Evidence for a Susceptibility Gene for Autism on Chromosome 2 and for Genetic Heterogeneity (original) (raw)
Abstract
Although there is considerable evidence for a strong genetic component to idiopathic autism, several genomewide screens for susceptibility genes have been performed with limited concordance of linked loci, reflecting either numerous genes of weak effect and/or sample heterogeneity. Because decreasing sample heterogeneity would increase the power to identify genes, the effect on evidence for linkage of restricting a sample of autism-affected relative pairs to those with delayed onset (at age >36 mo) of phrase speech (PSD, for phrase speech delay) was studied. In the second stage of a two-stage genome screen for susceptibility loci involving 95 families with two or more individuals with autism or related disorders, a maximal multipoint heterogeneity LOD score (HLOD) of 1.96 and a maximal multipoint nonparametric linkage (NPL) score of 2.39 was seen on chromosome 2q. Restricting the analysis to the subset of families (_n_=49) with two or more individuals having a narrow diagnosis of autism and PSD generated a maximal multipoint HLOD score of 2.99 and an NPL score of 3.32. The increased scores in the restricted sample, together with evidence for heterogeneity in the entire sample, indicate that the restricted sample comprises a population that is more genetically homogeneous, which could therefore increase the likelihood of positional cloning of susceptibility loci.
Autism/autistic disorder (MIM 209850) is a development disorder characterized by three classes of symptoms, including impairments in communication and reciprocal social interactions and repetitive or stereotyped behaviors and interests (Rapin and Katzman 1998). Twin studies have indicated that genetic factors play an important role in the etiology of autism (Folstein and Rutter 1977; Ritvo et al. 1985; Steffenburg et al. 1989; Bailey et al. 1995). The concordance rate for monozygotic twins is much higher than that of dizygotic twins. In addition, family studies indicate that the recurrence risk to siblings, estimated from multiple studies at 1%–3%, is profoundly higher than the risk to the general population, which is ∼.5–2/1,000 (Bolton et al. 1994; Szatmari et al. 1998). The mode of inheritance of autism appears complex, and latent-class analyses suggest that 3–10 genes may underlie the disorder (Pickles et al. 1995), although an interpretation of at least one genomewide linkage analysis has argued for >10 genes underlying the disorder (Risch et al. 1999).
Several genomewide screens for autism-susceptibility genes have recently been published (International Molecular Genetic Study of Autism Consortium [IMGSAC] 1998; Barrett et al. 1999; Philippe et al. 1999; Risch et al. 1999; see also Ashley-Koch et al. 1999; Auranen et al. 2000). In these screens, there was little overlap in the highest linkage peaks. The first screen, by the IMGSAC (1998), identified a peak on chromosome 7, where there appears to be the greatest concordance of findings (reviewed in Folstein and Mankoski 2000). However, even in those studies with some evidence for linkage to this region, there are discrepancies in peak localization. Furthermore, the evidence for linkage observed by the IMGSAC has decreased at this location with additional families (IMGSAC 2000). The next highest peak in the IMGSAC study and the highest peaks in other studies—for example, on chromosome 16p (IMGSAC 1998), 13 (Barrett et al. 1999), 2q, 7q, 16p, and 19p (Philippe et al. 1999), and 1p (Risch et al. 1999)—showed less reproducibility between studies. These issues have led to the suggestion that autism may involve extensive genetic heterogeneity (Lamb et al. 2000) and/or many interacting genes of weak effect (Risch et al. 1999). If so, it may prove very difficult, with the methods of linkage analysis and positional cloning, to use genomewide linkage analysis to identify susceptibility loci with the typical sample size of 50–200 families.
Regardless of the actual number of genes involved in autism, decreasing sample heterogeneity will increase the likelihood of identifying genes. Sample heterogeneity may be reduced by use of narrower inclusion criteria or by identication of traits that are shared among subgroups of families. Identifying traits that tend to be shared by family members—particularly by affected sibling pairs—is a means of identifying useful subgroups of families and may identify traits that have greater genetic components. Two studies, both with <50 sibships, have been published that identify Autism Diagnostic Interview (ADI) (Lord et al. 1994) components, which showed increased familiality. In one study (Spiker et al. 1994), familiality was observed in the repetitive behavior domain, and in the second study (MacLean et al. 1999), familiality was observed in impairments in nonverbal communication and verbal/nonverbal status. In a recent analysis of 136 sibling pairs with autism, strong evidence was found for concordance between affected sibling pairs for the severity of repetitive behaviors, the level of deficits in nonverbal communication, the presence of phrase speech, and the age at onset of phrase speech (J. M. Silverman, C. J. Smith, J. Schmeidler, E. Hollander, B. A. Lawlor, M. Fitzgerald, J. D. Buxbaum, K. Delaney, P. Galvin, unpublished data). Any of these clinical traits might be useful to subcategorize families for linkage analysis. Families in which affected members demonstrate greater deficits in these domains may be at greater genetic risk for autism and/or may be genetically more homogeneous.
On the basis of studies of traits in nonaffected members of autistic probands' families, the use of language has been proposed as a means to segregate families with autism for linkage (Folstein et al. 1999). More recently, because of the presence of a gene involved in language disorder on 7q (Fisher et al. 1998), near the peak of linkage to autism observed in several studies, it has been suggested that grouping families in subsets on the basis of language may increase evidence for linkage to this region (Folstein and Mankoski 2000). In one sample, restricting analyses to families with phrase-speech delay (PSD) and including as affected those parents who, through self-reporting, had delayed onset of speech, trouble learning to read, or persistent spelling difficulties, did in fact increase evidence for linkage to 7q (Folstein and CLSA 2000). On the basis of evidence for familiality of age at onset of phrase speech (MacLean et al. 1999; J. M. Silverman, C. J. Smith, J. Schmeidler, E. Hollander, B. A. Lawlor, M. Fitzgerald, J. D. Buxbaum, K. Delaney, P. Galvin, unpublished data), restricting analyses to families with PSD may be useful to decrease heterogeneity, thereby increasing the power to identify susceptibility loci throughout the genome.
In the current study, Autism Diagnostic Interview–Revised (ADI-R) (Lord et al. 1994) criteria were used to identify 95 families with affected relative pairs with autism, borderline autism, or Asperger disorder (table 1). To be considered affected with autism, an individual had to satisfy the prespecified cutoff scores in all three symptom areas of the ADI-R and had to present with evidence for onset of symptoms at age <36 mo (Lord et al. 1994). Individuals who failed to meet the ADI-R algorithm criteria for autism by no more than one point in the social domain and either, but not both, the communication or the repetitive-behavior domain were given a research diagnosis of borderline autism. A diagnosis of borderline autism was also given to individuals in which all three domains were above threshold but the onset criterion for autism was not met. A similar research category has been used in another study (IMGSAC 1998). A research diagnosis of Asperger disorder was given to those with neither autism nor borderline autism but who met DSM-IV criteria for Asperger disorder. At least one individual in each family had to satisfy ADI-R criteria for autism. In 84 families, a second individual also met ADI-R criteria for autism (families in class I), whereas in 8 families a second individual met criteria for borderline autism, and in 3 families a second individual met DSM-IV criteria for Asperger disorder (families in class II). Family studies suggest that genetic liability extends to other pervasive developmental disorders and to Asperger disorder, and it has been argued that including such individuals will not introduce significant genetic heterogeneity (IMGSAC 1998).
Table 1.
Demographics of Family Groups[Note]
| No. of Families with PSD/Total No. of Families | ||
|---|---|---|
| Class I | Classes I and II | |
| Families | 49/84 | 57/95 |
| Individuals genotyped | 231/383 | 273/439 |
| Autistic males | 82/134 | 90/145 |
| Autistic females | 16/35 | 16/35 |
| Borderline autistic males | 1/1 | 8/8 |
| Borderline autistic females | 0/0 | 1/1 |
| Males with Asperger disorder | 0/0 | 0/1 |
| Females with Asperger disorder | 0/0 | 0/2 |
| Sib pairs | 46/80 | 54/91 |
| Sib trios | 1/2 | 1/2 |
| First-cousin pairs | 2/2 | 2/2 |
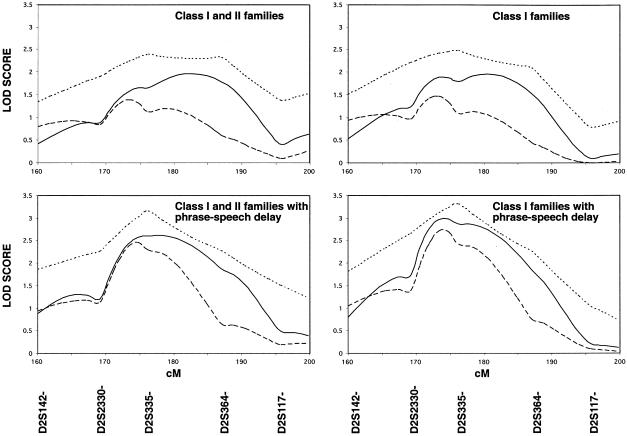
A genetic screen was performed in two stages. In the first stage, 35 families were genotyped with 382 ABI PRISM markers (Linkage Marker Set MD-10, Applied Biosystems) across the autosomes (∼10-cM density), using a microcapillary-based ABI PRISM 310 Genetic Analyzer. Using two-point analyses, we observed the strongest evidence for linkage on chromosome 2q, at marker D2S364 (heterogeneity LOD [HLOD] under dominant, 2.25) (table 2). An adjacent marker also provided evidence for linkage (D2S335, HLOD under dominant, 1.20). These markers, along with flanking markers, were then analyzed in the entire cohort (_n_=95). Evidence suggestive of linkage was observed in the entire sample on distal chromosome 2 by both parametric (maximal multipoint HLOD scores [MMHLS] of 1.96) and nonparametric (nonparametric linkage [NPL] score of 2.39) methods (fig. 1, top left, and table 3, bottom right). The estimate of families with linkage (α, table 3), as well as the divergence between LOD scores and HLOD scores (table 3), is suggestive of genetic heterogeneity in the sample.
Table 2.
Two-Point Linkage Results of Genome Scan[Note]
| Dominant | Recessive | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Chromosomeand Marker | Position(cM) | LOD | HLOD | α | LOD | HLOD | α | NPLScore | P |
| 1: | |||||||||
| D1S2842 | 273.5 | −.66 | 1.17 | .61 | −2.06 | 1.24 | .46 | 1.40 | .08 |
| 2: | |||||||||
| D2S319 | 7.6 | .02 | 1.20 | .66 | −3.41 | .55 | .31 | 1.69 | .04 |
| D2S112 | 141.6 | −3.57 | .39 | .38 | −4.74 | .44 | .28 | 1.04 | .14 |
| D2S335 | 175.9 | −1.48 | 1.20 | .58 | −5.98 | .54 | .26 | 1.59 | .05 |
| D2S364 | 186.2 | 1.80 | 2.25 | .82 | −1.28 | 1.65 | .46 | 2.45 | .01 |
| D2S325 | 204.5 | −3.26 | .67 | .47 | −5.52 | .65 | .29 | 1.52 | .06 |
| D2S2382 | 213.5 | −2.68 | .35 | .37 | −4.80 | .66 | .30 | 1.26 | .10 |
| 3: | |||||||||
| D3S1285 | 91.2 | −4.65 | .32 | .33 | −9.10 | .12 | .12 | 1.05 | .14 |
| D3S1278 | 129.7 | −5.50 | .45 | .34 | −6.76 | .64 | .27 | 1.44 | .08 |
| D3S1267 | 139.1 | −3.67 | .96 | .47 | −7.77 | .67 | .25 | 1.91 | .03 |
| D3S1292 | 146.6 | −9.74 | .39 | .27 | −10.67 | .86 | .25 | 1.24 | .10 |
| D3S1279 | 169.6 | −4.19 | .34 | .32 | −8.07 | .46 | .20 | 1.01 | .15 |
| 5: | |||||||||
| D5S406 | 11.9 | −.67 | 1.21 | .60 | −2.29 | 1.46 | .46 | 1.65 | .05 |
| D5S630 | 19.7 | −3.37 | .62 | .41 | −6.03 | .73 | .26 | 1.30 | .09 |
| 6: | |||||||||
| D6S309 | 14.1 | −2.73 | .56 | .44 | −4.44 | .85 | .32 | 1.65 | .05 |
| D6S264 | 179.1 | −2.64 | .56 | .16 | −3.56 | .90 | .39 | 1.12 | .13 |
| 7: | |||||||||
| D7S502 | 78.7 | −1.95 | .45 | .42 | −4.83 | .47 | .03 | 1.30 | .09 |
| D7S684 | 147.2 | −4.23 | .75 | .45 | −6.09 | .63 | .28 | 1.53 | .06 |
| 8: | |||||||||
| D8S550 | 21.3 | −4.18 | .74 | .46 | −6.69 | .64 | .27 | 1.59 | .05 |
| D8S270 | 103.7 | −4.08 | .31 | .33 | −6.97 | .38 | .22 | 1.12 | .13 |
| 9: | |||||||||
| D9S1817 | 59.3 | −2.37 | .44 | .43 | −6.94 | .10 | .12 | 1.04 | .14 |
| D9S175 | 70.3 | −2.21 | .51 | .46 | −5.82 | .31 | .21 | 1.25 | .10 |
| D9S283 | 94.9 | −1.75 | 1.09 | .56 | −3.98 | 1.09 | .38 | 1.72 | .04 |
| D9S158 | 161.7 | 1.05 | 1.66 | .80 | 1.16 | 1.89 | .74 | 1.41 | .07 |
| 11: | |||||||||
| D11S987 | 67.5 | −5.03 | .47 | .35 | −6.91 | .61 | .28 | 1.13 | .13 |
| D11S1314 | 73.6 | −3.12 | .65 | .43 | −5.35 | .74 | .30 | 1.44 | .07 |
| D11S937 | 80.0 | −4.27 | .73 | .43 | −7.51 | .36 | .21 | 1.19 | .11 |
| 17: | |||||||||
| D17S1868 | 64.2 | −1.39 | .69 | .51 | −3.33 | .87 | .37 | 1.10 | .13 |
| 19: | |||||||||
| D19S216 | 20.0 | −4.72 | .41 | .36 | −5.31 | .35 | .32 | 1.06 | .14 |
| D19S226 | 42.3 | −2.60 | .50 | .40 | −5.68 | .61 | .25 | 1.13 | .12 |
| 20: | |||||||||
| D20S115 | 21.2 | −2.56 | .54 | .47 | −4.89 | .52 | .29 | 1.04 | .15 |
Figure 1.
Multipoint analyses of chromosome 2q. Multipoint LOD analyses under dominant (solid lines) and recessive (dashed lines) modes of inheritance were performed (Vieland et al. 1992_b_; Durner et al. 1999), with penetrance set at 50% (Greenberg et al. 1998) and allowing for locus heterogeneity (HLOD) (Smith 1963; Durner and Greenberg 1992). This simple approach works well for complex traits, even if true modes of inheritance and numbers of genes contributing to the disease are not known (Vieland et al. 1992_a,_ 1993). All genotyped family members were included in the analyses, and any individual not receiving a research diagnosis of autism, borderline autism, or Asperger disorder was defined as phenotypically unknown. Disease-allele frequency was arbitrarily set at .1 for recessive inheritance and .006 for dominant inheritance. Multipoint linkage analyses using nonparametric estimates of sharing were also performed using NPL (dotted lines) with GENEHUNTER (Kruglyak et al. 1996).
Table 3.
Multipoint Analyses of Chromosome 2q[Note]
| With PSD | Total | |
|---|---|---|
| Families in Class I | ||
| 175.4 cM; 2.99 HLOD; α = .69 | 181.1 cM; 1.97 HLOD; α = .50 | |
| Dominant | 174.0 cM; 1.52 LOD | 183.3 cM; −3.49 LOD; |
| 174.4 cM; 2.71 HLOD; α = .50 | 174.0 cM; 1.47 HLOD; α = .27 | |
| Recessive | 174.4 cM; −1.88 LOD; | 172.6 cM; −13.21 LOD; |
| NPL | 176.8 cM; _Z_-score = 3.32; P = 3.8 × 10−4 | 176.8 cM; _Z_-score = 2.47; P = 6.2 × 10−3 |
| Families in Classes I and II | ||
| 179.0 cM; 2.62 HLOD; α = .61 | 183.3 cM; 1.96 HLOD; α = .48 | |
| Dominant | 181.2 cM; −.30 LOD | 183.3 cM; −4.54 LOD |
| 175.4 cM; 2.46 HLOD; α = .42 | 174.0 cM; 1.38 HLOD; α = .24 | |
| Recessive | 174.0 cM; −5.06 LOD; | 174.0 cM; −18.05 LOD; |
| NPL | 176.8 cM; _Z_-score = 3.17; P = 7.1 × 10−4 | 176.8 cM; _Z_-score = 2.39; P = 7.8 × 10−3 |
In an attempt to narrow diagnostic criteria to reduce heterogeneity, families in which there were two or more members with autism (families in class I) and onset of phrase speech at age >36 mo were examined for evidence of linkage. In the 49 families meeting these criteria, there was increased evidence for linkage (MMHLS of 2.99 and NPL score of 3.32), compared with the results for the entire cohort (fig. 1, bottom right, and table 3, top left).
Further, exploratory analyses were performed to identify the determinants in the family groups that contributed to the increased evidence for linkage. Linkage analysis only in families in which there were two individuals meeting ADI-R criteria for autism (families in class I, _n_=84), without a requirement for PSD, had little effect on observed MMHLS and NPL scores (fig. 1, top right, and table 3, top right), compared with analyses done on the entire sample. In contrast, linkage analysis in the 57 families in classes I and II in which the affected family members had PSD demonstrated more dramatic increases in MMHLS and NPL scores (fig. 1, bottom left, and table 3, bottom left), compared with analyses done in the entire sample. These data indicate that limiting analyses to families with PSD had a greater effect on the evidence for linkage than limiting the study to families in class I.
Evidence for heterogeneity decreased as more restrictive criteria were applied. Thus α, the estimate of families with linkage, increased from 48% to 69%, using multipoint HLOD analyses under the dominant model, and increased from 24% to 50%, using these analyses under the recessive model (table 3). With evidence for greater homogeneity in the families in class I with PSD, we performed traditional multipoint LOD score analyses in this sample (that is, we did not allow for heterogeneity). Under a dominant model, maximal LOD score was 1.52 at 166.4 cM (table 3, top left). At this position, the LOD score was −1.88 under a recessive model. In all other subsets of the data, LOD scores were below zero in both dominant and recessive models. The positive LOD score in the most restricted sample suggests that there is increased homogeneity in this subset of families.
To test for heterogeneity, we made use of the predivided sample test (Morton 1956). LOD or HLOD scores were compared between the entire sample, the most restricted sample, and all families excluded from the most restricted sample. For both the MMHLS and maximal multipoint LOD score analyses, these comparisons provided significant evidence for heterogeneity (χ2=6.31; _df_=1, _P_=.0120, and χ2=7.71, _df_=1; _P_=.0054, respectively).
The data in this study provide evidence suggesting linkage to autism for a region on chromosome 2. It has recently been reported that the IMGSAC, which had previously found its highest LOD score on chromosome 7q (IMGSAC 1998), has found that, after additional families were genotyped (for a total of 173), the highest MLS scores were on chromosomes 9 (MLS 2.61), 16 (MLS 2.55), and 2 (MLS 2.33) (IMGSAC 2000). The peak on chromosome 2 was localized to the same position as the peak we observed. In a recent scan involving 75 families, there is also weak evidence for linkage in the same region (∼180 cM) (Barrett et al. 1999). In addition, in an unpublished study involving 99 families, there was a peak (MLS 1.30 at 198.6 cM) on chromosome 2 that overlapped with our peak (Bass et al. 2000). Although slightly distal, in a scan involving 51 families, two-point MLS analysis yielded scores of 0.98 and 0.65 at markers D2S382 (at 210 cM) and D2S364 (at 231.5 cM) (Philippe et al. 1999). In the current analyses, there was increased evidence for linkage when the analyses were restricted to those families in which affected family members demonstrate PSD. Because the current analyses involved parametric analyses under two models as well as grouping the data in subsets, the results can only be taken as suggestive and require replication in a separate group of families before evidence for linkage will be compelling.
Several approaches have been considered in the autism research community as means to improve the chances of finding genes by positional cloning. These approaches include narrowing the definition of affectedness to define more genetically homogeneous groups and/or increasing information by defining individuals with a broader autism phenotype (Le Couteur et al. 1996; Piven et al. 1997) as affected. In the current study, the increase in LOD and HLOD scores in the restricted sample with PSD and the evidence for heterogeneity in the entire sample indicates that narrower inclusion criteria may result in decreased heterogeneity and may thereby increase power to detect linkage in autism. Because it is likely that autism-susceptibility genes result in a broader autism phenotype in unaffected family members, a further increase in power might be achieved in this restricted sample by including as affected those individuals with a broader autism phenotype.
In summary, the current results, when combined with results from other groups, indicate that there may be an autism-susceptibility gene on chromosome 2q. Furthermore, these results suggest that, if narrower inclusion criteria are used, it may be possible to define a genetically more-homogeneous group that could facilitate the positional cloning of putative autism-susceptibility genes.
Acknowledgments
We thank Dr. Susan Hodge for insights into the use of the predivided sample test. We also thank Cure Autism Now and the Autism Genetic Resource Exchange for providing DNA samples. These studies were supported by grants from the Seaver Autism Research Center (to K.L.D.) and Cure Autism Now (to J.D.B.).
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for autism/autistic disorder [MIM 209850]).
References
- Ashley-Koch A, Wolpert CM, Menold MM, Zaeem L, Basu S, Donnelly SL, Ravan SA, Powell CM, Qumsiyeh MB, Aylsworth AS, Vance JM, Gilbert JR, Wright HH, Abramson RK, DeLong GR, Cuccaro ML, Pericak-Vance MA (1999) Genetic studies of autistic disorder and chromosome 7. Genomics 61:227–236 [DOI] [PubMed] [Google Scholar]
- Auranen M, Nieminen T, Majuri S, Vanhala R, Peltonen L, Jarvela I (2000) Analysis of autism susceptibility gene loci on chromosomes 1p, 4p, 6q, 7q, 13q, 15q, 16p, 17q, 19q and 22q in Finnish multiplex families. Mol Psychiatry 5:320–322 [DOI] [PubMed] [Google Scholar]
- Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, Yuzda E, Rutter M (1995) Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med 25:63–77 [DOI] [PubMed] [Google Scholar]
- Barrett S, Beck JC, Bernier R, Bisson E, Braun TA, Casavant TL, Childress D, et al (1999) An autosomal genomic screen for autism: collaborative linkage study of autism. Am J Med Genet 88:609–615 [DOI] [PubMed] [Google Scholar]
- Bass MP, Menold MM, Joyner KL, Wolpert CM, Donnelly SL, Ravan SA, McClain C, vonWendt L, Gilbert JR, Wright HH, Abramson RK, DeLong GR, Cuccaro ML, Pericak-Vance MA (2000) Association analysis of GI candidate genes in autistic disorder. Am J Hum Genet 67 Suppl:S49 [Google Scholar]
- Bolton P, Macdonald H, Pickles A, Rios P, Goode S, Crowson M, Bailey A, Rutter M (1994) A case-control family history study of autism. J Child Psychol Psychiatry 35:877–900 [DOI] [PubMed] [Google Scholar]
- Durner M, Greenberg DA (1992) Effect of heterogeneity and assumed mode of inheritance on lod scores. Am J Med Genet 42:271–275 [DOI] [PubMed] [Google Scholar]
- Durner M, Vieland VJ, Greenberg DA (1999) Further evidence for the increased power of LOD scores compared with nonparametric methods. Am J Hum Genet 64:281–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher SE, Vargha-Khadem F, Watkins KE, Monaco AP, Pembrey ME (1998) Localisation of a gene implicated in a severe speech and language disorder. Nat Genet 18:168–170 [DOI] [PubMed] [Google Scholar]
- Folstein SE, for CLSA (2000) Autism LOD on chromosome 7q was increased subsetting the sample on language acquisition. Am J Hum Genet 67 Suppl:S49 [Google Scholar]
- Folstein SE, Mankoski RE (2000) Chromosome 7q: where autism meets language disorder? Am J Hum Genet 67:278–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folstein S, Rutter M (1977) Infantile autism: a genetic study of 21 twin pairs. J Child Psychol Psychiatry 18:297–321 [DOI] [PubMed] [Google Scholar]
- Folstein SE, Santangelo SL, Gilman SE, Piven J, Landa R, Lainhart J, Hein J, Wzorek M (1999) Predictors of cognitive test patterns in autism families. J Child Psychol Psychiatry 40:1117–1128 [PubMed] [Google Scholar]
- Greenberg DA, Abreu P, Hodge SE (1998) The power to detect linkage in complex disease by means of simple LOD-score analyses. Am J Hum Genet 63:870–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Molecular Genetic Study of Autism Consortium (1998) A full genome screen for autism with evidence for linkage to a region on chromosome 7q: International Molecular Genetic Study of Autism Consortium. Hum Mol Genet 7:571–578 [DOI] [PubMed] [Google Scholar]
- ——— (2000) Search for autism susceptibility loci: genome screen follow-up and fine mapping of a candidate region on chromosome 7q. Presented at Collegium Internationale Neuro-Psychopharamacologium 22nd Congress, Brussels, July 9–13 [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Lamb JA, Moore J, Bailey A, Monaco AP (2000) Autism: recent molecular genetic advances. Hum Mol Genet 9:861–868 [DOI] [PubMed] [Google Scholar]
- Le Couteur A, Bailey A, Goode S, Pickles A, Robertson S, Gottesman I, Rutter M (1996) A broader phenotype of autism: the clinical spectrum in twins. J Child Psychol Psychiatry 37:785–801 [DOI] [PubMed] [Google Scholar]
- Lord C, Rutter M, Le Couteur A (1994) Autism Diagnostic Interview-Revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Autism Dev Disord 24:659–685 [DOI] [PubMed] [Google Scholar]
- MacLean JE, Szatmari P, Jones MB, Bryson SE, Mahoney WJ, Bartolucci G, Tuff L (1999) Familial factors influence level of functioning in pervasive developmental disorder. J Am Acad Child Adolesc Psychiatry 38:746–753 [DOI] [PubMed] [Google Scholar]
- Morton NE (1956) The detection and estimation of linkage between the genes for elliptocytosis and the Rh blood type. Am J Hum Genet 8:80–96 [PMC free article] [PubMed] [Google Scholar]
- Philippe A, Martinez M, Guilloud-Bataille M, Gillberg C, Rastam M, Sponheim E, Coleman M, Zappella M, Aschauer H, Van Maldergem L, Penet C, Feingold J, Brice A, Leboyer M, van Malldergerme L (1999) Genome-wide scan for autism susceptibility genes: Paris Autism Research International Sibpair Study. Hum Mol Genet 8:805–812 [DOI] [PubMed] [Google Scholar]
- Pickles A, Bolton P, Macdonald H, Bailey A, Le Couteur A, Sim CH, Rutter M (1995) Latent-class analysis of recurrence risks for complex phenotypes with selection and measurement error: a twin and family history study of autism. Am J Hum Genet 57:717–726 [PMC free article] [PubMed] [Google Scholar]
- Piven J, Palmer P, Jacobi D, Childress D, Arndt S (1997) Broader autism phenotype: evidence from a family history study of multiple-incidence autism families. Am J Psychiatry 154:185–190 [DOI] [PubMed] [Google Scholar]
- Rapin I, Katzman R (1998) Neurobiology of autism. Ann Neurol 43:7–14 [DOI] [PubMed] [Google Scholar]
- Risch N, Spiker D, Lotspeich L, Nouri N, Hinds D, Hallmayer J, Kalaydjieva L, et al (1999) A genomic screen of autism: evidence for a multilocus etiology. Am J Hum Genet 65:493–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritvo ER, Freeman BJ, Mason-Brothers A, Mo A, Ritvo AM (1985) Concordance for the syndrome of autism in 40 pairs of afflicted twins. Am J Psychiatry 142:74–77 [DOI] [PubMed] [Google Scholar]
- Smith CAB (1963) Testing for heterogeneity of the recombination fraction values in human genetics. Ann Hum Genet 27:175–182 [DOI] [PubMed] [Google Scholar]
- Spiker D, Lotspeich L, Kraemer HC, Hallmayer J, McMahon W, Petersen PB, Nicholas P, Pingree C, Wiese-Slater S, Chiotti C, et al (1994) Genetics of autism: characteristics of affected and unaffected children from 37 multiplex families. Am J Med Genet 54:27–35 [DOI] [PubMed] [Google Scholar]
- Steffenburg S, Gillberg C, Hellgren L, Andersson L, Gillberg IC, Jakobsson G, Bohman M (1989) A twin study of autism in Denmark, Finland, Iceland, Norway and Sweden. J Child Psychol Psychiatry 30:405–416 [DOI] [PubMed] [Google Scholar]
- Szatmari P, Jones MB, Zwaigenbaum L, MacLean JE (1998) Genetics of autism: overview and new directions. J Autism Dev Disord 28:351–368 [DOI] [PubMed] [Google Scholar]
- Vieland VJ, Greenberg DA, Hodge SE (1993) Adequacy of single-locus approximations for linkage analyses of oligogenic traits: extension to multigenerational pedigree structures. Hum Hered 43:329–336 [DOI] [PubMed] [Google Scholar]
- Vieland V, Greenberg DA, Hodge SE, Ott J (1992_a_) Linkage analysis of two-locus diseases under single-locus and two-locus analysis models. Cytogenet Cell Genet 59:145–146 [DOI] [PubMed] [Google Scholar]
- Vieland VJ, Hodge SE, Greenberg DA (1992_b_) Adequacy of single-locus approximations for linkage analyses of oligogenic traits. Genet Epidemiol 9:45–59 [DOI] [PubMed] [Google Scholar]