Sardinia, and Sicily) inferred by Y-chromosome biallelic variability. Am J Phys Anthropol 121:270–279 10.1002/ajpa.10265 [DOI] [PubMed] [Google Scholar]
- Jobling MA, Tyler-Smith C (2003) The human Y chromosome: an evolutionary marker comes of age. Nat Rev Genet 4:598–612 10.1038/nrg1124 [DOI] [PubMed] [Google Scholar]
- Maca-Meyer N, Sanchez-Velasco P, Flores C, Larruga JM, Gonzales AM, Oterino A, Leyva-Cobian F (2003) Y chromosome and mithocondrial DNA characterization of Pasiegos, a human isolate from Cantabria (Spain). Ann Hum Genet 67:329–339 10.1046/j.1469-1809.2003.00045.x [DOI] [PubMed] [Google Scholar]
- Malaspina P, Cruciani F, Santolamazza P, Torroni A, Pangrazio A, Akar N, Bakalli V, Brdicka R, Jaruzelska J, Kozlov A, Malyarchuk B, Mehdi SQ, Michalodimitrakis E, Varesi L, Memmi MM, Vona G, Villems R, Parik J, Romano V, Stefan M, Stenico M, Terrenato L, Novelletto A, Scozzari R (2000) Patterns of male-specific inter-population divergence in Europe, West Asia and North Africa. Ann Hum Genet 64:395–412 10.1046/j.1469-1809.2000.6450395.x [DOI] [PubMed] [Google Scholar]
- Mathias N, Bayes M, Tyler-Smith C (1994) Highly informative compound haplotypes for the human Y chromosome. Hum Mol Genet 3:115–123 [DOI] [PubMed] [Google Scholar]
- Nasidze I, Sarkisian T, Kerimov A, Stoneking M (2003) Testing hypotheses of language replacement in the Caucasus: evidence from the Y-chromosome. Hum Genet 112:255–261 [DOI] [PubMed] [Google Scholar]
- Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York, pp 145–163 [Google Scholar]
- Passarino G, Cavalleri GL, Lin AA, Cavalli-Sforza LL, Borresen-Dale AL, Underhill PA (2002) Different genetic components in the Norwegian population revealed by the analysis of mtDNA and Y chromosome polymorphisms. Eur J Hum Genet 10:521–529 10.1038/sj.ejhg.5200834 [DOI] [PubMed] [Google Scholar]
- Passarino G, Underhill AP, Cavalli-Sforza L, Semino O, Pes MG, Carru C, Ferrucci L, Bonate M, Franceschi C, Deina L, Baggoi G, De Benedictis G (2001) Y chromosome binary markers to study the high prevalence of males in Sardinian centenarians and the genetic structure of the Sardinian population. Hum Hered 52:136–139 10.1159/000053368 [DOI] [PubMed] [Google Scholar]
- Perles C (2000) Greece, 30,000–20,000 bp. In: Roebroeks W, Mussi M, Svoboda J, Fennema K (eds) Hunters of the golden age. The Mid Upper Palaeolithic of Eurasia 30,000–20,000 BP: Analecta Praehistorica Leidensia. University of Leiden, Leiden, pp 375–398 [Google Scholar]
- Poloni ES, Semino O, Passarino G, Santachiara-Benerecetti AS, Dupanloup I, Langaney A, Excoffier L (1997) Human genetic affinities for Y-chromosome P49a,f/_Taq_I haplotypes show strong correspondence with linguistics. Am J Hum Genet 61:1015–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana-Murci L, Semino O, Poloni ES, Liu A, Van Gijn M, Passarino G, Brega A, Nasidze IS, Maccioni L, Cossu G, al-Zahery N, Kidd JR, Kidd KK, Santachiara-Benerecetti AS (1999) Y-chromosome specific YCAII, DYS19 and YAP polymorphisms in human populations: a comparative study. Ann Hum Genet 63:153–166 10.1046/j.1469-1809.1999.6320153.x [DOI] [PubMed] [Google Scholar]
- Richards M, Macaulay V, Hickey E, Vega E, Sykes B, Guida V, Rengo C, et al (2000) Tracing European founder lineages in the Near Eastern mtDNA pool. Am J Hum Genet 67:1251–1276 [PMC free article] [PubMed] [Google Scholar]
- Roewer L, Arnemann AJ, Spurr NK, Grzeschik KH, Epplen JT (1992) Simple repeated sequences on the Y chromosome are equally polymorphic as their autosomal counterparts. Hum Genet 89:389–394 [DOI] [PubMed] [Google Scholar]
- Roewer L, Kayser M, Dieltjes P, Nagy M, Bakker E, Krawczak M, de Knijff P (1996) Analysis of molecular variance (AMOVA) of Y-chromosome-specific microsatellites in two closely related human populations. Hum Mol Genet 5:1029–1033 10.1093/hmg/5.7.1029 [DOI] [PubMed] [Google Scholar]
- Sanchez JJ, Borsting C, Hallenberg C, Buchard A, Hernandez A, Morling N (2003) Multiplex PCR and minisequencing of SNPs: a model with 35 Y chromosome SNPs. Forensic Sci Int 137:74–84 10.1016/S0379-0738(03)00299-8 [DOI] [PubMed] [Google Scholar]
- Scozzari R, Cruciani F, Pangrazio A, Santolamazza P, Vona G, Moral P, Latini V, Varesi L, Memmi MM, Romano V, De Leo G, Gennarelli M, Jaruzelska J, Villems R, Parik J, Macaulay V, Torroni A (2001) Human Y-chromosome variation in the western Mediterranean area: implications for the peopling of the region. Hum Immunol 62:871–884 10.1016/S0198-8859(01)00286-5 [DOI] [PubMed] [Google Scholar]
- Semino O, Passarino G, Oefner PJ, Lin AA, Arbuzova S, Beckman LE, De Benedictis G, Francalacci P, Kouvatsi A, Limborska S, Marcikiae M, Mika A, Mika B, Primorac D, Santachiara-Benerecetti AS, Cavalli-Sforza LL, Underhill PA (2000) The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective. Science 290:1155–1159 10.1126/science.290.5494.1155 [DOI] [PubMed] [Google Scholar]
- Tambets K, Rootsi S, Kivisild T, Help H, Serk P, Loogväli EL, Tolk HV, et al (2004) The western and eastern roots of the Saami—the story of genetic “outliers” told by mtDNA and Y chromosomes. Am J Hum Genet 74:661–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MG, Bradman N, Flin HM (1999) High throughput analysis of 10 microsatellite and 11 diallelic polymorphisms on the human Y-chromosome. Hum Genet 105:577–581 10.1007/s004390051148 [DOI] [PubMed] [Google Scholar]
- Thomas MG, Skorecki K, Ben-Ami H, Parfitt T, Bradman N, Goldstein DB (1998) Origins of Old Testament priests. Nature 394:138–140 10.1038/28083 [DOI] [PubMed] [Google Scholar]
- Torroni A, Bandelt HJ, D’Urbano L, Lahermo P, Moral P, Sellitto D, Rengo C, Forster P, Savontaus ML, Bonne-Tamir B, Scozzari R (1998) mtDNA analysis reveals a major late Paleolithic population expansion from southwestern to northeastern Europe. Am J Hum Genet 62:1137–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Bandelt HJ, Macaulay V, Richards M, Cruciani F, Rengo C, Martinez-Cabrera V, et al (2001) A signal, from human mtDNA, of postglacial recolonization in Europe. Am J Hum Genet 69:844–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill PA, Passarino G, Lin AA, Shen P, Mirazon Lahr M, Foley RA, Oefner PJ, Cavalli-Sforza LL (2001) The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations. Ann Hum Genet 65:43–62 10.1046/j.1469-1809.2001.6510043.x [DOI] [PubMed] [Google Scholar]
- Underhill PA, Shen P, Lin AA, Jin L, Passarino G, Yang WH, Kauffman E, Bonne-Tamir B, Bertranpetit J, Francalacci P, Ibrahim M, Jenkins T, Kidd JR, Mehdi SQ, Seielstad MT, Wells RS, Piazza A, Davis RW, Feldman MW, Cavalli-Sforza LL, Oefner PJ (2000) Y chromosome sequence variation and the history of human populations. Nat Genet 26:358–361 10.1038/81685 [DOI] [PubMed] [Google Scholar]
- Wells RS, Yuldasheva N, Ruzibakiev R, Underhill PA, Evseeva I, Blue-Smith J, Jin L, et al (2001) The Eurasian heartland: a continental perspective on Y-chromosome diversity. Proc Natl Acad Sci USA 98:10244–10249 10.1073/pnas.171305098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Y Chromosome Consortium (2002) A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res 12:339–348 10.1101/gr.217602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Parra EJ, Sosnoski DM, Hiester K, Underhill PA, Shriver MD (2002) Melting curve SNP (McSNP) genotyping: a useful approach for diallelic genotyping in forensic sciense. J Forensic Sci 47:593–600 [PubMed] [Google Scholar]
- Zei G, Lisa A, Fiorani O, Magri C, Quintana-Murci L, Semino O, Santachiara-Benerecetti AS (2003) From surnames to the history of Y chromosomes: the Sardinian population as a paradigm. Eur J Hum Genet 11:802–807 10.1038/sj.ejhg.5201040 [DOI] [PubMed] [Google Scholar]
- Zhivotovsky LA (2001) Estimating divergence time with use of microsatellite genetic distances: impacts of population growth and gene flow. Mol Biol Evol 18:700–709 [DOI] [PubMed] [Google Scholar]
- Zhivotovsky LA, Underhill PA, Cinnioğlu C, Kayser M, Morar B, Kivisild T, Scozzari R, Cruciani F, Destro-Bisol G, Spedini G, Chambers GK, Herrera RJ, Yong KK, Gresham D, Tournev I, Feldman MW, Kalaydjieva L (2004) On the effective mutation rate at Y-chromosome STRs with application to human population divergence time. Am J Hum Genet 74:50–61 [DOI] [PMC free article] [PubMed] [Google Scholar]