Genomewide Screening Reveals High Levels of Insertional Polymorphism in the Human Endogenous Retrovirus Family HERV-K(HML2): Implications for Present-Day Activity (original) (raw)
Abstract
The published human genome sequence contains many thousands of endogenous retroviruses (HERVs) but all are defective, containing nonsense mutations or major deletions. Only the HERV-K(HML2) family has been active since the divergence of humans and chimpanzees; it contains many members that are human specific, as well as several that are insertionally polymorphic (an inserted element present only in some human individuals). Here we perform a genomewide survey of insertional polymorphism levels in this family by using the published human genome sequence and a diverse sample of 19 humans. We find that there are 113 human-specific HERV-K(HML2) elements in the human genome sequence, 8 of which are insertionally polymorphic (11 if we extrapolate to those within regions of the genome that were not suitable for amplification). The average rate of accumulation since the divergence with chimpanzees is thus approximately 3.8 × 10−4 per haploid genome per generation. Furthermore, we find that the number of polymorphic elements is not significantly different from that predicted by a standard population genetic model that assumes constant activity of the family until the present. This suggests to us that the HERV-K(HML2) family may be active in present-day humans. Active (replication-competent) elements are likely to have inserted very recently and to be present at low allele frequencies, and they may be causing disease in the individuals carrying them. This view of the family from a population perspective rather than a genome perspective will inform the current debate about a possible role of HERV-K(HML2) in human disease.
Endogenous retroviruses (ERVs) are derived from their exogenous counterparts via insertion (integration) into the germ line of their host. At insertion, each ERV element (provirus) consists of two identical, nontranslated long terminal repeats (LTRs) flanking an internal region that encodes proteins required for viral replication and assembly (5). ERVs become defective over time due to frameshift or nonsense mutations introduced during host DNA replication or via recombinational deletion of the internal region to leave a solo LTR structure (5, 31). Solo LTRs are approximately tenfold more abundant than their undeleted, full-length counterparts (31).
Within the published human genome sequence, there are over 98,000 human endogenous retroviruses (HERVs), but all are defective, containing nonsense mutations or major deletions. No replication-competent HERVs have been identified to date (26, 31, 33, 35), with only one (K113) with open reading frames for all genes (35), and thus their activity and infectivity is thought to have decreased substantially from levels occurring during earlier periods of primate evolution (1, 23, 34).
One possible exception to this trend is the HERV-K(HML2) family, which makes up less than 1% of HERV elements (27). This family has been active and infectious for much of the past 30 million years (2, 12, 20, 28, 35). It contains many members that inserted into the genome after the divergence of humans and chimpanzees approximately 6 million years ago, as well as several that are insertionally polymorphic (some human individuals have the insertion while other individuals have the empty, preinsertion site) (13, 21, 25, 35). Here we provide the first measures of the overall genomewide frequency of both human-specific and insertionally polymorphic elements in this HERV family. Full-length human-specific HERV-K(HML2) loci have been screened previously for insertional polymorphisms (21, 35), but this is not the case for the solo LTRs, which are much more abundant and can therefore provide substantially more data on the insertional history of an endogenous retrovirus family.
We also compare our observed level of insertional polymorphism to the value that we might expect if the HERV-K(HML2) family was still actively inserting at present. We generate this expectation by using a standard neutral population genetic model, whose two parameters are (i) an insertion rate that is calculated from the number of human-specific insertions in the published human genome sequence together with an estimate of the number of generations since the human-chimpanzee divergence and (ii) an estimate of the long-term population size in humans, as taken from the literature. The possibility that the family is active today is particularly important because it has been implicated in a range of human diseases.
MATERIALS AND METHODS
Screening.
Mining of HERV-K(HML2) elements was performed using build 31 of the human genome sequence as described previously (2). Human-specific solo LTRs, together with 500 bp of flanking sequence, were submitted to RepeatMasker (http://www.repeatmasker.org), and oligonucleotide primers were then designed against nonrepetitive DNA (when present) within the flanking regions. PCR amplification of a set of 19 diverse human DNA samples (Table 1) was performed under standard conditions, with typical annealing temperatures of 50 to 60°C. One set of reactions identified preinsertion loci and solo LTRs, and a second set was used to identify full-length elements. In the latter case, the 5′ flanking site primer was used in combination with a primer designed against a consensus leader sequence region of 12 full-length and human-specific HERV-K(HML2) elements.
TABLE 1.
Provenances of genomic DNA samples
| Sample | Sex | Ethnicity | Origin | Region or districta | Order no.b |
|---|---|---|---|---|---|
| 1 | Female | Biaka Pygmy | Central African Republic | Bagandu, SW CAR | NA10471 |
| 2 | Male | African | North of Sahara | NA17378 | |
| 3 | Female | Melanesian | Papua New Guinea | Bougainville | NA10539 |
| 4 | Female | Japanese | Japan | NA11589 | |
| 5 | Male | Mayan | Mexico | Yukatan | NA10975 |
| 6 | Female | Druze | Israel | The Galilee | NA11521 |
| 7 | Male | Adygei | Russia | Krasnodor, N. Caucasus | NA13619 |
| 8 | Male | Ami | Taiwan | E. Taiwan | NA13608 |
| 9 | Male | Atayal | Taiwan | Nanshi River, E. Taiwan | NA13597 |
| 10 | Female | Khmer | Cambodia | NA11373 | |
| 11 | Female | Mbuti Pygmy | Congo | Ituri Forest, NE DRC | NA10493 |
| 12 | Male | Russian | Russia | Zuevsky | NA13820 |
| 13 | Female | African | South of Sahara | NA17341 | |
| 14 | Male | Mbuti Pygmy | Congo | Ituri Forest, NE DRC | NA10492 |
| 15 | Male | Mbuti Pygmy | Congo | Ituri Forest, NE DRC | NA10494 |
| 16 | Male | Biaka Pygmy | Central African Republic | Bagandu, SW CAR | NA10469 |
| 17 | Male | Biaka Pygmy | Central African Republic | Bagandu, SW CAR | NA10470 |
| 18 | Female | Biaka Pygmy | Central African Republic | Bagandu, SW CAR | NA10472 |
| 19 | Female | Biaka Pygmy | Central African Republic | Bagandu, SW CAR | NA10473 |
Phylogenetic reconstruction.
From among all the HERV-K(HML2) elements extracted by our mining, we excluded the very old (and hence uninformative) insertions by selecting only the LTR sequences (from both solo LTRs and full-length elements) that were no more than 5% divergent from the 5′ LTR of a full-length element that appears to have inserted relatively recently (8c8, discussed below). This was performed using the program WATER (29). We then aligned the sequences by using CLUSTAL W (32) and, with such low sequence divergence, the resulting alignment was unambiguous (see Fig. S1 in the supplemental material). A maximum likelihood LTR phylogeny was then constructed for the selected LTR sequences by using the program PHYML (16) with the HKY+γ model of sequence evolution (parameter values estimated from the data). The phylogeny was rooted by using an element also present in the chimpanzee and gorilla genomes.
Calculation of the insertion rate (μ).
The average rate of insertion since the divergence of humans and chimpanzees was calculated by dividing the number of human-specific insertions by the number of generations in the human lineage since divergence, assuming an average generation time of 20 years (11, 14, 15).
The model and its parameters.
The program ms (19) generates samples drawn at random from a population obeying the Wright-Fisher model of genetic drift and an infinite-sites model of mutation (18). The infinite-sites model was used as it allows for an unlimited number of unique sites (in this case, loci) into which elements can insert and does not allow reversals to the preinsertion state. Briefly, the program performs the following functions. (i) It generates random genealogies for a specified number of samples, which in our case represent haploid genomes (a total of 39 representing the 19 human DNA samples plus the human genome sequence). (ii) Branch lengths are calculated (in terms of numbers of generations) using coalescent theory. (iii) Mutations, which in our case represent insertions, are randomly distributed onto these branches (following a Poisson distribution). (iv) The distribution of insertions among each sample is output from the program as a binary list (at each locus, 0 denotes a preinsertion site and 1 denotes an insertion). We then randomly selected one of these samples to represent the human genome sequence and calculated the number of loci that were represented by an inserted element in this sample but were insertionally polymorphic in the other 38 samples. We ran 1,000 simulations, and for each we incorporated free recombination by summing the results from 10,000 coalescent trees, on each of which the insertion rate was 0.0001 μ. It should be noted that we are considering here only insertions that are neutral, since insertions harmful to the host are likely to be lost rapidly from the host population as a result of selection.
RESULTS
Human-specific HERV-K(HML2) elements.
We mined the published human genome sequence for full-length elements and solo LTRs derived from the HERV-K(HML2) family. We then used the flanking regions to identify the orthologous location in the chimpanzee genome sequence (37) and determine which of the insertions were human specific. Excluding elements that had been copied via segmental duplication, we identified 113 human-specific insertions (15 of which were represented by full-length elements and 98 by solo LTRs) (Fig. 1). Humans and chimpanzees diverged approximately 6 million years ago, and the long-term average human generation time has been about 20 years (11, 14, 15). Thus, the average insertion rate (μ) has been approximately 3.8 × 10−4 per haploid genome per generation during this period. Most of the human-specific full-length elements inserted relatively recently, according to the low level of mutational divergence between their LTRs: the LTRs are identical at the time of insertion, and they diverge as they both accrue mutations independently during host replication. Of the full-length elements, the LTR divergence of 11 was below 0.5% and two had identical LTRs. A previous study (35) of another HERV-K(HML2) element with identical LTRs, which is not in the published human genome sequence, used the estimated rate of background mutation in the human genome to infer that such elements are, at most, a few hundred thousand years old.
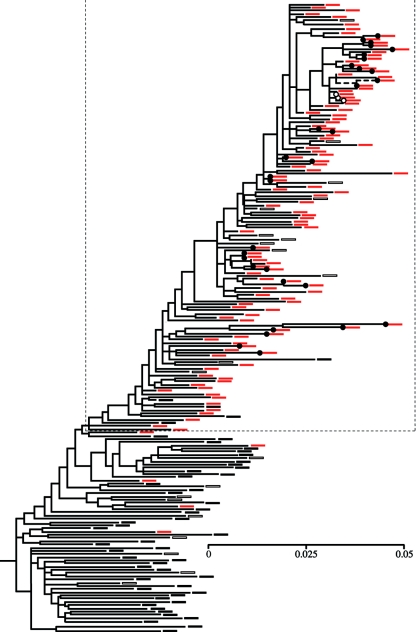
FIG. 1.
Maximum likelihood phylogeny of HERV-K(HML2) LTRs. Filled and open circles indicate LTRs from full-length and segmentally duplicated elements, respectively. Black boxes represent taxa present in both the chimpanzee and human genome sequences, whereas red boxes represent human-specific elements. Intermingling of the two classes is probably due to a variety of factors, such as gene conversion or ancestral polymorphism. Open boxes represent taxa whose distribution could not be determined directly (the chimpanzee genome project is incomplete), and their probable distribution was estimated from their position in the phylogeny. A dashed line indicates the placement of K113, which is absent from the published human genome sequence. The large boxed region (which excludes most non-human-specific elements) in shown in more detail in Fig. 2. Scale bar shows mean number of substitutions per site.
Detection of insertionally polymorphic HERV-K(HML2) elements.
We designed locus-specific primers against the flanking regions of 93 HERV-K(HML2) solo LTRs. Nineteen diverse human genomic DNA samples (Table 1) were then scored for the presence of either a preinsertion (empty) site or a solo LTR/full-length element by PCR amplification. The remaining five solo LTRs were not tested, as their basal phylogenetic location suggested that they were old and unlikely to be polymorphic.
Six of the 63 solo LTRs that were successfully amplified displayed insertional polymorphism (Fig. 2 and 3). Only one of these polymorphisms has been previously described (25). The status of 30 other solo LTRs could not be determined, as many are located in highly repetitive regions and gave multiple bands when amplified (unpublished results). Another two insertional polymorphisms are known from previous screening of the 15 full-length elements: 8c8, also known as K115 (35), and 154c11, also known as 11q22 (21). Thus, a total of 8 out of 78 tested elements (63 solo LTRs plus 15 full-length elements) in the published human genome are insertionally polymorphic (Fig. 3 and Table 2). Furthermore, assuming that the 30 untested loci are as likely to be polymorphic as those investigated successfully, another three loci (8/78 × 30) will be insertionally polymorphic; thus, a total of about 11 of the HERV-K(HML2) elements from the published sequence will display insertional polymorphism in our sample of human individuals. Most of the polymorphic loci are near the tip of the LTR phylogeny, indicating that the insertion events are likely to be relatively recent (Fig. 2). Also, the phylogeny shows many nodes near the tips, indicating recent insertional activity. The frequencies of the inserted elements range from 0.04 to 0.94, with a mean frequency of 0.61. Five of the six polymorphic loci that we examined displayed either preinsertion sites or solo LTRs, while only one, 859c12 (Fig. 3), displayed all three states including the full-length element. This suggests that in most cases solo LTR formation via recombinational deletion occurs rapidly and before the element reaches fixation. Indeed, only in the case of the element 8c8 (K115) is there no evidence of solo LTR formation, and this element has a low inserted allele frequency of only 0.04 within the human population (35).
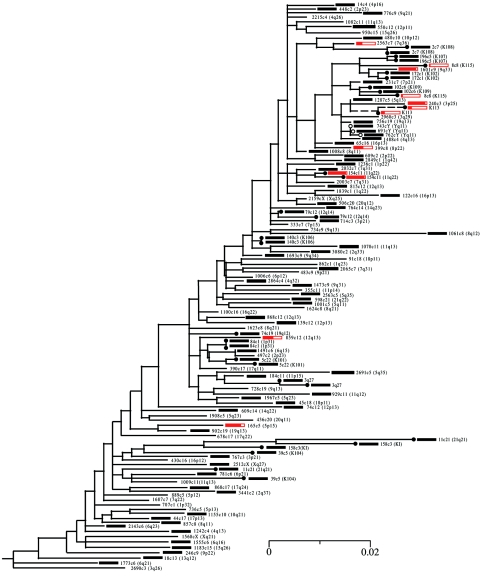
FIG. 2.
Elements screened for insertional polymorphism. Taxon names are followed by their genomic location in parentheses. Black boxes indicate elements homozygous for the insertion in all 19 individuals surveyed, whereas those in red display insertional polymorphism, with the filled region in each box being proportional to the frequency of the inserted element in the samples. The other (nonboxed) elements gave inconclusive results, usually because of their location in regions of highly repetitive DNA. Data from all full-length elements were taken from previous reports (21, 35). A nucleotide alignment of the surveyed solo LTRs, together with their flanking sequences, is shown in Fig. S1. Scale bar shows mean number of substitutions per site.
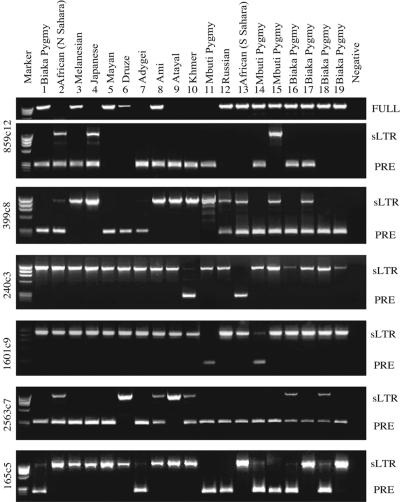
FIG. 3.
Detection of HERV-K(HML2) preinsertion sites. Amplification of solo LTR loci within the published human genome sequence showed a preinsertion site (PRE), a solo LTR (sLTR), or a full-length element (FULL) when tested against a panel of 19 individuals. The provenance of each individual is shown in Table 1, and allele frequencies are shown in Table 2. The identity of each band was confirmed by DNA sequencing. We rescreened the previously identified polymorphism 165c5, as the originally estimated frequencies were based largely on individuals from Russia (25).
TABLE 2.
Polymorphic HERV-K(HML2) insertions and their allele frequencies
| Name | Location | Allele frequencya | ||
|---|---|---|---|---|
| Preinsertion site | Solo LTR | Full-length element | ||
| 154c11 | 11q22 | 0.06 | 0.39 | 0.55 |
| 8c8 | 8p23 | 0.96 | 0.00 | 0.04 |
| 165c5 | 5p15 | 0.28 | 0.72 | 0.00 |
| 859c12 | 12q13 | 0.41 | 0.10 | 0.49 |
| 399c8 | 8p22 | 0.51 | 0.49 | 0.00 |
| 240c3 | 3p25 | 0.08 | 0.92 | 0.00 |
| 1609c9 | 9q33 | 0.08 | 0.92 | 0.00 |
| 2563c7 | 7q36 | 0.74 | 0.26 | 0.00 |
| Mean | 0.39 | 0.48 | 0.13 |
Modeling HERV-K(HML2) insertional polymorphism.
HERV-K(HML2) copying events lead to novel elements that are present initially only in a single host individual. The frequency of these elements in the host population may then increase as a result of genetic drift (we exclude here cases of co-option, which we assume are rare). If the HERV-K(HML2) family is still active and producing new copies at present, we expect to find elements that have inserted recently and are present in only a proportion of the human population. The actual number of insertionally polymorphic elements that we would expect, assuming the scenario of present-day activity, can be determined by using standard population genetic models.
We therefore calculated a frequency distribution (see Materials and Methods) for the expected number of loci in the published human genome sequence that would be insertionally polymorphic when compared to our sample of 19 individuals, assuming activity of the HERV-K(HML2) family until the present. Parameters used were our estimated insertion rate since the divergence from chimpanzees (μ = 3.8 × 10−4) and the previous estimate of long-term effective population size (Ne) of 10,000 (17, 36). Given that a polymorphism has a probability of 0.72 (78/108) of being detected in our survey (i.e., is in a region of the genome that can be amplified by PCR), the model predicts a mean of 10.6 polymorphic insertions, with 95% bounds of 5 and 18. Our observed value of eight polymorphic insertions is well within this range (P = 0.57). Moreover, the distribution of insertion frequencies is not significantly different from that predicted by the model (P is ≥0.25 [two-tailed] for the mean, variance, and skew). This result is robust to using other plausible parameter values. For example, our observed figure of eight polymorphic sites is not statistically different from the model's predictions if human generation time is 10 rather than 20 years or if the time since the human-chimpanzee divergence is 4.5 rather than 6 million years.
Note that the total number of polymorphic sites within the sample cannot be verified directly because we tested only those sites in the published human genome sequence that have an inserted element. Thus, after 1,000 replicates, the model predicts that there would be a mean of 63.4 polymorphic loci within our sample of 39 haploid genomes (of which 45.6 lie in amplifiable regions) but that only 14.5 (10.6 in amplifiable regions) of these would be present as inserted elements (as opposed to preinsertion sites) in any one haploid genome. Furthermore, although the model assumes neutrality, our inferences from it do not depend upon all insertions being neutral. Instead, we assume that elements with a negative effect on host fitness are lost from the host population and that we are thus observing the net rate of accumulation.
DISCUSSION
High level of insertional polymorphism in the HERV-K(HML2) family.
In the first genomewide survey of a HERV family, we find that approximately 10% of the human-specific loci tested (8/78) are insertionally polymorphic within our sample. From the observed allele frequencies in Table 2, we calculate that only 6% of individuals will be homozygous at all eight loci [calculated as Π(1−2_pq_), where p is the frequency of the insertion and q is the frequency of the preinsertion, and assuming Hardy-Weinberg equilibrium]. This is consistent with an infinite-sites mathematical model, which also predicts that 6% of the population will be homozygous at all sites (18). Gene flow between human subpopulations is relatively high, and so the assumption of random mating used to derive these expectations will not unduly bias our results (10, 30). Recently, Bennett et al. (3) examined the equivalent of an additional haploid genome for insertional polymorphisms and identified two HERV-K(HML2) sites as polymorphic when compared to the published human genome sequence. This is not significantly different from our result, as the model predicts that approximately 18% of individuals will be heterozygous at fewer than three sites.
Is the HERV-K(HML2) family active in present-day humans?
The high level of observed insertional polymorphism within the HERV-K(HML2) family indicates that a substantial number of insertions have occurred since the divergence of the human individuals investigated in this study. Furthermore, there are now several lines of evidence to suggest that the family may well still be active at present. First, there is the close match between the observed eight insertional polymorphisms and the number predicted using the population genetic model, which assumes continued activity until the present. However, we note that a recent cessation of activity would also be consistent with our data because there would still be insertionally polymorphic elements retained in the present-day human population. For example, in the model, stopping new insertions 650,000 years before the present predicts a mean of 2.9 polymorphic insertions, with 95% bounds of 0 and 7. Our observed value of eight polymorphic insertions falls outside these bounds, but it does not if the cessation was more recent (e.g., a cessation at 500,000 years ago gives a mean of 4.2, with 95% bounds of 1 and 9). Although we cannot exclude this possibility, we think it is unlikely. Also, the mean insertional rate has remained the same since the human-chimpanzee divergence: we found 440 HERV-K(HML2) elements (including solo LTRs) in the published human genome sequence that had inserted before the divergence of humans and chimpanzees (unpublished data). This gives a mean rate of 18 insertions per million years for the first 24 million years of the family's history, compared to 19 insertions per million years for the last 6 million years. The second line of evidence indicating continued activity is the phylogenetic pattern: most insertionally polymorphic elements, as well as many nodes, are near the tips of the phylogeny. Finally, the young age of some full-length elements, as determined by their LTRs having identical sequences, is compatible with continued activity.
Thus, we believe that the simplest explanation for our data is that the family is active at the present day. If we are correct, then a number of predictions can be made regarding HERV-K(HML2) polymorphism. The insertion rate for humans as a whole will be 2 Nμ, which suggests there are now approximately 4.5 × 106 new insertions occurring every generation (assuming a human population size [_N_] of 6 × 109), and the total number of polymorphic elements will be substantially higher than this figure. We have also shown previously that most HERV-K(HML2) insertions are the result of reinfection rather than retrotransposition within germ line cells (2), and thus the family is likely to be infectious as well as insertionally active. This reinfection may require movement only between cells of the same individual and does not necessarily require infectious transfer between individuals.
A model of HERV-K(HML2) evolution.
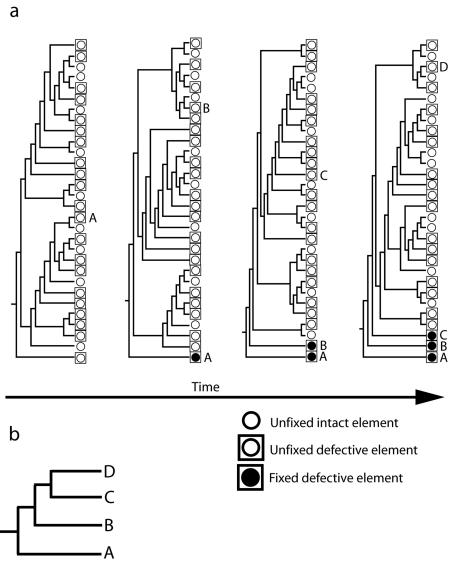
The absence of known, infectious members of the HERV-K(HML2) family and the lack of elements with a full coding potential within the published human genome sequence appears, initially, to contradict our conclusion that the family is likely to be active at present. Furthermore, the modeling presented above is based on insertions being neutral and therefore excludes any elements that never reach high allele frequencies due to negative selection acting on the host. Such elements are also ignored by our population genetic model. To take these factors into account, we propose the following scenario (shown in Fig. 4) for the evolution of the HERV-K(HML2) family. We suggest that there is (and has been for many millions of years) a large population of unfixed HERV-K(HML2) elements within the human germ line and that a subset of these elements is both active and infectious at any one time point. Because many of the active and infectious elements may be deleterious to their hosts, they are likely to be present only transiently and to rarely (due to negative selection) reach high allele frequencies in the population as a whole. Some of these elements then acquire, by chance, knockout mutations (for example, via recombinational deletion or frameshift mutations); it is these elements, now neutral and defective, which are able to reach high allele frequencies, and a few eventually become fixed. Thus, it is not surprising that the published human genome sequence [which contains most of the HERV-K(HML2) sequences characterized to date] contains no intact members; it is best regarded as a depository of old, defective elements that have drifted to fixation. This is because there is only a very small chance that any one individual or genome harbors one of the active and infectious members of the current HERV-K(HML2) population. We also note that recently inserted elements are less likely to have undergone the recombinational deletion events we observed for most of the solo LTR loci described here.
FIG. 4.
Proposed model of HERV-K(HML2) family evolution within humans. (a) At each time point, there is a large unfixed population of elements, a proportion of which are replication competent and infectious, whereas others are defective. Some of the subset of defective elements, but none of the replication-competent elements, eventually drift to fixation. The population of unfixed elements is continuously replenished by new insertions resulting from the replication of intact and unfixed elements. (b) Over time, the fixed and defective elements (i.e., A, B, and C) accumulate so that in any one genome all, or almost all, of the elements are defective, the intact and infectious elements being present only in a very small proportion of individuals.
Implications for disease.
HERV-K(HML2) elements commonly form viral particles in human cancer cells, and 60% of male patients with germ line cancers show specific immune reactions to HERV-K(HML2) antigens, compared to 4% of healthy individuals (4, 8, 22). Members of the family also encode an accessory protein, cORF or Rec, which can impair spermatogenesis in mice, possibly by binding to the transcription factor PLZF (promyelocytic leukemia zinc finger protein) (6). Impairment of spermatogenesis is thought to predispose humans to germ line tumors, and injection of cORF induces tumor formation in immunocompromised nude mice (6). Skepticism that HERV-K(HML2) elements are the cause, rather than just markers, of such tumors has been fuelled by the absence of known, active elements. From our study, we suggest that there may be many such active elements which, although rare in the general population, may well be causing disease in some of the individuals carrying them.
We consider that the rarity of novel HERV-K(HML2) insertions may explain why no active, disease-causing elements are known. Another type of retrotransposable element, long interspersed nuclear elements (LINEs), is much more active in humans, with a long-term accumulation rate of 4 × 10−3 elements per haploid genome per generation (7). Experimental work indicates that the actual frequency of novel LINEs among human individuals may be between 1 in 2 and 1 in 33 (9). However, despite this high level of activity, it appears that new insertions by LINEs are responsible for only approximately 1 out of every 1,000 disease-causing mutations in humans (24). Thus, disease-causing mutations caused by members of the HERV-K(HML2) family may well be sufficiently rare to have escaped detection to date.
Supplementary Material
[Supplemental material]
Acknowledgments
The work was supported by a grant from the Wellcome Trust.
We thank Vini Pereira and Aris Katzourakis for help with the mining of the human genome, Peter Kabat and Jonathan Ng for help with the molecular analysis, and Dick Hudson for advice on using ms.
Footnotes
REFERENCES
- 1.Bannert, N., and R. Kurth. 2004. Retroelements and the human genome: new perspectives on an old relation. Proc. Natl. Acad. Sci. USA 101**:**14572-14579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belshaw, R., V. Pereira, A. Katzourakis, G. Talbot, J. Pačes, A. Burt, and M. Tristem. 2004. Long-term reinfection of the human genome by endogenous retroviruses. Proc. Natl. Acad. Sci. USA 101**:**4894-4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett, E. A., L. E. Coleman, C. Tsui, W. S. Pittard, and S. E. Devine. 2004. Natural genetic variation caused by transposable elements in humans. Genetics 168**:**933-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bieda, K., A. Hoffmann, and K. Boller. 2001. Phenotypic heterogeneity of human endogenous retrovirus particles produced by teratocarcinoma cell lines. J. Gen. Virol. 82**:**591-596. [DOI] [PubMed] [Google Scholar]
- 5.Boeke, J. D., and J. P. Stoye. 1997. Retrotransposons, endogenous retroviruses, and the evolution of retroelements, p. 343-435. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. CSHL Press, New York, N.Y. [PubMed]
- 6.Boese, A., M. Sauter, U. Galli, B. Best, H. Herbst, J. Mayer, E. Kremmer, K. Roemer, and N. Mueller-Lantzsch. 2000. Human endogenous retrovirus protein cORF supports cell transformation and associates with the promyelocytic leukemia zinc finger protein. Oncogene 19**:**4328-4336. [DOI] [PubMed] [Google Scholar]
- 7.Boissinot, S., P. Chevret, and A. V. Furano. 2000. L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol. Biol. Evol. 17**:**915-928. [DOI] [PubMed] [Google Scholar]
- 8.Boller, K., O. Janssen, H. Schuldes, R. R. Tönjes, and R. Kurth. 1997. Characterization of the antibody response specific for the human endogenous retrovirus HTDV/HERV-K. J. Virol. 71**:**4581-4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brouha, B., J. Schustak, R. M. Badge, S. Lutz-Prigge, A. H. Farley, J. V. Moran, and H. H. Kazazian, Jr. 2003. Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl. Acad. Sci. USA 100**:**5280-5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cavalli-Sforza, L. L., P. Menozzi, and A. Piazza. 1994. The history and geography of human genes. Princeton University Press, Princeton, N.J.
- 11.Chen, F. C., and W. H. Li. 2001. Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am. J. Hum. Genet. 68**:**444-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costas, J. 2001. Evolutionary dynamics of the human endogenous retrovirus family HERV-K inferred from full-length proviral genomes. J. Mol. Evol. 53**:**237-243. [DOI] [PubMed] [Google Scholar]
- 13.Donner, H., R. R. Tonjes, R. E. Bontrop, R. Kurth, K. H. Usadel, and K. Badenhoop. 1999. Intronic sequence motifs of HLA-DQB1 are shared between humans, apes and Old World monkeys, but a retroviral LTR element (DQLTR3) is human specific. Tissue Antigens 53**:**551-558. [DOI] [PubMed] [Google Scholar]
- 14.Glazko, G. V., and M. Nei. 2003. Estimation of divergence times for major lineages of primate species. Mol. Biol. Evol. 20**:**424-434. [DOI] [PubMed] [Google Scholar]
- 15.Goodman, M., C. A. Porter, J. Czelusniak, S. L. Page, H. Schneider, J. Shoshani, G. Gunnell, and C. P. Groves. 1998. Toward a phylogenetic classification of primates based on DNA evidence complemented by fossil evidence. Mol. Phylogenet. Evol. 9**:**585-598. [DOI] [PubMed] [Google Scholar]
- 16.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52**:**696-704. [DOI] [PubMed] [Google Scholar]
- 17.Harpending, H. C., M. A. Batzer, M. Gurven, L. B. Jorde, A. R. Rogers, and S. T. Sherry. 1998. Genetic traces of ancient demography. Proc. Natl. Acad. Sci. USA 95**:**1961-1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartl, D. L., and A. G. Clark. 1997. Principles of population genetics. Sinauer Associates, Inc., Sunderland, Mass.
- 19.Hudson, R. R. 2002. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics 18**:**337-338. [DOI] [PubMed] [Google Scholar]
- 20.Hughes, J. F., and J. M. Coffin. 2001. Evidence for genomic rearrangements mediated by human endogenous retroviruses during primate evolution. Nat. Genet. 29**:**487-489. [DOI] [PubMed] [Google Scholar]
- 21.Hughes, J. F., and J. M. Coffin. 2004. Human endogenous retrovirus K solo-LTR formation and insertional polymorphisms: implications for human and viral evolution. Proc. Natl. Acad. Sci. USA 101**:**1668-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kleiman, A., N. Senyuta, A. Tryakin, M. Sauter, A. Karseladze, S. Tjulandin, V. Gurtsevitch, and N. Mueller-Lantzsch. 2004. HERV-K(HML2) GAG/ENV antibodies as indicator for therapy effect in patients with germ cell tumors. Int. J. Cancer 110**:**459-461. [DOI] [PubMed] [Google Scholar]
- 23.Lander, E. S., L. M. Linton, B. Birren, C. Nusbaum, M. C. Zody, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409**:**860-921. [DOI] [PubMed] [Google Scholar]
- 24.Lutz, S. M., B. J. Vincent, H. H. Kazazian, Jr., M. A. Batzer, and J. V. Moran. 2003. Allelic heterogeneity in LINE-1 retrotransposition activity. Am. J. Hum. Genet. 73**:**1431-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mamedov, I., Y. Lebedev, G. Hunsmann, E. Khusnutdinova, and E. Sverdlov. 2004. A rare event of insertion polymorphism of a HERV-K LTR in the human genome. Genomics 84**:**596-599. [DOI] [PubMed] [Google Scholar]
- 26.Mayer, J., M. Sauter, A. Racz, D. Scherer, N. Mueller-Lantzsch, and E. Meese. 1999. An almost-intact human endogenous retrovirus K on human chromosome 7. Nat. Genet. 21**:**257-258. [DOI] [PubMed] [Google Scholar]
- 27.Pačes, J., A. Pavlíček, and V. Pačes. 2002. HERVd: database of human endogenous retroviruses. Nucleic Acids Res. 30**:**205-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reus, K., J. Mayer, M. Sauter, H. Zischler, N. Müller-Lantzsch, and E. Meese. 2001. HERV-K(OLD): ancestor sequences of the human endogenous retrovirus family HERV-K(HML-2). J. Virol. 75**:**8917-8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rice, P., I. Longden, and A. Bleasby. 2000. EMBOSS: the European molecular biology open software suite. Trends Genet. 16**:**276-277. [DOI] [PubMed] [Google Scholar]
- 30.Rosenberg, N. A., J. K. Pritchard, J. L. Weber, H. M. Cann, K. K. Kidd, L. A. Zhivotovsky, and M. W. Feldman. 2002. Genetic structure of human populations. Science 298**:**2381-2385. [DOI] [PubMed] [Google Scholar]
- 31.Stoye, J. P. 2001. Endogenous retroviruses: still active after all these years? Curr. Biol. 11**:**R914-R916. [DOI] [PubMed] [Google Scholar]
- 32.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22**:**4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tönjes, R. R., F. Czauderna, and R. Kurth. 1999. Genome-wide screening, cloning, chromosomal assignment, and expression of full-length human endogenous retrovirus type K. J. Virol. 73**:**9187-9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tristem, M. 2000. Identification and characterization of novel human endogenous retrovirus families by phylogenetic screening of the human genome mapping project database. J. Virol. 74**:**3715-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turner, G., M. Barbulescu, M. Su, M. I. Jensen-Seaman, K. K. Kidd, and J. Lenz. 2001. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr. Biol. 11**:**1531-1535. [DOI] [PubMed] [Google Scholar]
- 36.Wall, J. D. 2003. Estimating ancestral population sizes and divergence times. Genetics 163**:**395-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Watanabe, H., A. Fujiyama, M. Hattori, T. D. Taylor, A. Toyoda, et al. 2004. DNA sequence and comparative analysis of chimpanzee chromosome 22. Nature 429**:**382-388. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental material]