Comparative genomic analysis of Vibrio cholerae: Genes that correlate with cholera endemic and pandemic disease (original) (raw)
Abstract
Historically, the first six recorded cholera pandemics occurred between 1817 and 1923 and were caused by Vibrio cholerae O1 serogroup strains of the classical biotype. Although strains of the El Tor biotype caused sporadic infections and cholera epidemics as early as 1910, it was not until 1961 that this biotype emerged to cause the 7th pandemic, eventually resulting in the global elimination of classical biotype strains as a cause of disease. The completed genome sequence of 7th pandemic El Tor O1 strain N16961 has provided an important tool to begin addressing questions about the evolution of V. cholerae as a human pathogen and environmental organism. To facilitate such studies, we constructed a V. cholerae genomic microarray that displays over 93% of the predicted genes of strain N16961 as spotted features. Hybridization of labeled genomic DNA from different strains to this microarray allowed us to compare the gene content of N16961 to that of other V. cholerae isolates. Surprisingly, the results reveal a high degree of conservation among the strains tested. However, genes unique to all pandemic strains as well as genes specific to 7th pandemic El Tor and related O139 serogroup strains were identified. These latter genes may encode gain-of-function traits specifically associated with displacement of the preexisting classical strains in South Asia and may also promote the establishment of endemic disease in previously cholera-free locations.
There have been seven pandemics of cholera, a severe diarrheal disease caused by Vibrio cholerae, a bacterium whose genomic sequence has recently been reported (1). The first six pandemics were caused by the classical biotype of V. cholerae, which spread from the Indian subcontinent to most other areas of the world between 1817 and 1923 (2–5). However, by 1900, cholera had disappeared from the Western hemisphere as an epidemic and endemic disease. A major epidemic in the Celebes Islands (present day Indonesia) was caused by an “El Tor” biotype strain in 1935, but cholera failed to become endemic in this locale and did not spread elsewhere in Asia at this time (2, 6). The 7th pandemic of cholera technically began in 1961, when pathogenic El Tor biotype strains established endemicity in Indonesia. These strains then rapidly spread to cause disease on the Asian mainland and in Africa, replacing classical strains as the cause of endemic cholera (7). In 1991, V. cholerae El Tor caused a massive epidemic in Lima, Peru. Since then, cholera has spread to virtually all neighboring countries and, as a result, is now endemic in much of Latin America. Molecular typing methods have suggested that isolates from the 1991 Latin American epidemic are clonal and closely related to Asian and African 7th pandemic strains (3, 7).
In 1992, a newly identified serogroup, O139, was recognized as the cause of epidemic cholera in South Asia (8). O139 strains initially displaced serogroup O1 El Tor strains as the main cause of cholera in India and Bangladesh. However, over the last few years, El Tor O1 strains have reestablished their prominence and now share this locale with O139 strains.
Much research has been focused on understanding the basis of V. cholerae pathogenicity and its pandemic potential. Numerous studies have highlighted the requirement of three essential components in pathogenic V. cholerae strains: the filamentous CTX bacteriophage (CTXφ) that encodes cholera toxin, the TCP pathogenicity island encoding the TCP pili, a colonization factor and receptor for CTXφ, and toxR, an essential virulence regulatory gene (7). Investigators have also sought to understand the evolution of 7th pandemic El Tor and O139 strains by using various molecular typing techniques, such as ribotyping, restriction fragment length polymorphisms, and sequence analysis of housekeeping and virulence genes (9–12). Data from these methods support the hypothesis that O139 strains evolved from recent 7th pandemic El Tor isolates (3, 13). Furthermore, although they are closely related, classical biotype strains and nonpathogenic El Tor environmental strains have evolved from a separate lineage from 7th pandemic El Tor isolates (14, 15).
In comparison to classical strains that caused the 6th pandemic, the El Tor strains responsible for the 7th pandemic show clear differences in regard to their epidemic and endemic behavior. It has been suggested that El Tor strains demonstrate increased persistence in aquatic ecosystems (16, 17). Furthermore, El Tor strains can be distinguished from classical strains by properties such as their hemolytic activity, agglutination reactions with erythrocytes, and polymyxin B resistance. However, there is no genetic evidence that these phenotypic properties correlate with the pandemic and endemic potential of V. cholerae strains, or with their ability to replace predecessor strains as observed during the 7th pandemic.
To begin to address these issues, we sought to identify genes unique to El Tor strains of the 7th pandemic. Here, we report the construction of a V. cholerae genomic microarray based on the sequenced O1 El Tor strain N16961 from the 7th pandemic of cholera (1). This array has been used to compare the gene content of classical, prepandemic El Tor, pandemic El Tor, and two nontoxigenic strains to that of strain N16961. Although these comparative genomic analyses revealed a high degree of genetic similarity among strains isolated over the past century, we were able to identify genes unique to pandemic El Tor and O139 strains. These genes potentially encode key properties that have led to the global success of 7th pandemic strains as agents of endemic and pandemic cholera.
Materials and Methods
Strains and Media.
V. cholerae strains used for this study are described in Table 1 and were obtained from either the American Type Culture Collection or laboratory stocks. All strains were grown in Luria–Bertani broth and stored as frozen stocks in Luria–Bertani broth with 20% glycerol.
Table 1.
Strains used for comparative analysis
| Strain | Origin | Year isolated | Biotype, serogroup | No. genes absent* |
|---|---|---|---|---|
| N16961 | Bangladesh | 1971 | El Tor, O1 (7th pandemic) | N/A |
| 2740-80 | Gulf Coast, United States | 1980 | environmental, nontoxigenic El Tor, O1 | 49 |
| NCTC 8457 | Saudi Arabia | 1910 | El Tor, O1 (prepandemic) | 39 |
| MAK 757 | Celebes Islands | 1937 | El Tor, O1 (prepandemic) | 49 |
| 569B | India | 1948 | Classical, O1 | 46 |
| O395 | India | 1965 | Classical, O1 | 36 |
| NIH 41 | India | 1940 | Classical, O1 | 48 |
| HK1 | Hong Kong | 1961 | El Tor, O1 (7th pandemic) | 0 |
| C6709 | Peru | 1991 | El Tor, O1 (7th pandemic) | 1 |
| MO10 | India | 1992 | El Tor, O139 | 47 |
Primer Design and PCR Amplification of ORFs.
PCR primers were designed by using PRIMER3 software (http://www.genome.wi.mit.edu/genome_software/other/primer3.html). The 5′ primer for each ORF began with the start codon and the 3′ primer began with the nucleotide preceding the stop codon. PCR amplification of each ORF was performed by using Takara Taq polymerase (Intergen, Purchase, NY) according to manufacturer's instructions and included 5% dimethyl sulfoxide. After an initial 5-min denaturation at 95°C, 30 PCR cycles were run as follows: 95°C for 30 s, 54°C for 30 s, and 72°C for 3 min. Products were purified by using the Multiscreen 96-well PCR unit according to the manufacturer's directions (Millipore). After elution with 3× SSC, Sarkosyl was added to a final concentration of 0.03%. Two-microliter aliquots of each purified product were run on 1% agarose gels to confirm the presence of a product migrating at the expected size for each ORF.
Array Printing.
Glass slides were coated with polylysine according to the protocol of Eisen and Brown (18). PCR products were spotted by using a GMS 417 arrayer (Affymetrix) or a Q arrayer (Genetix). DNA corresponding to the ORF of an unrelated gene, gfp from Aequorea victoria, was included to serve as a control for spotting and hybridization conditions.
Preparation and Hybridization of Fluorescently Labeled DNA.
Genomic DNA (gDNA) was prepared from each strain by using the Easy-DNA kit (Invitrogen) according to the manufacturer's instructions. For a 50-μl reaction, 2–3 μg of gDNA was combined with 3 μg of random hexamer, heated to >95°C for 5 min, and chilled on ice. Remaining components were added to final concentrations as follows: 0.05 mM dA/G/TTP, 0.5 mM Cy3 or Cy5-dCTP (NEN), 1× Eco Pol buffer, and 15 units of the Klenow fragment of Escherichia coli polymerase (NEB, Beverly, MA). The reaction was placed at 37°C for 2 h, and labeled DNA was purified by using the Qiagen (Chatsworth, CA) PCR purification kit according to manufacturer's instructions. Cy3- and Cy5-labeled DNAs were combined and precipitated according to standard protocols after addition of 50 μg/ml sheared salmon sperm DNA. DNA was recovered by centrifugation, and the pellet was washed with 70% EtOH and recentrifuged. The pellet was briefly air dried, then resuspended in hybridization buffer containing 50% formamide/6× SSC/5× Denhardt's solution/0.5% SDS/5 mM KH2PO4. Labeled DNA was heated to >95°C for 5 min and chilled on ice before use in hybridization.
Printed slides were crosslinked in a UV Stratalinker 2400 (Stratagene) at 1,000 × 100 μjoules after brief hydration over boiling dH2O. Crosslinked slides were washed briefly in 0.1% SDS, rinsed twice in dH2O, then placed in a boiling dH2O bath for 3–5 min. Slides were immersed in ice-cold 95% EtOH, dried by centrifugation, then prehybridized at 42°C for at least 1 h in 5× SSC/0.1% SDS/10 mg/ml BSA. Slides were washed twice in dH2O, twice in 95% EtOH, and dried by centrifugation. Hybridization was carried out under glass cover slips (VWR) in a sealed wet box at 42°C overnight. In at least one case for each test strain, the array was hybridized under the exact same conditions, except at 44°C. Following hybridization, slides were washed for 10 min in each of the following solutions: one time in 2× SSC/0.1% SDS heated to 55°C; one time in 0.2× SSC/0.1% SDS at room temperature two times in 0.2× SSC, and were then dried by centrifugation.
Data Generation and Analysis.
In a single array experiment, the genomic content from one of the test strains was compared with that of N16961. For each of the nine test strains, data were compiled from at least three array experiments. For each test strain, two independent gDNA preparations were used as template, and both Cy3 and Cy5-dCTP were used in independent labeling and hybridization experiments to account for any differences in DNA preparation or dye incorporation.
Hybridized, washed slides were scanned for Cy5 and Cy3 fluorescence intensities by using a ScanArray 5000 (Packard Instruments). Laser power and/or PMT were adjusted such that the two channels were balanced. The resulting files were analyzed using GENEPIX 3.0 software (Axon Instruments, Foster City, CA). Spots were excluded from analysis because of high local background fluorescence, slide abnormalities, or weak intensity as determined primarily by the background signal observed for hybridization to the gfp spot. For graphical representation of the results, gene absence or presence was converted to 1 for absent, 0 for present. cluster and treeview (19) were then used to compile and visualize the data (Fig. 1) by binary analysis.
Figure 1.
Representation of absent/present genes in nine test strains compared with N16961. Final absent/present calls for each gene were translated to binary code and analyzed using the cluster/treeview software of Eisen et al. (19). For each strain, a black line indicates the presence of a gene, whereas a white line indicates its absence. Black lines represent all genes from strain N16961, as this strain's sequence was used to construct the array.
Verification of Absent Genes.
Confirmation of absent genes included verification by Southern analysis, PCR analysis, or both methods. Standard 50-μl PCR reactions using Taq DNA polymerase were performed according to the manufacturer's instructions (Invitrogen). Southern analysis was performed by using standard protocols and the ECL method of probe labeling and detection (Amersham Pharmacia).
Results
Array Construction and Evaluation.
The V. cholerae microarray we constructed was composed of gene-length PCR products. Primer design was based on the initial release of the V. cholerae N16961 genomic sequence by TIGR, which reported 3,890 ORFs (compared with 3,885 ORFs in the final, published sequence). Of the 3,890 ORFs, gel electrophoresis revealed successful amplification of 3,632 ORFs, representing 93.5% of the genome. Seventy-two percent of PCR reactions classified as “failed” were from ORFs predicted to be less than 200 bp in length. Because it can be difficult to visualize short PCR products by agarose gel electrophoresis, it is possible that for some of these small ORFs, a significant amount of PCR product was actually generated and spotted on the array. In support of this possibility, significant signal was observed for some of the spots corresponding to these reactions when the array was hybridized with labeled N16961 DNA. In addition, a number of the “failed” reactions corresponding to ORFs of greater than 500 bp resulted in spots having significant fluorescent signal (greater than 5,000 units). On further inspection, many of the PCR reactions originally classified as “failed” actually produced weak bands that were barely visible by gel electrophoresis. Therefore, we believe that greater than 93.5% of the genome is truly represented by the array.
Classification of Genes from Test Strains as Absent or Present.
The background-corrected fluorescence ratio of N16961-labeled DNA divided by that of the test strain (N16961/test strain) was determined for each spot on the microarray by using the GENEPIX 3.0 software. Microarray results for ≈30 genes were chosen over a range of ratios, and the presence or absence of a gene was confirmed by PCR analysis using original ORF primers. This led to the following empirical metric used to report genes as absent or present for any given spot: if the N16961/test strain fluorescence ratio was ≥3.0, we had high confidence (90–100%) that the gene is absent in the test strain.
Conservation of the Genome Between N16961 and Test Strains.
Array analysis combined with PCR and Southern analysis identified 143 genes as absent from eight of the nine test strains that were compared with N16961 (Fig. 1, Table 1). Strain HK1 (an early 7th pandemic isolate) seems to be identical to strain N16961. Strain C6709 (a late 7th pandemic isolate) is missing only a single gene (described below). The remaining seven test strains are each missing between 36 and 49 genes (Table 1). Surprisingly, this represents only a ≈1% difference between the genomes of most test strains and the sequenced strain N16961. This is in contrast to results reported for Staphylococcus aureus and Helicobacter pylori (20, 21). Comparative studies using clinical isolates of these bacteria identified approximately a 12% difference between S. aureus clinical strains and the reference strain, and a 6% difference between two sequenced strains of H. pylori. Our results suggest a remarkable conservation of genomic information among the V. cholerae strains described here, despite their isolation over the past century.
For a number of cases, we found that the genes absent from test strains are clustered in the N16961 genome. These loci may correspond to multigene insertions or chromosomal “islands” (see below). However, the microarray data did not always indicate that every gene within that region was absent. In some cases, PCR and/or Southern analysis was conducted to confirm the presence or absence of genes within a suspected island insertion.
Genes we identified as absent fell into four main groups (Table 2). We were most interested in three of these groups (Fig. 2). First, we found genes that could differentiate classical biotype strains from El Tor biotype strains. These genes would be absent only from classical strains and present in all other strains selected for analysis (group I). Because the array was constructed by using the N16961 genome (a 7th pandemic isolate), we could not identify genes present only in classical strains and absent in all other strains. Although the “nonclassical” strains selected for this analysis include El Tor strains of diverse origins, all are TCP+ and therefore potentially pathogenic. Second, we identified genes present only in pandemic strains, including classical isolates (group II). Third, we found genes specific to 7th pandemic strains, including epidemic and endemic El Tor O1 strains and the closely related O139 strain MO10 (group III).
Table 2.
Classification of absent genes
| Group | Strains where genes are absent | No. of genes absent* |
|---|---|---|
| Group I | O395 | 7 |
| Genes present in El Tor but not classical strains. | 569B | 7 |
| NIH41 | 7 | |
| Group II | 2740-80 | 14 |
| Genes present only in strains able to cause epidemic disease (absent from environmental and prepandemic El Tor). | NCTC8457 MAK757 | 14 2 |
| Group III | 2740-80 | 22 |
| Genes present only in 7th pandemic strains HK1, | NCTC8457 | 22 |
| C6709, and MO10. | MAK757 | 22 |
| O395 | 22 | |
| 569B | 22 | |
| NIH41 | 22 | |
| Group IV | MO10 | 42 |
| Genes uniquely absent from a single strain. | NIH41 | 14 |
| MAK757 | 15 |
Figure 2.
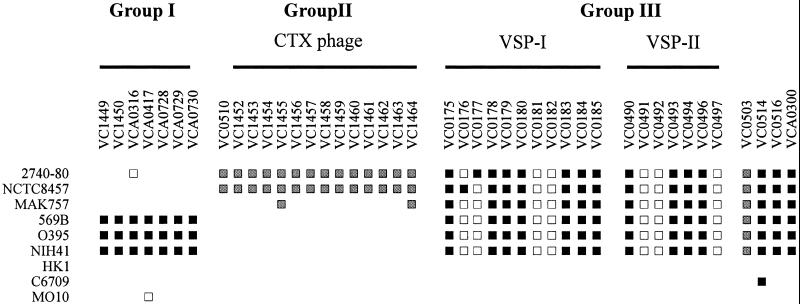
Group I–III genes identified by array, Southern, and PCR analysis. Absent genes were categorized based on criteria listed in Table 2. Black squares indicate genes identified as absent by array analysis and confirmed as absent by PCR or Southern analysis; gray squares indicate genes identified as absent by array analysis alone; and white squares indicate genes identified as absent by PCR or Southern analysis. The two regions of group III genes comprising newly identified Vibrio seventh pandemic islands are labeled VSP-I and VSP-II.
Group I: Genes Present in All El Tor Strains but Not Classical Strains.
Because classical and El Tor strains are believed to have evolved from separate lineages (12, 15), we sought to identify genes that would uniquely define strains of the El Tor biotype. Only seven genes were absent solely in classical strains but present in all other strains (Fig. 2). Interestingly, five of the seven are located on the small chromosome. Of these five, three are hypothetical proteins, for which we have no further information (VCA0728–VCA0730). The other two, VCA0316 and VCA0417, are identical genes predicted to encode a putative acetyltransferase. Both of these ORFs reside on the integron island, a gene capture system located on the small chromosome (22). PCR analysis using primers for neighboring genes combined with an ORF primer revealed that strain 2740-80 carries VCA0417 only, whereas strain MO10 carries only VCA0316. The conserved physical location of the VCA0417 and VCA0316 genes in these two strains suggests that each strain suffered an independent deletion of one copy of this particular gene. Strains NCTC8457, MAK757, HK1, and C6709 carry both copies of the gene.
The remaining two group I genes are located on the large chromosome in the region encoding the RTX toxin. Lin et al. previously showed that in classical strains, there is a deletion in this region that includes the rtxC gene (VC1450), which likely encodes an acylation enzyme that activates the toxin (23). Array analysis confirmed the absence of the rtxC gene in classical strains, as well as the absence of an adjoining gene predicted to encode a hypothetical protein (VC1449). In classical strains, the RTX deletion extends into the flanking genes that encode RtxA (VC1451) and RtxB (VC1448). However, there is sufficient sequence remaining for each of these two genes to provide hybridization on the array with labeled DNA from classical strains.
We were surprised that so few genes uniquely define the El Tor biotype. These data suggest that classical biotype strains may be derived from a primordial environmental strain that was more El Tor-like than previously thought.
Group II: Genes Present Only in Pandemic Strains.
This group represents genes that differentiate classical and 7th pandemic El Tor strains from environmental (2740-80) and prepandemic El Tor strains (MAK757 and NCTC8457, Fig. 2). NCTC8457 was isolated from individuals in an El Tor quarantine camp in 1910, and MAK757 was a clinical isolate from the Celebes Islands in 1935 (2, 6). Consistent with their isolation from human stools, both of these early El Tor, pre-7th pandemic strains were positive for TCP island genes (and thus likely encode the pilus colonization factor TCP). Strain 2740-80 is a nontoxigenic United States Gulf Coast environmental isolate (3, 5). Work by Goldberg and Murphy (24) has provided strong evidence that this strain is clonal with toxigenic CTX+ strains isolated from patients and Gulf Coast environmental sources. As expected, strain 2740-80 contains the TCP pathogenicity island but not the CTXφ genome. One additional absent gene was common to strain 2740-80 and NCTC8457: VC0510, whose product is similar to the E. coli RadC protein involved in DNA repair (25). Five absent genes were common to 2740-80 and MAK757, all encoding hypothetical proteins (data not shown).
Strain MAK757 seems to have all of the CTXφ genes except for VC1455 and VC1464, which are identical genes encoding the phage transcriptional regulator RstR. Waldor et al. have reported that RstR represses the transcription of rstA2, whose gene product is required for CTXφ replication (26). Although not identified by our array analysis, sequence divergence has been shown for RstR, consistent with its role in the formation of heteroimmune temperate phages (27, 28). We therefore predict that the CTXφ genome present in MAK757 possesses highly divergent copies of the rstR gene that are not detectable by microarray analysis. If so, this result is consistent with the independent emergence of MAK757 as a toxigenic clone distinct from earlier classical strains and the future 7th pandemic clone.
Group III: Genes Present Only in 7th Pandemic El Tor O1 Strains.
A total of 22 genes is missing from classical isolates (O395, 569B, and NIH41) and prepandemic TCP+ El Tor strains (NCTC8457 and MAK757). These genes are also absent from the environmental isolate 2740-80. Array analysis identified 15 of these genes, and Southern and PCR analysis assigned an additional 7 genes to this group (Fig. 2).
Seven genes identified by the array span a 16-kb region from VC0175 to VC0185, and Southern analysis confirmed that four additional genes within this region are also absent. This region therefore represents a block of 11 genes in 7th pandemic strains that has an uncharacteristically low GC content (40% vs. 47% for the entire genome), suggesting that it was likely acquired by horizontal transfer. Thus, we have designated this region “Vibrio seventh pandemic island-I” or VSP-I.
Seven of the genes in VSP-I encode hypothetical or conserved hypothetical proteins with no known function. The annotation of three other genes further suggests that this region may have been derived from a mobile genetic element. VC0185, which defines the 3′ end of the region, is predicted to encode a putative transposase, similar to that identified in the S. aureus transposon Tn554 (29). VC0175 shows similarity in its C-terminal domain to deoxycytidylate deaminase-related proteins. Deaminase proteins from a number of other species perform a variety of functions and can be involved in nucleotide scavenging or DNA uptake during competence (30, 31). Phage T2 and T4 also possess dCMP deaminase proteins (32, 33), although the similarity to VC0175 at the amino acid level is poor. VC0176 is homologous to the xre transcriptional regulator from the defective PBSX prophage of Bacillus subtilis (34) and contains a helix-turn-helix motif. Interestingly, VC0176 encodes a product belonging to a paralogous family that includes the lysogeny repressor protein for CTXφ, RstR. Thus, the product of VC0176 may serve as a regulator of genes within this region. Another gene in this region, VC0178, shows significant homology to a variety of phospholipases and is annotated as a “patatin-related protein.” Patatin is a major storage protein found in potato tubers and roots and has recently been shown to encode a novel phospholipase A activity (35).
We sequenced the chromosomal region corresponding to the “empty site” for insertion of the VSP-I island in the strains missing this island. The sequence was identical in these strains and shows that the flanking genes (VC0174 and VC0186) are intact. The junction lies within the intergenic region between VC0174 and VC0175 and encompasses 100 base pairs downstream of the VC0174 stop codon. This 100-bp region has a lower GC content than the rest of the genome (41%) and contains both inverted and direct repeats as well as a 14-bp palindromic sequence. The junction abuts the 3′ end of VC0185, which is transcribed on the negative DNA strand. Three additional nucleotides, predicted to encode an arginine residue, are inserted before the stop codon of VC0185 (data not shown).
Another region unique to 7th pandemic strains encompasses eight genes spanning VC0490–VC0497. Array analysis identified VC0490, VC0493, VC0494, and VC0496, and PCR analysis demonstrated that VC0491, VC0492, and VC0497 were also present only in 7th pandemic strains. Although PCR analysis of VC0495 was inconclusive, we believe that other methods will show it is a group III gene based on its location within this block of genes. Like VSP-I, the average GC content of genes VC490–VC0497 is lower than average (41% vs. 47% for the rest of the chromosome), suggesting that this region was also acquired by horizontal transfer. We therefore propose that this region likely represents a second chromosomal island, which we now designate as “Vibrio seventh pandemic island-II” or VSP-II. The boundaries of VSP-II are not precisely defined because of primer-related PCR technical problems. For example, we have not yet definitively confirmed the absence or presence of VC0498–VC0502 in nonepidemic/pandemic strains. If missing, the size of VSP-II would increase from 7.5 kb to ≈13.5 kb. Like VSP-I, VSP-II is composed of genes predicted to encode hypothetical and conserved hypothetical proteins. However, VC0497 is predicted to encode a transcriptional regulator, similar to a bacteriophage P4 protein (36).
Three other genes outside of VSP-I and VSP-II fit the group III pattern of distribution. Genes VC0514, encoding a methyl-accepting chemotaxis protein, and VC0516, encoding a putative phage integrase, were also located in regions of uncharacteristically low GC content. However, the absence of flanking genes that may contribute to a larger deletion awaits confirmation by another method. VC0514 is one of 42 methyl-accepting chemotaxis proteins identified by the sequencing project, all of which belong to a single paralogous family. VC0514 was the sole gene identified as absent from 7th pandemic strain C6709. Finally, a single, isolated gene located on the integron island (VCA0300) belongs to group III and is predicted to encode a chloramphenicol acetyltransferase.
Group IV: Genes Uniquely Absent from Individual Strains.
Based on array data, MO10 is missing two large regions of genes, which for simplicity are not shown in Fig. 2 (see Fig. 1). The first is from VC0241 to VC0270 and includes genes that encode enzymes involved in O antigen capsule and O139 lipopolysaccharide antigen synthesis. Because MO10 is an O139 serogroup strain and has an O antigen composition that differs from that of O1 strains, this result was expected.
The second region spans genes VC1761–VC1786. It includes gene VC1776, encoding a putative _N_-acetylneuraminate lyase, and VC1784, encoding the NanH neuraminidase. Both genes encode potential virulence factors, as cleavage of sialic acid residues from certain host gangliosides increases the sensitivity of host cells to cholera toxin (37). Nonetheless, PCR results verified that these two genes are absent in pandemic strain MO10. The missing region also includes genes VC1765 and VC1776, encoding a putative type I restriction enzyme and its cognate DNA methylase. In strain N16961, this region has a lower than average GC content. Together, these results suggest that this region might have originally been acquired by horizontal transfer (probably by the archaic precursor of both classical and El Tor strains) but was subsequently lost by strain MO10.
There is evidence of other potential strain-specific deletions. Classical strain NIH41 is apparently missing genes necessary for citrate metabolism, as 14 genes within a 20-gene region from VC0790 to VC0809 were called as absent by array analysis. Pre-7th pandemic El Tor strain MAK757 is missing a number of genes that likely encompass genes VC1791–VC1817. This region includes genes encoding a putative transcriptional regulator and a deoxyribodipyrimidine photolyase, likely involved in DNA repair. Microarray analysis also detected the previously reported deletion of the toxS gene in classical strain 569B (38, 39).
Conclusions
Microarray technology is a powerful new tool that allows global comparative analysis of gene content between different bacterial isolates of a given species. We have used this method to investigate the genetic similarity among strains of V. cholerae isolated from diverse geographical locales and over decades of time. Our analysis indicates that the nine test strains of V. cholerae we selected show a remarkably close degree of relatedness to strain N16961. Although the strains varied in biotype, serogroup, and year and site of isolation, as a group they lacked ≈1% of N16961 genes. Because of the asymmetric nature of microarray analysis, we cannot say whether the test strains carry more genes than N16961.
The close relatedness of the test strains to N16961 suggests that only a small group of V. cholerae strains might be capable of evolving to become human pathogens through the acquisition of the TCP pathogenicity island and the CTXφ genome. However, this conclusion assumes that other nonpathogenic (i.e., TCP− and CTX−) environmental isolates of V. cholerae (including non-O1 and non-O139 serogroup strains) will be quite different in gene content when compared with N16961. This analysis remains to be done. If such further analysis reveals that these nonpathogenic strains are as similar to N16961 as the test strains we selected, then we must conclude that virtually any strain of V. cholerae has the capacity to become a human pathogen simply by acquisition of the TCP island, the CTXφ, and perhaps the genes encoding a few other select properties (such as production of O1 or O139 O antigens). We are currently investigating this possibility by analyzing V. cholerae isolates representing environmentally well adapted, non-O1 and non-O139 strains. These isolates do not carry virulence genes usually associated with strains that cause cholera. Such studies should provide a near-complete evolutionary history of how the environmental organism V. cholerae can become a human pathogen.
It is important to appreciate that the genetic changes that have allowed V. cholerae to become a human pathogen may not be the sole determinants of its success as an endemic and pandemic pathogen. For example, at least one strain of pathogenic V. cholerae El Tor caused an epidemic before the beginning of the 7th pandemic (i.e., MAK757 in Indonesia in 1935), and another distinct El Tor clone caused sporadic cholera cases decades later along the Gulf Coast of the United States (2–6). Yet, these strains did not spread to cause significant disease outside the locales where they initially emerged. In contrast, 7th pandemic El Tor strains have flourished globally over four decades since their emergence, displacing resident classical strains in South Asia and establishing their endemic presence virtually everywhere they have been introduced (7).
Investigators using a variety of molecular typing methods have concluded that 7th pandemic strains represent a globally distributed clone that is closely related to the more recently emerged O139 clone (3, 10–15). Because our microarray was constructed based on the genomic sequence of the 7th pandemic strain N16961, we believed that comparative analysis would provide supporting evidence for the clonal nature of 7th pandemic strains. As predicted, we found no or only minor differences between N16961 and the 7th pandemic strains HK1 and C6709 as well as the O139 strain MO10. We further hypothesized that 7th pandemic strains would have unique gene content that might contribute to their apparent fitness as endemic and pandemic agents of cholera. This prediction was also supported by the microarray analysis, in that we identified genes specific to 7th pandemic isolates of V. cholerae. Most of these group III genes are located in two chromosomal gene clusters, which we have designated VSP-I and VSP-II (Fig. 2).
The VSP-I and VSP-II gene clusters have a lower than average GC content when compared with the rest of the V. cholerae chromosome, suggesting that these gene clusters represent chromosomal “islands” acquired by horizontal transfer. Although we cannot determine when these islands were acquired by V. cholerae, two possibilities seem most probable. The VSP-I and VSP-II islands might have been acquired by a fully pathogenic (i.e., TCP+, CTX+) pre-7th pandemic El Tor O1. Alternatively, a primordial, nonpathogenic El Tor O1 strain acquired the VSP islands first, followed by acquisition of the TCP and CTX genetic elements to complete its emergence as human pathogen.
We predict that the genes associated with the VSP-I and VSP-II islands are likely to encode some of the properties that are responsible for the unique characteristics of 7th pandemic clones. Although three other genes located outside this region (VC0514, VC0516, and VCA0300) show a similar group III pattern and could also contribute to these traits, the VSP islands encode far more group III genes and thus seem likely to be the origin of 7th pandemic properties. These might include adaptive properties that, for example, allow 7th pandemic strains to withstand nutrient deprivation or physical/chemical stresses and thus survive in aquatic environments more efficiently than pre-7th pandemic strains. Such environmental adaptation could also include genes that allow colonization of non-human hosts, such as phytoplankton, filamentous algae, or crustaceans.
Alternatively, the genes on the VSP islands might simply increase adaptation of V. cholerae to the human host in ways that may or may not manifest themselves as an increase in pathogenicity per se. For instance, human adaptation genes might encode functions that allow more efficient infection of humans by, for example, promoting bacterial resistance to stomach acid. These genes could also encode properties that allow more extensive or prolonged intestinal colonization, thus increasing the shedding of vibrios and seeding of environmental reservoirs.
If the genes of the VSP islands are, in fact, responsible for more efficient (or prolonged) infection of human hosts rather than improved survival in the aquatic ecosystems, we would favor a conclusion that has recently been unpopular in the field. The evolutionary success of the 7th pandemic clone of V. cholerae as an endemic and pandemic pathogen may be more related to its improved interaction with the human host than to its improved fitness within environmental reservoirs. Now that we have identified a number of unique genes that define the 7th pandemic clone, it will be possible to delete these genes systematically to address their potential roles in human infection and in promoting fitness of V. cholerae in environmental ecosystems.
Acknowledgments
We thank Catherine Lee, Su Chiang, and James Bina for critically reading the manuscript, and Christina Mills for generation of preliminary data. This work was funded by National Institutes of Health Grants AI18045 (to J.J.M.) and AI53535-02 (to TIGR).
References
- 1.Heidelberg J F, Elsen J A, Nelson W C, Clayton R J, Gwinn M L, Dodson R J, Haft D H, Hickey E K, Peterson J D, Umayam L, et al. Nature (London) 2000;406:477–484. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pollitzer R. Cholera. Geneva, Switzerland: World Health Organization Monograph; 1959. [Google Scholar]
- 3.Wachsmuth I K, Olsvik Ø, Evins G M, Popovic T. In: Vibrio cholerae and Cholera: Molecular to Global Perspectives. Wachsmuth I K, Blake P A, Olsvik Ø, editors. Washington, DC: Am. Soc. Microbiol.; 1994. pp. 357–370. [Google Scholar]
- 4.Blake P A. In: Vibrio cholerae and Cholera: Molecular to Global Perspectives. Wachsmuth I K, Blake P A, Olsvik Ø, editors. Washington, DC: Am. Soc. Microbiol.; 1994. pp. 293–295. [Google Scholar]
- 5.Blake P A. In: Vibrio cholerae and Cholera: Molecular to Global Perspectives. Wachsmuth I K, Blake P A, Olsvik Ø, editors. Washington, DC: Am. Soc. Microbiol.; 1994. pp. 309–319. [Google Scholar]
- 6.Barua D. In: Cholera. Barua D, Greenough W B III, editors. New York: Plenum; 1992. pp. 1–36. [Google Scholar]
- 7.Faruque S M, Albert M J, Mekalanos J J. Microbiol Mol Biol Rev. 1998;62:1301–1314. doi: 10.1128/mmbr.62.4.1301-1314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shimada T, Nair G B, Deb B C, Albert M J, Sack R B, Takeda Y. Lancet. 1993;341:1347. [Google Scholar]
- 9.Faruque S M, Asadulghani S M N, Alim A R M A, Albert M J, Nasirul Islam K M, Mekalanos J J. Infect Immun. 1998;66:5819–5825. doi: 10.1128/iai.66.12.5819-5825.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Byun R, Elbourne L D H, Lan R, Reeves P R. Infect Immun. 1999;67:1116–1124. doi: 10.1128/iai.67.3.1116-1124.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lan R, Reeves P R. Microbiology. 1998;144:1213–1221. doi: 10.1099/00221287-144-5-1213. [DOI] [PubMed] [Google Scholar]
- 12.Popovic T, Bopp C, Olsvik O, Wachsmuth I K. J Clin Microbiol. 1993;31:2474–2482. doi: 10.1128/jcm.31.9.2474-2482.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faruque S M, Alim A R M A, Roy S K, Khan F, Nair G B, Sack R B, Albert M J. J Clin Microbiol. 1994;32:1050–1053. doi: 10.1128/jcm.32.4.1050-1053.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaper J B, Bradford H B, Roberts N C, Falkow S. J Clin Microbiol. 1982;16:129–134. doi: 10.1128/jcm.16.1.129-134.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karaolis D K, Lan R, Reeves P R. J Bacteriol. 1995;177:3191–3198. doi: 10.1128/jb.177.11.3191-3198.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colwell R R, Huq A. In: Vibrio cholerae and Cholera: Molecular to Global Perspectives. Wachsmuth I K, Blake P A, Olsvik Ø, editors. Washington, DC: Am. Soc. Microbiol.; 1994. pp. 117–133. [Google Scholar]
- 17.Islam M S, Draser B S, Sack R B. J Diarrhoeal Dis Res. 1994;12:197–206. [PubMed] [Google Scholar]
- 18.Eisen M B, Brown P O. Methods Enzymol. 1999;303:179–205. doi: 10.1016/s0076-6879(99)03014-1. [DOI] [PubMed] [Google Scholar]
- 19.Eisen M B, Spellman P T, Brown P O, Botstein D. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fitzgerald J R, Sturdevant D E, Mackie S M, Gill S R, Musser J M. Proc Natl Acad Sci USA. 2001;98:8821–8826. doi: 10.1073/pnas.161098098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salama N, Guillemin K, McDaniel T K, Sherlock G, Tompkins L, Falkow S. Proc Natl Acad Sci USA. 2000;97:14668–14673. doi: 10.1073/pnas.97.26.14668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mazel D, Dychinco B, Webb V A, Davies J. Science. 1998;280:605–608. doi: 10.1126/science.280.5363.605. [DOI] [PubMed] [Google Scholar]
- 23.Lin W, Fullner K J, Clayton R J, Sexton J A, Rogers M B, Calia K E, Calderwood S B, Fraser C, Mekalanos J J. Proc Natl Acad Sci USA. 1999;96:1071–1076. doi: 10.1073/pnas.96.3.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goldberg S, Murphy J R. Infect Immun. 1983;42:224–230. doi: 10.1128/iai.42.1.224-230.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saveson C J, Lovett S T. Genetics. 1999;152:5–13. doi: 10.1093/genetics/152.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waldor M K, Rubin E J, Pearson G D, Kimsey H, Mekalanos J J. Mol Microbiol. 1997;24:917–926. doi: 10.1046/j.1365-2958.1997.3911758.x. [DOI] [PubMed] [Google Scholar]
- 27.Kimsey H H, Waldor M K. Proc Natl Acad Sci USA. 1998;95:7035–7039. doi: 10.1073/pnas.95.12.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis B D, Kimsey H H, Chang W, Waldor M K. J Bacteriol. 1999;181:6779–6787. doi: 10.1128/jb.181.21.6779-6787.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murphy E, Huwyler L, Do Carno de Freire Bastos M. EMBO J. 1985;4:3357–3365. doi: 10.1002/j.1460-2075.1985.tb04089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ribeiro G, Viveiros M, David H L, Costa J V. Microbiology. 1997;143:2701–2708. doi: 10.1099/00221287-143-8-2701. [DOI] [PubMed] [Google Scholar]
- 31.Richter G, Fischer M, Kreiger C, Eberhardt S, Luttgen H, Gerstenschlager I, Bacher A. J Bacteriol. 1997;179:2022–2028. doi: 10.1128/jb.179.6.2022-2028.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maley G F, Duceman B W, Wang A M, Martinez J, Maley F. J Biol Chem. 1990;265:47–51. [PubMed] [Google Scholar]
- 33.Maley G F, Guarina D U, Maley F. J Biol Chem. 1983;258:8290–8297. [PubMed] [Google Scholar]
- 34.Wood H E, Devine K M, McConnell D J. Gene. 1990;96:83–88. doi: 10.1016/0378-1119(90)90344-q. [DOI] [PubMed] [Google Scholar]
- 35.Hirschberg H J H B, Simons J-W F A, Dekker N, Egmond M R. Eur J Biochem. 2001;268:5037–5044. doi: 10.1046/j.0014-2956.2001.02411.x. [DOI] [PubMed] [Google Scholar]
- 36.Lin C S. Nucleic Acids Res. 1984;12:8667–8684. doi: 10.1093/nar/12.22.8667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Galen J E, Ketley J M, Fasano A, Richardson S H, Wasserman S S, Kaper J B. Infect Immun. 1992;60:406–415. doi: 10.1128/iai.60.2.406-415.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller V L, Taylor R K, Mekalanos J J. Cell. 1987;48:271–279. doi: 10.1016/0092-8674(87)90430-2. [DOI] [PubMed] [Google Scholar]
- 39.Miller V L, DiRita V J, Mekalanos J J. J Bacteriol. 1989;171:1288–1293. doi: 10.1128/jb.171.3.1288-1293.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]